Abstract
Parkinson’s disease can be caused by rare familial genetic mutations, but in most cases it is likely to result from an interaction between multiple genetic and environmental risk factors. Over recent years, many variants in a growing number of genes involved in the pathogenesis of Parkinson’s disease have been identified. Mutations in several genes have been shown to cause familial parkinsonism. In this review, we discuss 12 of them (SNCA, LRRK2, Parkin, PINK1, DJ1, ATP13A2, PLA2G6, FBXO7, UCHL1, GIGYF2, HTRA2, and EIF4G1). Additionally, six genes have been shown conclusively to be risk factors for sporadic Parkinson’s disease, and are also discussed (GBA, MAPT, BST1, PARK16, GAK, and HLA). Many more genes and genetic loci have been suggested, but need confirmation. There is evidence that pathways involved in the rare familial forms also play a role in the sporadic form, and that the respective genes might also be risk factors for sporadic Parkinson’s disease. The identification of genes involved in the development of Parkinson’s disease will improve our understanding of the underlying molecular mechanisms, and will hopefully lead to new drug targets and treatment strategies.
Keywords: Parkinson’s disease, genetics, SNCA, LRRK2, GBA, MAPT
Introduction
Parkinson’s disease is the second most common neurodegenerative disorder, after Alzheimer’s disease. An estimated 1%–2% of individuals over the age of 65 years are affected, and more than 4% of the population by the age of 85 years.1 Similar prevalence rates are found in different populations across the world. Because longevity is increasing in our society, these demographic age groups are growing, leading to increasing relevance of neurodegenerative diseases as a social and economic burden. Cardinal motor symptoms are tremor, rigidity, bradykinesia, and postural instability. Several nonmotor features are very common in Parkinson’s disease patients, including autonomic failure, cognitive impairment, depression, olfactory deficits, psychosis, and sleep disturbance. Parkinson’s disease is treated mainly with levodopa, which replaces the loss of dopamine. Complications can include the emergence of motor fluctuations and dyskinesias. Other therapeutic options include dopamine agonists, anticholinergics, monoamine oxidase inhibitors, and deep brain stimulation.2 A therapy protecting against loss of neurons is not available at present.3 The most frequently used diagnostic criteria are those established by the UK Parkinson’s Disease Society Brain Bank,4 which are based on the presence of cardinal motor features and the absence of atypical features, in addition to a positive response to levodopa treatment.2 In the early course of the disease, it is challenging to differentiate Parkinson’s disease from other parkinsonian syndromes, eg, progressive supranuclear palsy and multiple system atrophy.
During the course of the disease, dopaminergic cells are progressively lost in the substantia nigra pars compacta, resulting in a loss of dopamine in the striatal projection areas of these neurons, leading to the typical motor dysfunction, which becomes evident when approximately 80% of striatal dopamine and 50% of nigral neurons are lost.5 The hallmark of the pathology of Parkinson’s disease is loss of dopaminergic cells in the substantia nigra pars compacta and the presence of Lewy bodies and Lewy neurites. Lewy bodies are intracytoplasmic inclusions with a dense eosinophilic core, and are surrounded by a clearer halo on hematoxylin and eosin staining. Their principal component is the protein, alpha-synuclein, in a fibrillar form.6 It is still debated whether the Lewy body is neurotoxic or reflects an attempt to protect the cell.7 Lewy neurites also contain fibrillar alpha-synuclein.6 They are found in nerve cell processes, in high numbers in the CA2/3 region of the hippocampus, and in the substantia nigra.8 The development of neuropathological changes in Parkinson’s disease is thought to occur in different stages.9 The death of the dopaminergic neurons begins before the appearance of clinical symptoms and progresses throughout the course of the disease.10 In the first stages, the pathology is limited to the medulla oblongata and pontine tegmentum, including the dorsal motor nucleus of the vagus nerve, the locus coeruleus, and the olfactory bulb. Motor symptoms occur later, as the substantia nigra and other nuclei within the midbrain and forebrain, eg, the nucleus basalis of Meynert, become involved. In the late stages of the disease, the pathological changes advance to the mesocortex and neocortex.9
Parkinson’s disease is assumed to be multifactorial in most cases, caused by a combination of genetic and environmental risk factors. The most important “environmental” risk factor is aging. Age-dependent changes might initiate or maintain the process of neurodegeneration. In addition to that, a few toxic exposures (eg, MPTP, and certain pesticides) have been identified as selectively damaging dopaminergic neurons, but their relationship to sporadic Parkinson’s disease is still unclear. A few protective factors (caffeine, nicotine, and possibly nonsteroidal anti-inflammatory agents) have also been identified by epidemiologic studies.11 A family history is reported by approximately 10%–20% of patients.12 Genetic involvement has been substantiated in recent decades by the finding of several disease-causing genes and risk factors.
Genes causing monogenic forms of the disease are usually identified by linkage studies. This approach examines families with multiple affected and unaffected relatives, defining a rare genetic variant (pathogenic mutation) that is segregating with the disease. Genes that represent risk factors are mostly identified by association studies. Genetic risk variants are generally much more common in a population than high-penetrance disease-causing mutations, and if the frequency is higher in cases than in controls, this indicates that the variant confers a risk for a given disease. Because the effect strength of these genetic risk variants is usually low, association analyses need to include large cohorts of patients and controls. An unbiased approach for association analyses are genome-wide association studies. Linkage analyses have resulted in identification of several loci (PARK1-PARK13, PARK15, and EIF4G1). Association analyses have identified several risk factors (SNCA, LRRK2, GBA, MAPT, BST1, PARK16, GAK, and HLA).
Interestingly, the Parkinson’s disease-causing genes seem to fall into several classes. The dominant forms cause a phenotype closely resembling sporadic Parkinson’s disease. Some of the recessive forms present with an earlier age at onset and as pure parkinsonism, while others cause parkinsonism with additional clinical features (ATP13A2, PLA2G6, and FBXO7). In this paper we discuss the most important genes relevant to Parkinson’s disease (listed in Table 1) and their associated clinical features.
Table 1.
Summary of genes and loci underlying Parkinson’s disease
| Locus | Gene | Chromosomal location | Inheritance | Type of parkinsonism |
|---|---|---|---|---|
| PARK1/PARK4 | SNCA | 4q21 | AD + risk | LOPD/EOPD, dementia |
| PARK2 | Parkin | 6q25–q27 | AR | EOPD |
| PARK3 | Unknown | 2p13 | AD | LOPD |
| PARK5 | UCHL1 | 4p14 | AD | LOPD |
| PARK6 | PINK1 | 1p36 | AR | EOPD |
| PARK7 | DJ1 | 1p36 | AR | EOPD |
| PARK8 | LRRK2 | 12q12 | AD + risk | LOPD |
| PARK9 | ATP13 A2 | 1p36 | AR | EOPD, Kufor-Rakeb syndrome |
| PARK10 | Unknown | 1p32 | Unknown | LOPD |
| PARK11 | GIGYF2 | 2q37 | AD | LOPD |
| PARK12 | Unknown | Xq21–25 | X-linked | LOPD |
| PARK13 | HTRA2 | 2p12 | AD | LOPD |
| PARK14 | PLA2G6 | 22q13 | AR | EOPD, dystonia-parkinsonism |
| PARK15 | FBXO7 | 22q12–q13 | AR | EOPD, pallido-pyramidal syndrome |
| PARK16 | Unknown | 1q32 | Risk | LOPD |
| PARK17 | GAK | 4p16 | Risk | LOPD |
| PARK18 | HLA | 6p21 | Risk | LOPD |
| – | EIF4G1 | 3q27 | AD | LOPD |
| – | GBA | 1q21 | Risk | LOPD |
| – | MAPT | 17q21 | Risk | LOPD |
| – | BST1 | 4p15 | Risk | LOPD |
Abbreviations: AD, autosomal dominant; AR, autosomal recessive; EOPD, early-onset Parkinson’s disease; LOPD, late-onset Parkinson’s disease.
Autosomal dominant Parkinson’s disease genes
SNCA (PARK1/PARK4)
SNCA was the first causal Parkinson’s disease gene ever identified. Its mutations cause autosomal dominant Parkinson’s disease. The gene encodes the protein alpha-synuclein, which has been detected as the main component of Lewy bodies and Lewy neurites.6 The function of alpha-synuclein is still unknown. It binds to synaptic vesicles13 and may be involved in brain plasticity.14 The protein is expressed widely in the brain and localizes mostly to presynaptic nerve terminals.15 Natively unfolded, alpha-synuclein can oligomerize and form fibrils.16 These fibrillar moieties are the components of Lewy bodies in both familial and sporadic PD.6 It is controversial if these fibrils are toxic or protective to the cell. The discovery of the linkage of autosomal dominant Parkinson’s disease to chromosome 4q2117 was followed by identification of a pathogenic missense mutation in SNCA.18 To date, three missense mutations (A53T,18 A30P,19 and E46K20), duplications,21,22 and triplications23 of the entire gene have been identified. Duplications of the gene lead to a 1.5-fold increase of the protein, whereas triplications lead to a two-fold increase.24 These findings suggest that already the increased concentrations of wild-type alpha-synuclein protein appear to be toxic to neurons. A point mutation in alpha-synuclein might lead to an increased tendency to form aggregates,25 pointing to a gain-of-function hypothesis. Another hypothesis suggests that mutated SNCA leads to a compensatory overexpression of the wild-type allele. SNCA mutations are rare, accounting for less than 1% in Caucasian cases of Parkinson’s disease.26,27 Mutations have been identified in Caucasian and Asian families, and in some patients with apparently sporadic Parkinson’s disease.27,28 Some of the multiplications appear to have different genomic sizes in different families, suggesting that the mutations arose independently.29 The penetrance of duplication carriers is estimated to be around 40%.28,30 Accordingly, there are also reports of asymptomatic carriers.27,28,30,31 The clinical phenotype presents as levodopa-responsive Parkinson’s disease, with relatively early age at onset, rapid progression,32 a higher prevalence of dementia, and psychiatric and autonomic disturbances.27,33 Some patients show atypical clinical features, including myoclonus or multiple system atrophy.33 There is some evidence of genotype-phenotype correlation. Patients with SNCA duplications mostly show a typical late-onset Parkinson’s disease phenotype with slow progression and no atypical features. Patients with a SNCA triplication or the A53T missense mutation have an earlier age at onset (around 34 years), and carriers of a triplication, the A53T or the E46K missense mutation, present more frequently with dementia than duplication carriers or carriers of the A30P missense mutation.20–23,28,31,33,34 Pathological studies describe SNCA mutation carriers with typical brain stem32 or diffuse Lewy body disease.20,24 Common variants in SNCA have been found to be a risk factor in sporadic Parkinson’s disease. One signal represented by a dinucleotide repeat polymorphism (NACP-Rep1) is located in the promoter region;35 another signal maps to the 3′ end of the gene.36 SNCA was identified as a risk factor in all genome-wide association studies conducted so far.37–43 The variants associated with increased risk of Parkinson’s disease might lead to a slightly higher expression of alpha-synuclein.44
LRRK2 (PARK8)
Mutations in LRRK2 cause autosomal dominant Parkinson’s disease. The function of the encoded protein is unknown. It has been suggested to play a role in intracellular signaling pathways. The protein contains a kinase, a GTPase, and several protein–protein interaction domains.45 LRRK2 is expressed widely in the brain, and also in other organs.45,46 It is localized to membranes and vesicles, such as mitochondria, lysosomes, and endosomes.47 The locus was linked to chromosome 12q12,48 and subsequently several missense mutations were identified in the LRRK2 gene.45,46 Since then, many missense mutations have been identified in LRRK2, but the pathogenicity is only clearly established for six of them (ie, R1441C, R1441G, R1441H, Y1699C, G2019S, and I2020T).49 The mechanism by which LRRK2 mutations contribute to the development of Parkinson’s disease might be an increase in kinase activity,50 an impairment of the GTPase function,51 or an alteration in ability to dimerize,52 depending on which domain the mutation occurs in. Mutations in LRRK2 are the most common cause of familial Parkinson’s disease. The frequency in familial cases is 5%–15%, and in sporadic cases is around 1% in patients with Caucasian ancestry.53–55 One mutation, G2019S, is particular common, occurring in up to 2%–7% of Caucasian cases of familial Parkinson’s disease49,54,56,57 and in 1% of sporadic cases.49 In some populations, the frequency of G2019S is surprisingly high, being up to 20% in Ashkenazi Jews58 and 40% in North African Arabs.59 Occurrence of LRRK2 mutations in apparently sporadic patients and in healthy control individuals49,56 suggests a reduced penetrance. Estimations of the penetrance for G2019S range from 32% to 74%, depending on the familial background of the families analyzed.49,55,60 G2019S was also found in homozygosity, but with no difference in clinical phenotype compared with heterozygous carriers.57 This mutation is observed in a limited number of haplotypes, indicating that there are only a few founders.56,61 Similarly, the R1441G mutation was transmitted from a common founder in the Basque population,61 whereas the R1441C has several different founder haplotypes.54 Some mutations occur only in the Asian population, where the G2019S seems to be rare or absent.49,62 The clinical phenotype strongly resembles typical late-onset Parkinson’s disease, with an average age at onset of 59 years.45,53 Single cases cannot be distinguished from sporadic idiopathic Parkinson’s disease. On average, they seem to have a slightly more benign course of disease, slower progression, and lower frequency of dementia and psychiatric complications.49,53 One study reported that the predominant sign in LRRK2-related Parkinson’s disease is tremor.49 Pathological signs in LRRK2 mutation carriers are typical alpha-synuclein Lewy bodies in most cases, but some cases show atypical pathology, like diffuse Lewy body disease, tau pathology, neuronal loss without intracellular inclusions, and motor neuron disease.45,63 These different pathological changes can even be found among members of the same family, with the same pathogenic mutation.45 Two LRRK2 variants, ie, G2385R and R1628P, were identified as risk factors in the Asian population.64,65 These variants are very rare or absent in other populations. In a Caucasian and an Asian genome-wide association study, variation 5′ of the gene was identified as a risk factor in sporadic Parkinson’s disease.38,39
Autosomal recessive Parkinson’s disease genes
Parkin (PARK2)
Mutations in PARK2 (Parkin) cause autosomal recessive Parkinson’s disease. Parkin, the encoded protein, belongs to the ubiquitin E3 ligases.66 It interacts with ubiquitin-conjugating enzymes (E2s) to catalyze the attachment of ubiquitin to protein targets, thus tagging these proteins for destruction by the proteasome.66 Additionally, Parkin is involved in mitochondrial maintenance67,68 and might induce autophagy of dysfunctional mitochondria.69 Parkin is expressed in many tissues, including various brain regions and the substantia nigra.70 It is predominantly localized in the cytosol, but is also present in vesicles, the Golgi complex, the endoplasmic reticulum, and the outer mitochondrial membrane.71 The locus was first linked to chromosome 6q26,72 followed by the detection of mutations in the gene Parkin.70 Many mutations have been reported since then, including missense, nonsense, indels, exonic deletions, duplications, and triplications.73,74 Most of the exonic rearrangements have likely occurred as relatively recent new mutational events. Point mutations, on the other hand, are mostly transmitted from common founders.75 These mutations are involved in development of Parkinson’s disease probably by a loss-of-function mechanism.66 Patients with Parkinson’s disease and Parkin mutations have a mean age at onset of 32 years in the Caucasian population.74 Hence, Parkin mutations are the most common cause of early-onset Parkinson’s disease, occurring in up to 50% of those with age at onset under 25 years (and only 3%–7% in those with age at onset 30–45 years).73,74 However, cases with very early onset (<30 years) represent probably only 1%–2% of cases overall. Mutations were identified in familial as well as in sporadic patients and in populations of all ethnic origins. The penetrance for homozygous or compound heterozygous mutation carriers seems to be 100%. Patients with Parkin mutations present with levodopa-responsive Parkinson’s disease accompanied frequently by motor fluctuations and dyskinesias that often develop early in the course of treatment.74,76 A high percentage of patients show dystonia, usually in a lower extremity at the onset of disease.74 Additionally, psychiatric abnormalities can arise.76 The disease progression is slow. Missense mutations might lead to faster progression of the disease and a higher Unified Parkinson’s Disease Rating Scale motor score than truncating mutations. Missense mutations in functional domains of the gene can result in an earlier age at onset.77 Patients carrying mutations in Parkin rarely show the presence of Lewy bodies, although nigrostriatal cell loss is usually severe.78,79 Heterozygous mutations in Parkin may be a risk factor for Parkinson’s disease. However, several studies have reported controversial results.80,81 Further large-scale studies will be needed to elucidate the risk in heterozygous carriers.
PINK1 (PARK6)
Mutations in PINK1 also cause autosomal recessive Parkinson’s disease. The protein encoded by PINK1 is located in the mitochondrial membranes and expressed ubiquitously, including the brain.82 It is involved in the mitochondrial response to cellular and oxidative stress.83 By interaction with parkin, PINK1 might protect neurons against mitochondrial dysfunction and proteasome-induced apoptosis.84 The locus was first linked to chromosome 1p36,85 and then causal missense and nonsense mutations in the gene PINK1 were detected.83 Several missense, nonsense, frameshift mutations, and large deletions of multiple exons were identified thereafter.86–91 The mutations seem to impair the function of the protein.83 Mutations in PINK1 are a rare cause of early-onset Parkinson’s disease, accounting only for 2%–4% of early-onset cases in Caucasian populations86,91 and 4%–9% in Asian populations.87,90 The penetrance for homozygous and compound heterozygous mutation carriers seems to be 100%. The clinical phenotype is similar to that caused by mutations in Parkin.90 The early onset (24–47 years) is accompanied by slow disease progression and a good response to levodopa.86,87 Sometimes dementia occurs. Some patients show additional psychiatric disturbances, particularly anxiety and depression, which is only relatively rarely observed in Parkin-related cases.92 Occasionally, PINK1 mutations are found in late-onset Parkinson’s disease, restless legs syndrome with parkinsonism, and dopa-responsive dystonia.87–89 Pathological changes in a single reported case were those of typical Lewy body disease.93
DJ1 (PARK7)
Mutations in DJ1 are another, but very rare, cause of autosomal recessive Parkinson’s disease. The protein DJ1 is a sensor for oxidative stress and may mediate neuroprotection.94 It is translocated to the mitochondrial membrane upon presence of oxidative stress, and appears to act as an antioxidant.94 Nigral dopamine neurons are highly exposed to oxidative stress, highlighting the function of DJ1 as being particularly interesting in the pathogenesis of Parkinson’s disease. DJ1 is expressed in most tissues, including the brain, and has been found in neuronal and glial cells.95 The gene was originally mapped to chromosome 1p36.96 After the first discovery of a missense mutation and an exonic deletion,97 a few other missense and splice-site mutations, as well as deletions, were identified in homozygous or compound heterozygous states.98,99 One mutation, the L166P, leads to a less stable protein100 and to the reduction of antioxidative activity,101 implicating a loss-of-function mechanism for DJ1 mutations in general. Mutations in DJ1 seem to be very rare, accounting for only 1% of early-onset cases,98,99 but the penetrance for homozygous and compound heterozygous mutation carriers seems to be 100%. Similar to the other recessive Parkinson’s disease forms, the clinical picture includes an early onset (20–40 years), good response to levodopa, and slow progression. In one family, Parkinson’s disease was accompanied by dementia and amyotrophic lateral sclerosis.102 Some patients with DJ1 show psychiatric symptoms,103 short stature, and brachydactyly,104 suggesting that the phenotype can vary.
Autosomal recessive genes causing parkinsonism with atypical features
ATP13A2 (PARK9)
Mutations in ATP13A2 cause autosomal recessive parkinsonism with a complex phenotype. ATP13A2 encodes a lysosomal membrane protein with an ATPase domain, predominantly expressed in the brain.105 The exact function of this protein is still unknown. The locus was linked to chromosome 1p36, and the gene was subsequently identified as ATP13A2.105 Since then, several missense mutations, as well as splice-site and small deletions and insertions were identified.106,107 Some evidence indicates that the mutated protein is unstable and degraded.105 Mutation analyses in Caucasians show that these mutations are only a very rare cause of parkinsonism.108 The phenotype is atypical parkinsonism, also described as Kufor-Rakeb syndrome. Patients with mutations in ATP13A2 have a very early onset (11–16 years)109,110 and a rapid progression of parkinsonian symptoms. They are levodopa-responsive, have pyramidal signs, and show spasticity and supranuclear gaze palsy.109,110 Additionally, some also show facial, faucal, and finger minimyoclonus, visual hallucinations, and dystonic oculogyric spasm.110 A few cases have been reported with more typical early-onset Parkinson’s disease.
PLA2G6 (PARK14)
Mutations in PLA2G6 have been identified in autosomal recessive families. PLA2G6 encodes a calcium-independent phospholipase A2 enzyme that catalyzes hydrolysis of glycerophospholipids and is critical in cell membrane homeostasis.111 Two childhood-onset disorders are caused by mutations in PLA2G6, ie, infantile neuroaxonal dystrophy and neurodegeneration with brain iron accumulation or Karak syndrome.111 The pathology includes axonal swellings and spheroid bodies, and some show alpha-synuclein-positive Lewy bodies. The clinical phenotype is characterized by a young-onset progressive extrapyramidal–pyramidal syndrome with visual disturbances, early cerebellar signs, and a late-onset parkinsonian syndrome. By homozygosity mapping, missense mutations were identified in families with adult-onset levodopa-responsive dystonia-parkinsonism.112 Thereafter, several other mutations were identified in additional families, but the frequency in Parkinson’s disease seems to be very low.113 Parkinson’s disease patients with PLA2G6 mutations also show pyramidal signs, cognitive dysfunction, and levodopa-induced dyskinesias, but no cerebellar signs.112,114 Dystonia is not present in all patients, as well as iron accumulation in the brain.112,114
FBXO7 (PARK15)
Mutations in FBXO7 have been found in autosomal recessive families. The protein encoded by FBXO7 might serve as a molecular scaffold in the formation of protein complexes. It is also suggested to play a role in the ubiquitin-proteasome protein degradation pathway.115 The locus was originally linked to chromosome 22q1 in a family with juvenile Parkinson pyramidal syndrome. Subsequently, a missense mutation was identified in the FBXO7 gene.115 Additional cases with missense and splice site mutations were thereafter identified.116 The frequency of mutations in this gene seems to be very low.117 Patients with mutations in FBXO7 have early-onset, progressive parkinsonism and pyramidal tract signs, described as the pallido-pyramidal syndrome. Dystonia can be present in childhood, whereas pyramidal signs of the lower limbs can occur in later stages of the disease.116
Autosomal dominant genes and loci with unclear pathogenicity
UCHL1 (PARK5)
Mutations in UCHL1 have been suggested to cause autosomal dominant Parkinson’s disease. The encoded protein, UCHL1, is a component of the ubiquitin-proteasome system, removing abnormal and misfolded proteins, and generating free ubiquitin monomers.118 It is highly and specifically expressed in neurons,119 and is one of the most abundant proteins in the brain.118 It seems to be localized to synaptic vesicles. UCHL1 was sequenced in a candidate gene approach and one family was found to harbor a missense mutation (I93M).120 To date, no other pathogenic mutations of this gene have been identified. The I93M mutation leads to a 50% loss of catalytic hydrolase activity, suggesting a loss-of-function mechanism.120,121 Another hypothesis assumes a gain-of-function through alteration of the protein structure.121 There was an unaffected carrier in the affected family, suggesting reduced penetrance.120 This UCHL1 family presented with typical Parkinson’s disease, a good response to levodopa, and an age at onset of 49–51 years.120 There are no pathology data available. Later, a common missense variant (S18Y) was identified as being associated with risk of Parkinson’s disease.122 This S18Y mutation is thought to have a protective effect by decreasing ligase activity123 and increasing hydrolase activity.121 However, studies trying to replicate the association have yielded conflicting results.124–126 Because there was only one family with a potentially pathogenic mutation that was not fully segregating and no further evidence for UCHL1 as a genetic risk factor, its role in Parkinson’s disease has become very questionable.
GIGYF2 (PARK11)
The PARK11 locus was mapped by linkage to chromosome 2q37 with a dominant model in families with Parkinson’s disease.127 Later, mutations in the GIGYF2 gene were found to segregate with typical Parkinson’s disease in several small families.128 GIGYF2 encodes a component of the insulin-signaling pathway and is expressed in the brain.129 However, replication studies have failed to demonstrate the pathogenicity of these mutations,130,131 and the original PARK11 family used to define the locus has no mutation in GIGYF2.132 Therefore, it seems unlikely that GIGYF2 plays a role in susceptibility to Parkinson’s disease. In contrast with linkage approaches in single large families, the analysis of several families together is challenged by the heterogeneity of Parkinson’s disease. The fact that only small families were used in the GIGYF2-linkage might have additionally biased the analysis.
HTRA2 (PARK13)
Mutations in the HTRA2 gene were found in a small autosomal dominant family by a candidate gene approach.133 HTRA2 encodes for a serine protease that transfers from the mitochondria to the cytosol upon stimulation.134 One missense mutation (G399S) was found to segregate in a family with Parkinson’s disease. Another missense mutation (A141S) was more frequently detected in cases than in controls, indicating a risk factor. This A141S mutation leads to a decrease in protease function, suggesting a loss-of-function mechanism.133 But again, replication studies failed to demonstrate pathogenicity of the G399S mutation, which was also found in controls,135 and could not detect an association of A141S with risk for Parkinson’s disease.136
EIF4G1
A mutation in the gene EIF4G1 was found to be segregating with late-onset Parkinson’s disease in an autosomal dominant family.137 The encoded protein, EIF4G1, is a translation initiation factor and might be involved in stress response. Loss of EIF4G1 leads to impaired nutrient sensing and mitochondrial bioenergetics, while promoting autophagy. The same mutation was thereafter identified in several Caucasian families, but not in control individuals. These findings still need to be replicated.
PARK3, PARK10, PARK12
Several other loci have been mapped in families with Parkinson’s disease, but the genes have not yet been identified. PARK3, PARK10, and PARK12 were found by linkage analyses in families with typical late-onset Parkinson’s disease.127,138,139 The PARK3 locus includes the SPR gene. Polymorphisms in this gene have been found to be associated with age at onset in patients with Parkinson’s disease.140 Its protein is involved in the dopamine synthesis pathway. Future studies will be necessary to elucidate definitively the role of these loci in the pathogenesis of Parkinson’s disease.
Genetic risk factors
GBA
Homozygous or compound heterozygous mutations in the GBA gene cause Gaucher disease, and single heterozygous mutations have recently been found to be an important risk factor for Parkinson’s disease. Gaucher disease is a rare, recessively inherited, lipid storage disorder with multisystemic manifestation, including involvement of the liver, spleen, bone marrow, lungs, and nervous system.141 Some patients with Gaucher disease develop parkinsonism.142 The disease is characterized by accumulation of the glycolipid, glucosylceramide, in the lysosomal compartment of various cells. Relatives of patients with Gaucher disease have a higher prevalence of Parkinson’s disease,143 which led to the discovery that heterozygous carriers of mutations in GBA are at increased risk of Parkinson’s disease.144 The association of GBA mutations with Parkinson’s disease has been confirmed in a large meta-analysis involving many populations worldwide.145 GBA encodes glucosylceramidase, a lysosomal enzyme, which cleaves glucosylceramide to ceramide and glucose. Over 300 mutations causing Gaucher disease have been found in GBA, and are considered to be loss-of-function mutations. The mutations in Gaucher disease lead to accumulation of the substrate in the lysosome, but how heterozygous mutations contribute to the pathogenesis of Parkinson’s disease is still unclear. Because alpha-synuclein is at least in part degraded by the lysosome, decreased activity of GBA could lead to incomplete degradation and subsequent aggregation of alpha-synuclein. Likewise, misfolded GBA could impair lysosomal function and the autophagy pathway in general.146 Another hypothesis is based on the observation that alpha-synuclein binds to lipids in the plasma membrane147 and this binding influences the fibrillization capacity of alpha-synuclein. Actually, it has been shown that alpha-synuclein binds to glycosphingolipids containing glucosylceramide. Decreased activity of mutated GBA leads to accumulation of glucosylceramide, and might therefore influence lipid metabolism and aggregation of alpha-synuclein.146 The frequency of GBA mutations among Caucasian patients with Parkinson’s disease has been found to be 7%, as compared with 1% in control individuals. The frequency is much higher in Ashkenazi Jews, with 20% of Parkinson’s disease patients being carriers versus 4% in controls.145,148 In Caucasians, the two most common mutations are N370S and L444P, accounting for approximately 50% of all identified mutation carriers.145,148 In Asians, the L444P mutation is the most common mutation, and the N370S is only rarely found.149 In Ashkenazi Jews, the N370S is the most common mutation.145 The clinical phenotype resembles typical late-onset sporadic Parkinson’s disease with alpha-synuclein- positive Lewy body pathology and a higher frequency of diffuse neocortical Lewy body pathology.150 The onset is reported to be somewhat earlier, especially in carriers of the more severe mutations, eg, L444P.148 Parkinson’s disease patients with the GBA mutation often have more severe nonmotor symptoms, including cognitive changes.145 Bradykinesia as the onset symptom and levodopa-induced dyskinesias also seem to be more frequent.148 GBA mutations are also considered to be a risk factor for dementia with Lewy bodies.151
MAPT
MAPT is a known risk factor for progressive supranuclear palsy and corticobasal degeneration, and mutations have been found to cause one form of familial frontotemporal dementia.152–154 Many studies, including all published genome-wide association studies in Caucasian populations, have confirmed the MAPT locus as a risk factor for Parkinson’s disease.37–43,155,156 MAPT encodes the microtubule-associated protein tau, which plays a key role in the organization and integrity of the cytoskeleton.157 It is primarily expressed in neurons. Tau inclusions are found in several neurodegenerative diseases, referred to as “tauopathies”, including progressive supranuclear palsy, corticobasal degeneration, frontotemporal dementia, and Alzheimer’s disease. On the other hand, Parkinson’s disease, multiple system atrophy, and dementia with Lewy bodies, are referred to as “synucleinopathies”, due to the accumulation of alpha-synuclein. An interplay between the two pathophysiological mechanisms underlying these diseases has been repeatedly suggested.158 The MAPT locus is the longest region of linkage disequilibrium in Caucasians.159 There are two forms of this genomic region in Caucasians, ie, the H1/H2 haplotypes. The different haplotypes originate from a 900 kb inversion,160 which might have been introduced by the Neanderthals into the Caucasian genome.161 Because the H1 haplotype is associated with risk of Parkinson’s disease and other neurodegenerative disorders, the H2 haplotype might have been under positive selection since that time. This inversion is absent in the Asian population, which is why an Asian genome-wide association study did not identify MAPT as a risk factor. The H1 haplotype can be subdivided into subhaplotypes.162 Which of these subhaplotypes in particular is associated with Parkinson’s disease remains to be elucidated, but it seems that the risk variant increases expression of tau.
BST1
BST1 was identified as a risk factor in sporadic late-onset Parkinson’s disease in an Asian genome-wide association study.39 The encoded protein catalyzes formation of cyclic ADP-ribose and might play a role in Ca2+ homeostasis.163 The association with Parkinson’s disease was replicated in two Caucasian genome-wide association studies,42,43 but could not be replicated in one Caucasian38 and one Asian study.164 Allele frequencies differed markedly between the studies, and effect sizes seem to be lower in Caucasians. Further studies will be necessary to clarify the role of BST1 in Parkinson’s disease.
PARK16
A locus on chromosome 1q32 was identified to be associated with Parkinson’s disease in an Asian genome-wide association study.39 This region, designated PARK16, contains five genes. The locus was replicated in two Caucasian genome-wide association studies and several association studies.38,43,164 Potential disease-causing mutations were subsequently reported in families with typical Parkinson’s disease for two of the genes in this locus, ie, RAB7L1 and SLC41A1.165 The role of the PARK16 locus in monogenic families and as a risk factor needs to be determined further in future studies.
GAK (PARK17)
In a Caucasian genome-wide association study involving familial cases of Parkinson’s disease, a locus on chromosome 4p16 was identified.37 This locus was replicated in several other association studies as being significantly associated with Parkinson’s disease.41,43,156 GAK is one of the coding genes in this region. It encodes for a cell cycle regulator that is ubiquitously expressed.166 This protein might also be involved in clathrin-mediated endocytosis, like vesicle trafficking,167 which might be a link to alpha-synuclein. GAK is differentially expressed between Parkinson’s disease cases and controls in the substantia nigra.168
HLA (PARK18)
PARK18 was identified as a risk factor in a recent genome-wide association study.41 The locus contains the HLA region. HLA DRA and DRB proteins form the Class II HLA-DR antigens, which are expressed by antigen-presenting cells, including microglia in the brain. The genetic region is highly variable. Variation in the HLA-DR genes as a risk factor for Parkinson’s disease fits with the observations from previous studies implicating chronic inflammation and humoral immunity in the pathogenesis of Parkinson’s disease.169,170
Summary
Two genes (SNCA and LRRK2) have been conclusively linked to autosomal dominant Parkinson’s disease. These two genes also represent risk factors in the sporadic form of the disease. Three genes (Parkin, PINK1, and DJ1) have been shown to cause pure autosomal recessive Parkinson’s disease with early age at onset. Autosomal recessive parkinsonism in a complicated form is caused by mutations in three genes (ATP13A2, PLA2G, and FBXO7). There are more genes and loci suggested to be involved in monogenic Parkinson’s disease, but require replication before firm conclusions could be drawn (UCHL1, GIGYF2, HTRA2, EIF4G1, PARK3, PARK10, and PARK12). Six additional genes have been discussed as risk factors in sporadic Parkinson’s disease (GBA, MAPT, BST1, PARK16, GAK, and HLA). Many more variants have been found to be associated with Parkinson’s disease, most of which need further replication in independent studies.
Mendelian forms of Parkinson’s disease account for less than 10% of all Parkinson’s disease cases in most populations. The Ashkenazi Jewish population is an exception, because LRRK2 and GBA mutations are found at a higher frequency. Since the discovery that SNCA and LRRK2 not only cause the monogenic form of Parkinson’s disease, but also act as risk factors in the sporadic form, it has been suggested that different alterations in the same gene can be important in both forms. Other familial genes will be tested for association with risk and, in turn, known risk genes will be tested for causing the familial form of the disease.
The discovery of causal mutations in Parkinson’s disease has given researchers the opportunity to identify young presymptomatic mutation carriers who can be followed longitudinally to identify premotor changes using clinical, biochemical, and imaging methods. These biomarkers can be used to investigate the early phases of the disease in a general way, hopefully leading to earlier diagnoses and neuroprotective treatment strategies.
Many more susceptibility loci will likely be uncovered in the near future. A recent meta-analysis of five Caucasian genome-wide association studies revealed many additional risk genes.171 Future research methods will not only include traditional linkage analysis and genome-wide association studies, but also new methods, like meta-analysis of genome-wide association studies, gene–gene and gene–environment interaction studies, pathway analysis, and copy number variant analysis, as well as whole exome sequencing. It may also be promising to examine different and as yet unstudied populations. The identification of new genes involved in the pathogenesis of Parkinson’s disease is challenging because it has a late-in-life onset, which decreases the availability of large familial pedigrees. Furthermore, the underlying genetics are complex, including mutations with reduced penetrance and interaction of several risk factors. Replication studies have to consider that distinct populations differ in the composition of their risk factors, making comparisons across populations difficult.
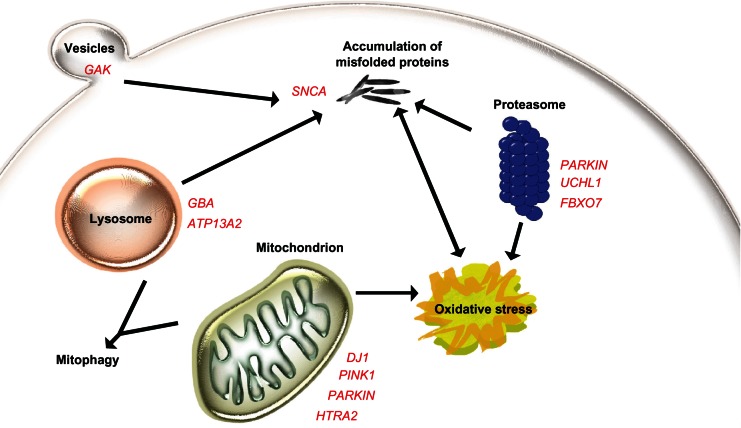
The genes identified so far are involved in pathways that might be of general interest in the development of Parkinson’s disease, in particular, those involving the mitochondria, like mitochondrial dysfunction, oxidative stress, mitochondrial fission and fusion, and mitophagy. Another broad field includes pathways involved in aberrant protein degradation, and proteasomal and lysosomal dysfunction (Figure 1). Although our knowledge about the pathogenesis of Parkinson’s disease is growing, the complete picture is not yet observable. How all the pathways and genes are interacting is still not completely understood. However, the discovery of new genes in conjunction with animal model studies will hopefully elucidate the pathogenesis. The standard treatment for Parkinson’s disease at the moment is effective in improving the motor symptoms of the disease, but few therapies treating nonmotor features are available, and there is currently no medication at all that prevents dopaminergic neurons from dying. Identifying the proteins and understanding the pathways involved in the development of Parkinson’s disease should lead to new drug targets, and might give rise to better treatment strategies.
Figure 1.
Model of the pathogenesis of Parkinson’s disease. The central step in Parkinson’s disease pathogenesis is proposed to be the accumulation of alpha-synuclein, while the regular function of alpha-synuclein (SNCA) might involve the formation of vesicles, like GAK. Mitochondrial maintenance and response to oxidative stress are to some extent regulated by DJ1, PINK1, parkin, and HTRA2. Five genes are involved in degradation pathways, either over the lysosome or the ubiquitin-proteasomal complex. If one component in these pathways is disturbed, this might lead to dysfunction of the other components.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.De Rijk MC, Launer LJ, Berger K, et al. Neurologic Diseases in the Elderly Research Group Prevalence of Parkinson’s disease in Europe: A collaborative study of population-based cohorts. Neurology. 2000;54(11 Suppl 5):S21–S23. [PubMed] [Google Scholar]
- 2.Samii A, Nutt JG, Ransom BR. Parkinson’s disease. Lancet. 2004;363(9423):1783–1793. doi: 10.1016/S0140-6736(04)16305-8. [DOI] [PubMed] [Google Scholar]
- 3.Jankovic J. An update on the treatment of Parkinson’s disease. Mt Sinai J Med. 2006;73(4):682–689. [PubMed] [Google Scholar]
- 4.Hughes AJ, Daniel SE, Kilford L, Lees AJ. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: A clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry. 1992;55(3):181–184. doi: 10.1136/jnnp.55.3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fearnley JM, Lees AJ. Ageing and Parkinson’s disease: Substantia nigra regional selectivity. Brain. 1991;114(Pt 5):2283–2301. doi: 10.1093/brain/114.5.2283. [DOI] [PubMed] [Google Scholar]
- 6.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha-synuclein in Lewy bodies. Nature. 1997;388(6645):839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 7.Cookson MR. Alpha-synuclein and neuronal cell death. Mol Neurodegener. 2009;4:9. doi: 10.1186/1750-1326-4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Love S. Neuropathological investigation of dementia: A guide for neurologists. J Neurol Neurosurg Psychiatry. 2005;76(Suppl 5):8–14. doi: 10.1136/jnnp.2005.080754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson’s disease-related pathology. Cell Tissue Res. 2004;318(1):121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 10.Halliday GM, McCann H. The progression of pathology in Parkinson’s disease. Ann N Y Acad Sci. 2010;1184:188–195. doi: 10.1111/j.1749-6632.2009.05118.x. [DOI] [PubMed] [Google Scholar]
- 11.Chade AR, Kasten M, Tanner CM. Nongenetic causes of Parkinson’s disease. J Neural Transm Suppl. 2006;70:147–151. doi: 10.1007/978-3-211-45295-0_23. [DOI] [PubMed] [Google Scholar]
- 12.Farrer MJ. Genetics of Parkinson disease: Paradigm shifts and future prospects. Nat Rev Genet. 2006;7(4):306–318. doi: 10.1038/nrg1831. [DOI] [PubMed] [Google Scholar]
- 13.Jensen PH, Nielsen MS, Jakes R, Dotti CG, Goedert M. Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson’s disease mutation. J Biol Chem. 1998;273(41):26292–26294. doi: 10.1074/jbc.273.41.26292. [DOI] [PubMed] [Google Scholar]
- 14.Abeliovich A, Schmitz Y, Farinas I, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25(1):239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 15.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: A neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8(8):2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer’s disease and learning, is natively unfolded. Biochemistry. 1996;35(43):13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 17.Polymeropoulos MH, Higgins JJ, Golbe LI, et al. Mapping of a gene for Parkinson’s disease to chromosome 4q21–q23. Science. 1996;274(5290):1197–1199. doi: 10.1126/science.274.5290.1197. [DOI] [PubMed] [Google Scholar]
- 18.Polymeropoulos MH, Lavedan C, Leroy E, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson’s disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 19.Kruger R, Kuhn W, Muller T, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson’s disease. Nat Genet. 1998;18(2):106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 20.Zarranz JJ, Alegre J, Gomez-Esteban JC, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55(2):164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 21.Chartier-Harlin MC, Kachergus J, Roumier C, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 22.Ibanez P, Bonnet AM, Debarges B, et al. Causal relation between alpha-synuclein gene duplication and familial Parkinson’s disease. Lancet. 2004;364(9440):1169–1171. doi: 10.1016/S0140-6736(04)17104-3. [DOI] [PubMed] [Google Scholar]
- 23.Singleton AB, Farrer M, Johnson J, et al. Alpha-synuclein locus triplication causes Parkinson’s disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 24.Farrer M, Kachergus J, Forno L, et al. Comparison of kindreds with parkinsonism and alpha-synuclein genomic multiplications. Ann Neurol. 2004;55(2):174–179. doi: 10.1002/ana.10846. [DOI] [PubMed] [Google Scholar]
- 25.Conway KA, Harper JD, Lansbury PT. Accelerated in vitro fibril formation by a mutant alpha-synuclein linked to early-onset Parkinson disease. Nat Med. 1998;4(11):1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 26.Berg D, Niwar M, Maass S, et al. Alpha-synuclein and Parkinson’s disease: Implications from the screening of more than 1,900 patients. Mov Disord. 2005;20(9):1191–1194. doi: 10.1002/mds.20504. [DOI] [PubMed] [Google Scholar]
- 27.Ahn TB, Kim SY, Kim JY, et al. Alpha-synuclein gene duplication is present in sporadic Parkinson disease. Neurology. 2008;70(1):43–49. doi: 10.1212/01.wnl.0000271080.53272.c7. [DOI] [PubMed] [Google Scholar]
- 28.Nishioka K, Ross OA, Ishii K, et al. Expanding the clinical phenotype of SNCA duplication carriers. Mov Disord. 2009;24(12):1811–1819. doi: 10.1002/mds.22682. [DOI] [PubMed] [Google Scholar]
- 29.Nuytemans K, Theuns J, Cruts M, Van Broeckhoven C. Genetic etiology of Parkinson disease associated with mutations in the SNCA, PARK2, PINK1, PARK7, and LRRK2 genes: A mutation update. Hum Mutat. 2010;31(7):763–780. doi: 10.1002/humu.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishioka K, Hayashi S, Farrer MJ, et al. Clinical heterogeneity of alpha-synuclein gene duplication in Parkinson’s disease. Ann Neurol. 2006;59(2):298–309. doi: 10.1002/ana.20753. [DOI] [PubMed] [Google Scholar]
- 31.Ibanez P, Lesage S, Janin S, et al. Alpha-synuclein gene rearrangements in dominantly inherited parkinsonism: Frequency, phenotype, and mechanisms. Arch Neurol. 2009;66(1):102–108. doi: 10.1001/archneurol.2008.555. [DOI] [PubMed] [Google Scholar]
- 32.Spira PJ, Sharpe DM, Halliday G, Cavanagh J, Nicholson GA. Clinical and pathological features of a Parkinsonian syndrome in a family with an Ala53Thr alpha-synuclein mutation. Ann Neurol. 2001;49(3):313–319. [PubMed] [Google Scholar]
- 33.Fuchs J, Nilsson C, Kachergus J, et al. Phenotypic variation in a large Swedish pedigree due to SNCA duplication and triplication. Neurology. 2007;68(12):916–922. doi: 10.1212/01.wnl.0000254458.17630.c5. [DOI] [PubMed] [Google Scholar]
- 34.Golbe LI, Di Iorio G, Bonavita V, Miller DC, Duvoisin RC. A large kindred with autosomal dominant Parkinson’s disease. Ann Neurol. 1990;27(3):276–282. doi: 10.1002/ana.410270309. [DOI] [PubMed] [Google Scholar]
- 35.Kruger R, Vieira-Saecker AM, Kuhn W, et al. Increased susceptibility to sporadic Parkinson’s disease by a certain combined alpha-synuclein/apolipoprotein E genotype. Ann Neurol. 1999;45(5):611–617. doi: 10.1002/1531-8249(199905)45:5<611::aid-ana9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 36.Mueller JC, Fuchs J, Hofer A, et al. Multiple regions of alpha-synuclein are associated with Parkinson’s disease. Ann Neurol. 2005;57(4):535–541. doi: 10.1002/ana.20438. [DOI] [PubMed] [Google Scholar]
- 37.Pankratz N, Wilk JB, Latourelle JC, et al. Genome-wide association study for susceptibility genes contributing to familial Parkinson disease. Hum Genet. 2009;124(6):593–605. doi: 10.1007/s00439-008-0582-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Simon-Sanchez J, Schulte C, Bras JM, et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41(12):1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Satake W, Nakabayashi Y, Mizuta I, et al. Genome-wide association study identifies common variants at four loci as genetic risk factors for Parkinson’s disease. Nat Genet. 2009;41(12):1303–1307. doi: 10.1038/ng.485. [DOI] [PubMed] [Google Scholar]
- 40.Edwards TL, Scott WK, Almonte C, et al. Genome-wide association study confirms SNPs in SNCA and the MAPT region as common risk factors for Parkinson disease. Ann Hum Genet. 2010;74(2):97–109. doi: 10.1111/j.1469-1809.2009.00560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamza TH, Zabetian CP, Tenesa A, et al. Common genetic variation in the HLA region is associated with late-onset sporadic Parkinson’s disease. Nat Genet. 2010;42(9):781–785. doi: 10.1038/ng.642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saad M, Lesage S, Saint-Pierre A, et al. Genome-wide association study confirms BST1 and suggests a locus on 12q24 as the risk loci for Parkinson’s disease in the European population. Hum Mol Genet. 2011;20(3):615–627. doi: 10.1093/hmg/ddq497. [DOI] [PubMed] [Google Scholar]
- 43.Spencer CC, Plagnol V, Strange A, et al. UK Parkinson’s Disease Consortium. Wellcome Trust Case Control Consortium 2 Dissection of the genetics of Parkinson’s disease identifies an additional association 5′ of SNCA and multiple associated haplotypes at 17q21. Hum Mol Genet. 2011;20(2):345–353. doi: 10.1093/hmg/ddq469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chiba-Falek O, Nussbaum RL. Effect of allelic variation at the NACP-Rep1 repeat upstream of the alpha-synuclein gene (SNCA) on transcription in a cell culture luciferase reporter system. Hum Mol Genet. 2001;10(26):3101–3109. doi: 10.1093/hmg/10.26.3101. [DOI] [PubMed] [Google Scholar]
- 45.Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44(4):601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 46.Paisan-Ruiz C, Jain S, Evans EW, et al. Cloning of the gene containing mutations that cause PARK8-linked Parkinson’s disease. Neuron. 2004;44(4):595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 47.Biskup S, Moore DJ, Celsi F, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60(5):557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- 48.Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson’s disease (PARK8) maps to chromosome 12p11.2–q13.1. Ann Neurol. 2002;51(3):296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 49.Healy DG, Falchi M, O’Sullivan SS, et al. Phenotype, genotype, and worldwide genetic penetrance of LRRK2-associated Parkinson’s disease: A case-control study. Lancet Neurol. 2008;7(7):583–590. doi: 10.1016/S1474-4422(08)70117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West AB, Moore DJ, Biskup S, et al. Parkinson’s disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci USA. 2005;102(46):16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun. 2007;357(3):668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sen S, Webber PJ, West AB. Dependence of leucine-rich repeat kinase 2 (LRRK2) kinase activity on dimerization. J Biol Chem. 2009;284(52):36346–36356. doi: 10.1074/jbc.M109.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Berg D, Schweitzer K, Leitner P, et al. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson’s disease*. Brain. 2005;128(Pt 12):3000–3001. doi: 10.1093/brain/awh666. [DOI] [PubMed] [Google Scholar]
- 54.Di Fonzo A, Tassorelli C, De Mari M, et al. Comprehensive analysis of the LRRK2 gene in sixty families with Parkinson’s disease. Eur J Hum Genet. 2006;14(3):322–331. doi: 10.1038/sj.ejhg.5201539. [DOI] [PubMed] [Google Scholar]
- 55.Latourelle JC, Sun M, Lew MF, et al. The Gly2019Ser mutation in LRRK2 is not fully penetrant in familial Parkinson’s disease: The GenePD study. BMC Med. 2008;6:32. doi: 10.1186/1741-7015-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kachergus J, Mata IF, Hulihan M, et al. Identification of a novel LRRK2 mutation linked to autosomal dominant parkinsonism: Evidence of a common founder across European populations. Am J Hum Genet. 2005;76(4):672–680. doi: 10.1086/429256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nichols WC, Pankratz N, Hernandez D, et al. Genetic screening for a single common LRRK2 mutation in familial Parkinson’s disease. Lancet. 2005;365(9457):410–412. doi: 10.1016/S0140-6736(05)17828-3. [DOI] [PubMed] [Google Scholar]
- 58.Ozelius LJ, Senthil G, Saunders-Pullman R, et al. LRRK2 G2019S as a cause of Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2006;354(4):424–425. doi: 10.1056/NEJMc055509. [DOI] [PubMed] [Google Scholar]
- 59.Lesage S, Durr A, Tazir M, et al. LRRK2 G2019S as a cause of Parkinson’s disease in North African Arabs. N Engl J Med. 2006;354(4):422–423. doi: 10.1056/NEJMc055540. [DOI] [PubMed] [Google Scholar]
- 60.Goldwurm S, Zini M, Mariani L, et al. Evaluation of LRRK2 G2019S penetrance: Relevance for genetic counseling in Parkinson disease. Neurology. 2007;68(14):1141–1143. doi: 10.1212/01.wnl.0000254483.19854.ef. [DOI] [PubMed] [Google Scholar]
- 61.Gonzalez-Fernandez MC, Lezcano E, Ross OA, et al. Lrrk2-associated parkinsonism is a major cause of disease in Northern Spain. Parkinsonism Relat Disord. 2007;13(8):509–515. doi: 10.1016/j.parkreldis.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 62.Tan EK, Shen H, Tan LC, et al. The G2019S LRRK2 mutation is uncommon in an Asian cohort of Parkinson’s disease patients. Neurosci Lett. 2005;384(3):327–329. doi: 10.1016/j.neulet.2005.04.103. [DOI] [PubMed] [Google Scholar]
- 63.Ross OA, Toft M, Whittle AJ, et al. Lrrk2 and Lewy body disease. Ann Neurol. 2006;59(2):388–393. doi: 10.1002/ana.20731. [DOI] [PubMed] [Google Scholar]
- 64.Di Fonzo A, Wu-Chou YH, Lu CS, et al. A common missense variant in the LRRK2 gene, Gly2385Arg, associated with Parkinson’s disease risk in Taiwan. Neurogenetics. 2006;7(3):133–138. doi: 10.1007/s10048-006-0041-5. [DOI] [PubMed] [Google Scholar]
- 65.Ross OA, Wu YR, Lee MC, et al. Analysis of Lrrk2 R1628P as a risk factor for Parkinson’s disease. Ann Neurol. 2008;64(1):88–92. doi: 10.1002/ana.21405. [DOI] [PubMed] [Google Scholar]
- 66.Shimura H, Hattori N, Kubo S, et al. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat Genet. 2000;25(3):302–305. doi: 10.1038/77060. [DOI] [PubMed] [Google Scholar]
- 67.Park J, Lee G, Chung J. The PINK1-Parkin pathway is involved in the regulation of mitochondrial remodeling process. Biochem Biophys Res Commun. 2009;378(3):518–523. doi: 10.1016/j.bbrc.2008.11.086. [DOI] [PubMed] [Google Scholar]
- 68.Kuroda Y, Mitsui T, Kunishige M, et al. Parkin enhances mitochondrial biogenesis in proliferating cells. Hum Mol Genet. 2006;15(6):883–895. doi: 10.1093/hmg/ddl006. [DOI] [PubMed] [Google Scholar]
- 69.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin-induced mitophagy in the pathogenesis of Parkinson disease. Autophagy. 2009;5(5):706–708. doi: 10.4161/auto.5.5.8505. [DOI] [PubMed] [Google Scholar]
- 70.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 71.Mouatt-Prigent A, Muriel MP, Gu WJ, et al. Ultrastructural localization of parkin in the rat brainstem, thalamus and basal ganglia. J Neural Transm. 2004;111(10–11):1209–1218. doi: 10.1007/s00702-004-0144-9. [DOI] [PubMed] [Google Scholar]
- 72.Matsumine H, Saito M, Shimoda-Matsubayashi S, et al. Localization of a gene for an autosomal recessive form of juvenile Parkinsonism to chromosome 6q25.2–27. Am J Hum Genet. 1997;60(3):588–596. [PMC free article] [PubMed] [Google Scholar]
- 73.Periquet M, Latouche M, Lohmann E, et al. Parkin mutations are frequent in patients with isolated early-onset parkinsonism. Brain. 2003;126(Pt 6):1271–1278. doi: 10.1093/brain/awg136. [DOI] [PubMed] [Google Scholar]
- 74.Lucking CB, Durr A, Bonifati V, et al. Association between early-onset Parkinson’s disease and mutations in the parkin gene. N Engl J Med. 2000;342(21):1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 75.Periquet M, Lucking C, Vaughan J, et al. Origin of the mutations in the parkin gene in Europe: Exon rearrangements are independent recurrent events, whereas point mutations may result from founder effects. Am J Hum Genet. 2001;68(3):617–626. doi: 10.1086/318791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Khan NL, Graham E, Critchley P, et al. Parkin disease: A phenotypic study of a large case series. Brain. 2003;126(Pt 6):1279–1292. doi: 10.1093/brain/awg142. [DOI] [PubMed] [Google Scholar]
- 77.Lohmann E, Periquet M, Bonifati V, et al. How much phenotypic variation can be attributed to parkin genotype? Ann Neurol. 2003;54(2):176–185. doi: 10.1002/ana.10613. [DOI] [PubMed] [Google Scholar]
- 78.Pramstaller PP, Schlossmacher MG, Jacques TS, et al. Lewy body Parkinson’s disease in a large pedigree with 77 Parkin mutation carriers. Ann Neurol. 2005;58(3):411–422. doi: 10.1002/ana.20587. [DOI] [PubMed] [Google Scholar]
- 79.Mori H, Kondo T, Yokochi M, et al. Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology. 1998;51(3):890–892. doi: 10.1212/wnl.51.3.890. [DOI] [PubMed] [Google Scholar]
- 80.Kay DM, Stevens CF, Hamza TH, et al. A comprehensive analysis of deletions, multiplications, and copy number variations in PARK2. Neurology. 2010;75(13):1189–1194. doi: 10.1212/WNL.0b013e3181f4d832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brooks J, Ding J, Simon-Sanchez J, Paisan-Ruiz C, Singleton AB, Scholz SW. Parkin and PINK1 mutations in early-onset Parkinson’s disease: Comprehensive screening in publicly available cases and control. J Med Genet. 2009;46(6):375–381. doi: 10.1136/jmg.2008.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gandhi S, Muqit MM, Stanyer L, et al. PINK1 protein in normal human brain and Parkinson’s disease. Brain. 2006;129(Pt 7):1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 83.Valente EM, Abou-Sleiman PM, Caputo V, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304(5674):1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 84.Yang Y, Gehrke S, Imai Y, et al. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc Natl Acad Sci USA. 2006;103(28):10793–10798. doi: 10.1073/pnas.0602493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valente EM, Bentivoglio AR, Dixon PH, et al. Localization of a novel locus for autosomal recessive early-onset parkinsonism, PARK6, on human chromosome 1p35–p36. Am J Hum Genet. 2001;68(4):895–900. doi: 10.1086/319522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Valente EM, Salvi S, Ialongo T, et al. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56(3):336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]
- 87.Tan EK, Yew K, Chua E, et al. PINK1 mutations in sporadic early-onset Parkinson’s disease. Mov Disord. 2006;21(6):789–793. doi: 10.1002/mds.20810. [DOI] [PubMed] [Google Scholar]
- 88.Gelmetti V, Ferraris A, Brusa L, et al. Late onset sporadic Parkinson’s disease caused by PINK1 mutations: Clinical and functional study. Mov Disord. 2008;23(6):881–885. doi: 10.1002/mds.21960. [DOI] [PubMed] [Google Scholar]
- 89.Leutenegger AL, Salih MA, Ibanez P, et al. Juvenile-onset Parkinsonism as a result of the first mutation in the adenosine triphosphate orientation domain of PINK1. Arch Neurol. 2006;63(9):1257–1261. doi: 10.1001/archneur.63.9.1257. [DOI] [PubMed] [Google Scholar]
- 90.Li Y, Tomiyama H, Sato K, et al. Clinicogenetic study of PINK1 mutations in autosomal recessive early-onset parkinsonism. Neurology. 2005;64(11):1955–1957. doi: 10.1212/01.WNL.0000164009.36740.4E. [DOI] [PubMed] [Google Scholar]
- 91.Bonifati V, Rohe CF, Breedveld GJ, et al. Early-onset parkinsonism associated with PINK1 mutations: Frequency, genotypes, and phenotypes. Neurology. 2005;65(1):87–95. doi: 10.1212/01.wnl.0000167546.39375.82. [DOI] [PubMed] [Google Scholar]
- 92.Ephraty L, Porat O, Israeli D, et al. Neuropsychiatric and cognitive features in autosomal-recessive early parkinsonism due to PINK1 mutations. Mov Disord. 2007;22(4):566–569. doi: 10.1002/mds.21319. [DOI] [PubMed] [Google Scholar]
- 93.Samaranch L, Lorenzo-Betancor O, Arbelo JM, et al. PINK1-linked parkinsonism is associated with Lewy body pathology. Brain. 2010;133(Pt 4):1128–1142. doi: 10.1093/brain/awq051. [DOI] [PubMed] [Google Scholar]
- 94.Canet-Aviles RM, Wilson MA, Miller DW, et al. The Parkinson’s disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc Natl Acad Sci USA. 2004;101(24):9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bandopadhyay R, Kingsbury AE, Cookson MR, et al. The expression of DJ-1 (PARK7) in normal human CNS and idiopathic Parkinson’s disease. Brain. 2004;127(Pt 2):420–430. doi: 10.1093/brain/awh054. [DOI] [PubMed] [Google Scholar]
- 96.Van Duijn CM, Dekker MC, Bonifati V, et al. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am J Hum Genet. 2001;69(3):629–634. doi: 10.1086/322996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bonifati V, Rizzu P, Squitieri F, et al. DJ-1( PARK7), a novel gene for autosomal recessive, early onset parkinsonism. Neurol Sci. 2003;24(3):159–160. doi: 10.1007/s10072-003-0108-0. [DOI] [PubMed] [Google Scholar]
- 98.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson’s disease. Ann Neurol. 2003;54(3):283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 99.Hague S, Rogaeva E, Hernandez D, et al. Early-onset Parkinson’s disease caused by a compound heterozygous DJ-1 mutation. Ann Neurol. 2003;54(2):271–274. doi: 10.1002/ana.10663. [DOI] [PubMed] [Google Scholar]
- 100.Moore DJ, Zhang L, Dawson TM, Dawson VL. A missense mutation (L166P) in DJ-1, linked to familial Parkinson’s disease, confers reduced protein stability and impairs homo-oligomerization. J Neurochem. 2003;87(6):1558–1567. doi: 10.1111/j.1471-4159.2003.02265.x. [DOI] [PubMed] [Google Scholar]
- 101.Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson’s disease patients. Biochem Biophys Res Commun. 2004;320(2):389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- 102.Annesi G, Savettieri G, Pugliese P, et al. DJ-1 mutations and parkinsonism-dementia-amyotrophic lateral sclerosis complex. Ann Neurol. 2005;58(5):803–807. doi: 10.1002/ana.20666. [DOI] [PubMed] [Google Scholar]
- 103.Dekker MC, van Swieten JC, Houwing-Duistermaat JJ, et al. A clinical-genetic study of Parkinson’s disease in a genetically isolated community. J Neurol. 2003;250(9):1056–1062. doi: 10.1007/s00415-003-0151-z. [DOI] [PubMed] [Google Scholar]
- 104.Dekker MC, Galjaard RJ, Snijders PJ, Heutink P, Oostra BA, van Duijn CM. Brachydactyly and short stature in a kindred with early-onset parkinsonism. Am J Med Genet A. 2004;130A(1):102–104. doi: 10.1002/ajmg.a.30021. [DOI] [PubMed] [Google Scholar]
- 105.Ramirez A, Heimbach A, Grundemann J, et al. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat Genet. 2006;38(10):1184–1191. doi: 10.1038/ng1884. [DOI] [PubMed] [Google Scholar]
- 106.Di Fonzo A, Chien HF, Socal M, et al. ATP13A2 missense mutations in juvenile parkinsonism and young onset Parkinson disease. Neurology. 2007;68(19):1557–1562. doi: 10.1212/01.wnl.0000260963.08711.08. [DOI] [PubMed] [Google Scholar]
- 107.Schneider SA, Paisan-Ruiz C, Quinn NP, et al. ATP13A2 mutations (PARK9) cause neurodegeneration with brain iron accumulation. Mov Disord. 2010;25(8):979–984. doi: 10.1002/mds.22947. [DOI] [PubMed] [Google Scholar]
- 108.Djarmati A, Hagenah J, Reetz K, et al. ATP13A2 variants in early-onset Parkinson’s disease patients and controls. Mov Disord. 2009;24(14):2104–2111. doi: 10.1002/mds.22728. [DOI] [PubMed] [Google Scholar]
- 109.Najim al-Din AS, Wriekat A, Mubaidin A, Dasouki M, Hiari M. Pallido-pyramidal degeneration, supranuclear upgaze paresis and dementia: Kufor-Rakeb syndrome. Acta Neurol Scand. 1994;89(5):347–352. doi: 10.1111/j.1600-0404.1994.tb02645.x. [DOI] [PubMed] [Google Scholar]
- 110.Williams DR, Hadeed A, al-Din AS, Wreikat AL, Lees AJ. Kufor Rakeb disease: Autosomal recessive, levodopa-responsive parkinsonism with pyramidal degeneration, supranuclear gaze palsy, and dementia. Mov Disord. 2005;20(10):1264–1271. doi: 10.1002/mds.20511. [DOI] [PubMed] [Google Scholar]
- 111.Morgan NV, Westaway SK, Morton JE, et al. PLA2G6, encoding a phospholipase A2, is mutated in neurodegenerative disorders with high brain iron. Nat Genet. 2006;38(7):752–754. doi: 10.1038/ng1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Paisan-Ruiz C, Bhatia KP, Li A, et al. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann Neurol. 2009;65(1):19–23. doi: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tan EK, Ho P, Tan L, Prakash KM, Zhao Y. PLA2G6 mutations and Parkinson’s disease. Ann Neurol. 2010;67(1):148. doi: 10.1002/ana.21663. [DOI] [PubMed] [Google Scholar]
- 114.Yoshino H, Tomiyama H, Tachibana N, et al. Phenotypic spectrum of patients with PLA2G6 mutation and PARK14-linked parkinsonism. Neurology. 2010;75(15):1356–1361. doi: 10.1212/WNL.0b013e3181f73649. [DOI] [PubMed] [Google Scholar]
- 115.Shojaee S, Sina F, Banihosseini SS, et al. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am J Hum Genet. 2008;82(6):1375–1384. doi: 10.1016/j.ajhg.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Di Fonzo A, Dekker MC, Montagna P, et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72(3):240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 117.Luo LZ, Xu Q, Guo JF, et al. FBXO7 gene mutations may be rare in Chinese early-onset Parkinsonism patients. Neurosci Lett. 2010;482(2):86–89. doi: 10.1016/j.neulet.2010.06.083. [DOI] [PubMed] [Google Scholar]
- 118.Wilkinson KD, Lee KM, Deshpande S, Duerksen-Hughes P, Boss JM, Pohl J. The neuron-specific protein PGP 9.5 is a ubiquitin carboxyl-terminal hydrolase. Science. 1989;246(4930):670–673. doi: 10.1126/science.2530630. [DOI] [PubMed] [Google Scholar]
- 119.Doran JF, Jackson P, Kynoch PA, Thompson RJ. Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis. J Neurochem. 1983;40(6):1542–1547. doi: 10.1111/j.1471-4159.1983.tb08124.x. [DOI] [PubMed] [Google Scholar]
- 120.Leroy E, Boyer R, Auburger G, et al. The ubiquitin pathway in Parkinson’s disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- 121.Nishikawa K, Li H, Kawamura R, et al. Alterations of structure and hydrolase activity of parkinsonism-associated human ubiquitin carboxyl-terminal hydrolase L1 variants. Biochem Biophys Res Commun. 2003;304(1):176–183. doi: 10.1016/s0006-291x(03)00555-2. [DOI] [PubMed] [Google Scholar]
- 122.Maraganore DM, Farrer MJ, Hardy JA, Lincoln SJ, McDonnell SK, Rocca WA. Case-control study of the ubiquitin carboxy-terminal hydrolase L1 gene in Parkinson’s disease. Neurology. 1999;53(8):1858–1860. doi: 10.1212/wnl.53.8.1858. [DOI] [PubMed] [Google Scholar]
- 123.Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT., Jr The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson’s disease susceptibility. Cell. 2002;111(2):209–218. doi: 10.1016/s0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- 124.Hutter CM, Samii A, Factor SA, et al. Lack of evidence for an association between UCHL1 S18Y and Parkinson’s disease. Eur J Neurol. 2008;15(2):134–139. doi: 10.1111/j.1468-1331.2007.02012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ragland M, Hutter C, Zabetian C, Edwards K. Association between the ubiquitin carboxyl-terminal esterase L1 gene (UCHL1) S18Y variant and Parkinson’s disease: A HuGE review and meta-analysis. Am J Epidemiol. 2009;170(11):1344–1357. doi: 10.1093/aje/kwp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tan EK, Lu CS, Peng R, et al. Analysis of the UCHL1 genetic variant in Parkinson’s disease among Chinese. Neurobiol Aging. 2010;31(12):2194–2196. doi: 10.1016/j.neurobiolaging.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 127.Pankratz N, Nichols WC, Uniacke SK, et al. Genome screen to identify susceptibility genes for Parkinson disease in a sample without parkin mutations. Am J Hum Genet. 2002;71(1):124–135. doi: 10.1086/341282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lautier C, Goldwurm S, Durr A, et al. Mutations in the GIGYF2 (TNRC15) gene at the PARK11 locus in familial Parkinson disease. Am J Hum Genet. 2008;82(4):822–833. doi: 10.1016/j.ajhg.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Giovannone B, Lee E, Laviola L, Giorgino F, Cleveland KA, Smith RJ. Two novel proteins that are linked to insulin-like growth factor (IGF-I) receptors by the Grb10 adapter and modulate IGF-I signaling. J Biol Chem. 2003;278(34):31564–31573. doi: 10.1074/jbc.M211572200. [DOI] [PubMed] [Google Scholar]
- 130.Bras J, Simon-Sanchez J, Federoff M, et al. Lack of replication of association between GIGYF2 variants and Parkinson disease. Hum Mol Genet. 2009;18(2):341–346. doi: 10.1093/hmg/ddn340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zimprich A, Schulte C, Reinthaler E, et al. PARK11 gene (GIGYF2) variants Asn56Ser and Asn457Thr are not pathogenic for Parkinson’s disease. Parkinsonism Relat Disord. 2009;15(7):532–534. doi: 10.1016/j.parkreldis.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 132.Nichols WC, Kissell DK, Pankratz N, et al. Variation in GIGYF2 is not associated with Parkinson disease. Neurology. 2009;72(22):1886–1892. doi: 10.1212/01.wnl.0000346517.98982.1b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Strauss KM, Martins LM, Plun-Favreau H, et al. Loss of function mutations in the gene encoding Omi/HtrA2 in Parkinson’s disease. Hum Mol Genet. 2005;14(15):2099–2111. doi: 10.1093/hmg/ddi215. [DOI] [PubMed] [Google Scholar]
- 134.Hegde R, Srinivasula SM, Zhang Z, et al. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J Biol Chem. 2002;277(1):432–438. doi: 10.1074/jbc.M109721200. [DOI] [PubMed] [Google Scholar]
- 135.Simon-Sanchez J, Singleton AB. Sequencing analysis of OMI/HTRA2 shows previously reported pathogenic mutations in neurologically normal controls. Hum Mol Genet. 2008;17(13):1988–1993. doi: 10.1093/hmg/ddn096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kruger R, Sharma M, Riess O, et al. A large-scale genetic association study to evaluate the contribution of Omi/HtrA2 (PARK13) to Parkinson’s disease. Neurobiol Aging. 2011;32(3):548.e9–548.e18. doi: 10.1016/j.neurobiolaging.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chartier-Harlin MC, Dachsel J, Hulihan M, et al. EIF4G1 mutations in familial parkinsonism. Parkinsonism Relat Disord. 2009;15(Suppl 2):145–146. [Google Scholar]
- 138.Gasser T, Muller-Myhsok B, Wszolek ZK, et al. A susceptibility locus for Parkinson’s disease maps to chromosome 2p13. Nat Genet. 1998;18(3):262–265. doi: 10.1038/ng0398-262. [DOI] [PubMed] [Google Scholar]
- 139.Hicks AA, Petursson H, Jonsson T, et al. A susceptibility gene for late-onset idiopathic Parkinson’s disease. Ann Neurol. 2002;52(5):549–555. doi: 10.1002/ana.10324. [DOI] [PubMed] [Google Scholar]
- 140.Sharma M, Mueller JC, Zimprich A, et al. The sepiapterin reductase gene region reveals association in the PARK3 locus: Analysis of familial and sporadic Parkinson’s disease in European populations. J Med Genet. 2006;43(7):557–562. doi: 10.1136/jmg.2005.039149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hruska KS, LaMarca ME, Scott CR, Sidransky E. Gaucher disease: Mutation and polymorphism spectrum in the glucocerebrosidase gene (GBA) Hum Mutat. 2008;29(5):567–583. doi: 10.1002/humu.20676. [DOI] [PubMed] [Google Scholar]
- 142.Neudorfer O, Giladi N, Elstein D, et al. Occurrence of Parkinson’s syndrome in type I Gaucher disease. QJM. 1996;89(9):691–694. doi: 10.1093/qjmed/89.9.691. [DOI] [PubMed] [Google Scholar]
- 143.Halperin A, Elstein D, Zimran A. Increased incidence of Parkinson disease among relatives of patients with Gaucher disease. Blood Cells Mol Dis. 2006;36(3):426–428. doi: 10.1016/j.bcmd.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 144.Aharon-Peretz J, Rosenbaum H, Gershoni-Baruch R. Mutations in the glucocerebrosidase gene and Parkinson’s disease in Ashkenazi Jews. N Engl J Med. 2004;351(19):1972–1977. doi: 10.1056/NEJMoa033277. [DOI] [PubMed] [Google Scholar]
- 145.Sidransky E, Nalls MA, Aasly JO, et al. Multicenter analysis of glucocerebrosidase mutations in Parkinson’s disease. N Engl J Med. 2009;361(17):1651–1661. doi: 10.1056/NEJMoa0901281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Velayati A, Yu WH, Sidransky E. The role of glucocerebrosidase mutations in Parkinson disease and Lewy body disorders. Curr Neurol Neurosci Rep. 2010;10(3):190–198. doi: 10.1007/s11910-010-0102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jo E, McLaurin J, Yip CM, St George-Hyslop P, Fraser PE. Alpha-synuclein membrane interactions and lipid specificity. J Biol Chem. 2000;275(44):34328–34334. doi: 10.1074/jbc.M004345200. [DOI] [PubMed] [Google Scholar]
- 148.Lesage S, Anheim M, Condroyer C, et al. Large-scale screening of the Gaucher’s disease-related glucocerebrosidase gene in Europeans with Parkinson’s disease. Hum Mol Genet. 2011;20(1):202–210. doi: 10.1093/hmg/ddq454. [DOI] [PubMed] [Google Scholar]
- 149.Wu YR, Chen CM, Chao CY, et al. Glucocerebrosidase gene mutation is a risk factor for early onset of Parkinson disease among Taiwanese. J Neurol Neurosurg Psychiatry. 2007;78(9):977–979. doi: 10.1136/jnnp.2006.105940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Neumann J, Bras J, Deas E, et al. Glucocerebrosidase mutations in clinical and pathologically proven Parkinson’s disease. Brain. 2009;132(Pt 7):1783–1794. doi: 10.1093/brain/awp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Goker-Alpan O, Giasson BI, Eblan MJ, et al. Glucocerebrosidase mutations are an important risk factor for Lewy body disorders. Neurology. 2006;67(5):908–910. doi: 10.1212/01.wnl.0000230215.41296.18. [DOI] [PubMed] [Google Scholar]
- 152.Conrad C, Andreadis A, Trojanowski JQ, et al. Genetic evidence for the involvement of tau in progressive supranuclear palsy. Ann Neurol. 1997;41(2):277–281. doi: 10.1002/ana.410410222. [DOI] [PubMed] [Google Scholar]
- 153.Houlden H, Baker M, Morris HR, et al. Corticobasal degeneration and progressive supranuclear palsy share a common tau haplotype. Neurology. 2001;56(12):1702–1706. doi: 10.1212/wnl.56.12.1702. [DOI] [PubMed] [Google Scholar]
- 154.Poorkaj P, Bird TD, Wijsman E, et al. Tau is a candidate gene for chromosome 17 frontotemporal dementia. Ann Neurol. 1998;43(6):815–825. doi: 10.1002/ana.410430617. [DOI] [PubMed] [Google Scholar]
- 155.Pastor P, Ezquerra M, Munoz E, et al. Significant association between the tau gene A0/A0 genotype and Parkinson’s disease. Ann Neurol. 2000;47(2):242–245. [PubMed] [Google Scholar]
- 156.Rhodes SL, Sinsheimer JS, Bordelon Y, Bronstein JM, Ritz B. R eplication of GWAS associations for GAK and MAPT in Parkinson’s disease. Ann Hum Genet. 2011;75(2):195–200. doi: 10.1111/j.1469-1809.2010.00616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Paglini G, Peris L, Mascotti F, Quiroga S, Caceres A. Tau protein function in axonal formation. Neurochem Res. 2000;25(1):37–42. doi: 10.1023/a:1007531230651. [DOI] [PubMed] [Google Scholar]
- 158.Galpern WR, Lang AE. Interface between tauopathies and synucleinopathies: A tale of two proteins. Ann Neurol. 2006;59(3):449–458. doi: 10.1002/ana.20819. [DOI] [PubMed] [Google Scholar]
- 159.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307(5712):1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 160.Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet. 2005;37(2):129–137. doi: 10.1038/ng1508. [DOI] [PubMed] [Google Scholar]
- 161.Hardy J, Pittman A, Myers A, et al. Evidence suggesting that Homo neanderthalensis contributed the H2 MAPT haplotype to Homo sapiens. Biochem Soc Trans. 2005;33(Pt 4):582–585. doi: 10.1042/BST0330582. [DOI] [PubMed] [Google Scholar]
- 162.Pastor P, Ezquerra M, Perez JC, et al. Novel haplotypes in 17q21 are associated with progressive supranuclear palsy. Ann Neurol. 2004;56(2):249–258. doi: 10.1002/ana.20178. [DOI] [PubMed] [Google Scholar]
- 163.Yamamoto-Katayama S, Ariyoshi M, Ishihara K, Hirano T, Jingami H, Morikawa K. Crystallographic studies on human BST-1/CD157 with ADP-ribosyl cyclase and NAD glycohydrolase activities. J Mol Biol. 2002;316(3):711–723. doi: 10.1006/jmbi.2001.5386. [DOI] [PubMed] [Google Scholar]
- 164.Tan EK, Kwok HH, Tan LC, et al. Analysis of GWAS-linked loci in Parkinson disease reaffirms PARK16 as a susceptibility locus. Neurology. 2010;75(6):508–512. doi: 10.1212/WNL.0b013e3181eccfcd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Tucci A, Nalls MA, Houlden H, et al. Genetic variability at the PARK16 locus. Eur J Hum Genet. 2010;18(12):1356–1359. doi: 10.1038/ejhg.2010.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Kimura SH, Tsuruga H, Yabuta N, Endo Y, Nojima H. Structure, expression, and chromosomal localization of human GAK. Genomics. 1997;44(2):179–187. doi: 10.1006/geno.1997.4873. [DOI] [PubMed] [Google Scholar]
- 167.Ungewickell EJ, Hinrichsen L. Endocytosis: Clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19(4):417–425. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 168.Grunblatt E, Mandel S, Jacob-Hirsch J, et al. Gene expression profiling of parkinsonian substantia nigra pars compacta; alterations in ubiquitin-proteasome, heat shock protein, iron and oxidative stress regulated proteins, cell adhesion/cellular matrix and vesicle trafficking genes. J Neural Transm. 2004;111(12):1543–1573. doi: 10.1007/s00702-004-0212-1. [DOI] [PubMed] [Google Scholar]
- 169.Wersinger C, Sidhu A. An inflammatory pathomechanism for Parkinson’s disease? Curr Med Chem. 2006;13(5):591–602. doi: 10.2174/092986706776055760. [DOI] [PubMed] [Google Scholar]
- 170.Orr CF, Rowe DB, Mizuno Y, Mori H, Halliday GM. A possible role for humoral immunity in the pathogenesis of Parkinson’s disease. Brain. 2005;128(Pt 11):2665–2674. doi: 10.1093/brain/awh625. [DOI] [PubMed] [Google Scholar]
- 171.Nalls MA, Plagnol V, Hernandez DG, et al. International Parkinson Disease Genomics Consortium Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: A meta-analysis of genome-wide association studies. Lancet. 2011;377(9766):641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]