Abstract
Safety is the foremost issue in all human cell therapies, but human induced pluripotent stem cells (iPSCs) currently lack a useful safety indicator. Studies in chimeric mice have demonstrated that certain lines of iPSCs are tumorigenic; however a similar screen has not been developed for human iPSCs. Here, we show that in vitro cartilage tissue engineering is an excellent tool for screening human iPSC lines for tumorigenic potential. Although all human embryonic stem cells (ESCs) and most iPSC lines tested formed cartilage safely, certain human iPSCs displayed a pro-oncogenic state, as indicated by the presence of secretory tumors during cartilage differentiation in vitro. We observed five abnormal iPSC clones amoungst 21 lines derived from five different reprogramming methods using three cellular origins. We conclude that in vitro cartilage tissue engineering is a useful approach to identify abnormal human iPSC lines.
Human induced pluripotent stem cell (iPSC) technology has great potential for cell therapy1,2; however a risk of tumorigenicity has been identified. Indeed, incidents of tumor formation have been observed in chimeric mice derived from mouse iPSCs due to the presence of the pro-oncogene, c-MYC, in reprogramming3,4,5. In the undifferentiated state, the pro-oncogenic nature of iPSC lines is not entirely predictable as global gene expression patterns and their epigenetic state in iPSCs are similar to that in embryonic stem cells (ESCs)6,7. Like ESCs, when iPSCs are transplanted into animals, they will form teratomas; however, teratoma formation cannot predict the pro-oncogenicity of select iPSCs. Currently, chimera formation in mouse is a reliable approach to identify pro-oncogenic iPSCs, but this method cannot be practiced using human iPSCs. In addition, tests in animal models can be time consuming. For example teratoma formation, which is not a test for oncogenic potential can take between one and ten months8. Therefore, a quick and reliable surrogate test for oncogenicity in human iPSCs needs to be developed.
The in vitro differentiation of iPSCs may provide an alternative approach to evaluating tumorigenic risk. We have focused on using cartilage tissue engineering as a differentiation model for ESCs, since functional 3D cartilage tissues can be formed in vitro with simple and robust protocols9,10. Unique and uniform morphology of cartilage tissue helps to identify any abnormal cell growth or differentiation by histological analysis. Here, we report that cartilage tissue engineering using iPSCs is an excellent method for identifying cell lines, which display abnormality during differentiation.
Results
Human ESCs and iPSCs differentiate into the chondrocyte lineage
Previously, we have demonstrated cartilage tissue engineering as a differentiation model for mouse ESCs. In this study, we modified this method for human pluripotent stem cells. For chondrogenic differentiation, high-cell density micro-mass culture was treated with low concentration of FBS, Insulin-Transferrin-Selenium-X supplement, BMP-2, TGF-β1, and ascorbic acid, as previously described11. We performed chondrogenic differentiation experiments using 6 human ESC lines and 21 human iPSC lines derived from 5 different methods: i) retrovirus 4F (OSKM) with EOS reporter12, ii) retrovirus 4F (OSKM)[1], iii) retrovirus 3F (OSK, without c-MYC)4, iv) retrovirus L-Myc (OSKL, L-MYC instead of c-MYC)5 and v) integration-free episomal 6F (OSKLp53LIN)13 and 3 cellular origins, i) infant fibroblasts, ii) adult fibroblasts and iii) adult dental pulp (Supplementary Table 1).
Human ESCs formed cartilage-like tissues in vitro and in vivo
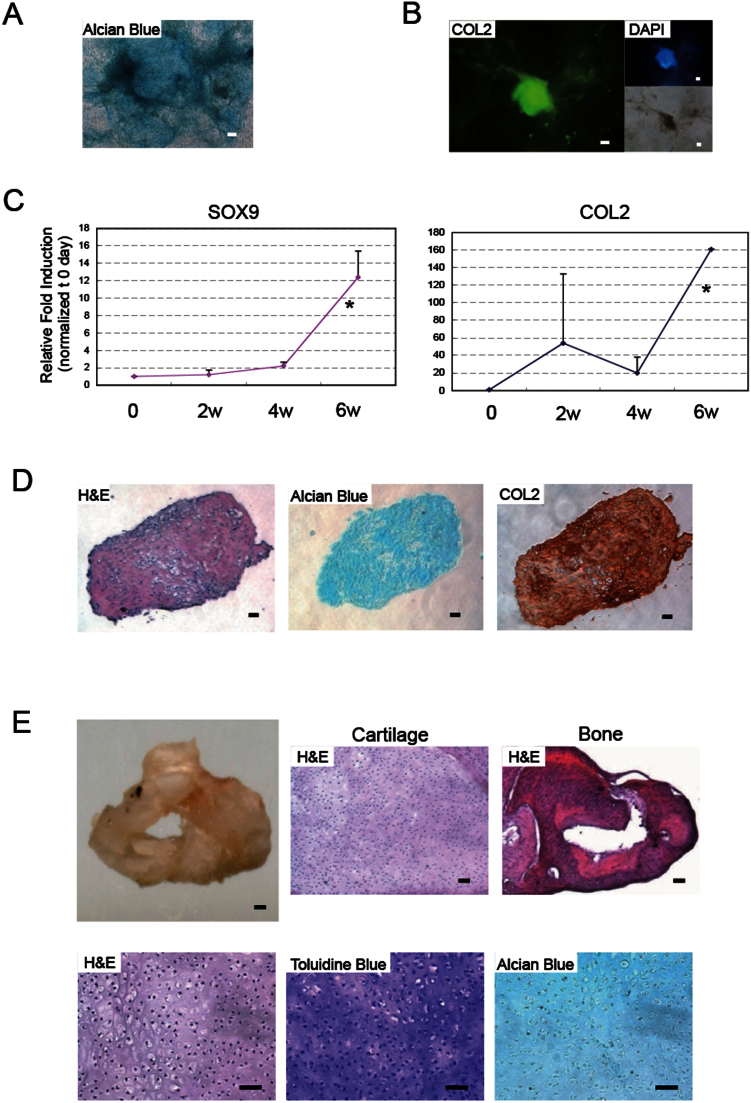
Following micro-mass chondrogenic differentiation, human ESCs formed aggregates, and expressed cartilage matrix proteoglycans as detected by Alcian Blue staining and collagen 2 (COL2) as detected by immunohistochemistry (Figure 1A and B). Differentiated ESCs expressed chondrocyte-related genes, SOX9 and COL2 (Figure 1C). Following 6 weeks of culture, human ESCs formed the phenotypic hallmarks of cartilage, as indicated by histological (H&E, and Alcian Blue) and immunohistochemical (COL2) staining (Figure 1D).
Figure 1. Human ESCs form cartilage.
(A) Chondrogenic differentiation of ESCs stained by Alcian Blue following 2 weeks of culture. (B) Expression of the chondrocyte marker, COL2 following 2 weeks of differentiation culture. Immunofluorescence of COL2 (left), DAPI (right, top) and brightfield (right, bottom). Scale bars represent 50 μm. (C) Expression of chondrocyte-related genes by real-time PCR: SOX9 (left), AGGRECAN (middle), COL 2 (right). Data is expressed as mean ± SEM (N = 3) per lane. *indicates significance; P < 0.05. (D) Histological section of ESC-derived aggregates following 6 weeks of differentiation: H&E (left), Alcian Blue (middle), COL2 immunostaining (right). (E) Transplantation of aggregates. ESC-derived hard tissue (top, left); scale bar represents 100 μm. Cartilage (top, middle) and bone (top, right) stained by H&E. Cartilage (bottom) at high magnification. H&E (left). Toluidine Blue (middle) and Acian Blue (right) stain cartilage purple and blue respectively. Scale bars represent 50 μm throughout.
In order to characterize the ESC-derived cartilage in vivo, transplants were performed subcutaneously into SCID mice. Six months after transplantation, not only was cartilage found, as indicated by Toluidine Blue and Alcian Blue staining, but so was bone, as indicated by Methylene Blue staining following transplantation (Figure 1E. Supplementry Figure 1). We attribute this to endochondral ossification, a natural process whereby cartilage is replaced by bone tissue following hypertrophy and degradation. These results indicated that human ESCs could generate cartilage safely in vitro and in vivo.
Certain human iPSCs formed tumors during cartilage differentiation in vitro
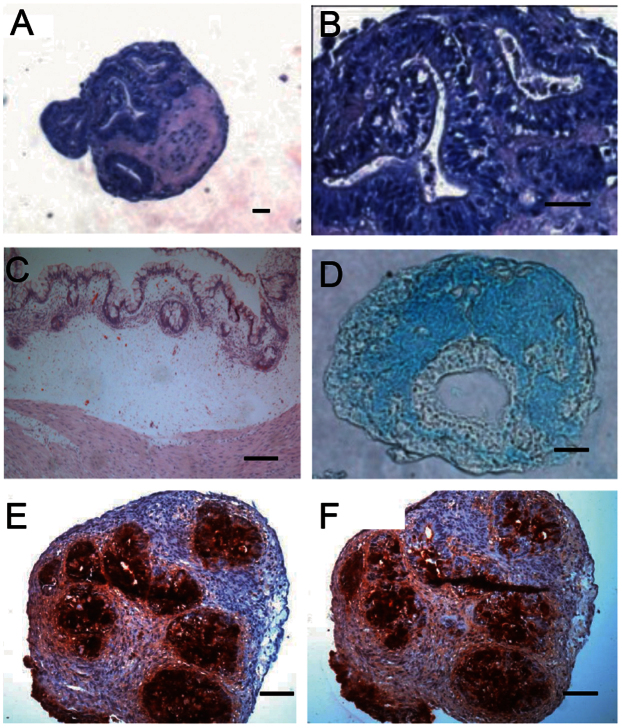
Similar to human ESCs, all iPSC lines differentiated into the chondrocyte lineage as indicated by Alcian Blue and COL2 staining (Supplementry Figure 2). Although certain human iPSC lines could form cartilage following 6 weeks of culture, a few iPSC lines diplay glandular epithelial cysts complete with columnar epithelium following cartilage differentiation (Figure 2). This glandular epithelium region displayed a lack of cartilage matrix/proteoglycan and expressed carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) (Figure 2), tumor markers of adenocarcinoma prevalent in colorectal or gastric cancer14. Following transplantation into immuno-compromised mice, this iPSC-derived tissue formed glandular epithelial tumors in vivo (Figure 2). When cultured in an undifferentiated state, these iPSC lines did not display any abnormalities in cell morphology, plating efficiency, cell expansion karyotype karyotype (Supplementary Figure 3) or teratoma forming ability (Supplementary Figure 5). These results indicated that certain iPSC lines might have pro-oncogenic potential, which is revealed following differentiation into cartilage.
Figure 2. Glandular tumor formation during cartilage formation in certain iPSC line.
(A, B) Low and high magnification of 6 week in vitro differentiated iPSCs aggregate stained with H&E. (C) H&E staining of in vivo glandular tumor three months after transplantation into SCID mice, (D–F) In vitro differentiated aggregates stained with alcian blue (D) or glandular tumor markers CEA (E) and CA19-9 (F). Scale bars represent 50 μm.
Identify abnormal differentiation of human iPSC clones through in vitro cartilage tissue engineering
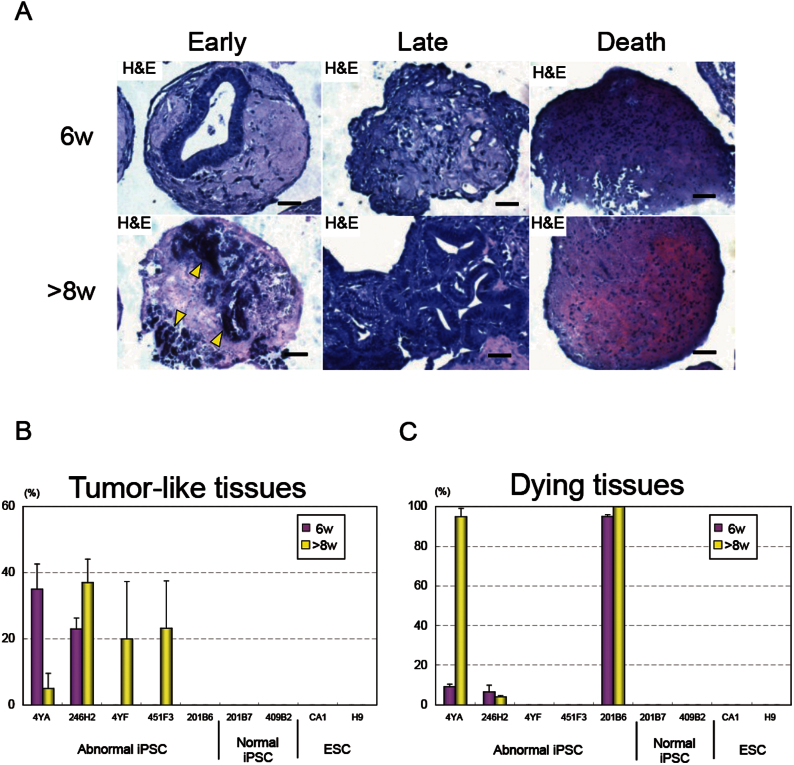
Using in vitro cartilage tissue engineering, we could identify abnormal iPSC clones. As a typical case, one such abnormal line (BJ-EOS-4YA) displayed glandular epithelial cysts in approximately 35% of micro-mass aggregates following six weeks of diffrentiation into cartilage (Figure 3A, Supplementary Table 2). This, and one other line (246H2) were identified as “early” tumorigenic lines, because the abnormality appeared at a relatively early stage during cartilage differentiation (Figure 3B, Table 1, Supplementry Figure 4).
Figure 3. Identification abnormal iPSC clones through in vitro cartilage tissue engineering.
(A) Three types of abnormal differentiation patterns, i) tumor formation and subsequent death (left), ii) late blooming tumor formation (middle), iii) direct death (right), following cartilage differentiation; Histological section of iPSC-derived aggregates with H&E staining at 6 (top) and >8 weeks (bottom). Scale bars represent 50 μm throughout. (B) Percentage of aggregates containing tumor-like tissues following 6, >8 weeks of cartilage differentiation: Mean ± SEM (N = 3) per lane. (C) Percentage of aggregates with dying tissue following 6, >8 weeks of cartilage differentiation: Mean ± SEM (N = 3) per lane.
Table 1. Abnormal Differentiation in Human iPSCs.
| Early | 2/21 (9.5%) |
| Late | 2/21 (9.5%) |
| Death | 1/21 (4.8%) |
We also identified the other two “late” tumorigenic lines (4YF, 451F3) that formed secretory tumors later (>8 weeks) within the cartilage template (Figure 3A, 3B, Table 1, Supplementary Figure 4, Supplementary Table 2). These late tumor-forming iPSC lines formed teratomas in vivo (Supplementary Figure 5). One final line (201B6) displayed many apoptotic cells following differentiation (Fig. 3A, 3C, Table 1, Supplementary Figure 6, Supplementary Table 3). Aggregates from this “direct cell death” line contained small apoptotic nuclei as indicated by TUNEL staining at 6 and 8 weeks of differentiation (Figure 3A, Supplementary Figure 7).
Exploring the predominant genes in each type of abnormal clone
We next investigated gene expression at 4 weeks of differentiation in early tumorigenic lines. Two tumorigenic lines (4YA, 246H2) showed higher expression levels of the endogenous proto-oncogenes, c-MYC and LIN28B compared with the other lines (Supplementary Figure 8). To contrast, the late tumorigenic lines showed residual expression of the pluripotency genes, endogenous OCT4 and NANOG at 2 weeks of differentiation (Supplementary Figure 9). In the death line, where we observed apoptotic cells within the cartilage template, the inflammatory cytokines, IL6 and TNFα were expressed at much higher expression levels in these iPSC clone (Supplementary Figure 10).
Reprogramming method and cell origin affect tumor-forming propensity
When comparing reprogramming methods, the use of proto-oncogenes, c-MYC and LIN28, as a reprogramming factors showed the higher risk of pro-oncogenicity (4/13 [30%]), regardless of retroviral or episomal transduction (Table 2). Similar to previous reports of mouse iPSC-derived chimera5, L-MYC and without c-MYC decreased the tumor formation in human (0/3, 1/5 [20%], respectively). Although integration-free iPSC lines theoretically should present a low tumor risk due to lack of genomic damage13, one such iPSC line (451F3) still showed late tumor forming capacity. These results imply that the reprogramming method, in particular the combination of reprogramming factors, is important for preventing the risk of iPSC tumor forming capacity.
Table 2. Differences among Combinations of Reprogramming Factors.
| Method | Abnormality |
|---|---|
| EOS-4F | 2/3 (66.6%) Early (1); Late (1) |
| Retro-4F | 1/5 (20%) Death (1) |
| w/o MYC | 1/5 (20%) Early (1) |
| L-MYC | 0/3 (0%) |
| Episo-6F | 1/5 (20%) Late (1) |
| ESC | 0/6 (0%) |
One iPSC line (246H2) displayed tumors regardless of the absence c-MYC. Interestingly, this iPSC line was generated from the same infant fibroblast cell line (BJ) as two other pro-oncogenic iPSC lines (Table 3, Supplementary Table 1). These results imply that the tumor-forming propensity may also depend on the cell origin in addition to the reprogramming factors.
Table 3. Abnormal Differentiation by Among Cell Origins.
| Origin | Abnormality |
|---|---|
| Infant | 3/6 (50%) Early (2); Late (1) |
| Adult | 1/12 (8.3%) Death (1) |
| Dental Pulp | 1/3 (33.3%) Late (1) |
Discussion
Evaluating the safety of iPSCs is a foremost issue in cell therapy. As the reprogramming factor c-MYC or LIN28 are oncogenes found in many human cancers15,16, there remains a risk of oncogenic transformation. However, this risk is unclear as a useful model system to evaluate this property has been lacking thus far.
In this study, we found that certain human iPSC clones are in a pro-oncogenic state that is revealed by ectopic presence of secretory tumor tissue during in vitro cartilage differentiation. Our results imply that even though ESCs and iPSCs display nearly identical cell behaviour in the undifferentiated state, their pro-oncogenic differences might only become revealed by differentiation. Here, cartilage differentiation may provide a special environment for revealing tumorigenicity, as we have not observed tumorigenesis using other cell differentiation paradigms (data not shown). Since we observe that ESCs and most iPSC lines generate cartilage safely in vitro and in vivo, cartilage differentiation may be an excellent system for recognizing a pro-oncogenic state in select human iPSC lines.
In this study we evaluated 21 human iPSC lines derived using five different reprogramming methods and three cellular origins. We observed that most lines could form cartilage normally (57.1%), however five iPSC lines clearly showed abnormal differentiation following differentiation into cartilage. We observed two different abnormal tissue formations. Four lines displayed epithelial tumor formation, while one displayed a cell death phenotype.
Through the timing of tumor and/or dying tissue observation, we could clarify three types of abnormal differentiation: early or late tumor formation, and cell death. In the early tumor-forming clones, we observed tumors following six weeks of differentiation. We could not identify any abnormalities in the late tumor forming lines at six weeks; these tumors usually occurred two weeks later. However, if we treated differentiation cultures with the p53 inhibitor pifithrin, tumors would occur earlier (Supplementary Figure 4). Because pifithrin increased the penetrance of tumor formation, we used it routinely to increase the sensitivity of our interogation, making sure that we did not miss iPSC clones that were mildly tumorigenic.
For each type of abnormal differentiation, a pattern of gene expression was prevalent. Early tumorigenic clones showed high expression levels for the oncogenes, c-MYC and LIN28 following four weeks of differentiation. It has been reported that the activation of c-MYC and LIN28 promotes oncogenic transformation and is associated with induced inflammation and/or apoptosis16,17,18,19. Although c-MYC's involvement is easy to reconcile, as this gene is used in reprogramming and could have been “epigenetically awakened” during differentiation, the involvement of LIN28 is less clear. LIN28 can be used to functionally replace c-MYC in the reprogramming cocktail2. LIN28 also appears to be transcriptional targets of c-MYC in early development20,21.
In contrast, the late tumorigenic clones showed high expression levels of the pluripotency genes, OCT4 and NANOG following two weeks of differentiation. As these genes counteract differentiation, this might explain the delayed appearance of tumor formation in these clones. Such changes in gene expression were not observed in the ESCs lines examined, nor in normal iPSC clones meaning that in the future it may be possible to simply screen for abnormal lines using gene expression profiling following cartilage differentiation.
When comparing reprogramming methods, the use of proto-oncogenes, c-MYC and LIN28, as reprogramming factors showed a higher risk of pro-oncogenic state (4/13 [30%]), regardless of retroviral and episomal transduction. In addition to the selected reprogramming factors, cell source also appeared to affect this tumor-forming propensity, as three of the four tumorigenic lines were derived from the BJ fibroblast line. Although we were short of iPSC clone numbers to make firm conclusions, our results suggest that reprogramming methods and origin will have an impact on their risks of human iPSCs.
In summary, we have demonstrated a new way to test the oncogenic risk of human iPSCs using in vitro cartilage tissue engineering. It has been reported that human iPSCs show unexpected differentiation in various lineage promotion approaches22,23,24. This in vitro cartilage tissue engineering model has an advantage that it displays abnormal differentiation and/or tumor formation in vitro without having to use animal transplantation experiments. As undifferentiated iPSCs naturally give rise to teratomas when transplanted into animals, and differentiated cells suffer from immunological rejection due to xenograft, it is difficult to use an in vivo approach to identify abnormal cell lines. Our results here show that human iPSCs need to be tested for their differentiation propensity using a suitable differentiation model before cell therapy should be undertaken.
Methods
Chondrogenic differentiation
Human iPSCs were generated in University of Toronto and Kyoto University according to their respective University Ethics committees. The use of human ESCs and iPSCs were approved by the University of Calgary's Research Ethics Board.
Human ESCs and iPSCs were differentiated into chondrocytes using micro-mass culture. Briefly, ESCs and iPSCs were cultured at high-density, ~1.0 × 105 cells per 10 μL × 7 spots in 6 cm culture dish for 3 hours. After incubation, medium was added to each dish without dissociating the cell drops. For differentiating pluripotent stem cells into chondrocytes, the differentiation medium contained: DMEM (Gibco), 1% non-essential amino acids (Invitrogen), 50 U/ml penicillin and 50 μg/ml streptomycin (Invitrogen), 0.1 mM 2-mercaptoethanol (Invitrogen), 1% ITS (Invitrogen), 1% FBS (Gibco), 10 ng/ml TGF-β1 (PeproTech), 10 ng/ml BMP-2 (PeproTech), and 50 μg/ml Ascorbic acid (Sigma). The medium was changed every 2 days. After 5 days, aggregates were physically separated and transferred to suspension culture in 6 cm petri dishes containing differentiation media 1% ITS, 1% FBS, 10 ng/ml BMP-2 and 50 μg/ml ascorbic acid.
Alcian blue staining
Proteoglycans secreted by chondrocytes were stained with Alcian Blue. Cells were fixed in 4% paraformaldehyde for 15 min and washed twice with PBS-T (0.05% Tween-20 in PBS). They were then stained with Alcian Blue (Wako) for 30 min, followed by de-staining for 3 min in 0.1 N HCl, and washed twice with PBS-T before visualization.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 15 min and washed twice with PBS-T. Cell permeabilization was performed with PBS containing 0.1% Triton-X, followed by two washes with PBS-T, and a blocking step with PBS containing 3% BSA for 1 hour at room temperature. Primary antibody, COL2 (Santa Cruz, 1:200) was added and incubated overnight at 4°C. Cells were washed three times with PBS-T, incubated with an Alexa Fluor 488 conjugated secondary antibody (1:500) and DAPI (1:500) for 2 hours at room temperature. Unbound secondary antibody was removed with three washes in PBS-T. Fluorescent images were captured using an IX70 fluorescent microscope (Olympus, Japan) equipped with a CCD camera (Q color5, Olympus, Qcapture Pro 6.0).
RNA isolation and quantitative real-time RT-PCR
Total RNA was isolated from cell culture using the RNeasy Mini Kit (Qiagen) according to the manufacturer's instructions with on-column DNase I digestion. The amount of total RNA was measured using a Bio Photometer (Eppendorf). Five hundred nanograms of total RNA were used as a template for cDNA synthesis with the Super Script III First-Strand Synthesis System (Invitrogen).
Amplified products were used to derive standard curves for quantitative real-time RT-PCR. Real-time RT-PCR was performed in an iCycler iQ system using a SYBR green PCR master mix (BioRad), with the following cycle conditions: an initial denaturation step of 95°C for 3 min, followed by 45 cycles of denaturation at 95°C for 30 sec, annealing at 58°C for 30 sec and extension at 72°C for 30 sec. Primers were designed based on the mouse sequence, and specificity was checked using BLAST (NCBI). Primer sequences are described in Supplementary Table 4.
In vitro aggregate assay
The aggregates at 6, 8 and 10 weeks of differentiation were fixed in 4% paraformaldehyde. After dehydration in ascending concentrations of ethanol and xylene, the specimens were embedded in paraffin. The paraffin sections (5 μm) were then deparaffinized, hydrated, and stained with hematoxylin and eosin (H&E), Alcian Blue (Wako), COL 2, CEA, and CA19-9 (Santa Cruz).
In vivo implant assay
To determine chondrogenic potential in vivo, aggregates were injected into SCID mice. ESCs were differentiated into the chondrocyte lineage in vitro for 30 days; aggregates were then transplanted into SCID mice subcutaneously with a 25-gauge syringe. SCID mice were obtained from Charles River Canada and housed in the single-barrier animal facility of the Faculty of Medicine at the University of Calgary. After 6 months, the animals were euthanized and tissue at the point of injection was dissected. Tissue was fixed in 4% paraformaldehyde and decalcified using Cal-EX (Fisher) for a week. After dehydration in ascending concentrations of ethanol and xylene, the specimens were embedded in paraffin. Six μm paraffin sections were mounted and then deparaffinized, hydrated, and stained with H&E, Alcian Blue, Toluidine Blue (Wako), and Methylene Blue. Methylene Blue consisted of methylene blue (Sigma) and 0.03% basic fuchsin (Sigma).
TUNEL staining
To identify apoptotic cells, TUNEL staining was performed using the in situ cell apoptosis detection kit (Roche). Sections were fixed with 4% paraformaldehyde for 30 min. Terminal deoxynucleotidyl transferase (TdT) and biotin-11-dUTP reactions were performed for 1 hour at 37°C.
Tumor observation
To identify the amount of oncogenic transformation following cartilage differentiation, the number of aggregates with secretory tumor tissue was counted based on histomorphology, and was divided by the total number of aggregates to determine the percentage. Secretory tumor tissue was defined as the glandular epithelium and mucous gland cysts. Such secretory tumors were not observed in ESC-derived aggregates. Abnormal lines were defined has having >5% tumor observation.
Cell death and dying tissue
To identify the amount of dying tissue following cartilage differentiation, the number of aggregates with dead tissue was counted by histomorphology, and was divided by the total number of aggregates to determine the percentage. Dying tissue was defined as non/small-nuclear cells within cartilage or the degradation of mucous glands. The abnormal clones were defined as having a greater than 5% observation.
Statistical analysis
All charts contain the mean of N = 3 with error bars of S.E.M. Treatment groups, or cells were compared across time points using a two-way ANOVA in GraphPad Prism version 5.0c. Each time point was then analyzed using Bonferroni post-tests to compare each cell type for each time point; P values <0.05 were significant. One-way ANOVA was also performed.
Author Contributions
A.Y. and D.E.R. designed research. A.Y. and S.L. performed research. A.Y., S.L., K.W., B.T. and G.M. analysed data. K.W., A.H., K.T., J.E. and S.Y. provided iPSC clones. A.Y., B.T., D.E.R. wrote the main manuscript. All authors reviewed the manuscript.
Supplementary Material
Supplementary Dataset 1
Acknowledgments
This work was supported by a Canadian Institutes of Health Regenerative Medicine Team Grant and by a grant from the Funding Program for World-Leading Innovative Research and Development on Science and Technology (First Program) of the Japanese Society for the Promotion of Science. A.Y. is supported by postdoctoral fellowships from the Alberta Innovates Health Solutions and the Japanese Society for the Promotion of Science Postdoctoral Fellow for Research Abroad.
References
- Takahashi K. et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872 (2007). [DOI] [PubMed] [Google Scholar]
- Yu J. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920 (2007). [DOI] [PubMed] [Google Scholar]
- Okita K., Ichisaka T. & Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature 448, 313–317 (2007). [DOI] [PubMed] [Google Scholar]
- Nakagawa M. et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 26, 101–106 (2008). [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Takizawa N., Narita M., Ichisaka T. & Yamanaka S. Promotion of direct reprogramming by transformation-deficient Myc. Proc. Natl. Acad. Sci. USA. 107, 14152–14157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther M. G. et al. Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7, 249–257 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman A. M. & Cooper J. B. Lab-specific gene expression signatures in pluripotent stem cells. Cell Stem Cell 7, 258–262 (2010). [DOI] [PubMed] [Google Scholar]
- Miura K. et al. Variation in the safety of induced pluripotent stem cell lines. Nat. Biotechnol. 27, 743–745 (2009). [DOI] [PubMed] [Google Scholar]
- Yamashita A., Nishikawa S. & Rancourt D. E. Identification of five developmental processes during chondrogenic differentiation of embryonic stem cells. PLoS One 5, e10998 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medvedev S. P. et al. Human Induced Pluripotent Stem Cells Derived from Fetal Neural Stem Cells Successfully Undergo Directed Differentiation into Cartilage. Stem Cells Dev. 20, 1099–1012 (2011). [DOI] [PubMed] [Google Scholar]
- Yamashita A., Krawetz R. & Rancourt D. E. Loss of discordant cells during micro-mass differentiation of embryonic stem cells into the chondrocyte lineage. Cell Death Differ. 16, 278–286 (2009). [DOI] [PubMed] [Google Scholar]
- Hotta A. et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat. Methods. 6, 370–376 (2009). [DOI] [PubMed] [Google Scholar]
- Okita K. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods 8, 409–412 (2011). [DOI] [PubMed] [Google Scholar]
- Takahashi Y. et al. The usefulness of CEA and/or CA19-9 in monitoring for recurrence in gastric cancer patients: a prospective clinical study. Gastric Cancer 6, 142–145 (2003). [DOI] [PubMed] [Google Scholar]
- Peng S., Maihle N. J. & Huang Y. Pluripotency factors Lin28 and Oct4 identify a subpopulation of stem cell–like cells in ovarian cancer. Oncogene 29, 2153–2159 (2010). [DOI] [PubMed] [Google Scholar]
- Dang C. V. et al. The c-Myc target gene network. Semin. Cancer Biol. 16, 253–264 (2006). [DOI] [PubMed] [Google Scholar]
- Viswanathan S. R. et al. Lin28 promotes transformation and is associated with advanced human malignancies. Nat. Genet. 41, 843–848 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T. et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460, 1140–1144 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D., Hirsch H. A. & Struhl K. An epigenetic switch involving NF-kappaB, Lin28, LET7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139, 693–706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T. C. et al. Lin-28B transactivation is necessary for Myc-mediated let-7 repression and proliferation. Proc. Natl. Acad. Sci. USA. 106, 3384–9 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangi-Garimella S. et al. Raf kinase inhibitory protein suppresses a metastasis signaling cascade involving LIN28 and let-7. EMBO J. 28, 347–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q. et al. Hemangioblastic derivatives from human induced pluripotent stem cells exhibit limited expansion and early senescence. Stem Cells 28, 704–712 (2010). [DOI] [PubMed] [Google Scholar]
- Hu B. Y. et al. Neural differentiation of human induced pluripotent stem cells follows developmental principles but with variable potency. Proc. Natl. Acad. Sci. USA. 107, 4335–4340 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajiwara M. et al. Donor-dependent variations in hepatic differentiation from human-induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 109, 12538–12543 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Dataset 1