Abstract
Introduction
Epigenetic events are, along with genetic alteration, important in the development and progression of cancer. Promoter hypermethylation causes gene silencing and is thought to be an early event in carcinogenesis. The role of promoter hypermethylation in male breast cancer has not yet been studied.
Methods
In a group of 108 male breast cancers, the methylation status of 25 genes was studied using methylation-specific multiplex ligation-dependent probe amplification. Methylation of more than 15% was regarded indicative for promoter hypermethylation. Methylation status was correlated with clinicopathological features, with patients' outcome and with 28 female breast cancer cases.
Results
Promoter hypermethylation of the genes MSH6, WT1, PAX5, CDH13, GATA5 and PAX6 was seen in more than 50% of the cases, but was uncommon or absent in normal male breast tissue. High overall methylation status was correlated with high grade (P = 0.003) and was an independent predictor of poor survival (P = 0.048; hazard ratio 2.5). ESR1 and GSTP1 hypermethylation were associated with high mitotic count (P = 0.037 and P = 0.002, respectively) and high grade (both P = 0.001). No correlation with survival was seen for individual genes. Compared with female breast cancers (logistic regression), promoter hypermethylation was less common in a variety of genes, particularly ESR1 (P = 0.005), BRCA1 (P = 0.010) and BRCA2 (P < 0.001). The most frequently hypermethylated genes (MSH6, CDH13, PAX5, PAX6 and WT1) were similar for male and female breast cancer.
Conclusion
Promoter hypermethylation is common in male breast cancer and high methylation status correlates with aggressive phenotype and poor survival. ESR1 and GSTP1 promoter hypermethylation seem to be involved in development and/or progression of high-grade male breast cancer. Although female and male breast cancer share a set of commonly methylated genes, many of the studied genes are less frequently methylated in male breast cancer, pointing towards possible differences between male and female breast carcinogenesis.
Introduction
Along with genetic alterations, epigenetic events are important in cancer development and progression. Hypermethylation of CpG islands in promoter regions (further denoted methylation) is the most well characterized epigenetic change and is a common mechanism for silencing tumor suppressor genes [1]. Methylation is reversible and therefore is an attractive therapeutic target, and can serve as a marker for therapy response and prognosis [2].
Methylation is very common in virtually all cancer types but can also be a physiological event, as in genomic imprinting [3]. Methylation is involved in the development of female breast cancer, with frequent methylation of PAX6, BRCA2, PAX5, WT1, CDH13 and MSH6 in ductal carcinoma in situ and invasive ductal cancer [4]. On the contrary, methylation is less common in estrogen receptor (ER)-negative, lymph node-negative and BRCA1-associated female breast cancer [5]. Methylation is thought to be an early event in carcinogenesis of female breast cancer, and the methylation status of specific genes may therefore be useful as a potential screening target in clinical practice [4,6].
Most of the diagnostic and therapeutic algorithms for male breast cancer have been extrapolated from female breast cancer although we and others have already demonstrated that there seem to be important differences between the two. Male breast cancers are more often hormone-positive while HER2-amplified and basal-like breast cancers are rare in men [7-10]. Different genes and mechanisms of oncogene activation also play a role in the carcinogenesis of male breast cancer: high-level amplification is less common, but whole chromosome arm gains are more often seen in male breast cancer [11]. Because of its general importance in carcinogenesis, methylation is probably also important in the development of male breast cancer, but this has not yet been studied.
Several techniques are available to assess methylation. The methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) technique allows simultaneous evaluation of the methylation status of a variety of genes in one PCR reaction. With this high-throughput approach, which shows good correlations with other methylation-specific techniques, a reliable general view of methylation in several important tumor suppressor genes can be obtained [12,13].
In this study we investigated the role of methylation of several important tumor suppressor genes in male breast cancer using MS-MLPA. We correlated methylation patterns with clinicopathological features and prognosis. The results were also compared with a group of female breast cancers.
Materials and methods
Patients: specimens and clinical information
One hundred and ten consecutive cases of surgical breast specimens of invasive male breast cancer from 1986 to 2010 were collected from four different pathology laboratories in the Netherlands (St Antonius Hospital Nieuwegein, Diakonessenhuis Utrecht, University Medical Center Utrecht and Laboratory for Pathology East Netherlands) as described in more detail previously [10]. H & E slides were reviewed by three experienced observers (PJvD, RK and AHJV-M) to confirm the diagnosis and to type and grade according to current standards.
Pathology reports were used to retrieve information on age, tumor size and lymph node status. The mean age of these patients was 66 years (range 32 to 89 years). The tumor size ranged from 0.8 to 5.5 cm (average 2.2 cm). In 86% of cases the lymph node status was known, and 55% of these patients had lymph node metastases. The majority of cases were diagnosed (according to the World Health Organization) as invasive ductal carcinoma (90%). The remaining cases were lobular (n = 3), mixed type (ductal/lobular) (n = 2), invasive cribriform (n = 1), papillary (n = 1), mucinous (n = 2), invasive micropapillary (n = 1) or adenoid cystic carcinomas (n = 1). According to the modified Bloom and Richardson score [14] most tumors were grade 2 (41%) or grade 3 (36%). Mitotic activity was assessed as described previously [15] with a mean mitotic index per 2 mm2 of 11 (range 0 to 56). For all cases, the hormone receptor and HER2 status were reassessed as described previously [10]. Tissue microarray slides were used for immunohistochemical stainings for ER and progesterone receptor (PR), and chromogenic in situ hybridization for HER2 assessment. Most tumors were ER-positive (102/110, 93%) and PR-positive (71/110; 65%), and HER2 amplification was rare (4/110, 4%).
In addition, normal male breast tissue was obtained from 10 autopsies. These patients had no history of a breast tumor. The subareolar region was resected and, after fixation in 4% formalin, was dissected and embedded in paraffin. From these blocks, 4 μm sections were cut and stained for H & E; if ducts were present, the areas richest in ducts were dissected for DNA isolation. Anonymous use of redundant tissue for research purposes is part of the standard treatment agreement with patients in our hospital [16]. Ethical approval was not required.
DNA extraction and MS-MLPA analysis
Representative tumor areas were identified on H & E-stained slides and corresponding areas (at least 1 cm2) were dissected from 8 μm paraffin slides using a scalpel. DNA was extracted by overnight incubation in proteinase K (10 mg/ml; Roche, Almere, the Netherlands) at 56°C. MS-MLPA was performed according to the manufacturers' instructions (MRC Holland, Amsterdam, the Netherlands), using a Veriti® 96-well thermal cycler (Applied Biosystems, Foster City, CA, USA). The ME002-B1 kit (MRC Holland), containing 25 tumor suppressor genes (Table 1), was used as previously [4]. Two different CpG probes were available for the genes MGMT and RB1.
Table 1.
Genes in the MS-MLPA kit and frequencies of promoter hypermethylation in male breast cancer patients
| Gene | Hypermethylation | Chromosome | Gene name |
|---|---|---|---|
| MSH6 | 104 (96%) | 02p16.3 | mutS homologue 6 |
| WT1 | 91 (84%) | 11p13 | Wilms tumor 1 |
| PAX5 | 85 (79%) | 09p13.2 | Paired box 5 |
| CDH13 | 83 (77%) | 16q24.1 | Cadherin 13, H-cadherin |
| GATA5 | 60 (56%) | 20q13.33 | GATA binding protein 5 |
| PAX6 | 57 (53%) | 11p13 | Paired box 6 |
| GSTP1 | 47 (44%) | 11q13.1 | Glutathione S-transferase p1 |
| THBS1 | 21 (19%) | 15q14 | Thrombospondin 1 |
| BRCA2 | 18 (17%) | 13q13.1 | Breast cancer gene 2 |
| CD44 | 17 (16%) | 11p13 | CD44 molecule (Indian blood group) |
| TP73 | 14 (13%) | 01p36.32 | Tumor protein p73 |
| TP53 | 12 (11%) | 17p13.1 | Tumor protein p53 |
| ESR1 | 9 (8%) | 06q25.1 | Estrogen receptor 1 |
| CADM1 | 9 (8%) | 11q23.2 | Cell adhesion molecule 1 |
| MGMT | 8 (7%) | 10q26.3 | O-6-Methylguanine-DNA methyltransferase |
| STK11 | 8 (7%) | 19p13.3 | Serine/threonine kinase 11 |
| RARB | 5 (5%) | 03p24.2 | Retinoic acid receptor beta |
| PTEN | 5 (5%) | 10q23.31 | Phosphatase and tensin homologue |
| PYCARD | 5 (5%) | 16p11.2 | PYD and CARD domain containing (TMS1) |
| RB1 | 3 (3%) | 13q14.2 | Retinoblastoma 1 |
| BRCA1 | 2 (2%) | 17q21.31 | Breast cancer gene 1 |
| CDKN2A | 2 (2%) | 09p21.3 | Cyclin-dependent kinase inhibitor 2A (p14-ARF) |
| VHL | 2 (2%) | 03p25.3 | von Hippel-Lindau |
| ATM | 1 (1%) | 11q22.3 | Ataxia telangiectasia mutated |
| CHFR | 1 (1%) | 12q24.33 | Checkpoint with forkhead and ring finger domains |
Genes contained in the ME002-B1 MS-MLPA kit (MRC Holland, Amsterdam, the Netherlands) and frequencies of promoter hypermethylation (> 15%) in 108 male breast cancer patients.
The principle of MS-MLPA has been described elsewhere in more detail [17]. In short, MS-MLPA kits contain probes for methylation quantification, which are similar to those in conventional multiplex ligation-dependent probe amplification except that the sequence detected by the MS-MLPA probes contains a restriction site for the methylation-sensitive HhaI enzyme. After DNA denaturation and overnight incubation with the probe mix, the samples are divided into two tubes, one of which is incubated with HhaI. In this tube, unmethylated DNA is digested and not exponentially amplified by PCR. Because methylated DNA is prevented from being digested by HhaI, these probes are ligated and therefore amplified by PCR. The ratio between probes incubated with and without HhaI gives an estimation of the methylation status. Appropriate negative and positive (Sssi-methylated DNA) controls were taken along with each MS-MLPA run. The PCR products were separated by electrophoresis on an ABI 3730 capillary sequencer (Applied Biosystems).
Methylation analysis was carried out with Genescan v4.1 (Applied Biosystems) and Coffalyser v9.4 (MRC Holland) software. Relative probe peaks were calculated by dividing the signal of each probe by the signal of every reference probe in one sample (intra-sample normalization). For the final methylation status, the ratio of relative probe peaks of the undigested sample (without HhaI) and the corresponding digested sample (with HhaI) were calculated for each probe. In case two CpG loci were present for one gene (MGMT and RB1), the mean methylation status was calculated for further analysis.
A promoter methylation ratio > 0.15 (corresponding to > 15% methylation) was regarded as indicative for promoter hypermethylation, based on cell-line experiments and previous experience [4,18]. The cumulative methylation index (CMI) was calculated as the sum of the methylation percentage of all genes [5].
Comparison with female breast cancer
A previously described group of female breast cancers was used to compare promoter hypermethylation in male and female breast cancer [4]. This group consists of 33 patients with invasive ductal carcinoma and a mean age of 55 years (range 32 to 81 years). The tumor size ranged from 0.5 to 6.5 cm (average 2.1 cm). The mean mitotic activity was 14 per 2 mm2 and, according to the modified Bloom and Richardson score, most tumors were grade 2 (10/33, 30%) or grade 3 (17/33, 52%). ER-positivity was common (27/31, 87%) and 71% of the tumors were PR-positive (22/31, 71%). HER2 amplification was seen in two cases (2/31, 6%). The same tumor suppressor kit (ME002-B1; MRC Holland) was used.
Statistical analysis
Statistical calculations were performed using SPSS for Windows v15.0 (SPSS Inc., Chicago, IL, USA), regarding two-sided P < 0.05 as significant. Correlations between promoter hypermethylation (> 15% methylation) and clinicopathological characteristics were calculated with analysis of variance for continuous variables and with the Pearson chi-square test (or Fisher's exact test when appropriate) for categorical variables. The following clinicopathological features were dichotomized: age (> 50 years), tumor size (> 2.0 cm), mitotic count (> 8) and histological grade (1/2 vs. 3). The Mann-Whitney test was used to calculate differences in CMI and clinicopathological features. Correlation between the number of methylated genes and clinicopathological features was calculated with Spearman's rho. Unsupervised hierarchical clustering using the statistical program R [19] was performed to analyze relevant clusters and co-methylation. Absolute methylation percentages were used and all cases with methylation < 5% were pooled together. Logistic regression analysis was performed to compare methylation in male and female breast cancer, taking significant clinicopathological differences between the two groups into account. Backward stepwise method was used until the most predictive variables remained.
Survival data were obtained from the Integral Cancer Registration the Netherlands (IKNL). Outcome data were available for 101 cases with a mean follow-up of 5.7 years. Survival analysis was therefore based on 5-year survival rates. For univariate survival analysis, Kaplan-Meier curves were plotted and analyzed with the log-rank test. Multivariate survival analysis was performed with Cox regression (enter and remove limits 0.10). The CMI and number of methylated genes were dichotomized for survival analysis according to the most predictive threshold.
Results
Methylation status by MS-MLPA
In two male breast cancer cases the amount of DNA was insufficient, leaving 108 cases for further analysis. The methylation status of the 25 analyzed tumor suppressor genes is presented in Table 1. All cases except one showed methylation (15% cutoff) of at least one gene, with an average of six genes (range 0 to 26). Methylation was very common for MSH6 (96%), WT1 (83%), PAX5 (79%) and CDH13 (77%). On the contrary, methylation was very rare in RB1 (3%), BRCA1 (2%), CDKN2A (2%), VHL (2%), ATM (1%) and CHFR (1%). The mean CMI was 364 (range 129 to 904).
In male breast tissue derived from autopsies, gynecomastia was seen in three cases. The other seven cases harbored normal male breast ducts. Methylation was seen in the genes MSH6 (4/10, 40%), ESR1 (2/10, 20%), PAX5 (1/10, 10%) and CDH13 (1/10, 10%). No methylation was found in all of the other genes. The mean CMI in these cases was also low at 16 (range 11 to 27).
Correlation with clinicopathological features
Higher CMI was correlated with high mitotic count (P = 0.046) and high grade (P = 0.003). The number of methylated genes was significantly higher in grade 3 cancers (P = 0.034), and correlated with a high mean mitotic count (P = 0.021). Two individual genes were associated with a more aggressive phenotype: the mean mitotic count was higher in tumors with ESR1 (10 vs. 16; P = 0.037) and GSTP1 (8 vs. 14; P = 0.002) methylation. Both genes were also associated with high grade (both P = 0.001). For ESR1 eight out of nine methylated tumors were grade 3, and for GSTP1 25 out of 47 methylated tumors were grade 3. Finally, tumors with MGMT methylation had a mean tumor size of 3.2 cm, which was significantly larger compared with tumors without MGMT methylation (2.1 cm; P = 0.002). No association was seen between any genes on the one hand and age, lymph node, PR and HER2 status on the other.
Cluster analysis
Hierarchical cluster analysis revealed three groups of clustered genes (Figure 1). One group consisted of the genes WT1, CDH13, MSH6, PAX5, GSTP1, GATA5 and PAX6, seven genes in which methylation was very common. Indeed, in 15% of cases all these genes showed methylation. The second cluster was formed by genes with intermediate methylation rates (5 to 19%). In the third group the remaining genes clustered together. Methylation was rare (< 8%) in these genes. Regarding all of the patients, male breast cancer cases were not divided into clear distinctive clusters. At least four different groups could be identified and these clusters displayed no distinct clinicopathological features. One case did not fit into any of the groups. This grade 3 male breast cancer case showed a high methylation ratio in nearly all genes.
Figure 1.
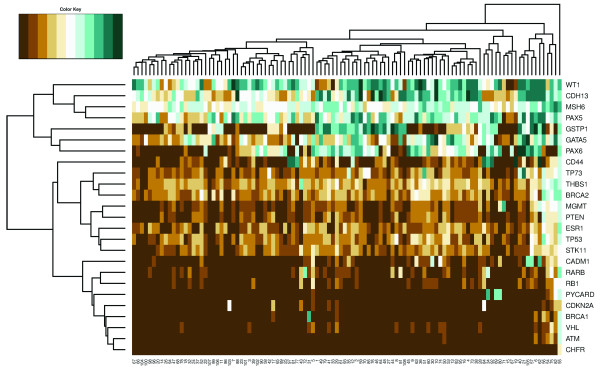
Hierarchical clustering analysis of male breast cancer patients. Unsupervised hierarchical clustering of absolute methylation percentages in 25 genes in 108 male breast cancer patients. One gene cluster consisted of WT1, CDH13, MSH6, PAX5, GSTP1, GATA5 and PAX6, seven genes in which methylation was very common. The second cluster was formed by genes with intermediate methylation rates (5 to 19%), and in the third cluster the remaining genes with little methylation (< 8%) grouped together. No clear distinctive clusters of male breast cancer cases were found.
Comparison with female breast cancer
Because breast cancer is a heterogeneous disease, only luminal-type male breast cancer and luminal-type female breast cancer (defined by ER and/or PR expression) were compared. In this approach, age was the only clinicopathological feature that was significantly different between the two groups. Male breast cancer patients were significantly older (66 years vs. 54 years; P < 0.001).
Figure 2 illustrates the methylation status of the 25 studied genes in luminal-type male (n = 95) and luminal-type female (n = 28) breast cancer. Methylation was much less frequent in male breast cancer in a variety of genes. Particularly, ESR1, BRCA1 and BRCA2 were less often methylated compared with female breast cancer and were strong independent predictors of gender in logistic regression analysis (P = 0.005, P = 0.010 and P < 0.001, respectively). The genes CD44 (P = 0.050), RARB (P = 0.026), ATM (P = 0.017) and STK11 (P = 0.040) also showed less frequent methylation in male breast cancer. On the other hand, the high frequency of methylation in MSH6, PAX5, PAX6 and CDH13 was shared between male and female breast cancer.
Figure 2.
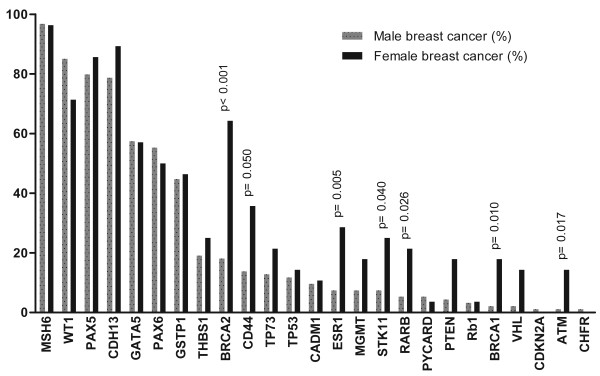
Methylation status of the 25 studied genes in luminal-type breast cancer. Promoter hypermethylation (> 15% methylation) of the 25 studied genes in luminal-type male (n = 95) and luminal-type female (n = 28) breast cancer.
Only age was taken into account during logistic regression analysis using gender as the determinant, because no other clinicopathological feature was significantly different between the two groups. When leaving out age and using the Pearson chi-square test, methylation in PTEN and VHL was also significantly less common in male breast cancer (P = 0.029 and P = 0.025, respectively). None of the studied genes was more frequently methylated in male breast cancer.
Survival analysis
Grade 3 (P = 0.027), high mitotic count (> 8; P = 0.015) and large tumor size (> 2.0 cm; P = 0.036) were correlated with decreased 5-year survival as expected. No individual methylated gene was significantly correlated with patients' outcome, although tumors with GATA5 methylation showed a trend towards decreased 5-year survival (64% vs. 82%; P = 0.083). When the number of methylated genes was dichotomized using a threshold of six methylated genes, however, the group with six or more methylated genes had significantly decreased survival compared with tumors with less than six methylated genes (P = 0.022; Figure 3), but was not a significant independent prognostic factor in Cox regression (P = 0.057). Tumors with high CMI (> 350) also had decreased survival (P = 0.033; Figure 3) and high CMI was an independent prognosticator in Cox regression (P = 0.048; hazard ratio 2.5).
Figure 3.
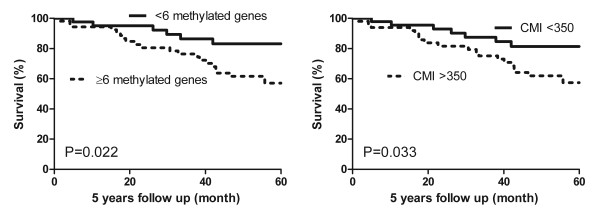
Five-year survival according to number of methylated genes and cumulative methylation index. Five-year survival with corresponding P values (log rank) according to high number of methylated genes (≥ 6) and high cumulative methylation index (CMI > 350).
Discussion
Promoter hypermethylation is an important gene-silencing mechanism thought to be an early event in carcinogenesis [1]. Understanding the epigenetic role in male breast cancer is important to gain further insight into male breast carcinogenesis and for the identification of potential biomarkers for diagnosis and treatment [20,21]. Epigenetic changes in male breast cancer had not yet been studied and therefore we investigated promoter hypermethylation in a large group of 108 patients with this rare disease using the high-throughput MS-MLPA approach, enabling evaluation of the methylation status of a variety of genes in one PCR.
Not surprisingly, methylation does occur in male breast cancer. The genes MSH6, WT1, PAX5, CDH13, GATA5 and PAX6 showed promoter hypermethylation in more than 50% of cases, indicating that these genes are probably often involved in male breast carcinogenesis. These genes are required for normal development of several organ systems and/or play a role in DNA repair, cell adhesion, cell growth and migration, although the function of some of these genes is still poorly understood [22-26]. Loss of function of both alleles leads to complete knockdown of these genes, which may facilitate malignant transformation. Methylation, with aberrant silencing of one of these alleles, could be the initiating event, the second hit or both [27]. MSH6 methylation was also quite common in the normal male breast, although at a lower frequency than our group of male breast cancer cases. The other commonly methylated genes in male breast cancer were not found to be methylated in our 10 cases of normal male breast tissue, confirming the important role of methylation in the development of male breast cancer.
In male breast cancer, methylation was very rare in BRCA1, CDKN2A, VHL, ATM and CHFR (< 2%) - indicating that methylation of these genes does not seem to play a prominent role in male breast carcinogenesis.
Male breast cancer with an aggressive phenotype harbored an increased number of methylated genes and had a higher CMI. In addition, tumors with six or more methylated genes or high CMI had a worse outcome. High CMI was even an independent predictor of poor survival when corrected for grade, mitotic count and tumor size. This indicates that accumulation of methylated genes and an overall higher methylation status seem to be important in the development of more aggressive male breast cancer with poor survival. The hallmark of high-grade breast cancer is genetic instability [28], which in male breast cancer seems to include accumulation of methylated genes. A similar trend was noted in female breast cancer, and female breast cancer patients with an increasing number of methylated genes also have an unfavorable outcome [4,29,30].
Two single genes were identified in which methylation was correlated with high mitotic count and high grade: ESR1 and GSTP1. High-grade breast cancer is believed to arise from high-grade precursor lesions by gaining different genetic and epigenetic changes compared with low-grade breast cancer [31,32]. ESR1 and GSTP1 methylation could be important in the development of these high-grade male breast cancers. GSTP1 belongs to a family of metabolic enzymes and is involved in the detoxification of carcinogens and chemotherapeutic agents by conjugating them with glutathione [33]. In female breast cancer, GSTP1 hypermethylation is correlated with high-grade ductal carcinoma in situ and high-grade invasive breast cancer, presence of lymph node metastasis and poor outcome [4,30,34,35]. ER, encoded by ESR1, is an important factor in breast cancer, because studies in females have shown that patients with hormone-negative tumors do not benefit from endocrine therapy [36]. In the present study we could not demonstrate a relation between ESR1 methylation and ER expression, although this needs to be interpreted with caution since only seven out of 108 cases were ER-negative in the present study. Another recent study also concluded that the relation between ESR1 methylation and protein expression is weak and unlikely to represent a predominant mechanism of ER silencing [37]. There was also no relation between methylation and expression of TWIST as shown by us, so this may not be unusual [38]. Larger series of ER-negative male breast cancer cases will be needed to further explore this relationship. Similar to female breast cancer, methylation of ESR1 seems to be a biomarker for high malignant male breast cancer. Indeed, in female breast cancer ESR1 promoter hypermethylation has been correlated with poor prognosis [39]. ESR1 methylation and GSTP1 methylation were not significantly correlated with poor survival in our group of male breast cancer and therefore do not seem to be useful prognostic biomarkers in male breast cancer.
Compared with female breast cancer, methylation was less common in male breast cancer in several of the studied genes, particularly ESR1, BRCA1 and BRCA2. BRCA1 and BRCA2 promoter hypermethylation was encountered in, respectively, 2% and 18% of the male breast cancers, but was seen in 18% and 64% of the female breast cancers, using the same approach and similar cutoff criteria. These results points towards possible important differences between female and male breast carcinogenesis with regard to methylation. BRCA1 methylation is more common in relatively young, premenopausal women [40], which could explain the higher incidence in female breast cancer since the male breast cancer patients were significantly older than the female breast cancer patients. However, in the present study we corrected for age in logistic regression, so gender-specific differences also seem to play a role here. Differences in genetic predisposition may also influence the epigenetic profile of these tumors and could be responsible for some of the differences found in promoter hypermethylation between male and female breast cancer. Approximately 10% of men with breast cancer are known to have a genetic predisposition, and especially BRCA2 mutations seem to be important [41]. Unfortunately no data regarding BRCA germline mutations were available for both cohorts, but it seems probable that there is a higher rate of hidden BRCA2 mutation carriers in the male breast cancer group. This higher rate of BRCA2 mutation carriers may well explain the lower rate of BRCA2 promoter hypermethylation in the male breast cancer group compared with female breast cancers [42]. Interestingly, genes with frequent methylation in male breast cancer (MSH6, CDH13, PAX5, PAX6 and WT1) were also very commonly methylated in female breast cancer.
The methylation status of both groups was obtained using the same technique. However, the male breast cancer cases were microdissected by a scalpel and the female breast cancer cases by laser microdissection. Although the latter method is more precise we do not think this may have influenced our results. The male breast cancer tumors were quite large and rich in tumor cells and could therefore be well harvested for DNA isolation based on scalpel dissection. Besides, multiplex ligation-dependent probe amplification is relatively insensitive to tumor cell content [43].
Conclusion
Methylation seems to be important in the development of male breast cancer. More than 50% of the tumors showed methylation in MSH6, WT1, PAX5, CDH13, GATA5 and PAX6. The accumulation of methylated genes and an overall high methylation status was correlated with a more aggressive phenotype and poor survival. ESR1 and GSTP1 were the only single genes associated with mitotically active and high-grade male breast cancers. Compared with female breast cancer, methylation occurred less often in male breast cancer. On the other hand, the most frequently methylated genes were shared between male and female breast cancer. Our results point towards differences in carcinogenesis between male and female breast cancer, hidden behind similarities.
Abbreviations
ATM: ataxia telangiectasia mutated; BRCA1: breast cancer gene 1; BRCA2: breast cancer gene 2; CADM1: cell adhesion molecule 1; CDH13: cadherin 1; CDKN2A: cyclin-dependent kinase inhibitor 2A; CHFR: checkpoint with forkhead and ring finger domains; CMI: cumulative methylation index; ER: estrogen receptor; ESR1: estrogen receptor 1; GATA5: GATA binding protein 5; GSTP1: glutathione S-transferase p1; H & E: hematoxylin and eosin; HER2: human epidermal growth receptor 2; MGMT: O-6-methylguanine-DNA methyltransferase; MSH6: mutS homologue 6; MS-MLPA: methylation-specific multiplex ligation-dependent probe amplification; PAX5: paired box 5; PAX6: paired box 6; PCR: polymerase chain reaction; PR: progesterone receptor; PTEN: phosphatase and tensin homologue; PYCARD: PYD and CARD domain containing; RARB: retinoic acid receptor beta; RB1: retinoblastoma 1; STK11: serine/threonine kinase 11; THBS1: thrombospondin 1; TP53: tumor protein p53; TP73: tumor protein p73; VHL: von Hippel-Lindau; WT1: Wilms tumor 1.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PJvD, RK and AHJV-M conceived the experiments. RK, MCHH, JJO and PCdB were involved in collecting male breast cancer cases. RK and CBM carried out the experiments and analyzed data. All authors were involved in writing the paper and had final approval of the submitted and published versions.
Contributor Information
Robert Kornegoor, Email: R.Kornegoor@umcutrecht.nl.
Cathy B Moelans, Email: C.B.Moelans@umcutrecht.nl.
Anoek HJ Verschuur-Maes, Email: A.Maes@umcutrecht.nl.
Marieke CH Hogenes, Email: m.hogenes@laborpath.nl.
Peter C de Bruin, Email: p.bruin@antoniusziekenhuis.nl.
Joost J Oudejans, Email: joudejan@diakhuis.nl.
Paul J van Diest, Email: p.j.vandiest@umcutrecht.nl.
References
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000;14:168–174. doi: 10.1016/S0168-9525(99)01971-X. [DOI] [PubMed] [Google Scholar]
- Jovanovic J, Ronneberg JA, Tost J, Kristensen V. The epigenetics of breast cancer. Mol Oncol. 2010;14:242–254. doi: 10.1016/j.molonc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surani MA. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;14:309–312. doi: 10.1016/S0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- Moelans CB, Verschuur-Maes AH, van Diest PJ. Frequent promoter hypermethylation of BRCA2, CDH13, MSH6, PAX5, PAX6 and WT1 in ductal carcinoma in situ and invasive breast cancer. J Pathol. 2011;14:222–231. doi: 10.1002/path.2930. [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk KP, Fackler MJ, Sukumar S, van Gils CH, van Laar T, van der Wall E, Vooijs M, van Diest PJ. Methylation is less abundant in BRCA1-associated compared with sporadic breast cancer. Ann Oncol. 2008;14:1870–1874. doi: 10.1093/annonc/mdn409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoque MO, Prencipe M, Poeta ML, Barbano R, Valori VM, Copetti M, Gallo AP, Brait M, Maiello E, Apicella A, Rossiello R, Zito F, Stefania T, Paradiso A, Carella M, Dallapiccola B, Murgo R, Carosi I, Bisceglia M, Fazio VM, Sidransky D, Parrella P. Changes in CpG islands promoter methylation patterns during ductal breast carcinoma progression. Cancer Epidemiol Biomarkers Prev. 2009;14:2694–2700. doi: 10.1158/1055-9965.EPI-08-0821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson WF, Jatoi I, Tse J, Rosenberg PS. Male breast cancer: a population-based comparison with female breast cancer. J Clin Oncol. 2010;14:232–239. doi: 10.1200/JCO.2009.23.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano SH, Cohen DS, Buzdar AU, Perkins G, Hortobagyi GN. Breast carcinoma in men: a population-based study. Cancer. 2004;14:51–57. doi: 10.1002/cncr.20312. [DOI] [PubMed] [Google Scholar]
- Muir D, Kanthan R, Kanthan SC. Male versus female breast cancers. A population-based comparative immunohistochemical analysis. Arch Pathol Lab Med. 2003;14:36–41. doi: 10.5858/2003-127-36-MVFB. [DOI] [PubMed] [Google Scholar]
- Kornegoor R, Verschuur-Maes AH, Buerger H, Hogenes MC, de Bruin PC, Oudejans JJ, van der Groep P, Hinrichs B, van Diest PJ. Molecular subtyping of male breast cancer by immunohistochemistry. Mod Pathol. 2012;14:398–404. doi: 10.1038/modpathol.2011.174. [DOI] [PubMed] [Google Scholar]
- Johansson I, Nilsson C, Berglund P, Strand C, Jönsson G, Staaf J, Ringnér M, Nevanlinna H, Barkardottir RB, Borg A, Olsson H, Luts L, Fjällskog ML, Hedenfalk I. High-resolution genomic profiling of male breast cancer reveals differences hidden behind the similarities with female breast cancer. Breast Cancer Res Treat. 2010;14:747–760. doi: 10.1007/s10549-010-1262-8. [DOI] [PubMed] [Google Scholar]
- Suijkerbuijk KP, Pan X, van der Wall E, van Diest PJ, Vooijs M. Comparison of different promoter methylation assays in breast cancer. Anal Cell Pathol (Amst) 2010;14:133–141. doi: 10.3233/ACP-CLO-2010-0542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dikow N, Nygren AO, Schouten JP, Hartmann C, Kramer N, Janssen B, Zschocke J. Quantification of the methylation status of the PWS/AS imprinted region: comparison of two approaches based on bisulfite sequencing and methylation-sensitive MLPA. Mol Cell Probes. 2007;14:208–215. doi: 10.1016/j.mcp.2006.12.002. [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;14:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- van Diest PJ, Baak JP, Matze-Cok P, Wisse-Brekelmans EC, van Galen CM, Kurver PH, Bellot SM, Fijnheer J, van Gorp LH, Kwee WS, Los J, Peterse JL, Ruitenberg HM, Schapers RFM, Schipper MEI, Somsen JG, Willig AWPM, Ariens ATh. Reproducibility of mitosis counting in 2,469 breast cancer specimens: results from the Multicenter Morphometric Mammary Carcinoma Project. Hum Pathol. 1992;14:603–607. doi: 10.1016/0046-8177(92)90313-R. [DOI] [PubMed] [Google Scholar]
- van Diest PJ. No consent should be needed for using leftover body material for scientific purposes. For. BMJ. 2002;14:648–651. doi: 10.1136/bmj.325.7365.648. [DOI] [PubMed] [Google Scholar]
- Nygren AO, Ameziane N, Duarte HM, Vijzelaar RN, Waisfisz Q, Hess CJ, Schouten JP, Errami A. Methylation-specific MLPA (MS-MLPA): simultaneous detection of CpG methylation and copy number changes of up to 40 sequences. Nucleic Acids Res. 2005;14:e128. doi: 10.1093/nar/gni127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gylling A, Abdel-Rahman WM, Juhola M, Nuorva K, Hautala E, Jarvinen HJ, Mecklin JP, Aarnio M, Peltomaki P. Is gastric cancer part of the tumour spectrum of hereditary non-polyposis colorectal cancer? A molecular genetic study. Gut. 2007;14:926–933. doi: 10.1136/gut.2006.114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Statistical program R. http://r-project.org
- Laird PW. The power and the promise of DNA methylation markers. Nat Rev Cancer. 2003;14:253–266. doi: 10.1038/nrc1045. [DOI] [PubMed] [Google Scholar]
- Mulero-Navarro S, Esteller M. Epigenetic biomarkers for human cancer: the time is now. Crit Rev Oncol Hematol. 2008;14:1–11. doi: 10.1016/j.critrevonc.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Andreeva AV, Kutuzov MA. Cadherin 13 in cancer. Genes Chromosomes Cancer. 2010;14:775–790. doi: 10.1002/gcc.20787. [DOI] [PubMed] [Google Scholar]
- Hohenstein P, Hastie ND. The many facets of the Wilms' tumour gene, WT1. Hum Mol Genet. 2006;14:R196–R201. doi: 10.1093/hmg/ddl196. [DOI] [PubMed] [Google Scholar]
- Martin SA, Lord CJ, Ashworth A. Therapeutic targeting of the DNA mismatch repair pathway. Clin Cancer Res. 2010;14:5107–5113. doi: 10.1158/1078-0432.CCR-10-0821. [DOI] [PubMed] [Google Scholar]
- Palmisano WA, Crume KP, Grimes MJ, Winters SA, Toyota M, Esteller M, Joste N, Baylin SB, Belinsky SA. Aberrant promoter methylation of the transcription factor genes PAX5 alpha and beta in human cancers. Cancer Res. 2003;14:4620–4625. [PubMed] [Google Scholar]
- Zheng R, Blobel GA. GATA transcription factors and cancer. Genes Cancer. 2010;14:1178–1188. doi: 10.1177/1947601911404223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;14:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Al-Kuraya K, Schraml P, Torhorst J, Tapia C, Zaharieva B, Novotny H, Spichtin H, Maurer R, Mirlacher M, Köchli O, Zuber M, Dieterich H, Mross F, Wilber K, Simon R, Sauter G. Prognostic relevance of gene amplifications and coamplifications in breast cancer. Cancer Res. 2004;14:8534–8540. doi: 10.1158/0008-5472.CAN-04-1945. [DOI] [PubMed] [Google Scholar]
- Sharma G, Mirza S, Yang YH, Parshad R, Hazrah P, Datta Gupta S, Ralhan R. Prognostic relevance of promoter hypermethylation of multiple genes in breast cancer patients. Cell Oncol. 2009;14:487–500. doi: 10.3233/CLO-2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Shen J, Gammon MD, Zhang YJ, Wang Q, Gonzalez K, Xu X, Bradshaw PT, Teitelbaum SL, Garbowski G, Hibshoosh H, Neugut AI, Chen J, Santella RM. Prognostic significance of gene-specific promoter hypermethylation in breast cancer patients. Breast Cancer Res Treat. 2012;14:197–205. doi: 10.1007/s10549-011-1712-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buerger H, Mommers EC, Littmann R, Simon R, Diallo R, Poremba C, Dockhorn-Dworniczak B, van Diest PJ, Boecker W. Ductal invasive G2 and G3 carcinomas of the breast are the end stages of at least two different lines of genetic evolution. J Pathol. 2001;14:165–170. doi: 10.1002/path.875. [DOI] [PubMed] [Google Scholar]
- Hwang ES, DeVries S, Chew KL, Moore DH, Kerlikowske K, Thor A, Ljung BM, Waldman FM. Patterns of chromosomal alterations in breast ductal carcinoma in situ. Clin Cancer Res. 2004;14:5160–5167. doi: 10.1158/1078-0432.CCR-04-0165. [DOI] [PubMed] [Google Scholar]
- Coles BF, Kadlubar FF. Detoxification of electrophilic compounds by glutathione S-transferase catalysis: determinants of individual response to chemical carcinogens and chemotherapeutic drugs? Biofactors. 2003;14:115–130. doi: 10.1002/biof.5520170112. [DOI] [PubMed] [Google Scholar]
- Arai T, Miyoshi Y, Kim SJ, Taguchi T, Tamaki Y, Noguchi S. Association of GSTP1 CpG islands hypermethylation with poor prognosis in human breast cancers. Breast Cancer Res Treat. 2006;14:169–176. doi: 10.1007/s10549-006-9241-9. [DOI] [PubMed] [Google Scholar]
- Lasabova Z, Tilandyova P, Kajo K, Zubor P, Burjanivova T, Danko J, Plank L. Hypermethylation of the GSTP1 promoter region in breast cancer is associated with prognostic clinicopathological parameters. Neoplasma. 2010;14:35–40. doi: 10.4149/neo_2010_01_035. [DOI] [PubMed] [Google Scholar]
- Hayashi SI, Eguchi H, Tanimoto K, Yoshida T, Omoto Y, Inoue A, Yoshida N, Yamaguchi Y. The expression and function of estrogen receptor alpha and beta in human breast cancer and its clinical application. Endocr Relat Cancer. 2003;14:193–202. doi: 10.1677/erc.0.0100193. [DOI] [PubMed] [Google Scholar]
- Gaudet MM, Campan M, Figueroa JD, Yang XR, Lissowska J, Peplonska B, Brinton LA, Rimm DL, Laird PW, Garcia-Closas M, Sherman ME. DNA hypermethylation of ESR1 and PGR in breast cancer: pathologic and epidemiologic associations. Cancer Epidemiol Biomarkers Prev. 2009;14:3036–3043. doi: 10.1158/1055-9965.EPI-09-0678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gort EH, Suijkerbuijk KP, Roothaan SM, Raman V, Vooijs M, van der Wall E, van Diest PJ. Methylation of the TWIST1 promoter, TWIST1 mRNA levels, and immunohistochemical expression of TWIST1 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2008;14:3325–3330. doi: 10.1158/1055-9965.EPI-08-0472. [DOI] [PubMed] [Google Scholar]
- Ramos EA, Camargo AA, Braun K, Slowik R, Cavalli IJ, Ribeiro EM, Pedrosa Fde O, de Souza EM, Costa FF, Klassen G. Simultaneous CXCL12 and ESR1 CpG island hypermethylation correlates with poor prognosis in sporadic breast cancer. BMC Cancer. 2010;14:23. doi: 10.1186/1471-2407-10-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Gammon MD, Zhang Y, Bestor TH, Zeisel SH, Wetmur JG, Wallenstein S, Bradshaw PT, Garbowski G, Teitelbaum SL, Neugut AI, Santella RM, Chen J. BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat. 2009;14:397–404. doi: 10.1007/s10549-008-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korde LA, Zujewski JA, Kamin L, Giordano S, Domchek S, Anderson WF, Bartlett JM, Gelmon K, Nahleh Z, Bergh J, Cutuli B, Pruneri G, McCaskill-Stevens W, Gralow J, Hortobagyi G, Cardoso F. Multidisciplinary meeting on male breast cancer: summary and research recommendations. J Clin Oncol. 2010;14:2114–2122. doi: 10.1200/JCO.2009.25.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin AM, Spearman AD, Tseng SY, Sweet K, Toland AE. Methylation not a frequent 'second hit' in tumors with germline BRCA mutations. Fam Cancer. 2009;14:339–346. doi: 10.1007/s10689-009-9240-1. [DOI] [PubMed] [Google Scholar]
- Moelans CB, de Weger RA, Ezendam C, van Diest PJ. HER-2/neu amplification testing in breast cancer by multiplex ligation-dependent probe amplification: influence of manual- and laser microdissection. BMC Cancer. 2009;14:4. doi: 10.1186/1471-2407-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


