Abstract
Background:
The tyrosine kinase receptor HER4 is a member of the epidermal growth factor receptor (EGFR) family. It plays diverse roles in cancer development and cancer progression and can both exert oncogenic and tumour-suppressive activities. Alternatively spliced isoforms of HER4 are critical to the different signalling possibilities of HER4.
Methods:
We use a splice-switching oligonucleotide (SSO) to direct the alternative splicing of HER4 from the CYT1 to the CYT2 isoform in HER4-expressing breast cancer cells.
Results:
Treatment with a target-specific SSO was accompanied by a decreased growth of the cells (P<0.0001). In addition, the SSO treatment induced a decreased activity of Akt. We confirmed the SSO-dependent switching of the HER4 isoform CYT1 to CYT2 expression in a xenografted mouse tumour model driven by subcutaneously injected MCF7 cells. We hence demonstrated the feasibility of SSO-directed splice-switching activity in vivo. Furthermore, the SSO treatment efficiently decreased the growth of the xenografted tumour (P=0.0014).
Conclusion:
An SSO directing the splicing of HER4 towards the CYT2 isoform has an inhibitory effect of cancer cell growth in vitro and in vivo. These results may pave the way for the development of new anticancer drugs in HER4-deregulated cancers in humans.
Keywords: HER4, Erbb4, alternative splicing, splice-switching oligonucleotide (SSO), EGFR family, breast cancer
Human epidermal growth factor (EGF) receptor 4 (HER4/ErbB4) belongs to the EGF receptor (EGFR/HER1/ErbB1) family, a family of transmembrane tyrosine kinase receptors and their ligands. Besides EGFR and HER4, the family includes HER2 (ErbB2/Neu) and HER3 (ErbB3) and at least 12 ligands. The receptors play various roles in the growth and maintenance of numerous tissues, among others by regulating cell division, apoptosis, differentiation, and migration. Dysregulation of the EGFR family is involved in the development and growth of numerous epithelial cancers (Yarden and Sliwkowski, 2001; Hynes and Macdonald, 2009). The oncogenic nature of EGFR and HER2 has led to clinical use of specific inhibitors of these two receptors for the treatment of different cancers (Gschwind et al, 2004). In addition, HER4 has been found to be oncogenic. Thus, the receptor is overexpressed in a number of carcinomas of the central nervous system such as medullablastomas, ependymomas, glioblastomas, and pilocytic astrocytomas (Carpenter, 2003; Hollmen and Elenius, 2010). The expression of HER4 is associated with a late tumour stage in colorectal cancer and with distant metastasis and decreased overall survival in oral squamous cell carcinomas (Xia et al, 1999; Lee et al, 2002; Hollmen and Elenius, 2010). It has therefore been proposed that HER4 may be a potential candidate for cancer treatment. On the other hand, HER4 has also in some cases been shown to be an indicator of a well-differentiated phenotype and a good prognosis. Thus, HER4 expression is associated with a better survival in breast, bladder, and cervix cancer (Memon et al, 2004; Lee et al, 2005; Uberall et al, 2008); and HER4 expression is often absent in aggressive cancers of the pancreas, larynx, and prostate (Thybusch-Bernhardt et al, 2001; Saglam et al, 2007; Uberall et al, 2008).
The recent findings that HER4 is the most frequently mutated kinase in metastatic melanomas (19%) and that gene amplification of HER4 is frequent in gastric cancer (67%) make HER4 an attractive target for cancer therapy (Prickett et al, 2009; Shi et al, 2012). On this background, there is a need for a better understanding of the conditions behind the receptor's oncogenic and tumour-suppressing functions. The HER4-activating ligands and dimerisation partners present in the specific cellular environment contribute to the variation in signalling (Mill et al., 2011b) as does the cellular localisation of HER4 (Junttila et al, 2005; Sundvall et al, 2007). However, another important reason for the variable functions of HER4 is that the expression of different, alternatively spliced isoforms is associated with different biological functions.
By alternative splicing, HER4 is expressed in a number of isoforms. In the juxtamembraneous area of HER4, inclusion of either exon 15b or exon 16 results in the JM-a or JM-b isoforms, respectively (Elenius et al, 1997; Rio et al, 2000; Cheng et al, 2003). The JM-a isoform and not the JM-b isoform can be processed to release a soluble HER4 intracellular domain (ICD) that localises to the cytoplasm, nucleus, and mitochondria (Ni et al, 2001; Naresh et al, 2006). The HER4 ICD localisation in the nucleus is associated with poor survival (Junttila et al, 2005); and HER4 ICD can induce apoptosis when associated with mitochondria (Naresh et al, 2006).
Further complexity in the HER4 structure arises from alternative splicing in the cytoplasmic region of HER4, which gives rise to the CYT1 and CYT2 isoforms. The skipping of exon 26 results in a receptor (CYT2) lacking 16 of the amino acids of the full-length receptor (CYT1) (Elenius et al, 1999).
The 16 amino acid sequence, which is unique to CYT1, encompasses a PPXY motif that comprises the only direct phosphoinositide 3-kinase (PI3K) interaction site of HER4. This indicates that CYT1, but not CYT2, is capable of directly binding and activating PI3K (Elenius et al, 1999). Hence, through this interaction, only CYT1 can directly influence PI3K-mediated survival and inhibit apoptosis (Kainulainen et al, 2000). In accordance with this finding, CYT1 was found to have a more proliferative phenotype than CYT2 in medulla blastoma cells because of its ability to escape apoptosis through activation of the PI3K/Akt pathway (Ferretti et al, 2006). Besides having an inhibitory effect on apoptosis, CYT1 has also been found to induce chemotaxis (Kainulainen et al, 2000) and to increase proliferation and decrease differentiation in rat adrenal gland pheochromocytoma cells (Veikkolainen et al, 2011). These results point to CYT1 as the main contributor to the oncogenic activities of HER4. However, results have also been obtained that suggest that CYT1 and CYT2 have the opposite effects. In mammary epithelial cells, the cleavable 80 kD cytoplasmic domain of CYT1 was found to decrease growth and induce a more differentiated phenotype, whereas the cleavable 80 kD cytoplasmic domain of CYT2 increased growth and induced no differentiation (Muraoka-Cook et al, 2009). The CYT1-unique PPXY motif directs ubiquitination and degradation as well as inhibits CYT1 from entering the cell nucleus to the same extent as CYT2, and HER4 nuclear localisation is associated with poor survival (Komuro et al, 2003; Aqeilan et al, 2005; Junttila et al, 2005; Omerovic et al, 2007; Sundvall et al, 2007; Zeng et al, 2007; Sundvall et al, 2008; Zeng et al, 2009). Even though cell line studies give no clear indications as to which of the two cytoplasmic isoforms is the most oncogenic, the isoform status of HER4 undoubtedly contributes to the outcome of HER4 activity.
Clinical data on the prognostic value of CYT1 and CYT2 expression are sparse. However, in medulla blastoma patients the CYT1/CYT2 ratio was found to be higher in aggressive tumours than in less aggressive tumours (Ferretti et al, 2006), and in ovarian cancer, the CYT1 variant but not the CYT2 variant was associated with poor survival (Paatero et al, 2013). These data suggest that CYT1 is the more tumourigenic cytoplasmic isoform of HER4.
An evaluation of the potential of HER4 as a target for cancer therapy should take into consideration whether it is desirable to target the total pool of HER4 isoforms or only specific isoforms. An antibody specific for the JM-a isoform of HER4 has been found to inhibit the growth of breast cancer cells (Hollmen et al, 2009); however, apart from this, no studies have explored the effect of inhibiting specific splicing isoforms of HER4 on cancer cell growth.
Exon skipping to favour the expression of one splice variant over another can be done with splice-switching oligonucleotides (SSOs) (reviewed by Sazani and Kole, 2003). By hybridisation to pre-mRNA, SSOs sterically block sequences important for alternative splicing. This prevents exon recognition by the splicing machinery and causes the exon to be skipped (Sierakowska et al, 1996). Modifications to the oligonucleotide backbone such as 2′-O-methyl, 2′-O-methoxyethyl, and locked nucleic acids (LNAs) prevent SSO-induced antisense downregulation via RNaseH and also stabilise the oligoes towards nucleolytic degradation. These modifications also usually increase the binding affinity of the pre-mRNA target, and the increased competitiveness with spliceosome may account for the higher efficacy in vivo (Sazani and Kole, 2003; Roberts et al, 2006). The splice-switching capacities of SSOs have been demonstrated in several cell culture studies (Cartegni and Krainer, 2003; Scaffidi and Misteli, 2005; Williams and Kole, 2006; Khoo et al, 2007). More recently, successful SSO-directed splice-switching activity in mouse models has opened up the possibility that SSOs can have therapeutic applications in human diseases in which modulation of alternative splicing is desirable. Most advanced are the clinical trials on patients suffering from Duchenne muscular dystrophy, where SSO-mediated restoration of the aberrant splicing of the dystrophin pre-mRNA has shown some efficacy (van Deutekom et al, 2007; Graziewicz et al, 2008; Svasti et al, 2009; Bauman et al, 2010).
The purpose of our study was to investigate the possibility of targeting only one cytoplasmic isoform of HER4 and to further study the different functions of CYT1 and CYT2. We have investigated the ability of an SSO molecule to direct HER4 mRNA splicing towards the CYT2 isoform at the expense of the CYT1 isoform and demonstrate that a particular SSO can effectively decrease the CYT1/CYT2 ratio. The alteration of HER4 isoform expression is accompanied by a decreased growth of breast cancer cells. This is further demonstrated in a xenograft mouse model in which the SSO-induced switch in alternative splicing towards the CYT2 isoform of HER4 in the inoculated tumour causes decreased tumour growth.
Materials and methods
Cell lines
The human breast cancer cell lines MCF7 and T47D were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). The cells were tested for growth properties, morphology, mycoplasma contamination, and species by ATCC. Cells were cultured at 37 °C in a humidified air atmosphere with 5% CO2. The MCF7 cells were grown in DMEM medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin and T47D cells were grown in RPMI medium supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% hepes, and 1% pyruvate (all media and supplements were from Life Technologies Carlsbad, CA, USA).
Oligonucleotide treatment
A 15-mer-LNA modified oligoribonucleotide SSOe26 (5′-(l)T (m)U (l)A (m)C (m)U (l)C (m)C (m)U (l)G (m)A (m)C (l)A (m)U (l)G (l)G-3′ (l=LNA, m=methylated)) antisense to the 5′ splice site of HER4 exon 26 was used. A scrambled oligonucleotide, SSOsc, containing the same nucleotides as SSOe26 at scrambled positions and modifications at the same positions as in SSOe26, was used as a negative control (5′-(l)G (m)T (l)T (m)C (m)A (l)G (m)T (m)C (l)G (m)C (m)A (l)T (m)A (l)C (l)T-3′). Both oligonucleotides were synthesised and purified by RiboTask ApS, Langeskov, Denmark.
Cells were plated in serum-free medium 24 h before oligonucleotide treatment. The cells were treated with oligonucleotides complexed with Lipofectamine 2000 (Life Technologies) according to the manufacturer's directions at the concentrations indicated in the figures. Analyses were carried out 24 h after transfection for quantitative reverse transcription PCR (Q–PCR) and western blotting or continuously for cell proliferation assays.
RNA isolation and Q–PCR
The RNA was purified according to the manufacturer's directions (Qiagen ‘Total RNA isolation kit' (Qiagen, Chatsworth, CA, USA)) and resuspended in RNase-free water. The amount of purified RNA was quantified by UV spectroscopy (A260 nm=1 corresponds to 40 μg ml−1 RNA). Complementary DNA (cDNA) synthesis for the total HER4 assay was obtained from 0.1 μg of RNA and 0.4 μg of RNA was used for cDNA synthesis for the CYT1 and CYT2 assays. For the reverse transcription (RT) reaction was used: 1 × PCR buffer (Life Technologies, Carlsbad, CA, USA), 6.3 mmol l−1 MgCl2 (Applied Biosystems), 25 mmol l−1 each of dATP/dTTP/dGTP and dCTP (Pharmasia), 2.5 mmol l−1 16-mer oligo dT primer (DNA Technology), 20 U RNase inhibitor (Life Technologies), and 50 U reverse transcriptase (Life Technologies). Temperature profile was as follows: 42 °C for 30 min; 96 °C for 5 min; 4 °C end.
cDNA (1 μl) was used for Q-PCR using SYBR green Master (Roche, Basel, Switzerland) on a LightCycler 480 at: 95 °C for 10 min; 50 cycles of 95 °C for 10 s, specific annealing temperature for 10 s, 72 °C for 5 s; 99 °C for 1 s; 59 °C for 15 s; 95 °C for 1 s; and cooling to 40 °C. The forward and reverse primers were HER4: forward: 5′-ACAGCAGTACCGAGCCTTTGCG-3′ reverse: 5′-GCCACTACCACGTAGCCTGTGAC-3′. CYT1: forward: 5′- GGATGAAGAGGATTTGGAAG -3′ reverse: 5′- TCCTGACATGGGGGTGTA-3′. CYT2: forward: 5′- GAATAGGAACCAGTTTGTATACCG-3′ reverse: 5′- ACAGCAGGAGTCATCAAAAATC-3′. Annealing temperatures were HER4: 64 °C; CYT1 and CYT2: 56 °C.
Western blotting
Cells were collected in scraping buffer and pelleted by centrifugation. Cell pellets were incubated on ice for 15 min in RIPA buffer (50 mℳ Tris-HCL pH 7.4, 150 mℳ NaCl, 1% NP-40, 0.25% Na-deoxycholate, 1 mℳ PMSF, 1 mℳ Orthovanadate, 0.5 ng ml−1 Aprotinin, 0.5 ng ml−1 Chymostatin, 0.5 ng ml−1 Leupeptin, and 0.5 ng ml−1 Pepstatin) and homogenised with gentle vortexing and cleared by centrifugation. The protein concentration was determined with BCA reagent (Pierce Chemical, Rockford, IL, USA). Equal amounts of protein (30 μg) were resolved by 8–12% SDS–PAGE. The resolved proteins were transferred onto PVDF membrane, and blocked with 5% (w/v) nonfat dry milk in TBS-T solution (25 mℳ Tris base, pH 7.5, 150 mℳ NaCl, 0.05% (w/v) Tween-20). After washing, the blots were incubated with specific primary and secondary antibodies according to the data sheets provided by the manufacturers. Immunoreactive bands were detected by ECL reagents (GE Healthcare, Little Chalfont, UK). Antibodies used for western blotting were: total HER4 (Epitomics, Burlingame, CA, USA), β-actin (Sigma, St Louis, MO, USA), p-Akt (Ser473; Cell Signaling Technology), total-Akt (Cell Signaling Technology, Danvers, MA, USA), p-44/42 MAPK (Thr202/Tyr204; Cell Signaling Technology, Danvers, MA, USA), and total-44/42 MAPK (Cell Signaling Technology).
Cell proliferation assay
Cell proliferation was measured using Roche xCELLigence system.
The xCELLigence system measures electrical impedance across interdigitated micro-electrodes integrated on the bottom of tissue culture E-Plates. The cell index (CI) is derived as a relative change in measured electrical impedance to represent cell status.
A total of 50 μl of cell culture medium in each well of an E-plate 96 was used for determination of background impedance. A total of 100 μl of cell suspension was subsequently added (5000 cells per well). Cells were incubated for 30 min at room temperature before placing the E-plate in the Real-Time Cell Analyzer (RTCA) station. Impedance was measured every 1 h. At 24 h post cell seeding, 50 μl of media containing Lipofectamine 2000 or oligonucleotides complexed with Lipofectamine 2000 was added. Cell index was measured using the software provided by the manufacturer and normalised to the time just after treatment.
Tumour xenografts
At 4 days before cell inoculation, a β-estradiol pellet (0.5 mg, 60-day release; Innovative Research of America, Sarasota, FL, USA) was implanted subcutaneously in the neck region of the mouse to support tumour growth. MCF7 cells (4 × 106 cells in 200 μl of PBS) were injected subcutaneously into the right flank of each mouse (9 weeks old, female, nu/nu BALB/c (C.Cg/AnBomTac-Foxn1nu; Taconic, Ejby, Denmark)). Isoflurane (Abbott, Copenhagen, Denmark) was used for anaesthesia for implantation of pellets and inoculation of tumour cells. Tumours were allowed to grow until visible (1 week; average volume 83 mm3 (tumour volume=½ (length × width2)) before they were randomly allocated into two groups with approximately the same total tumour volume in each group (6 mice in each group). Treatment was performed with 400 μg SSOe26 or SSOsc in 150 μl PBS intraperitoneally three times per week. Tumour sizes were measured at the days of treatment by caliper measurements of the largest longitudinal (length) and transverse (width) diameter; the tumour volume was then calculated by the formula given above. At the end of the experiment, the mice were killed and the tumours were removed and snap-frozen for RNA isolation. The tumours were mechanically homogenised and RNA was extracted as described above. Animal experiments were conducted with approval from The Danish Experimental Animal Inspectorate.
Statistics
Differences in mRNA expressions were analysed using t-tests.
Cell growth and in vivo tumour growth were analysed using univariate ANOVA models for repeated measurements on the log-transformed data.
Results
SSOe26 shifts the splicing equilibrium of the CYT1 and CYT2 isoforms of HER4
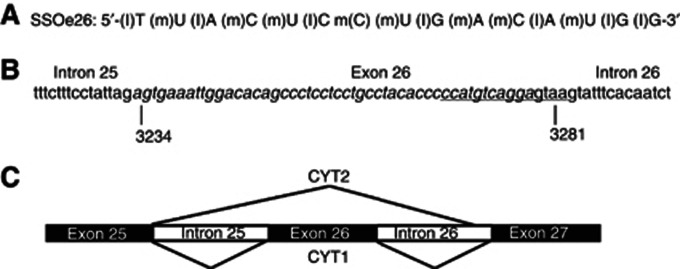
A 15-mer LNA-modified oligoribonucleotide, SSOe26 (SSO exon 26; Figure 1A), was designed to anneal to the 5′ splice site of exon 26 of the HER4 pre-mRNA (Figure 1B). The nucleotide sequence of SSOe26 was scrambled to create a control oligo, SSOsc (SSO scrambled). By annealing to the 5′ splice site of exon 26, SSOe26 makes the splice site inaccessible for the splicing machinery, which results in exon skipping and thereby expression of the CYT2 isoform (Figure 1C).
Figure 1.
Sequence and annealing position of SSOe26. (A) Sequence and backbone modifications of SSOe26. (l)=LNA, (m)=2′-O-methyl. (B) Part of the HER4 pre-mRNA sequence showing the 3′-end of intron 25, exon 26 (italic) and the 5′-end of intron 26. The sequence of 15 nucleotides annealing with SSOe26 is underlined. The numbers indicate the nucleotide number of the first and the last nucleotide in exon 26 according to NM_005235.2, NCBI. (C) Alternative splicing of exon 26 in the HER4 pre-mRNA. Inclusion of exon 26 in the HER4 pre-mRNA generates the CYT1 isoform. Skipping of exon 26 generates the CYT2 isoform.
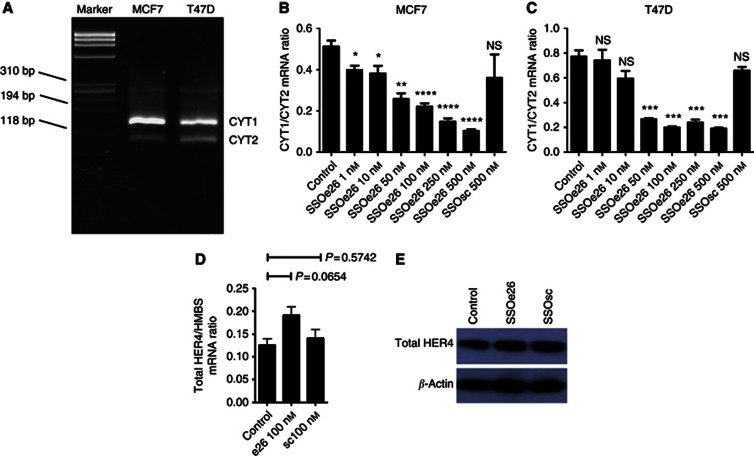
Both MCF7 and T47D, two breast cancer cell lines, express HER4. To determine the relative CYT1 and CYT2 expression in these two cell lines, a semiquantitative PCR assay was performed. For this assay one set of primers anneal to either site of exon 26 to give PCR products of different size depending on isoform expression. The isoforms can be visualised upon gel electrophoresis. This assay revealed that in both cell lines, the CYT1 variant is the predominant cytoplasmic isoform of HER4 (Figure 2A). The MCF7 and T47D cells were then transfected with SSOe26 at increasing concentrations as indicated in Figures 2B and C. Cells were harvested 24 h after transfection, and CYT1 and CYT2 mRNA expression was quantified by Q-PCR. A dose-dependent decrease in the ratio of the CYT1/CYT2 isoform expression was induced by SSOe26 treatment (Figures 2B and C). For the MCF7 cells, a decrease was seen with as little as 1 nℳ of SSOe26 (P=0.0158), and with 100 nℳ SSOe26, the CYT1/CYT2 ratio was reduced to approximately half of the ratio of untreated cells (mean ratio for control cells: 0.51±0.027 (s.e.m.); mean ratio for SSOe26-treated cells: 0.22±0.015 (s.e.m.)). For the T47D, 50 nℳ of SSOe26 was necessary to give a significant decrease in the CYT1/CYT2 ratio (P=0.0005). Treatment with SSOsc at the highest dose used for SSOe26 (500 nℳ) did not change the CYT1/CYT2 ratio significantly (P=0.3894 for MCF7 cells and P=0.1135 for T47D cells).
Figure 2.
The effect of SSOe26 and SSOsc on HER4 CYT1/CYT2 mRNA ratio and on total HER4 mRNA and protein expression. (A) Agarose gel electrophoresis depicting the mRNA expression of the CYT1 and CYT2 isoforms in MCF7 and T47D cells. The CYT1 amplicon contains additional 45 amino acids compared with CYT2 and, hence, CYT1 is more retarded in the gel. Bands representing CYT1 and CYT2 are indicated in the figure. A third of ∼300 bp represents an artefact as a result of the smaller products annealing together (Elenius et al, 1999). (B) The mRNA expression ratio of CYT1 and CYT2 in MCF7 cells treated with increasing concentrations of SSOe26. The cells were treated with fresh media containing transfection reagent (control) or indicated amounts of SSOe26 or SSOsc formulated with transfection reagent as indicated. Bars represent means of four individual samples, error bars are s.e.m. Asterisks indicate the level of significance (*P⩽0.05; **P⩽0.01; ***P⩽0.001; and ****P⩽0.0001). (C) The mRNA expression ratio of CYT1 and CYT2 in T47D cells treated with increasing concentrations of SSOe26. The cells were treated with fresh media containing transfection reagent (control) or indicated amounts of SSOe26 or SSOsc formulated with transfection reagent as indicated. Bars represent means of four individual samples, error bars are s.e.m. Asterisks indicate the level of significance (***P⩽0.001). (D) Total HER4 mRNA expression relative to the mRNA expression of the HMBS household gene. The cells were treated with fresh media containing transfection reagent (control) or 100 nℳ of SSOe26 or 100 nℳ of SSOsc formulated with transfection reagent as indicated. Bars represent means of four individual samples, error bars are s.e.m. (E) Total HER4 protein expression. The cells were treated with fresh media containing transfection (control) reagent or 100 nℳ of SSOe26 or 100 nℳ of SSOsc formulated with transfection reagent as indicated. Stripping and reblotting with β-actin demonstrates equal loading.
To ensure that binding of SSOe26 did not cause degradation of the HER4 pre-mRNA, total HER4 mRNA expression was quantified by Q-PCR in MCF7 cells (Figure 2D). The SSOe26 did not decrease mRNA expression of HER4. On the contrary, a small, although not significant (P=0.0654), increase was observed. The SSOsc had no effect on HER4 mRNA expression. Also, on protein level, neither SSOe26 nor SSOsc induced a change in the HER4 expression as demonstrated by western blotting (Figure 2E).
Together, these results demonstrate that SSOe26 successfully induces splice switching of HER4 to decrease the ratio of CYT1/CYT2 isoform expression without inhibiting total HER4 expression.
SSOe26 decreases breast cancer cell growth
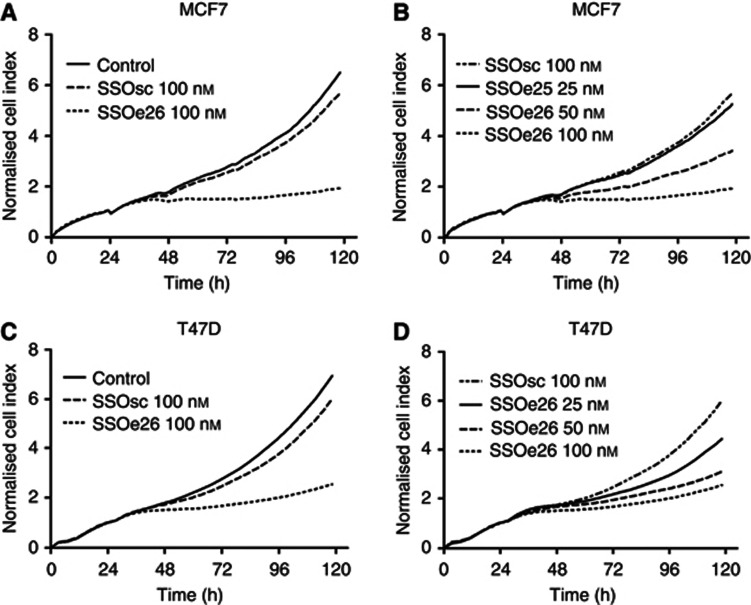
The effect of an SSO-induced decrease in the CYT1/CYT2 isoform ratio was assessed by real-time measurements of MCF7 and T47D cell growth. Compared with the control cells, splice switching of HER4 with SSOe26 caused an almost complete growth arrest (P<0.0001; Figures 3A and C). The SSOsc had a negligible effect on cell growth. These results from a real-time growth experiment were reproduced using a calorimetric method to investigate cell growth (MTS assay, results not shown).
Figure 3.
Growth inhibition of MCF7 and T47D cells by SSOe26 treatment. Proliferation was measured as the cell index using a Real-Time Cell Analyzer (RTCA) station. (A) MCF7 cells were treated with fresh media containing transfection reagent (control) or SSOe26 (100 nℳ) or SSOsc (100 nℳ) formulated with transfection reagent. Shown are the mean of triplicates. (B) Growth inhibition of MCF7 cells by increasing concentrations of SSOe26. Cells were treated with 25, 50 and 100 nℳ SSOe26 or 100 nℳ of SSOsc as indicated. Shown are the mean of triplicates. (C) T47D cells were treated with fresh media containing transfection reagent (control) or SSOe26 (100 nℳ) or SSOsc (100 nℳ) formulated with transfection reagent. Shown are the mean of triplicates. (D) Growth inhibition of T47D cells by increasing concentrations of SSOe26. Cells were treated with 25, 50 and 100 nℳ SSOe26 or 100 nℳ of SSOsc as indicated. Shown are the mean of triplicates.
The SSOe26-induced inhibition of cell growth was dose dependent and decreased with decreasing concentrations of SSOe26 (Figures 3B and D).
SSOe26 alters the activity of signalling proteins and induces apoptosis
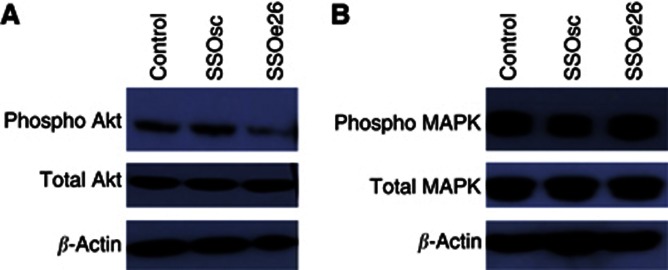
Two of the main pathways activated by HER4 are the MAPK pathway and the PI3K/Akt pathway, the latter specific for CYT1. We investigated how splice switching with SSOe26 influences the activity of these pathways. Phosphorylation of Akt in response to SSOe26 treatment was examined by western blotting and resulted in decreased phosphorylation of Akt compared with cells treated with SSOsc (Figure 4A). The total amount of Akt protein was unchanged upon treatment (Figure 4A). The phosphorylation status and the total protein content of MAPK as a result of SSOe26 treatment were also examined by western blotting. We detected no change in either the phosphorylation status or the total protein expression of this protein (Figure 4 B).
Figure 4.
Western blotting of total protein extracts from MCF7 cells treated with fresh media containing transfection reagent (control) or 100 nℳ of SSOsc or 100 nℳ of SSOe26 formulated with transfection reagent as indicated. Cells were harvested 24 h after treatment. Stripping and reblotting with β-actin demonstrates equal loading. (A) Protein expression of phosphorylated Akt and total Akt. (B) Protein expression of phosphorylated MAPK and total MAPK.
SSOe26 induces splice switching and decreases tumour growth in mouse xenografts
After observing a splice-switching activity of SSOe26 in cell culture studies, the ability of SSOe26 to induce splice switching in vivo was tested. The MCF7 breast cancer cells were planted subcutaneously onto the right flank of immune-deficient mice and SSOe26 or SSOsc was hereafter injected intraperitoneally three times per week. After 15 days of treatment, the tumours were removed and CYT1 and CYT2 mRNA expression was quantified by Q-PCR.
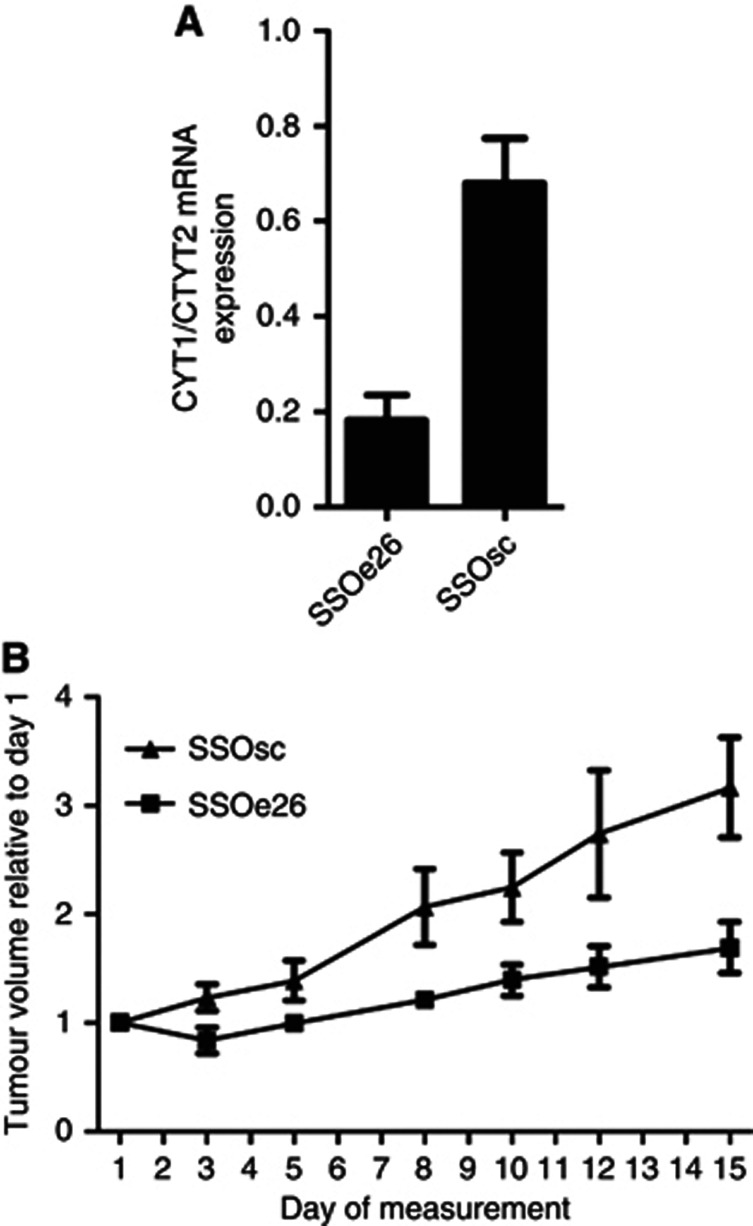
In accordance with the cell culture experiments, tumours from mice receiving SSOe26 had a substantially lower CYT1/CYT2 mRNA ratio than tumours from mice receiving the control oligo (P<0.0001), which demonstrates the splice-switching activity of SSOe26 in vivo (Figure 5A).
Figure 5.
Tumour growth of a xenograft mouse model. Mice received 400 μg of SSOe26 or SSOsc intraperitoneally three times per week for 15 days. (A) The mRNA expression ratio of CYT1 and CYT2 in tumours removed from mice treated with SSOe26 or SSOsc as indicated. Shown are mean of six mice and error bars indicate s.e.m. (B) The tumour size was manually measured three times per week for 15 days. Size was calculated as ½ × (length × with2). Shown are mean of six mice. Error bars indicate s.e.m.
The effect of SSOe26 on the growth of the tumour xenografts was assessed by manually measuring the tumour size three times/week. The tumours of SSOe26-treated mice showed a significantly slower growth than the tumours of SSOsc-treated mice (P=0.0014; Figure 5B).
The in vivo results clearly demonstrate the ability of SSOe26 to reach the subcutaneous location of the xenograft tumour from the intraperitoneal cavity site of injection and to induce splice-switching activity in this setting. The decreased tumour growth indicates that CYT2 has less proliferative potential than CYT1 in these tumours.
Discussion
Divergent data exist on the role of HER4 in tumour growth and development. In some settings, HER4 exerts tumourigenic phenotype characteristics in accordance with the biology of its family members EGFR and HER2; however, in other settings, tumour HER4 expression is an indicator of a better survival of cancer patients (Hollmen and Elenius, 2010). The existence of alternatively spliced isoforms of the receptor most likely contributes to the contradictory data on HER4 because the alternatively spliced isoforms have been reported to exert different biological effects (Veikkolainen et al, 2011). The HER4 has been described as a possible new specific target for cancer therapy in numerous connections (Kurppa and Elenius, 2009; Settleman, 2009; Rudloff and Samuels, 2010). For this purpose, targeting only one isoform of HER4 may be preferable. We investigated the potential of using SSO to modulate the splicing of HER4 pre-mRNA into its CYT1 and CYT2 isoforms. The purpose of this was to investigate the possibility of targeting only one cytoplasmic isoform of HER4 and to further study the different functions of CYT1 and CYT2.
The use of an SSO that targets the junction of intron 26 and exon 26 resulted in a successful modulation of the HER4 pre-mRNA that decreased the CYT1/CYT2 ratio both in vitro and in vivo. The ability of the SSO to successfully change the expression of HER4 cytoplasmic isoforms makes SSO molecules a useful tool for investigating the function of the CYT1 and CYT2 splice variants. The majority of studies on differences between CYT1 and CYT2 functions rely on an exogenous expression of the full-length HER4 or the ICD of the HER4, which leads to excessive overexpression of the gene. In contrast, the SSO functions in settings where expression levels and background environments are physiologically relevant, which makes this an attractive alternative to overexpression.
One of the purposes of backbone modifications of the SSO is to prevent mRNA degradation by RNA interference. In our studies, the mRNA expression level was, indeed, conserved. On the contrary, we observed a slight increase in the total HER4 mRNA level. This increase could be a result of regulatory mechanisms responding to the binding to the mRNA and/or response to regulations of HER4 isoform expression patterns. The change in mRNA expression was not, however, translated to the protein level.
The SSO-induced decrease in the CYT1/CYT2 ratio was accompanied by a decrease in the growth of the HER4-expressing MCF7 and T47D breast cancer cells. This finding was supported by the ability of the SSO to inhibit tumour growth in the xenograft mouse model. It has previously been found that downregulation of endogenous HER4 in MCF7 breast cancer cells reduces cell growth in vitro and in vivo (Tang et al, 1999). Our results indicate that it is the downregulation of the CYT1 isoform of HER4 that gives rise to this growth inhibition. The CYT1, but not CYT2, directly couples to the PI3K/Akt pathway, and CYT1, but not CYT2, can execute an inhibitory effect on apoptosis via the PI3K/Akt pathway (Kainulainen et al, 2000). We did, indeed, see a decrease in Akt activity in cells upon switching from the CYT1 towards the CYT2 isoform. The downstream targets of Akt are numerous. In medulla blastoma cells, CYT1 overexpression has a PI3k/Akt-dependent inhibitory effect on apoptose upon serum starvation or etopside treatment (Ferretti et al, 2006). Also, CYT1 has been found to induce chemotaxis (Kainulainen et al, 2000). In rat adrenal gland pheochromocytoma cells, CYT1 expression promoted less differentiation and increased cell proliferation compared with CYT2 (Veikkolainen et al., 2011). Together with our present results, these findings add to the picture of CYT1 as a more carcinogenic isoform than CYT2, and they suggest that targeting only this isoform of HER4 is therapeutically most relevant.
However, results have also been obtained that suggest that CYT1 and CYT2 have the opposite effects. In mammary epithelial cells, overexpression of the cleavable 80 kD cytoplasmic domain of CYT1 was found to decrease growth and to induce a more differentiated phenotype, whereas the cleavable 80 kD cytoplasmic domain of CYT2 increased growth and induced no differentiation (Muraoka-Cook et al, 2009). In addition, HER4 nuclear localisation is associated with poor survival and CYT2 has been found to enter the nucleus more readily than CYT1 (Junttila et al, 2005; Sundvall et al, 2007; Zeng et al, 2007).
The contradictory results regarding the roles of CYT1 and CYT2 demonstrate that the divergent roles of HER4 do not come down to cytoplasmic isoform expression alone. Besides isoform expression, HER4 functions are influenced by the activating ligand, the dimerisation partner, and the localisation of cleavage products. That the activating ligand is important also in regard to HER4 cytoplasmic isoform expression was demonstrated in Madin-Darby Canine Kidney (MDCK) II cells in which HB-EGF had a greater proliferative effect than NRG1β in CYT2-expressing cells, whereas NRG1β had a greater proliferative effect than HB-EGF in CYT1-expressing cells (Zeng et al, 2007). With regard to the importance of the dimerisation partner, it has recently been found that HER2 expression is important for HER4 to possess an oncogenic phenotype (Mill et al, 2011b). In addition, a constitutively dimerised variant of HER4 possessed a proliferative potential different from that of ligand activation of a wild-type receptor in prostate cancer cell lines (Mill et al, 2011a).
This study is the first to selectively target only one cytoplasmic splice variant of HER4. Despite the contradictory results on CYT1 and CYT2 functions, there is no doubt that the cytoplasmic isoforms play a role in the divergent roles of HER4 and that in some cases, it will be attractive to target only one of these isoforms. Targeting only one isoform will potentially result in fewer side effects as other isoforms of HER4 can still exert their functions. Example of conditions in which targeting of CYT1 could be attractive is medulla blastoma where the CYT1/CYT2 ratio was found to be higher in aggressive than in less aggressive tumours and in ovarian cancer in which the CYT1 but not the CYT2 isoform was associated with poor survival (Ferretti et al, 2006). Under the conditions of our experiments, we also found that the decrease in the CYT1/CYT2 ratio inhibited the growth of MCF7 breast cancer cells and mice in xenografted tumours. If this effect can also be achieved in humans, SSO targeted to HER4 may represent a novel strategy in cancer treatment in the future.
Acknowledgments
This work was supported by the Danish Cancer Society.
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- Aqeilan RI, Donati V, Palamarchuk A, Trapasso F, Kaou M, Pekarsky Y, Sudol M, Croce CM. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005;65 (15:6764–6772. doi: 10.1158/0008-5472.CAN-05-1150. [DOI] [PubMed] [Google Scholar]
- Bauman JA, Li SD, Yang A, Huang L, Kole R. Anti-tumor activity of splice-switching oligonucleotides. Nucleic Acids Res. 2010;38 (22:8348–8356. doi: 10.1093/nar/gkq731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter G. ErbB-4: mechanism of action and biology. Exp Cell Res. 2003;284 (1:66–77. doi: 10.1016/s0014-4827(02)00100-3. [DOI] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Correction of disease-associated exon skipping by synthetic exon-specific activators. Nat Struct Biol. 2003;10 (2:120–125. doi: 10.1038/nsb887. [DOI] [PubMed] [Google Scholar]
- Cheng QC, Tikhomirov O, Zhou W, Carpenter G. Ectodomain cleavage of ErbB-4: characterization of the cleavage site and m80 fragment. J Biol Chem. 2003;278 (40:38421–38427. doi: 10.1074/jbc.M302111200. [DOI] [PubMed] [Google Scholar]
- Elenius K, Choi CJ, Paul S, Santiestevan E, Nishi E, Klagsbrun M. Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene. 1999;18 (16:2607–2615. doi: 10.1038/sj.onc.1202612. [DOI] [PubMed] [Google Scholar]
- Elenius K, Corfas G, Paul S, Choi CJ, Rio C, Plowman GD, Klagsbrun M. A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J Biol Chem. 1997;272 (42:26761–26768. doi: 10.1074/jbc.272.42.26761. [DOI] [PubMed] [Google Scholar]
- Ferretti E, Di ML, Gessi M, Mattei T, Greco A, Po A, De SE, Giangaspero F, Riccardi R, Di RC, Pazzaglia S, Maroder M, Alimandi M, Screpanti I, Gulino A. Alternative splicing of the ErbB-4 cytoplasmic domain and its regulation by hedgehog signaling identify distinct medulloblastoma subsets. Oncogene. 2006;25 (55:7267–7273. doi: 10.1038/sj.onc.1209716. [DOI] [PubMed] [Google Scholar]
- Graziewicz MA, Tarrant TK, Buckley B, Roberts J, Fulton L, Hansen H, Orum H, Kole R, Sazani P. An endogenous TNF-alpha antagonist induced by splice-switching oligonucleotides reduces inflammation in hepatitis and arthritis mouse models. Mol Ther. 2008;16 (7:1316–1322. doi: 10.1038/mt.2008.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gschwind A, Fischer OM, Ullrich A. The discovery of receptor tyrosine kinases: targets for cancer therapy. Nat Rev Cancer. 2004;4 (5:361–370. doi: 10.1038/nrc1360. [DOI] [PubMed] [Google Scholar]
- Hollmen M, Elenius K. Potential of ErbB4 antibodies for cancer therapy. Future Oncol. 2010;6 (1:37–53. doi: 10.2217/fon.09.144. [DOI] [PubMed] [Google Scholar]
- Hollmen M, Maatta JA, Bald L, Sliwkowski MX, Elenius K. Suppression of breast cancer cell growth by a monoclonal antibody targeting cleavable ErbB4 isoforms. Oncogene. 2009;28 (10:1309–1319. doi: 10.1038/onc.2008.481. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Macdonald G. ErbB receptors and signaling pathways in cancer. Curr Opin Cell Biol. 2009;21 (2:177–184. doi: 10.1016/j.ceb.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Junttila TT, Sundvall M, Lundin M, Lundin J, Tanner M, Harkonen P, Joensuu H, Isola J, Elenius K. Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 2005;65 (4:1384–1393. doi: 10.1158/0008-5472.CAN-04-3150. [DOI] [PubMed] [Google Scholar]
- Kainulainen V, Sundvall M, Maatta JA, Santiestevan E, Klagsbrun M, Elenius K. A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J Biol Chem. 2000;275 (12:8641–8649. doi: 10.1074/jbc.275.12.8641. [DOI] [PubMed] [Google Scholar]
- Khoo B, Roca X, Chew SL, Krainer AR. Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB. BMC Mol Biol. 2007;8:3. doi: 10.1186/1471-2199-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Nagai M, Navin NE, Sudol M. WW domain-containing protein YAP associates with ErbB-4 and acts as a co-transcriptional activator for the carboxyl-terminal fragment of ErbB-4 that translocates to the nucleus. J Biol Chem. 2003;278 (35:33334–33341. doi: 10.1074/jbc.M305597200. [DOI] [PubMed] [Google Scholar]
- Kurppa K, Elenius K. Mutated ERBB4: a novel drug target in metastatic melanoma. Pigment Cell Melanoma Res. 2009;22 (6:708–710. doi: 10.1111/j.1755-148X.2009.00635.x. [DOI] [PubMed] [Google Scholar]
- Lee CM, Shrieve DC, Zempolich KA, Lee RJ, Hammond E, Handrahan DL, Gaffney DK. Correlation between human epidermal growth factor receptor family (EGFR, HER2, HER3, HER4), phosphorylated Akt (P-Akt), and clinical outcomes after radiation therapy in carcinoma of the cervix. Gynecol Oncol. 2005;99 (2:415–421. doi: 10.1016/j.ygyno.2005.05.045. [DOI] [PubMed] [Google Scholar]
- Lee JC, Wang ST, Chow NH, Yang HB. Investigation of the prognostic value of coexpressed erbB family members for the survival of colorectal cancer patients after curative surgery. Eur J Cancer. 2002;38 (8:1065–1071. doi: 10.1016/s0959-8049(02)00004-7. [DOI] [PubMed] [Google Scholar]
- Memon AA, Sorensen BS, Melgard P, Fokdal L, Thykjaer T, Nexo E. Expression of HER3, HER4 and their ligand heregulin-4 is associated with better survival in bladder cancer patients. Br J Cancer. 2004;91 (12:2034–2041. doi: 10.1038/sj.bjc.6602251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill CP, Gettinger KL, Riese DJ. Ligand stimulation of ErbB4 and a constitutively-active ErbB4 mutant result in different biological responses in human pancreatic tumor cell lines. Exp Cell Res. 2011a;317 (4:392–404. doi: 10.1016/j.yexcr.2010.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mill CP, Zordan MD, Rothenberg SM, Settleman J, Leary JF, Riese DJ. ErbB2 Is Necessary for ErbB4 Ligands to Stimulate Oncogenic Activities in Models of Human Breast Cancer. Genes Cancer. 2011b;2 (8:792–804. doi: 10.1177/1947601911431080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraoka-Cook RS, Sandahl MA, Strunk KE, Miraglia LC, Husted C, Hunter DM, Elenius K, Chodosh LA, Earp HS., III ErbB4 splice variants Cyt1 and Cyt2 differ by 16 amino acids and exert opposing effects on the mammary epithelium in vivo. Mol Cell Biol. 2009;29 (18:4935–4948. doi: 10.1128/MCB.01705-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, Tovey S, Cooke TG, Bartlett JM, Jones FE. The ERBB4/HER4 intracellular domain 4ICD is a BH3-only protein promoting apoptosis of breast cancer cells. Cancer Res. 2006;66 (12:6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294 (5549:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- Omerovic J, Santangelo L, Puggioni EM, Marrocco J, Dall'Armi C, Palumbo C, Belleudi F, Di ML, Frati L, Torrisi MR, Cesareni G, Gulino A, Alimandi M. The E3 ligase Aip4/Itch ubiquitinates and targets ErbB-4 for degradation. FASEB J. 2007;21 (11:2849–2862. doi: 10.1096/fj.06-7925com. [DOI] [PubMed] [Google Scholar]
- Paatero I, Lassus H, Junttila TT, Kaskinen M, Butzow R, Elenius K. CYT-1 isoform of ErbB4 is an independent prognostic factor in serous ovarian cancer and selectively promotes ovarian cancer cell growth in vitro. Gynecol Oncol. 2013;129 (1:179–187. doi: 10.1016/j.ygyno.2012.12.044. [DOI] [PubMed] [Google Scholar]
- Prickett TD, Agrawal NS, Wei X, Yates KE, Lin JC, Wunderlich JR, Cronin JC, Cruz P, Rosenberg SA, Samuels Y. Analysis of the tyrosine kinome in melanoma reveals recurrent mutations in ERBB4. Nat Genet. 2009;41 (10:1127–1132. doi: 10.1038/ng.438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rio C, Buxbaum JD, Peschon JJ, Corfas G. Tumor necrosis factor-alpha-converting enzyme is required for cleavage of erbB4/HER4. J Biol Chem. 2000;275 (14:10379–10387. doi: 10.1074/jbc.275.14.10379. [DOI] [PubMed] [Google Scholar]
- Roberts J, Palma E, Sazani P, Orum H, Cho M, Kole R. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol Ther. 2006;14 (4:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]
- Rudloff U, Samuels Y. A growing family: adding mutated Erbb4 as a novel cancer target. Cell Cycle. 2010;9 (8:1487–1503. doi: 10.4161/cc.9.8.11239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglam O, Shah V, Worsham MJ. Molecular differentiation of early and late stage laryngeal squamous cell carcinoma: an exploratory analysis. Diagn Mol Pathol. 2007;16 (4:218–221. doi: 10.1097/PDM.0b013e3180d0aab5. [DOI] [PubMed] [Google Scholar]
- Sazani P, Kole R. Therapeutic potential of antisense oligonucleotides as modulators of alternative splicing. J Clin Invest. 2003;112 (4:481–486. doi: 10.1172/JCI19547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaffidi P, Misteli T. Reversal of the cellular phenotype in the premature aging disease Hutchinson-Gilford progeria syndrome. Nat Med. 2005;11 (4:440–445. doi: 10.1038/nm1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settleman J. A therapeutic opportunity in melanoma: ErbB4 makes a mark on skin. Cancer Cell. 2009;16 (4:278–279. doi: 10.1016/j.ccr.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Shi J, Yao D, Liu W, Wang N, Lv H, He N, Shi B, Hou P, Ji M. Frequent gene amplification predicts poor prognosis in gastric cancer. Int J Mol Sci. 2012;13 (4:4714–4726. doi: 10.3390/ijms13044714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sierakowska H, Sambade MJ, Agrawal S, Kole R. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA. 1996;93 (23:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M, Korhonen A, Paatero I, Gaudio E, Melino G, Croce CM, Aqeilan RI, Elenius K. Isoform-specific monoubiquitination, endocytosis, and degradation of alternatively spliced ErbB4 isoforms. Proc Natl Acad Sci USA. 2008;105 (11:4162–4167. doi: 10.1073/pnas.0708333105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundvall M, Peri L, Maatta JA, Tvorogov D, Paatero I, Savisalo M, Silvennoinen O, Yarden Y, Elenius K. Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene. 2007;26 (48:6905–6914. doi: 10.1038/sj.onc.1210501. [DOI] [PubMed] [Google Scholar]
- Svasti S, Suwanmanee T, Fucharoen S, Moulton HM, Nelson MH, Maeda N, Smithies O, Kole R. RNA repair restores hemoglobin expression in IVS2-654 thalassemic mice. Proc Natl Acad Sci USA. 2009;106 (4:1205–1210. doi: 10.1073/pnas.0812436106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang CK, Concepcion XZ, Milan M, Gong X, Montgomery E, Lippman ME. Ribozyme-mediated down-regulation of ErbB-4 in estrogen receptor-positive breast cancer cells inhibits proliferation both in vitro and in vivo. Cancer Res. 1999;59 (20:5315–5322. [PubMed] [Google Scholar]
- Thybusch-Bernhardt A, Beckmann S, Juhl H. Comparative analysis of the EGF-receptor family in pancreatic cancer: expression of HER-4 correlates with a favourable tumor stage. Int J Surg Investig. 2001;2 (5:393–400. [PubMed] [Google Scholar]
- Uberall I, Kolar Z, Trojanec R, Berkovcova J, Hajduch M. The status and role of ErbB receptors in human cancer. Exp Mol Pathol. 2008;84 (2:79–89. doi: 10.1016/j.yexmp.2007.12.002. [DOI] [PubMed] [Google Scholar]
- van Deutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, den Dunnen JT, Koop K, van der Kooi AJ, Goemans NM, de Kimpe SJ, Ekhart PF, Venneker EH, Platenburg GJ, Verschuuren JJ, van Ommen GJ. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357 (26:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- Veikkolainen V, Vaparanta K, Halkilahti K, Iljin K, Sundvall M, Elenius K. Function of ERBB4 is determined by alternative splicing. Cell Cycle. 2011;10 (16:2647–2657. doi: 10.4161/cc.10.16.17194. [DOI] [PubMed] [Google Scholar]
- Williams T, Kole R. Analysis of prostate-specific membrane antigen splice variants in LNCap cells. Oligonucleotides. 2006;16 (2:186–195. doi: 10.1089/oli.2006.16.186. [DOI] [PubMed] [Google Scholar]
- Xia W, Lau YK, Zhang HZ, Xiao FY, Johnston DA, Liu AR, Li L, Katz RL, Hung MC. Combination of EGFR, HER-2/neu, and HER-3 is a stronger predictor for the outcome of oral squamous cell carcinoma than any individual family members. Clin Cancer Res. 1999;5 (12:4164–4174. [PubMed] [Google Scholar]
- Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2 (2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- Zeng F, Xu J, Harris RC. Nedd4 mediates ErbB4 JM-a/CYT-1 ICD ubiquitination and degradation in MDCK II cells. FASEB J. 2009;23 (6:1935–1945. doi: 10.1096/fj.08-121947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng F, Zhang MZ, Singh AB, Zent R, Harris RC. ErbB4 isoforms selectively regulate growth factor induced Madin-Darby canine kidney cell tubulogenesis. Mol Biol Cell. 2007;18 (11:4446–4456. doi: 10.1091/mbc.E07-03-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]