Abstract
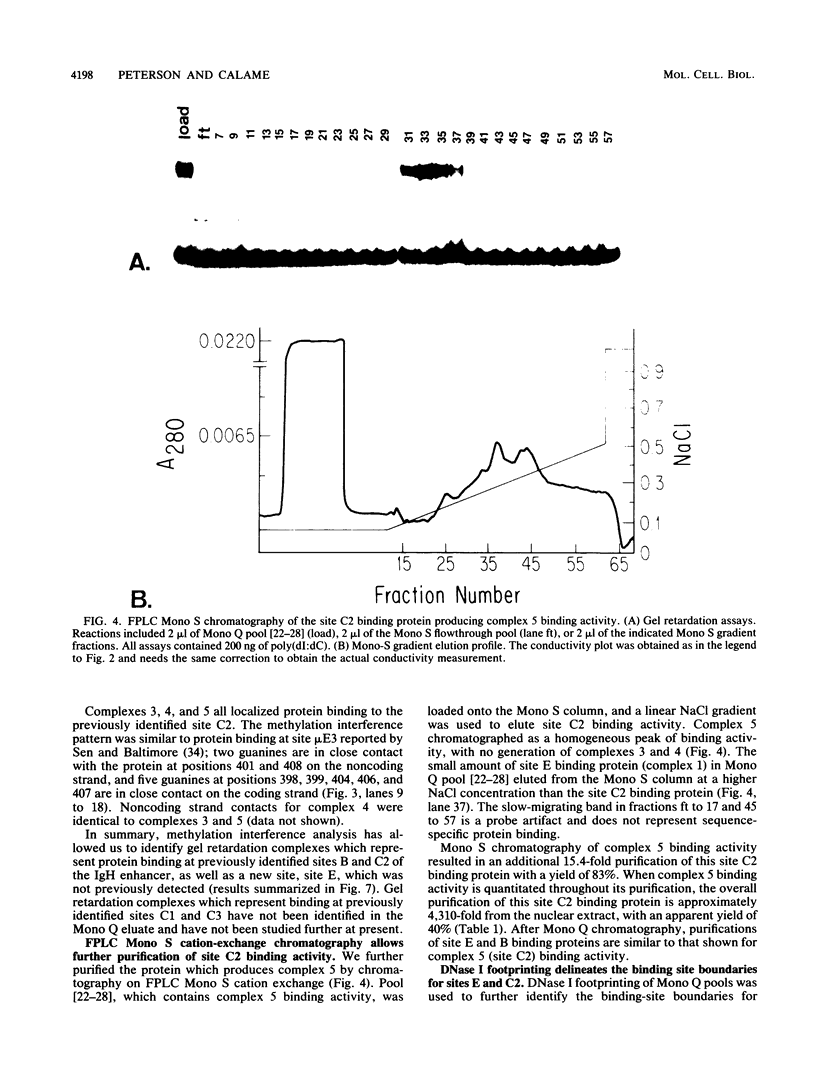
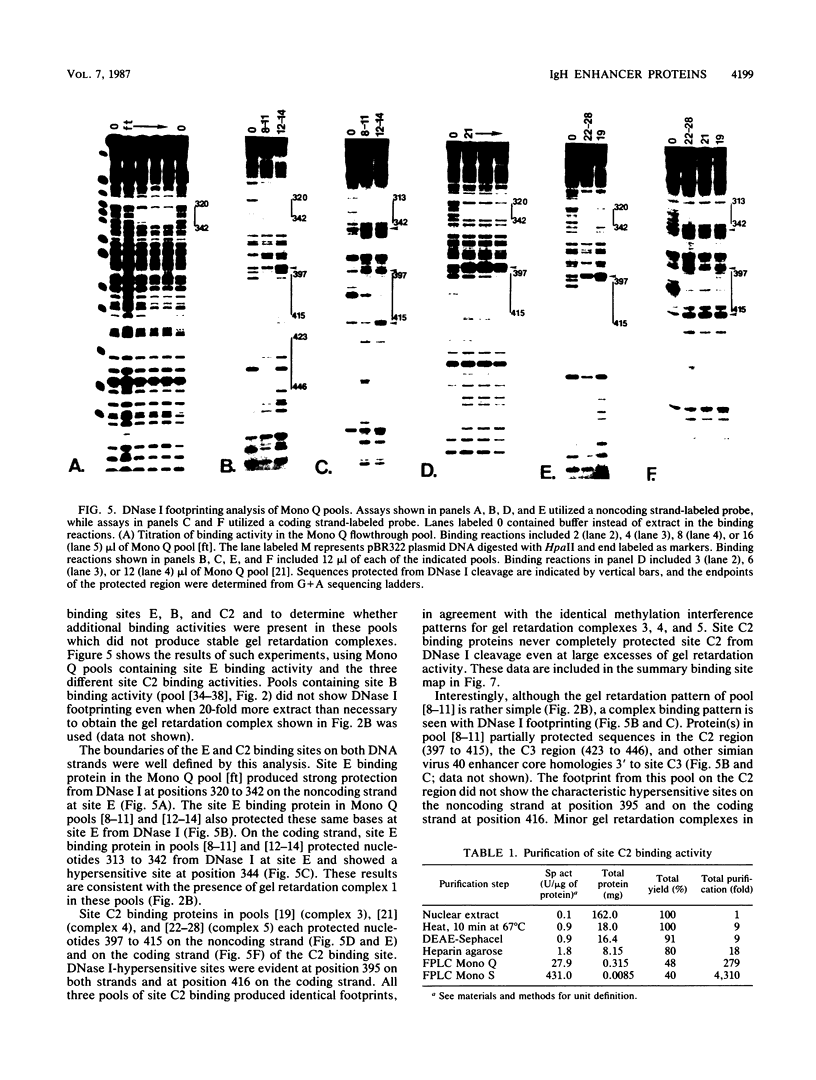
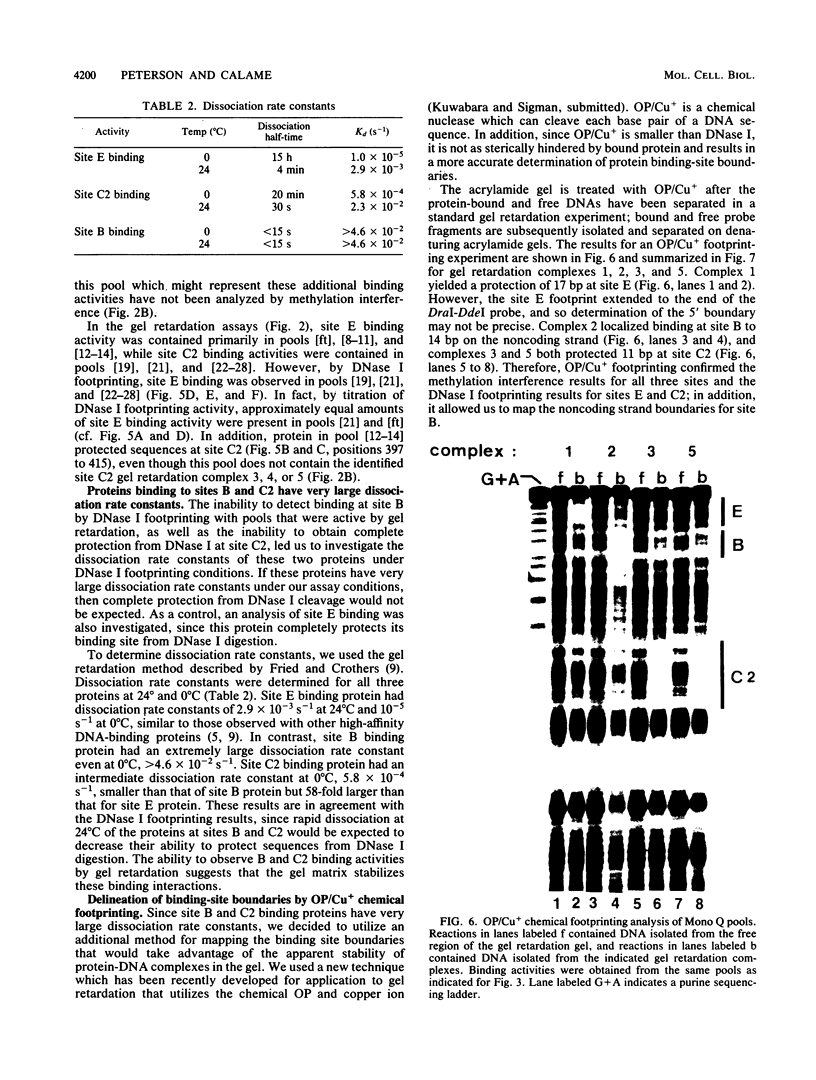
We have begun to purify and characterize several proteins which bind to the mouse immunoglobulin heavy-chain enhancer to understand the molecular interactions important for enhancer activity. Three proteins which bind to different sites on the immunoglobulin heavy-chain enhancer have been chromatographically separated and partially purified. One protein binds a site which has not been reported previously and does not bind to other reported protein-binding sites on the immunoglobulin heavy-chain enhancer. Binding-site boundaries for the three partially purified proteins have been precisely mapped by methylation interference, DNase I footprinting, and orthophenanthroline/copper chemical nuclease footprinting. We have also characterized these three proteins with respect to dissociation rate constants.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Augereau P., Chambon P. The mouse immunoglobulin heavy-chain enhancer: effect on transcription in vitro and binding of proteins present in HeLa and lymphoid B cell extracts. EMBO J. 1986 Aug;5(8):1791–1797. doi: 10.1002/j.1460-2075.1986.tb04428.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Olson L., Schaffner W. A lymphocyte-specific cellular enhancer is located downstream of the joining region in immunoglobulin heavy chain genes. Cell. 1983 Jul;33(3):729–740. doi: 10.1016/0092-8674(83)90015-6. [DOI] [PubMed] [Google Scholar]
- Benoist C., Chambon P. In vivo sequence requirements of the SV40 early promotor region. Nature. 1981 Mar 26;290(5804):304–310. doi: 10.1038/290304a0. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chodosh L. A., Carthew R. W., Sharp P. A. A single polypeptide possesses the binding and transcription activities of the adenovirus major late transcription factor. Mol Cell Biol. 1986 Dec;6(12):4723–4733. doi: 10.1128/mcb.6.12.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983 Mar 11;11(5):1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi A., Church G. M., Tonegawa S., Gilbert W. B lineage--specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985 Jan 11;227(4683):134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Ohno S., Yasumitsu H., Taniguchi T. Delimitation and properties of DNA sequences required for the regulated expression of human interferon-beta gene. Cell. 1985 Jun;41(2):489–496. doi: 10.1016/s0092-8674(85)80022-2. [DOI] [PubMed] [Google Scholar]
- Gillies S. D., Morrison S. L., Oi V. T., Tonegawa S. A tissue-specific transcription enhancer element is located in the major intron of a rearranged immunoglobulin heavy chain gene. Cell. 1983 Jul;33(3):717–728. doi: 10.1016/0092-8674(83)90014-4. [DOI] [PubMed] [Google Scholar]
- Gilman M. Z., Wilson R. N., Weinberg R. A. Multiple protein-binding sites in the 5'-flanking region regulate c-fos expression. Mol Cell Biol. 1986 Dec;6(12):4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez N., Wiggs J., Chamberlin M. J. A simple procedure for resolution of Escherichia coli RNA polymerase holoenzyme from core polymerase. Arch Biochem Biophys. 1977 Aug;182(2):404–408. doi: 10.1016/0003-9861(77)90521-5. [DOI] [PubMed] [Google Scholar]
- Goodbourn S., Zinn K., Maniatis T. Human beta-interferon gene expression is regulated by an inducible enhancer element. Cell. 1985 Jun;41(2):509–520. doi: 10.1016/s0092-8674(85)80024-6. [DOI] [PubMed] [Google Scholar]
- Graves B. J., Johnson P. F., McKnight S. L. Homologous recognition of a promoter domain common to the MSV LTR and the HSV tk gene. Cell. 1986 Feb 28;44(4):565–576. doi: 10.1016/0092-8674(86)90266-7. [DOI] [PubMed] [Google Scholar]
- Gruss P., Dhar R., Khoury G. Simian virus 40 tandem repeated sequences as an element of the early promoter. Proc Natl Acad Sci U S A. 1981 Feb;78(2):943–947. doi: 10.1073/pnas.78.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadesch T., Zervos P., Ruezinsky D. Functional analysis of the murine IgH enhancer: evidence for negative control of cell-type specificity. Nucleic Acids Res. 1986 Oct 24;14(20):8209–8221. doi: 10.1093/nar/14.20.8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga J. T., Tjian R. Affinity purification of sequence-specific DNA binding proteins. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5889–5893. doi: 10.1073/pnas.83.16.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury G., Gruss P. Enhancer elements. Cell. 1983 Jun;33(2):313–314. doi: 10.1016/0092-8674(83)90410-5. [DOI] [PubMed] [Google Scholar]
- Landolfi N. F., Capra J. D., Tucker P. W. Interaction of cell-type-specific nuclear proteins with immunoglobulin VH promoter region sequences. Nature. 1986 Oct 9;323(6088):548–551. doi: 10.1038/323548a0. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mercola M., Goverman J., Mirell C., Calame K. Immunoglobulin heavy-chain enhancer requires one or more tissue-specific factors. Science. 1985 Jan 18;227(4684):266–270. doi: 10.1126/science.3917575. [DOI] [PubMed] [Google Scholar]
- Mercola M., Wang X. F., Olsen J., Calame K. Transcriptional enhancer elements in the mouse immunoglobulin heavy chain locus. Science. 1983 Aug 12;221(4611):663–665. doi: 10.1126/science.6306772. [DOI] [PubMed] [Google Scholar]
- Moreau P., Hen R., Wasylyk B., Everett R., Gaub M. P., Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981 Nov 25;9(22):6047–6068. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuberger M. S. Expression and regulation of immunoglobulin heavy chain gene transfected into lymphoid cells. EMBO J. 1983;2(8):1373–1378. doi: 10.1002/j.1460-2075.1983.tb01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson C. L., Orth K., Calame K. L. Binding in vitro of multiple cellular proteins to immunoglobulin heavy-chain enhancer DNA. Mol Cell Biol. 1986 Dec;6(12):4168–4178. doi: 10.1128/mcb.6.12.4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riggs A. D., Bourgeois S., Cohn M. The lac repressor-operator interaction. 3. Kinetic studies. J Mol Biol. 1970 Nov 14;53(3):401–417. doi: 10.1016/0022-2836(70)90074-4. [DOI] [PubMed] [Google Scholar]
- Rosenfeld P. J., Kelly T. J. Purification of nuclear factor I by DNA recognition site affinity chromatography. J Biol Chem. 1986 Jan 25;261(3):1398–1408. [PubMed] [Google Scholar]
- Sassone-Corsi P., Wildeman A., Chambon P. A trans-acting factor is responsible for the simian virus 40 enhancer activity in vitro. Nature. 1985 Feb 7;313(6002):458–463. doi: 10.1038/313458a0. [DOI] [PubMed] [Google Scholar]
- Schlokat U., Bohmann D., Schöler H., Gruss P. Nuclear factors binding specific sequences within the immunoglobulin enhancer interact differentially with other enhancer elements. EMBO J. 1986 Dec 1;5(12):3251–3258. doi: 10.1002/j.1460-2075.1986.tb04636.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholer H., Haslinger A., Heguy A., Holtgreve H., Karin M. In vivo competition between a metallothionein regulatory element and the SV40 enhancer. Science. 1986 Apr 4;232(4746):76–80. doi: 10.1126/science.3006253. [DOI] [PubMed] [Google Scholar]
- Schöler H. R., Gruss P. Cell type-specific transcriptional enhancement in vitro requires the presence of trans-acting factors. EMBO J. 1985 Nov;4(11):3005–3013. doi: 10.1002/j.1460-2075.1985.tb04036.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöler H. R., Gruss P. Specific interaction between enhancer-containing molecules and cellular components. Cell. 1984 Feb;36(2):403–411. doi: 10.1016/0092-8674(84)90233-2. [DOI] [PubMed] [Google Scholar]
- Sen R., Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986 Aug 29;46(5):705–716. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- Staudt L. M., Singh H., Sen R., Wirth T., Sharp P. A., Baltimore D. A lymphoid-specific protein binding to the octamer motif of immunoglobulin genes. Nature. 1986 Oct 16;323(6089):640–643. doi: 10.1038/323640a0. [DOI] [PubMed] [Google Scholar]
- Storb U., Wilson R., Selsing E., Walfield A. Rearranged and germline immunoglobulin kappa genes: different states of DNase I sensitivity of constant kappa genes in immunocompetent and nonimmune cells. Biochemistry. 1981 Feb 17;20(4):990–996. doi: 10.1021/bi00507a053. [DOI] [PubMed] [Google Scholar]
- Treisman R., Maniatis T. Simian virus 40 enhancer increases number of RNA polymerase II molecules on linked DNA. Nature. 1985 May 2;315(6014):73–75. doi: 10.1038/315072a0. [DOI] [PubMed] [Google Scholar]
- Wasylyk C., Wasylyk B. The immunoglobulin heavy-chain B-lymphocyte enhancer efficiently stimulates transcription in non-lymphoid cells. EMBO J. 1986 Mar;5(3):553–560. doi: 10.1002/j.1460-2075.1986.tb04246.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger J., Baltimore D., Sharp P. A. Distinct factors bind to apparently homologous sequences in the immunoglobulin heavy-chain enhancer. 1986 Aug 28-Sep 3Nature. 322(6082):846–848. doi: 10.1038/322846a0. [DOI] [PubMed] [Google Scholar]
- Zenke M., Grundström T., Matthes H., Wintzerith M., Schatz C., Wildeman A., Chambon P. Multiple sequence motifs are involved in SV40 enhancer function. EMBO J. 1986 Feb;5(2):387–397. doi: 10.1002/j.1460-2075.1986.tb04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Villiers J., Schaffner W. A small segment of polyoma virus DNA enhances the expression of a cloned beta-globin gene over a distance of 1400 base pairs. Nucleic Acids Res. 1981 Dec 11;9(23):6251–6264. doi: 10.1093/nar/9.23.6251. [DOI] [PMC free article] [PubMed] [Google Scholar]