Abstract
The apical-most region of cell-to-cell contact in a vertebrate epithelium is the tight junction (TJ) complex. It is composed of bicellular TJs (bTJs) that bridge two adjacent epithelial cells and tricellular TJs (tTJs) that are points of contact between three adjoining epithelial cells. Tricellulin (TRIC) is a transmembrane TJ protein of vertebrates that is found in the tTJ complex. Full-length cDNA encoding rainbow trout TRIC was cloned and sequenced. In silico analysis of rainbow trout TRIC revealed a tetraspannin protein with several putative posttranslational modification sites. TRIC mRNA was broadly expressed in rainbow trout tissues and exhibited moderately greater abundance in the gill. In a primary cultured gill epithelium, TRIC localized to tTJs and TRIC protein abundance increased in association with corticosteroid-induced reductions in paracellular permeability. Sodium caprate was used to compromise cultured gill epithelium integrity by disrupting the tTJ complex. Sodium caprate treatment caused a reversible reduction in transepithelial resistance, caused an increase in paracellular permeability (as measured by [3H]PEG-4000 flux), and displaced TRIC from tTJs while leaving bTJs intact. Data from this study support the view that tTJs and the TJ protein TRIC 1) play a role in maintaining gill epithelium integrity and 2) contribute to the regulation of gill epithelium permeability.
Keywords: tight junction, pavement cell, paracellular permeability, sodium caprate, cortisol
the junctional complex that bridges adjacent vertebrate epithelial cells typically comprises a tripartite arrangement of elements that include lateral desmosomes and adherens junctions and apico-lateral tight junctions (TJs; Ref. 19). Bicellular TJs (bTJs) link the membranes of two neighboring epithelial cells and run alongside each other around the cell periphery. bTJs form the majority of the paracellular interface between epithelial cells and the apical (outer) environment (41). In contrast, tricellular TJs (tTJs) run vertically in an apical-to-basal direction at contact points of three adjacent epithelial cells (51). The tTJ is currently thought to be necessary for intercellular occlusion at barrier-weak points where three adjacent bTJs meet (26, 41). bTJs and tTJs consist of transmembrane TJ proteins with 1) extracellular loops that link within the extracellular cleft to control the properties of the paracellular pathway and 2) intracellular domains that connect with scaffolding TJ proteins, which, in turn, connect to the cell cytoskeleton (23). Transmembrane proteins of the claudin superfamily localize exclusively to bTJs and considerable evidence supports the view that bTJ claudin composition is chiefly responsible for tissue- and cell-specific paracellular permeability properties in vertebrate epithelia (24, 55). In contrast, the protein composition of tTJs and their contribution to epithelium paracellular permeability properties have received less attention.
Members of the TJ-associated MARVEL domain-containing subfamily of proteins (TAMPs) include the TJ proteins occludin, tricellulin, and MarvelD3 (46). Occludin and MarvelD3 localize to bTJs exclusively, while tricellulin (TRIC) is the only TAMP described so far that localizes to both bTJs and tTJs (22, 26, 34, 42, 44, 52). At the tTJ, TRIC is distributed as vertically oriented short rods, and knockdown of TRIC has been reported to significantly reduce transepithelial resistance (TER) and increase paracellular marker flux across Eph4 mammary epithelia (26). Mutations in TRIC gene were reported to cause disrupted barrier formation between cochlear and vestibular epithelia in the inner ear, leading to nonsyndromic deafness in humans (13), and overexpression of TRIC has been reported to change the conductive properties of Madin-Darby canine kidney II epithelia (35). In this latter case, when TRIC was overexpressed in tTJs, a reduction in macromolecule permeability was observed whereas TRIC overexpression in bTJs as well as tTJs decreased paracellular Na+ and Cl− permeability as well as macromolecule permeability (35). More recently, it has also been shown that using sodium caprate to displace TRIC from the tTJ complex in epithelia composed of HT-29/B6 cells resulted in a decrease in TER and increase in paracellular tracer flux (36). Therefore, there is strong evidence supporting an integral role for TRIC in the maintenance of epithelial integrity in a variety of vertebrate tissues.
The gill epithelium of teleost fishes plays an important role in the ability of these organisms to maintain salt and water balance (18). The gill epithelium interfaces directly with the surrounding aquatic medium, and its permeability properties contribute significantly to ionoregulatory homeostasis. Although the varying properties of the gill epithelium TJ complex have long been implicated in the regulation of ion movement across the gill, studies aimed at investigating the molecular composition and properties of the fish gill TJ complex have only recently been initiated (for review, see Ref. 12). In this regard, the bulk of work conducted thus far has focused on members of the claudin superfamily (e.g., Refs. 1, 2, 4, 7, 16, 17, 29, 37, 38, 44, 49, 53, 56). However, a number of studies have also revealed the potential importance of occludin (a member of the TAMP subfamily) in teleost fish gills (5, 6, 7, 8, 9, 10, 11, 12, 29). Occludin is found in gill pavement cells as well as gill mitochondria-rich cells (ionocytes), it is responsive to osmoregulatory hormones and environmental change, and it has been shown that increased occludin abundance is associated with decreased paracellular permeability (5, 6, 7, 8, 9, 10, 11, 29). It has also been shown that knockdown of occludin caused an increase in gill epithelium paracellular permeability (11). Nevertheless, virtually nothing is known about other members of the TAMP subfamily in fishes and nothing is known about the properties of the tTJ complex in the gill epithelium or other teleost fish tissues.
In view of the above, the goal of the current study was to consider the importance of the tTJ complex in the fish gill epithelium by investigating the potential role that the tTJ protein TRIC may play in this domain. The important contribution that TRIC appears to play in maintaining the integrity of a wide variety of other vertebrate epithelia (see Refs. 21, 22, 26, 34, 36, 41, 47, 50, 52) strongly supports the hypothesis that TRIC may contribute to the maintenance of gill epithelium integrity in teleost fishes.
MATERIALS AND METHODS
Experimental animals.
Rainbow trout (Oncorhynchus mykiss) were obtained from a local supplier (Humber Springs Trout Hatchery, Orangeville ON, Canada) and held in 600-liter opaque polyethylene tanks supplied with flow-through dechlorinated freshwater (approximate composition in μM: 590 [Na+], 920 [Cl−], 760 [Ca2+], and 43 [K+], pH 7.35). Fish (stocking density of 8–10 g/l) were held under a constant photoperiod of 12:12-h light-dark and were fed ad libitum once daily with commercial trout pellets (Martin Profishent, Elmira, ON, Canada). Fish culture and all experimental procedures were conducted according to a protocol approved by the York University Animal Care Committee and conformed to the guidelines of the Canadian Council on Animal Care.
Total RNA extraction and cDNA synthesis.
Total RNA was isolated from trout tissues using TRIzol Reagent (Invitrogen) according to manufacturer's instructions. Total RNA yield from each sample was determined using a Multiskan Spectrum UV/Vis microplate spectrophotometer (Thermo Fisher Scientific, Nepean, ON, Canada). A fixed quantity of RNA (2 μg) was treated with DNase I (Amplifications Grade; Invitrogen) and used for cDNA synthesis. First-strand cDNA was synthesized using SuperScriptTM III Reverse Transcriptase and Oligo(dT)12–18 primers (Invitrogen).
Identification, cloning, gene assembly, and analysis of rainbow trout TRIC.
Expressed sequence tags (ESTs) from the rainbow trout genome that were similar to mouse TRIC (GenBank: ABA18656.1) and Atlantic salmon MARVEL-domain containing protein 2 (Gene ID: 100380371; Ref. 39) were sought using the National Center for Biotechnology Information (NCBI) database BLAST search engine. Newly identified ESTs were confirmed to be protein encoding using a reverse χBLAST. A reading frame was established using nBLAST alignment and ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/). Two sets of primers were designed based on the predicted TRIC-encoding EST sequence using Primer3 software (v. 0.4.0). Two putative TRIC fragments were amplified using RT-PCR. Amplicon size was verified using agarose gel electrophoresis. Amplicons were then isolated and purified for sequencing using a QIAquick gel extraction kit (Qiagen, Mississauga, ON, Canada). Sequencing was conducted at the York University Core Molecular Biology Facility (Department of Biology, York University, ON, Canada). A full coding sequence for rainbow trout TRIC was confirmed using a BLAST search and submitted to GenBank (accession no. KC603902). Once amplicon identity was confirmed through sequencing at Core Molecular Facility at York University, more primer sets were designed to sequence across the gene to assemble the ESTs into one full coding sequence of rainbow trout TRIC (combinations of forward and reverse primers from different primer sets were used to ensure the integrity of the assembled gene). ClustalX software was used to align and assemble multiple sequenced segments. The phylogenetic relationship between rainbow trout TRIC and TRICs of other vertebrate species was established by building a phylogenetic tree using Geneious software (v. 5.3.4). Amino acid and nucleotide sequences of TRIC from different vertebrates were aligned using ClustalX software. In silico analysis of the rainbow trout TRIC amino acid sequence was performed using EXPASY PROSITE (posttranslational modifications and protein domains), ProtParam (protein weight and stability parameters such as predicted half-life), PSIPRED (secondary structure), and ProtScale and TMHMM (hydrophobicity scale and transmembrane domains). Final rainbow trout TRIC topography was visualized using TOPO2 software.
Rainbow trout TRIC mRNA expression profile.
Rainbow trout were net-captured, anesthetized in 0.5 g/l tricaine methasulfonate (MS-222; Syndel Laboratories), and killed by spinal transection. Discrete tissues were removed, quick frozen in liquid nitrogen, and stored at −80°C until analysis. The following tissues were examined: brain, eye, gill, oesophagus, pyloric caeca, anterior intestine, middle intestine, posterior intestine, kidney, and skin. Total RNA was extracted and cDNA was synthesized as described previously. TRIC transcript presence and abundance were determined by quantitative real-time PCR (qRT-PCR) using SYBR Green I Supermix (Bio-Rad Laboratories, Mississauga, ON, Canada) and a Chromo4 Detection System (CFB-3240; Bio-Rad Laboratories). Primers for rainbow trout TRIC (forward: 5′-GTCACATCCCCAAACCAGTC-3′ and reverse: 5′-GTCCAGCTCGTCAAACTTCC-3′, predicted amplicon size ∼170 bp) were designed using the CDS as outlined previously. The following reaction conditions were used: one cycle denaturation (95°C, 4 min), followed by 40 cycles of 1) denaturation (95°C, 30 s), 2) annealing (51–61°C, 30 s), and 3) extension (72°C, 30 s). To ensure that a single PCR product was synthesized during reactions, a dissociation curve analysis was carried out after each qRT-PCR run. TRIC mRNA abundance was normalized to β-actin using primers and conditions previously described (10). The use of β-actin for normalization of TRIC transcript abundance between different rainbow trout tissues was validated by statistically comparing β-actin threshold cycle values between tissues to confirm that no statistically significant changes occurred.
Preparation and culture of a trout gill epithelium.
Primary cultured gill epithelia (composed of gill pavement cells) were generated using methods originally reported by Wood et al. (58) and described in detail by Kelly et al. (32). Briefly, trout gill cells were isolated by trypsination and initially cultured in 25-cm2 flasks with Leibovitz's culture medium supplemented with 6% fetal bovine serum (L15). Flasks were held in an air atmosphere at 18°C. At confluence (∼ 6 days in culture), cells were harvested from flasks by trypsination and seeded into cell culture inserts (polyethylene terephthalate filters, 0.9-cm2 growth area, 0.4-μm pore size, and 1.6 × 106 pore/cm2 pore density; BD Falcon TM; BD Biosciences, Mississauga, ON, Canada). Inserts were housed in companion cell culture plates (Falcon BD), and the apical and basolateral compartments of the culture system contained L15 media.
Western blot analysis of TRIC in a cultured trout gill epithelium.
TRIC protein abundance was examined in cultured gill epithelia by Western blot analysis according to methods previously described by Chasiotis et al. (10). In short, cells were rinsed with ice-cold PBS, incubated in lysis buffer containing protease inhibitor cocktail, and homogenized by repeatedly passing through a syringe needle. Homogenized samples were centrifuged to remove debris. The supernatant was collected and protein concentration was determined using a Bradford assay (Sigma-Aldrich). A fixed amount of protein from each sample was preheated in homogenization buffer and subjected to a 12% SDS-PAGE. Semidry transfer was performed to transfer protein samples from the gel to a polyvinylidene difluoride membrane. The membrane was then blocked with skimmed milk solution and incubated with a rabbit anti-TRIC (COOH terminus) polyclonal antibody (Invitrogen), followed by a horseradish peroxidase-conjugated goat anti-rabbit antibody (Bio-Rad Laboratories). Antigen reactivity was then examined using an enhanced Chemiluminescence Plus Western blotting system (GE Healthcare BioSciences, Baie d′Urfé, QC, Canada).
Immunolocalization of TRIC in a cultured trout gill epithelium.
Procedures for the immunolocalization of TRIC in cultured gill epithelia were conducted according to Chasiotis et al. (6). In brief, cultured gill epithelia were fixed in 3% paraformaldehyde, permeabilized with methanol, washed with Triton X in phosphate PBS, and blocked with antibody-dilution buffer. TRIC was detected using a rabbit anti-TRIC (COOH terminus) polyclonal antibody (Invitrogen). A mouse anti-cingulin polyclonal antibody (catalog no. 37-4300; Invitrogen) was used as a bTJ marker. After being washed with PBS, preparations were incubated with TRITC-labeled goat-anti-rabbit and FITC-labeled goat anti-mouse antibodies (1:500 in antibody-dilution buffer each; Jackson ImmunoResearch Laboratories, West Grove, PA). After incubation with primary and secondary antibodies, epithelia were mounted on slides using mounting medium containing 4′,6-diamidino-2-phenylindole. Images were captured using differential interference contrast and laser-scanning confocal microscopy with an Olympus BX-51 in conjunction with a Fluoview unit and MellesGriot green and red argon lasers (Olympus). As a negative control, epithelia were prepared as previously described in the absence of primary antibody.
Effect of cortisol on TER, [3H]PEG-4000 flux, and TRIC abundance in a cultured trout gill epithelium.
Cultured trout gill epithelia (prepared as described previously) were treated with 500 ng/ml cortisol (hydrocortisone 21-hemisuccinate sodium salt; Sigma-Aldrich) by addition of hormone to the basolateral side of cell culture inserts immediately following seeding. TER was measured using chopstick electrodes (STX-2) connected to a custom-modified EVOM epithelial voltohmmeter (World Precision Instruments, Sarasota, FL). All TER measurements are expressed as ohms per centimeter squared after correcting for TER measured across “blank” culture inserts. Paracellular permeability across cultured epithelia was determined by measuring [3H]polyethylene glycol (molecular mass 4,000 Da; PEG-4000; PerkinElmer, Woodbridge, ON, Canada) flux according to methods and calculations previously described (58).
TRIC mRNA abundance was determined in cultured gill epithelia by qRT-PCR using methods for total RNA extraction, cDNA synthesis, and qRT-PCR as previously outlined. TRIC mRNA abundance was normalized with rainbow trout elongation factor-1α (EF-1α). Primers for rainbow trout EF-1α (forward: 5′-GGCAAGTCAACCACCACAG-3′ and reverse: 5′-GATACCACGCTCCCTCTCAG-3′, predicted amplicon size ∼159 bp) were designed using GenBank accession no. AF498320.1 and the following PCR reaction conditions were used: one cycle denaturation (95°C, 4 min), followed by 40 cycles of 1) denaturation (95°C, 30 s); 2) annealing (51–61°C, 30 s); and (3) extension (72°C, 30 s). The use of EF-1α for normalization in qRT-PCR experiments was validated by statistically comparing EF-1α threshold cycle values between experimental groups to confirm that no significant changes occurred.
TRIC protein abundance was examined using Western blot analysis as previously outlined. Following TRIC detection, membranes were stripped and incubated with mouse monoclonal anti-actin antibody (Developmental Studies Hybridoma Bank, Iowa City, IA). TRIC protein abundance in cultured epithelia was normalized with actin. The use of actin for normalization was validated by statistically comparing actin abundance between experimental groups to confirm that no significant changes occurred. TRIC and actin protein abundance was quantified using a Molecular Imager Gel Doc XR+ System and Quantity One 1D analysis software (Bio-Rad Laboratories).
Effect of sodium caprate on TER, [3H]PEG-4000 flux, and TRIC localization in a cultured trout gill epithelium.
With the use of mature cultured gill epithelia that exhibited a plateau in TER (∼ 5 days after seeding cell culture inserts), sodium caprate (sodium decanoate, Sigma-Aldrich Canada) was added to apical media (3 mM) for a period of 2 h, after which sodium caprate-containing media were rinsed out (i.e., both apical and basolateral media were changed) and epithelia were allowed to recover for 3 h. During the course of the experiment, TER was monitored as previously described. [3H]PEG-4000 flux was examined during the period of sodium caprate exposure according to methods outlined previously. Control and sodium caprate-treated gill epithelia samples were fixed for immunocytochemical analysis at the end of the 2-h sodium caprate exposure period. Samples were processed for TRIC and cingulin immunohistochemistry as previously described.
Statistical analysis.
All data are expressed as mean values ± SE (n), where n represents the number of fish or, when cultured gill epithelia were used, the number of cell culture inserts. Either a Student's t-test or one-way or two-way ANOVA followed by a Student-Newman-Keuls test was used to determine significant differences (P < 0.05) between groups as appropriate. All statistical analyses were conducted using SigmaStat 3.5 software (Systat Software, San Jose, CA).
RESULTS
Characteristics of TRIC in rainbow trout.
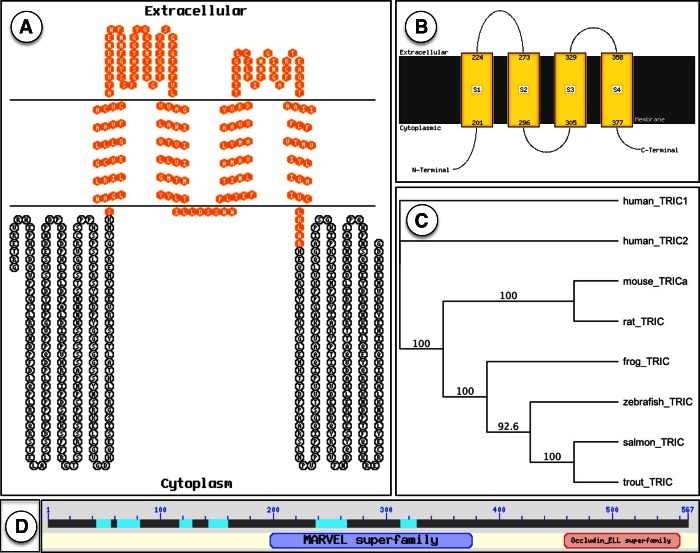
A single full coding sequence of rainbow trout TRIC was obtained by assembly of ESTs using NCBI EST databases. Extensive BLAST searches coupled with the use of multiple primer sets spanning the assembled TRIC gene indicated the presence of one single isoform of rainbow trout TRIC. Aligning rainbow trout TRIC nucleotide sequence to existing vertebrate TRIC protein sequences using a χBLAST engine revealed 98% similarity to Atlantic salmon MARVEL domain-containing protein 2 (=TRIC; accession no. NP001167127.1) and 55% sequence similarity to mouse tricellulin isoform a (accession number ABA18656.1), as illustrated with a bootstrapped phylogenetic tree (Fig. 1C). To confirm the presence of a continuous message encoding for TRIC in trout tissue, primers were designed to amplify regions within and across ESTs that were used to build the gene in silico using stock trout gill cDNA. Assembled sequence identity was confirmed by performing BLAST and χBLAST searches using the full coding sequence of TRIC. Rainbow trout TRIC (predicted ∼64 kDa) demonstrated more amino acid and nucleotide sequence similarity to closely related vertebrates. For example, trout TRIC demonstrated more sequence similarity to Atlantic salmon TRIC (98%) than to zebrafish TRIC (60%, NP001119878.1), frog (Xenopus tropicalis) TRIC (57%, NP001017292.1), or mammalian TRICs (55%, mouse, rat, and human; Fig. 1C). In silico analysis of encoded protein (using PSIPRED, ProtScale, and TMHMM databases) revealed a tetraspannin protein containing four transmembrane α-helices, two extracellular loops, and intracellular NH2 and COOH termini. ProtParam database was used to determine the primary structure of the protein, which was at least 567 residues with theoretical pI = 6.49 and possesses an estimated half-life of ∼30 h in an in vitro epithelial system. In silico analysis of the full rainbow trout TRIC sequence using EXPASY PROSITE database revealed a MARVEL domain and a COOH-terminal occludin-ELL homology domain, with sequence similarity to a variety of vertebrate TRIC homologues (visualized using TOPO2 software in Fig. 1A) as well as a proline-rich region (residues 45–76) and several putative PTM sites (such as protein kinase C, tyrosine kinase, and casein kinase II phosphorylation sites as well as several possible N-myristoylation sites along the amino acid sequence of the protein). Proline regions of many structural proteins have been related to a binding function or protein-protein interactions (57).
Fig. 1.
A: amino acid sequence of rainbow trout tricellulin (TRIC) with MARVEL domain highlighted in orange. B: membrane topology of amino acid sequence of TRIC. C: phylogenetic tree relating vertebrate TRIC amino acid sequences. Diagram illustrates evolutionary relatedness between human, mouse, rat, frog, zebrafish, Atlantic salmon, and rainbow trout TRIC amino acid sequences, with shorter human TRIC isoform 2 used as an outgroup (D) protein foldout generated by χBLAST search in the NCBI database; positions of the MARVEL and occludin-ELL superfamily domains are indicated.
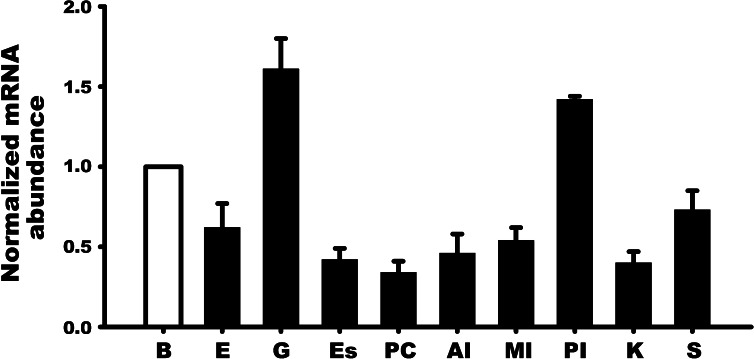
TRIC was observed in all tissues examined in this study. TRIC mRNA abundance was relatively similar in most cases, but in the gill and to some extent the posterior intestine, TRIC mRNA abundance was comparatively higher than other tissues (Fig. 2).
Fig. 2.
Expression profile of TRIC mRNA in discrete tissues of rainbow trout as determined by quantitative real-time PCR analysis. TRIC mRNA abundance was normalized with β-actin and expressed relative to the brain tissue assigned a value of 1.0. Data are expressed as mean values ± SE (n = 4). B, brain; E, eye; G, gill; Es, esophagus; PC, pyloric caeca; AI, anterior intestine; MI, mid-intestine; PI, posterior intestine; K, kidney; S, skin.
Western blot analysis and immunolocalization of TRIC in a cultured trout gill epithelium.
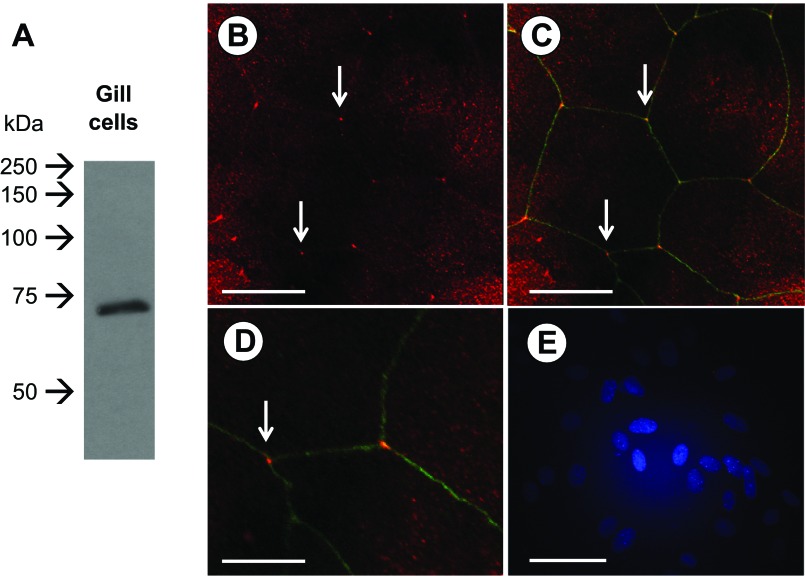
Western blot analysis of TRIC in gill tissue revealed a single immunoreactive band that resolved at ∼70 kDa (Fig. 3A). This same antibody localized to regions of tricellular contact in a cultured gill epithelium (Fig. 3, B–D). This was clearly delineated by cingulin immunoreactivity in regions of bicellular contact (Fig. 3, C and D). However, although TRIC immunoreactivity was strongest at regions of tricellular contact, faint bicellular immunoreactivity could also be observed (Fig. 3B). Junction staining was absent when primary antibodies were omitted from immunocytochemistry procedures (Fig. 3E).
Fig. 3.
Immunodetection and localization of TRIC in gill epithelia by Western blot analysis (A) and immunofluorescence (B–D). In A, a single immunoreactive band can be seen that resolved at ∼70 kDa. In B, TRIC can be seen (in red) localizing to areas of tricellular contact in gill epithelial cells (indicated by white arrows) and this is highlighted further (C and D) by cingulin immunoreactivity (in green), which localizes along areas of bicellular contact between gill epithelial cells. Negative control (primary antibody omitted) is shown in E where 4′,6-diamidino-2-phenylindole staining of nuclei can be seen in blue. Scale bars = 50 μm (B, C, and E) and 25 μm (D).
Effect of cortisol on permeability and TRIC abundance in a cultured trout gill epithelium.
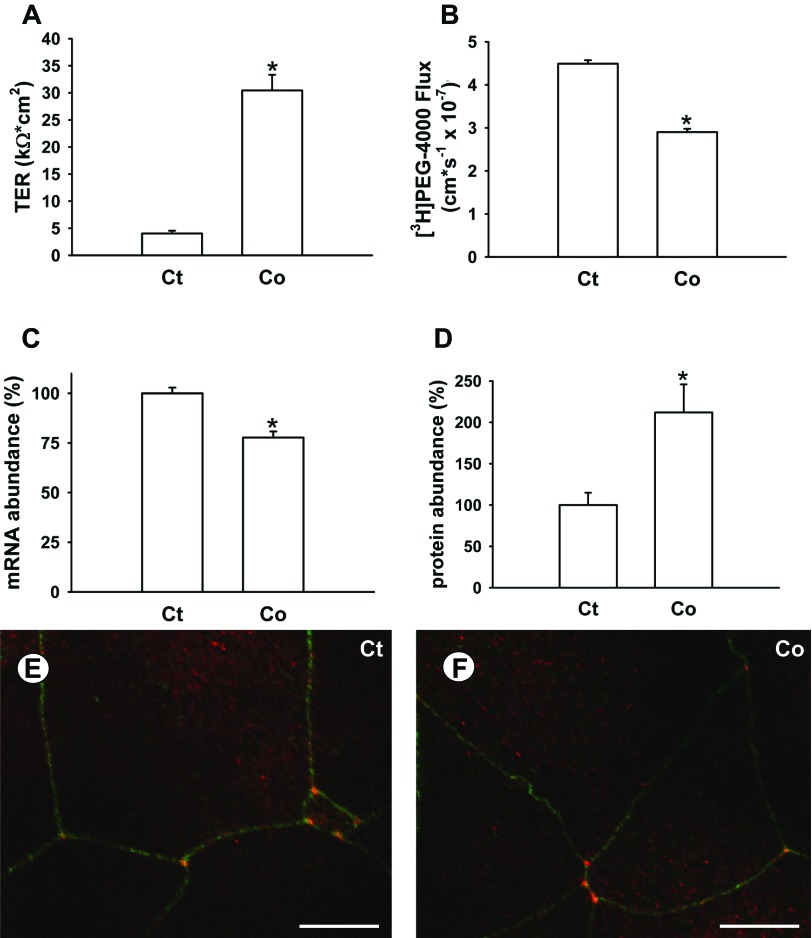
Cortisol treatment of cultured gill epithelia increased TER (Fig. 4A) and decreased [3H]PEG-4000 flux (Fig. 4B). TRIC mRNA abundance decreased slightly but significantly in response to cortisol treatment (Fig. 4C). In contrast, TRIC protein abundance exhibited a marked and significant increase in response to cortisol treatment (Fig. 4D). Cortisol treatment did not affect the localization of TRIC in the gill epithelium (Fig. 4, E and F).
Fig. 4.
Effect of cortisol (500 ng/ml) on transepithelial resistance (TER; A), [3H]PEG-4000 flux (B), TRIC mRNA (C), and protein abundance (D), as well as immunolocalization of TRIC in control (Ct; E) and cortisol-treated (Co; F) cultured gill epithelia derived from rainbow trout. Data are expressed as mean values ± SE (n = 12–20). *P < 0.05, significant difference between Ct and Co epithelia. Scale bar = 25 μm in E and F.
Effect of sodium caprate on permeability and TRIC immunolocalization in a cultured trout gill epithelium.
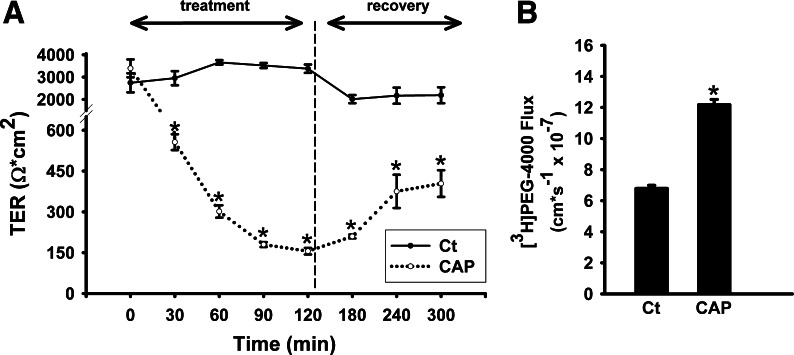
Sodium caprate treatment (2 h, 3 mM) caused a gradual decrease in TER from 3,390 ± 399 Ω·cm2 at 0-h exposure to 556 ± 29 Ω·cm2 after 1-h exposure, and down to 155 ± 12 Ω·cm2 at 2-h exposure (Fig. 5A). The gradual decrease in TER was accompanied by an approximate doubling of PEG-4000 flux from control values of 6.79 ± 0.20 cm/s × 10−7 to 12.18 ± 0.34 cm/s × 10−7 following 2 h of sodium caprate exposure (Fig. 5B). When sodium caprate was removed from cultured gill epithelia by rinsing and replacing media, epithelia began to recover as indicated by TER measurements that increased ∼2.6-fold over 3 h (Fig. 5A).
Fig. 5.
Effect of sodium caprate (CAP) on TER (A) and [3H]PEG-4000 flux (B) in cultured rainbow trout gill epithelia. CAP was added to apical media (final concentration of 3 mM) at 0 min and TER monitored for 120 min. Following this, medium bathing CAP treated epithelia were removed and replaced (after being rinsed) with Ct medium (indicated by vertical dashed line) to allow epithelia to recover. [3H]PEG-4000 flux was measured during the 120-min CAP treatment. Data are expressed as mean values ± SE (n = 10). *P < 0.05, significant difference between Ct and CAP-treated preparations.
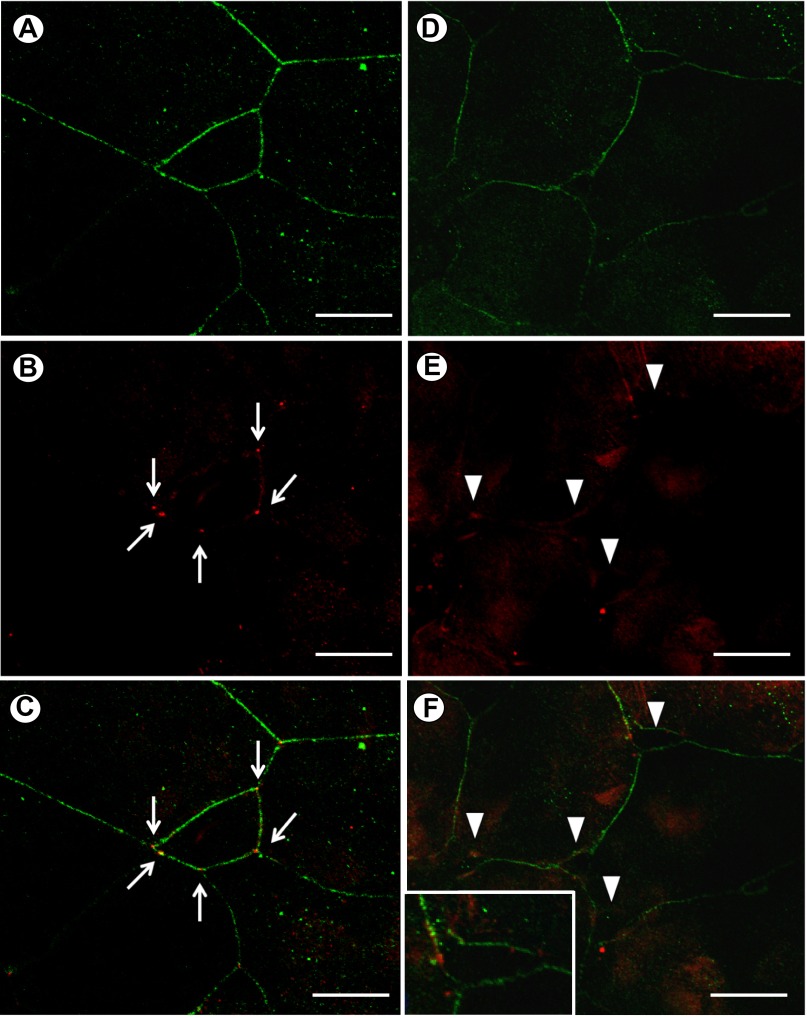
Following 2 h of exposure to sodium caprate, immunocytochemical observations revealed that regions of tricellular contact exhibited compromised integrity (Fig. 6). Specifically, instead of meeting at a single point of contact among three cells (see Fig. 6, A–C), regions of tricellular contact had become “unzipped” (see Fig. 6, D–F). Furthermore, although cingulin continued to stain the periphery of gill cells, no distinct point of TRIC immunoreactivity could be observed in sodium caprate treated cells (Fig. 6E). Instead, only faint diffuse staining could be observed along the cell periphery at regions that were formerly tricellular contact points.
Fig. 6.
Immunolocalization of TRIC and cingulin (CING) in control (A–C) and sodium CAP-treated (D–F) epithelia collected 120 min following the addition of CAP (final concentration of 3 mM) to apical media of cultured rainbow trout gill epithelia. In A, CING (in green) of control epithelia can be seen around the periphery of gill cells while distinct localization of TRIC (in red) can be seen at intact tricellular contact points (indicated by white arrows) in B and in C where CING and TRIC images are merged. In D, CING of CAP-treated epithelia can still be seen around the periphery of the cells, but in E, TRIC can no longer be seen at tricellular contact points because these have been disrupted (indicated by arrowheads). In F and its higher magnification (inset), a merged image of CING and TRIC reveals disrupted tricellular contact points and intact bicellular contact between gill epithelial cells. Scale bars in A–F = 50 μm.
DISCUSSION
Overview.
The current study supports the hypothesis that TRIC plays a significant role in maintaining the integrity of the gill epithelium of fishes as well as contributes to the regulation of paracellular permeability across this tissue. TRIC exhibited a distinct pattern of localization at regions of tricellular contact in a cultured gill epithelium, which is consistent with the presence of TRIC at the gill epithelium tTJ. When TRIC is displaced from the tTJ using sodium caprate, the integrity of the gill epithelium paracellular pathway (as measured by TER and [3H]PEG-4000 flux) is compromised. Furthermore, when the paracellular permeability of the gill epithelium is reduced in response to cortisol treatment, TRIC protein abundance is significantly elevated. In fishes, the role of cortisol as a stress hormone as well as an osmoregulatory hormone is well documented. Therefore, a cortisol-induced elevation in TRIC abundance at barrier weak points of tricellular contact in gill tissue would be advantageous as it could participate in the maintenance of gill epithelium integrity under stressful conditions and/or when osmoregulatory strategies require. Therefore, this study provides us with a first look at the potential importance of the tTJ complex in fish epithelia as well as the first functional insight into TRIC in the gill epithelium of fishes. In addition, this work also offers further insight into the molecular physiology of fish gill TJs and the endocrine control of gill epithelium permeability.
Characteristics of rainbow trout TRIC.
In silico analysis indicates that rainbow trout TRIC is encoded by a single exon, resulting in a nucleotide sequence with 98% sequence similarity to the gene encoding for Atlantic salmon MARVEL domain-containing protein 2. Protein sequence similarity to TRIC as described in other vertebrates (human, mouse, rat, frog, and zebrafish) ranged from 55 to 60%. These observations are consistent with those made for trout occludin, which exhibited 46–48% amino acid sequence identity to mammalian occludins, 50% amino acid sequence identity with frog occludin, and 63% identity to zebrafish occludin (10). Further information on rainbow trout TRIC as determined by in silico analysis of the deduced amino acid sequence characterized trout TRIC as a tetraspannin transmembrane protein with 1) MAL and related protein for vesicle trafficking and membrane link (MARVEL), and 2) occludin-ELL homology domains, both of which are characteristic of other vertebrate TRIC proteins. This is also characteristic of other members of the vertebrate TAMP subfamily such as occludin and marvelD3 (45; NCBI protein BLAST results). Analysis of the primary and secondary structure of TRIC using PSIPRED and TMHMM bioinformatic databases indicated that transmembrane elements of TRIC consist of α-helices. Amino acid sequence analysis indicated ample space for possible posttranslational modifications such as phosphorylation and myristoylation. TAMPs have been reported to be regulated through phosphorylation, and TRIC specifically is known to be phosphorylated during its transit from the cytosolic to the membrane-bound pool (15, 33, 52).
Distribution of TRIC in rainbow trout tissues.
An expression profile of mRNA encoding for TRIC revealed its presence in all rainbow trout tissues examined in this study. Furthermore, TRIC mRNA abundance was similar in most regions examined, although the gill (and to a lesser extent the esophagus) exhibited moderately elevated levels of transcript. Broad expression and relatively constant abundance are not unexpected for a protein that localizes to regions of tricellular contact in epithelial tissue as all epithelia will have these. In fishes, another member of the TAMP subfamily, occludin, has also been found in a broad array of tissues (7, 10). In addition, broad TRIC expression in trout was comparable to reports in mammals (26, 33, 35, 47, 50), where it has been found in testis, small intestine, kidney, lung, liver, brain, inner ear, retina, epidermal cells, and pancreas. Furthermore, cytosolic TJ proteins such as zonula occludens 1 (ZO-1) also show broad and consistent expression patterns in fish tissues (8). Although ZO-1 is not a member of the TAMP subfamily, it is, like TRIC, an integral component of all epithelial TJs and as such exhibits a broad expression pattern not unlike that seen for TRIC. In contrast, claudins exhibit expression patterns in fishes that often reveal the absence of protein in discrete tissues (e.g., Refs. 1, 2, 8, 14, 40, 53). In a recent study, a broad expression pattern for a partial clone of TRIC in Atlantic salmon has also been described, although transcript abundance seemed to vary between tissues in this species (54).
Immunoblotting and localization of TRIC in a trout gill epithelium.
Western blot analysis of TRIC in cultured gill epithelial cells revealed a single immunoreactive band that resolved at ∼70 kDa (Fig. 3A). This is consistent with mammalian TRIC, which was reported to fall within a similar size range, between 66 and 72 kDa (26). Localization of TRIC revealed prominent immunoreactivity in regions of the cultured gill epithelium where three cells converged (i.e., regions of tricellular contact). This strongly supports the idea that TRIC localizes to the tTJ complex of the gill epithelium. TRIC is conventionally reported to be a tTJ marker in many vertebrate epithelia (26, 27, 35, 36, 43, 46, 47). However, faint bTJ TRIC immunoreactivity is also present in select epithelia (26). In the cultured gill epithelium, very faint bTJ immunoreactivity could sometimes also be observed. This would suggest that although TRIC appears to be prominent in the tTJ of the gill epithelium, it might also be weakly involved in the bTJ complex of the fish gill epithelium.
Effect of cortisol on gill epithelium permeability and TRIC abundance.
Cortisol reduced the permeability of cultured trout gill epithelia as indicated by elevated TER (Fig. 4A). This was at least partly attributable to a decrease in paracellular permeability as the flux rate of the paracellular permeability marker PEG-4000 was significantly decreased in the presence of cortisol (Fig. 4B). The “tightening” effect of cortisol on primary cultured gill epithelia has previously been documented in trout (10, 30, 59, 60) as well as cultured gill epithelia derived from other fish species (7, 31). In cultured gill epithelia, corticosteroid-induced reductions in paracellular permeability are associated with elevated protein and/or transcript levels of bTJ proteins such as occludin (10, 29), various claudins (4, 7, 29), and the cytosolic scaffolding TJ protein ZO-1 (29). In the current study, TRIC protein also significantly elevated in response to cortisol treatment (Fig. 4D) and this would suggest that cortisol can reduce paracellular permeability and/or improve the integrity of the gill epithelium by altering the molecular physiology of both bTJ and tTJ complexes. Curiously, transcript abundance of TRIC marginally (but significantly) declined in response to cortisol treatment (Fig. 4C). Given that Western blot analysis reflects a measure of how much functional (or at least properly folded) TRIC the cells have accumulated, increased TRIC abundance in cortisol-treated epithelia indicates that these cells have accumulated more TRIC than control epithelia following ∼7 days of hormone exposure. In contrast, reduced TRIC mRNA abundance in cortisol-treated preparations could suggest that at the same point in time, TRIC was being transcribed and translated at a lower rate compared with controls. In silico analysis of rainbow trout TRIC indicates that it has a comparatively long half-life of 30 h in an in vitro system. Therefore, it is not unreasonable to suggest that following 7 days of cortisol treatment, transcription of TRIC may be downregulated while stable TRIC protein has accumulated at the tTJ. Such divergent correlation of mRNA and protein abundance has been studied in a genomic context in prokaryotes and eukaryotes and is postulated to be a consequence of mRNA-regulatory mechanisms (3, 20, 28). Another possibility is that an increased presence of a TRIC cofactor such as lipolysis-stimulated lipoprotein receptor (LSR) may have increased TRIC stability (25, 43). However, nothing is known about LSR in fishes and this idea will require further study.
Effect of sodium caprate on gill epithelium permeability and TRIC localization.
A drug absorption enhancer by design, sodium caprate has been reported to cause a rapid and reversible increase in the permeability of cultured epithelial models (36, 48). In addition, sodium caprate has recently been reported to compromise the integrity of a human intestinal cell line (HT-29/B6) by displacing TRIC from the tTJ (36). Treatment of trout gill epithelia with sodium caprate reversibly decreased TER and increased paracellular permeability (Fig. 5), which is consistent with observations in other models (36, 48). Furthermore, sodium caprate treatment also appeared to cause gill epithelial cells to detach from one another at tricellular regions and associated with this was a displacement of TRIC (Fig. 6). This would suggest that sodium caprate treatment results in TRIC movement away from tTJs in the gill epithelium. However, despite sodium caprate-induced TRIC displacement from areas of tricellular contact in the gill epithelium, cingulin immunoreactivity remained along the cell periphery in these “unzipped” areas as well as in regions of bicellular contact, where bTJs remained intact (Fig. 6). This provides evidence that the sodium caprate-induced changes observed in TER and PEG-4000 flux can be at least partly attributable to a compromised tTJ.
Perspectives and Significance
The importance of the gill as a barrier to passive solute movement is broadly acknowledged to play a key role in the ability of fishes to maintain salt and water balance in an aquatic setting. In turn, the significance of the gill TJ complex as a vital component of the gill barrier, either to impede solute movement or to act as a selectively permeable secretory pathway, has been recognized for some time (for review see Ref. 12). However, gill TJs have only received attention from a bicellular perspective, as we are unaware of any attempt to examine the importance of the tTJ complex or the contribution that its molecular architecture may make to gill epithelium integrity. Greater opportunity to consider the tTJ complex has arisen due to the discovery of tTJ proteins that include TRIC (26), LSR (43), and the LSR-related proteins immunoglobulin-like domain-containing receptor 1 and 2 (ILDR-1 and ILDR-2; Ref. 25), and the current work offers a first insight into the potential importance of the tTJ complex and its molecular components in the gill epithelium of fishes. This sets the stage for future studies that will significantly expand our understanding of what roles the tTJ complex and its proteins may play in regulating (or contributing to the regulation of) the barrier properties of the gill epithelium. Future studies that broaden our knowledge of how tTJ proteins such as TRIC may contribute to gill function in accord with altered environmental conditions such as salinity change, or among heterogeneous gill cell types such as mitochondria rich cells, accessory cells, as well as pavement cells, will be of particular interest.
GRANTS
This work was supported by a Natural Sciences and Engineering Research Council Discovery Grant and Discovery Accelerator Supplement (to S. P. Kelly). D. Kolosov was the recipient of an Ontario Graduate Scholarship.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: D.K. and S.P.K. conception and design of research; D.K. and S.P.K. performed experiments; D.K. and S.P.K. analyzed data; D.K. and S.P.K. interpreted results of experiments; D.K. and S.P.K. prepared figures; D.K. and S.P.K. drafted manuscript; D.K. and S.P.K. edited and revised manuscript; D.K. and S.P.K. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank Dr. Susanne Krug for helpful discussions and advice on work with caprate, as well as Dr. Helen Chasiotis and Phuong Bui for insightful discussion. The mouse monoclonal anti-actin antibody (JLA20) was developed by Jim Jung-Chin Lin and obtained from the Developmental Studies Hybridoma Bank developed under National Institute of Child Health and Human Development and maintained by the Department of Biology, University of Iowa.
REFERENCES
- 1. Bagherie-Lachidan M, Wright SI, Kelly SP. Claudin-3 tight junction proteins in Tetraodon nigroviridis: cloning, tissue-specific expression, and a role in hydromineral balance. Am J Physiol Regul Integr Comp Physiol 294: R1638–R1647, 2008 [DOI] [PubMed] [Google Scholar]
- 2. Bagherie-Lachidan M, Wright SI, Kelly SP. Claudin-8 and -27 tight junction proteins in puffer fish Tetraodon nigroviridis acclimated to freshwater and seawater. J Comp Physiol B 179: 419–431, 2009 [DOI] [PubMed] [Google Scholar]
- 3. Bevilacqua A, Ghisolfi L, Franzi S, Maresca G, Gherzi R, Capaccioli S, Nicolin A, Canti G. Stabilization of cellular mRNAs and up-regulation of proteins by oligoribonucleotides homologous to the Bcl2 adenine-uridine rich element motif. Mol Pharmacol 71: 531–538, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Bui P, Bagherie-Lachidan M, Kelly SP. Cortisol differentially alters claudin isoforms in cultured puffer fish gill epithelia. Mol Cell Endocrinol 317: 120–126, 2010 [DOI] [PubMed] [Google Scholar]
- 5. Chasiotis H, Kelly SP. Occludin immunolocalization and protein expression in goldfish. J Exp Biol 211: 1524–1534, 2008 [DOI] [PubMed] [Google Scholar]
- 6. Chasiotis H, Kelly SP. Permeability properties and occludin expression in a primary cultured model gill epithelium from the stenohaline freshwater goldfish. J Comp Physiol B 181: 487–500, 2011 [DOI] [PubMed] [Google Scholar]
- 7. Chasiotis H, Kelly SP. Effect of cortisol on permeability and tight junction protein transcript abundance in primary cultured gill epithelia from stenohaline goldfish and euryhaline trout. Gen Comp Endocrinol 172: 494–504, 2011 [DOI] [PubMed] [Google Scholar]
- 8. Chasiotis H, Kelly SP. Effects of elevated circulating cortisol levels on hydromineral status and gill tight junction protein abundance in the stenohaline goldfish. Gen Comp Endocrinol 175: 277–283, 2012 [DOI] [PubMed] [Google Scholar]
- 9. Chasiotis H, Effendi JC, Kelly SP. Occludin expression in goldfish held in ion-poor water. J Comp Physiol B 179: 145–154, 2008 [DOI] [PubMed] [Google Scholar]
- 10. Chasiotis H, Wood CM, Kelly SP. Cortisol reduces paracellular permeability and increases occludin abundance in cultured trout gill epithelia. Mol Cell Endocrinol 323: 232–238, 2010 [DOI] [PubMed] [Google Scholar]
- 11. Chasiotis H, Kolosov D, Kelly SP. Permeability properties of the teleost gill epithelium under ion-poor conditions. Am J Physiol Regul Integr Comp Physiol 302: R727–R739, 2012 [DOI] [PubMed] [Google Scholar]
- 12. Chasiotis H, Kolosov D, Bui P, Kelly SP. Tight junctions, tight junction proteins and paracellular permeability across the gill epithelium of fishes: a review. Respir Physiol Neurobiol 184: 269–281, 2012 [DOI] [PubMed] [Google Scholar]
- 13. Chishti MS, Bhatti A, Tamim S, Lee K, McDonald ML, Leal SM, Ahmad W. Splice-site mutations in the TRIC gene underlie autosomal recessive nonsyndromic hearing impairment in Pakistani families. J Hum Genet 53: 101–105, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clelland ES, Kelly SP. Tight junction proteins in zebrafish ovarian follicles: stage specific mRNA abundance and response to 17β-estradiol, human chorionic gonadotropin, and maturation inducing hormone. Gen Comp Endocrinol 168: 388–400, 2010 [DOI] [PubMed] [Google Scholar]
- 15. Dörfel MJ, Huber O. Modulation of tight junction structure and function by kinases and phosphatases targeting occludin. J Biomed Biotech 2012: 807356, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duffy NM, Bui P, Bagherie-Lachidan M, Kelly SP. Epithelial remodeling and claudin mRNA abundance in the gill and kidney of puffer fish (Tetraodon biocellatus) acclimated to altered environmental ion levels. J Comp Physiol B 181: 219–238, 2010 [DOI] [PubMed] [Google Scholar]
- 17. Engelund MB, Yu ASL, Li J, Madsen SS, Faergeman NJ, Tipsmark CK. Functional characterization and localization of a gill-specific claudin isoform in Atlantic salmon. Am J Physiol Regul Integr Comp Physiol 302: R300–R311, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Evans DH, Piermarini PM, Choe KP. The multifunctional fish gill: Dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85: 97–177, 2005 [DOI] [PubMed] [Google Scholar]
- 19. Farquhar MG, Palade GE. Junctional complexes in various epithelia. J Cell Biol 17: 375–412, 1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fournier ML, Paulson A, Pavelka N, Mosley AL, Gaudenz K, Bradford WD, Glynn E, Li H, Sardiu ME, Fleharty B, Seidel C, Florens L, Washburn MP. Delayed correlation of mRNA and protein expression in rapamycin-treated cells and a role for Ggc1 in cellular sensitivity to rapamycin. Mol Cell Proteomics 9: 271–284, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fukumatsu M, Ogawa M, Arakawa S, Suzuki M, Nakayama K, Shimizu S, Kim M, Mimuro H, Sasakawa C. Shigella targets epithelial tricellular junctions and uses a noncanonical clathrin-dependent endocytic pathway to spread between cells. Cell Host Microbe 11: 325–336, 2012 [DOI] [PubMed] [Google Scholar]
- 22. Furuse M, Oda Y, Higashi T, Iwamoto N, Masuda S. Lipolysis-stimulated lipoprotein receptor: a novel membrane protein of tricellular tight junctions. Ann NY Acad Sci 1257: 54–58, 2012 [DOI] [PubMed] [Google Scholar]
- 23. González-Mariscal L, Betanzos A, Nava P, Jaramillo BE. Tight junction proteins. Prog Biophys Mol Biol 81: 1–44, 2003 [DOI] [PubMed] [Google Scholar]
- 24. Günzel D, Fromm M. Claudins and other tight junction proteins. Compr Physiol 2: 1819–1852, 2012 [DOI] [PubMed] [Google Scholar]
- 25. Higashi T, Tokuda S, Kitajiri SI, Masuda S, Nakamura H, Oda Y, Furuse M. Analysis of the angulin family consisting of LSR, ILDR1 and ILDR2: tricellulin recruitment, epithelial barrier function and implication in deafness pathogenesis. J Cell Sci 126: 966–977, 2013 [DOI] [PubMed] [Google Scholar]
- 26. Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. J Cell Biol 171: 939–945, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S. Loss of occludin affects tricellular localization of tricellulin. Mol Biol Cell 19: 4687–4693, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jayapal KP, Philp RJ, Kok YJ, Yap MG, Sherman DH, Griffin TJ, Hu WS. Uncovering genes with divergent mRNA-protein dynamics in Streptomyces coelicolor. PLoS One 3: 2097, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kelly SP, Chasiotis H. Glucocorticoid and mineralocorticoid receptors regulate paracellular permeability in a primary cultured gill epithelium. J Exp Biol 214: 2308–2318, 2011 [DOI] [PubMed] [Google Scholar]
- 30. Kelly SP, Wood CM. Effect of cortisol on the physiology of cultured pavement cell epithelia from freshwater trout gills. Am J Physiol Regul Integr Comp Physiol 281: R811–R820, 2001 [DOI] [PubMed] [Google Scholar]
- 31. Kelly SP, Wood CM. Cultured gill epithelia from freshwater tilapia (Oreochromis niloticus): effect of cortisol and homologous serum supplements from stressed and unstressed fish. J Membr Biol 190: 29–42, 2002 [DOI] [PubMed] [Google Scholar]
- 32. Kelly SP, Fletcher M, Part P, Wood CM. Procedures for the preparation and culture of “reconstructed” rainbow trout branchial epithelia. Methods Cell Sci 22: 153–163, 2000 [DOI] [PubMed] [Google Scholar]
- 33. Kojima T, Fuchimoto J, Yamaguchi H, Ito T, Takasawa A, Ninomiya T, Kikuchi S, Ogasawara N, Ohkuni T, Masaki T, Hirata K, Himi T, Sawada N. c-Jun N-terminal kinase is largely involved in the regulation of tricellular tight junctions via tricellulin in human pancreatic duct epithelial cells. J Cell Physiol 225: 720–733, 2010 [DOI] [PubMed] [Google Scholar]
- 34. Korompay A, Borka K, Lotz G, Somorácz Á, Törzsök P, Erdélyi-Belle B, Kenessey I, Baranyai Z, Zsoldos F, Kupcsulik P, Bodoky G, Schaff Z, Kiss A. Tricellulin expression in normal and neoplastic human pancreas. Histopathology 60: E76–E86, 2012 [DOI] [PubMed] [Google Scholar]
- 35. Krug SM, Amasheh S, Richter JF, Milatz S, Günzel D, Westphal JK, Huber O, Schulzke JD, Fromm M. Tricellulin forms a barrier to macromolecules in tricellular tight junctions without affecting ion permeability. Mol Biol Cell 20: 3713–3724, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Krug SM, Amasheh M, Dittmann I, Christoffel I, Fromm M, Amasheh S. Sodium caprate as an enhancer of macromolecule permeation across tricellular tight junctions of intestinal cells. Biomaterials 34: 275–282, 2013 [DOI] [PubMed] [Google Scholar]
- 37. Kumai Y, Bahubeshi A, Steele S, Perry SF. Strategies for maintaining Na+ balance in zebrafish (Danio rerio) during prolonged exposure to acidic water. Comp Physiol Biochem A Mol Integr Physiol 160: 52–62, 2011 [DOI] [PubMed] [Google Scholar]
- 38. Kwong RWM, Kumai Y, Perry SF. Evidence for a role of tight junctions in regulating sodium permeability in zebrafish (Danio rerio) acclimated to ion-poor water. J Comp Physiol B 183: 203–213, 2012 [DOI] [PubMed] [Google Scholar]
- 39. Leong JS, Jantzen SG, Schalburg von KR, Cooper GA, Messmer AM, Liao NY, Munro S, Moore R, Holt RA, Jones SJ, Davidson WS, Koop BF. Salmo salar and Esox lucius full-length cDNA sequences reveal changes in evolutionary pressures on a post-tetraploidization genome. BMC Genomics 11: 279, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Loh YH, Christoffels A, Brenner S, Hunziker W, Venkatesh B. Extensive expansion of the claudin gene family in the teleost fish, Fugu rubripes. Genome Res 14: 1248–1257, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mariano C, Sasaki H, Brites D, Brito MA. A look at tricellulin and its role in tight junction formation and maintenance. Eur J Cell Biol 90: 787–796, 2011 [DOI] [PubMed] [Google Scholar]
- 42. Masuda R, Semba S, Mizuuchi E, Yanagihara K, Yokozaki H. Negative regulation of the tight junction protein tricellulin by snail-induced epithelial-mesenchymal transition in gastric carcinoma cells. Pathobiology 77: 106–113, 2010 [DOI] [PubMed] [Google Scholar]
- 43. Masuda S, Oda Y, Sasaki H, Ikenouchi J, Higashi T, Akashi M, Nishi E, Furuse M. LSR defines cell corners for tricellular tight junction formation in epithelial cells. J Cell Sci 124: 548–555, 2011 [DOI] [PubMed] [Google Scholar]
- 44. Ohkuni T, Kojima T, Ogasawara N, Masaki T, Ninomiya T, Kikuchi S, Go M, Takano KI, Himi T, Sawada N. Expression and localization of tricellulin in human nasal epithelial cells in vivo and in vitro. Med Mol Morphol 42: 204–211, 2009 [DOI] [PubMed] [Google Scholar]
- 45. Pinto PI, Matsumura H, Thorne MA, Power DM, Terauchi R, Reinhardt R, Canário AV. Gill transcriptome response to changes in environmental calcium in the green spotted puffer fish. BMC Genomics 11: 476, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Raleigh DR, Marchiando AM, Zhang Y, Shen L, Sasaki H, Wang Y, Long M, Turner JR. Tight junction-associated MARVEL proteins marveld3, tricellulin, and occludin have distinct but overlapping functions. Mol Biol Cell 21: 1200–1213, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Riazuddin S, Ahmed ZM, Fanning AS, Lagziel A, Kitajiri SI, Ramzan K, Khan SN, Chattaraj P, Friedman PL, Anderson JM, Belyantseva IA, Forge A, Riazuddin S, Friedman TB. Tricellulin is a tight-junction protein necessary for hearing. Am J Hum Genet 79: 1040–1051, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rosenthal R, Heydt MS, Amasheh M, Stein C, Fromm M, Amasheh S. Analysis of absorption enhancers in epithelial cell models. Ann NY Acad Sci 1258: 86–92, 2012 [DOI] [PubMed] [Google Scholar]
- 49. Sandbichler AM, Egg M, Schwerte T, Pelster B. Claudin 28b and F-actin are involved in rainbow trout gill pavement cell tight junction remodeling under osmotic stress. J Exp Biol 214: 1473–1487, 2011 [DOI] [PubMed] [Google Scholar]
- 50. Schlüter H, Moll I, Wolburg H, Franke WW. The different structures containing tight junction proteins in epidermal and other stratified epithelial cells, including squamous cell metaplasia. Eur J Cell Biol 86: 645–655, 2007 [DOI] [PubMed] [Google Scholar]
- 51. Staehelin LA. Further observations on the fine structure of freeze-cleaved tight junctions. J Cell Sci 13: 763–786, 1973 [DOI] [PubMed] [Google Scholar]
- 52. Takasawa A, Kojima T, Ninomiya T, Tsujiwaki M, Murata M, Tanaka S, Sawada N. Behavior of tricellulin during destruction and formation of tight junctions under various extracellular calcium conditions. Cell Tiss Res 351: 73–84, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tipsmark CK, Kiilerich P, Nilsen TO, Ebbesson LOE, Stefansson SO, Madsen SS. Branchial expression patterns of claudin isoforms in Atlantic salmon during seawater acclimation and smoltification. Am J Physiol Regul Integr Comp Physiol 294: R1563–R1574, 2008 [DOI] [PubMed] [Google Scholar]
- 54. Tipsmark CK, Madsen SS. Tricellulin, occludin and claudin-3 expression in salmon intestine and kidney during salinity adaptation. Comp Biochem Physiol A 162: 378–385, 2012 [DOI] [PubMed] [Google Scholar]
- 55. Van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annu Rev Physiol 68: 403–429, 2006 [DOI] [PubMed] [Google Scholar]
- 56. Whitehead A, Roach JL, Zhang S, Galvez F. Genomic mechanisms of evolved physiological plasticity in killifish distributed along and environmental salinity gradient. Proc Natl Acad Sci USA 108: 6193–6198, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J 297: 249–260, 1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wood CM, Gilmour KM, Pärt P. Passive and active transport properties of a gill model, the cultured branchial epithelium of the freshwater rainbow trout (Oncorhynchus mykiss). Comp Biochem Physiol A Mol Integr Physiol 119: 87–96, 1998 [DOI] [PubMed] [Google Scholar]
- 59. Wood CM, Kelly SP, Zhou B, Fletcher M, O'Donnell MJ, Eletti B, Pärt P. Cultured gill epithelia as models for the freshwater fish gill. BBA - Biomembranes 1566, 72–83, 2002. [DOI] [PubMed] [Google Scholar]
- 60. Zhou B, Kelly SP, Ianowski JP, Wood CM. Effects of cortisol and prolactin on Na+ and Cl− transport in cultured branchial epithelia from FW rainbow trout. Am J Physiol Regul Integr Comp Physiol 285: R1305–R1316, 2003 [DOI] [PubMed] [Google Scholar]