Video abstract
Video
Keywords: chronic lymphocytic leukemia, chromosomal abnormality, miRNA deregulation
Abstract
Chronic lymphocytic leukemia is the most common leukemia in adults. By cytogenetic investigations major subgroups of the disease can be identified that reflect different routes of tumor development. Of these chromosomal deviations, trisomy 12 and deletions of parts of either the long arm of chromosome 13, the long arm of chromosome 11, or the short arm of chromosome 17 are most commonly detected. In some of these aberrations the molecular target has been identified as eg, ataxia telangiectasia mutated (ATM) in case of deletions of chromosomal region 11q22~23 and the genes encoding microRNAs miR-15a/16-1 as likely targets of deletions of chromosomal band 13q14.3. Of note, these aberrations do not characterize independent subgroups but often coexist within the metaphases of one tumor. Generally, complex aberrations are associated with a worse prognosis than simple karyotypic alterations. Due to smaller sizes of the missing segment the detection of recurrent deletions is not always possible by means of classical cytogenetics but requires more advanced techniques as in particular fluorescence in situ hybridization (FISH). Nevertheless, at this time it is not recommended to replace classical cytogenetics by FISH because this would miss additional information given by complex or secondary karyotypic alterations. However, the results of cytogenetic analyses allow the stratification of prognostic and predictive groups of the disease. Of these, the group characterized by deletions involving TP53 is clinically most relevant. In the future refined methods as eg, array-based comparative genomic hybridization will supplement the existing techniques to characterize CLL.
Chronic lymphocytic leukemia: the most common leukemia in Western countries
Chronic lymphocytic leukemia (CLL) is characterized by the presence of small, monomorphic, round to slightly irregular B lymphocytes in the peripheral blood, bone marrow, lymph nodes, liver, and spleen. Nonleukemic disease presenting with the tissue morphology and immunophenotype of CLL are termed small lymphocytic lymphoma (SLL).1 The disease is more common in males. Patients are often asymptomatic at the time of diagnosis but the continuing accumulation of CLL lymphocytes leads to development of symptoms, disease-related complications, and a need for therapy. CLL causes approximately 4,500 deaths per year in the United States alone.2,3
There are approximately 15,000 new cases of CLL reported annually in the United States3 and CLL is generally the most common leukemia of the adult population in Western countries with an incidence of two to six cases per 100,000 persons/year. Incidence appears to be correlated with age, with more than ten cases per 100,000 in those aged over 65 years. Moreover, approximately 6% of elderly individuals present with a clonal B-cell population that immunophenotypically and cytogenetically resembles CLL.4 This latter condition is called monoclonal B-cell lymphocytosis (MBL). MBL is known to progress to CLL in approximately 2% of cases, and CLL may generally be preceded by MBL. Thus, MBL may sometimes be more appropriately considered to be early or low stage CLL, and in the past patients have been diagnosed as having CLL when they would only fulfill the diagnostic criteria for MBL. There is not only a progression from MBL to CLL, but CLL is also able to undergo further clinicopathologic transformation to become an aggressive lymphoma with a predominance of diffuse large B-cell lymphomas. This transformation is referred to as Richter syndrome.
Although the f irst description of CLL dates back 150 years there is still little known about the etiology of the disease. Pathogenetically, the immunophenotype of the resting B cells characterizing CLL (CD5, CD23, CD27, and low level of surface immunoglobulin [Ig]) is different from that of normal B cells. CLL cells either have somatically mutated or unmutated IgV genes, suggesting that the corresponding tumor precursor cells originated from an antigen-dependent or -independent developmental stage, respectively.4
Cytogenetic findings: methods, and pathogenetic and clinical implications
Generally, there is little doubt that CLL is a monoclonal disease which makes it similar to most other benign or malignant tumors. This is supported by the results of cytogenetic studies. Techniques that appear to be for specific in vitro stimulation of the tumor cells in CLL by an immunostimulatory CpG oligonucleotide plus interleukin 25 represent a major step forward in understanding the pathogenesis of the disease. By applying these techniques, metaphases can be obtained in the vast majority of cases. Clonal chromosome abnormalities have been detected in more than 80% of the cases studied through the use of classical cytogenetics.6 The percentage of cases with at least one abnormalitiy increases when fluorescence in situ hybridization (FISH) is applied using appropriate probes to detect the most frequent abnormalities in addition to classical cytogenetics, mainly due to the detection of deletions under the detection limit made possible through the analysis of chromosomal bands.3 There is no doubt that most cytogenetic changes seen in CLL are causally related to the disease and its progression, and cytogenetic analyses in CLL have generally turned out to be good for providing prognoses and predictions. Analyses are usually carried out by classical as well as molecular cytogenetic methods. There has been debate as to whether FISH can replace classical cytogenetics in the genetic analysis of CLL, but the larger spectrum of aberrations that can be detected through classical cytogenetics seems to make this indispensable at present.6
In contrast to most other leukemias and lymphomas, typical recurrent chromosomal translocations are very rare in CLL, if they exist at all. Translocations involving chromosomal band 14q32 including the t(11;14)(q13;q32) (Figure 1) typically seen in Mantle cell lymphoma (MCL) have repeatedly have been reported, but distinguishing MCL from CLL can be complicated due to overlapping morphologic features and similar clinical manifestations.7 Hence, it may be speculated that these cases would be diagnosed as MCL if strict diagnostic criteria were followed.
Figure 1.
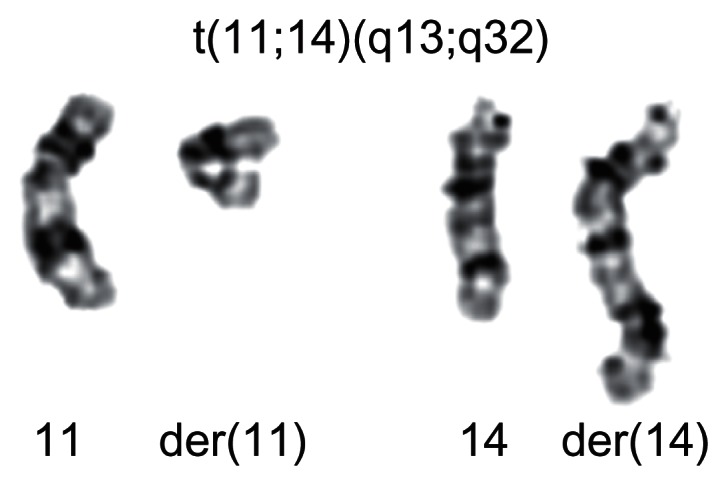
The translocation t(11;14)(q13;q32) is a nonrandom chromosomal rearrangement in Mantle cell lymphoma.
However, the identification of four main cytogenetic changes of in CLL was possible due to several recurrent and common gains and losses of chromosomal material: these are trisomy 12 (16%), and deletions of chromosomal regions 13q14 (55%), 11q (18%), and 17p (7%).4 Chromosomal deviations can occur either as the sole chromosome abnormality or coexist with other changes of the number or structure of the chromosomes, which leads to more complex changes of the karyotype. This also holds true for the most frequent cytogenetic abnormalities, which allows the independent subgroups to be distinguished. It is noted that these abnormalities are not mutually exclusive and so can occur together (Figure 2).
Figure 2.
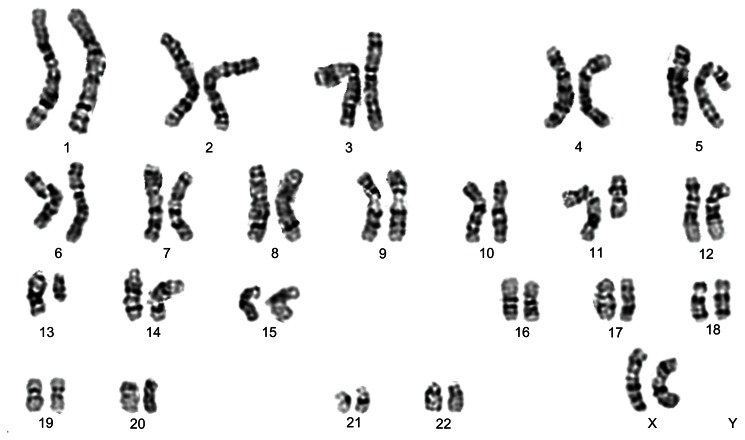
The frequent karyotype abnormalities in CLL are not mutually exclusive. Representative G-banded karyotype of a CLL showing deletions of the long arm of chromosome 11 as well as of chromosome 13: 46,XX,del(11)(q21),del(13)(q14).
Abbreviation: CCL, chronic lymphocytic leukemia.
Deletions of the long arm of chromosome 13 involving 13q14: microRNAs impairing apoptosis?
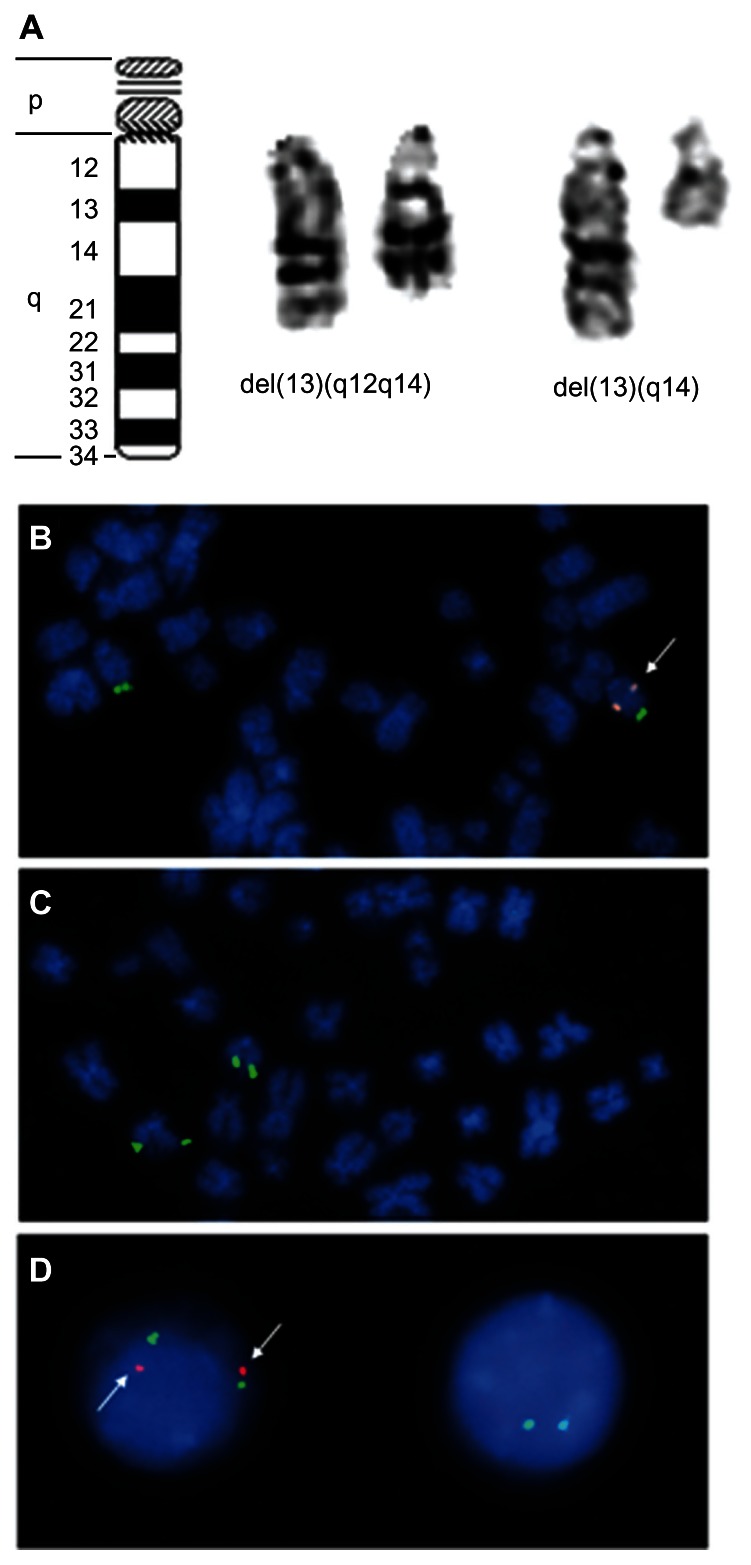
The most frequent chromosomal abnormalities observed in CLL are deletions of the long arm of chromosome 13 involving 13q14, which occurs in more than half of all CLL cases.8–10 The size of the deletion varies from those comprising almost the entire long arm of the chromosome to those detectable only by FISH (Figure 3A–D). Most of these deletions are monoallelic, but some are biallelic. In a study by Hernández et al,11 roughly 20% of the deletions that were detected as the sole abnormality at the time of first diagnosis were found to be biallelic. This deletion is found at the same frequency in MBL.12 The 13q14 deletion is not confined to CLL, as deletions of the same minimal deleted region (MDR) appear to also occur in subsets of multiple myelomas and mature T-cell lymphomas. Moreover, deletions comprising band 13q14 have been found in several benign and malignant solid tumors as lipomas and thyroid adenomas.13–18 In CLL the minimal deleted region contains sequences encoding two apparently sterile transcripts: DLEU2 (deleted in leukemia-2) and DLEU1.19,20 Intron 4 of DLEU2 encodes two microRNAs, miR-15a and miR-16-1, which are processed from its primary transcripts, and so share DLEU2’s transcriptional regulation and seem to interact with pathways governing apoptosis.21 Downregulation of DLEU2 and miR-15a/16-1 expression compared to normal B cells has been described for CLL without 13q14 deletions as well,22 suggesting an epigenetic mechanism is suppressing the DLEU2/miR-15a/16-1 cluster in cases lacking 13q14 deletion.23 miRNAs miR-15a/16-1 may have a pathogenetic function, and both seem to be involved in keeping the balance between proliferation and apoptosis. Nevertheless, their action as a posttranscriptional repressor of the BCL2 (B-cell lymphoma 2) expression is still a matter of an debate.24–26
Figure 3.
Deletions of chromosome 13 are the most frequent karyotype alterations in CLL. (A, left to right): Ideogram of chromosome 13, example of an interstitial deletion of the long arm of chromosome 13 in CLL, and example of a terminal deletion of chromosome 13 in CLL. (B) Part of a metaphase with monoallelic deletion of chromosome 13 as detected by FISH, the orange (spectrum: orange)-labeled probe detects the minimal deleted region, the green (spectrum: green)-labeled probe detects a terminal DNA segment of the long arm of chromosome 13 to facilitate its identification. Chromosomes are counterstained by DAPI. (C) Part of a metaphase with diallelic deletion of chromosome 13 as detected by FISH. For probes and counterstaining see (B). (D, left to right): nucleus of a normal cell without 13q-deletion and nucleus of a cell with biallelic deletion. For probes and counterstaining see (B).
Abbreviations: CCL, chronic lymphocytic leukemia; DAPI, 4′6-diamidino-2-phenylindole; FISH, fluorescent in situ hybridization.
Deletions of chromosome 13 are associated with quite a favorable prognosis with no detectable impact if these occur as the sole genetic abnormality and if they are mono- or biallelic.11,27 It is interesting to note that the absence of del(13) (q14) seems to protect against the transformation of CLL into diffuse large B-cell lymphoma, as the absence of del(13)(q14) characterizes patient subgroups at high risk of developing Richter syndrome.28 Some recent reports suggest that the size of the deletions influences prognosis. In particular, codeletion of the retinoblastoma gene (RB1) locus seems to be relevant.29
Trisomy 12: gene dosage effects and several oncogenes
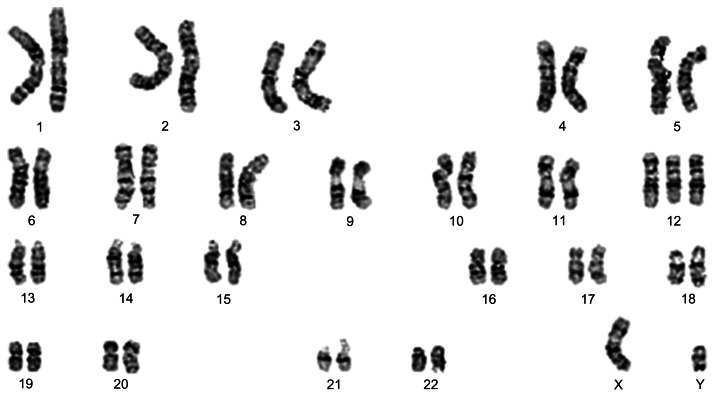
Deletions of chromosome 13 trisomy 12 (Figure 4) appear to be similar to frequent karyotypic alterations in CLL that are not confined to this type of tumor, such as a cytogenetic subgroup of uterine leiomyomas characterized by trisomy 12 as the sole karyotypic abnormality.30 The molecular mechanisms by which trisomy 12 supports tumorigenic transformation in CLL are unknown. However, it is assumed that trisomy 12 may cause an elevated gene dosage of a candidate proto-oncogene. Several genes with oncogenic potential have been assigned to chromosome 12, such as HMGA2 (high-mobility group AT-hook 2)31 which encodes a so-called architectural transcription factor implicated in mechanisms of benign and malignant tumorigenesis. Further examples are MDM2 (murine double minute 2), which is known to be amplified in some malignant solid tumors encoding a major repressor of p53, and GLI1 (glioma-associated oncogene homologue 1). Overall, the prognosis of CLL with trisomy as the only aberration is less favorable than that of CLL with deletions of the long arm of chromosome 13, but better than that of cases with either deletions of chromosome 11 or 17.
Figure 4.
Trisomy 12 is the second most frequent karyotypic abnormality seen in CLL. Representative G-banded karyotype of a CLL with trisomy 12 as the sole karyotypic deviation. Karyotype: 47,XY,+12.
Abbreviation: CCL, chronic lymphocytic leukemia.
Deletions of the long arm of chromosome 11 involving 11q22-q23: ATM deficiency destabilizing the tumor genome
Deletions of chromosomal region 11q22-q23 (Figure 5A) can be detected in almost 20% of cases, which makes them the second most frequent karyotypic change in CLL. These deletions often lead to a hemizygous loss of the ataxia telangiectasia (ATM) gene.32 Mechanistically, ATM is able to signal DNA damage and to mediate p53 activation which, as a major player assuring genome stability, can ultimately trigger apoptosis or oncogene-induced senescence if the damage is not repaired. Therefore, damaged cells are not adequately removed and ATM deficiency causes genomic instability.
Figure 5.
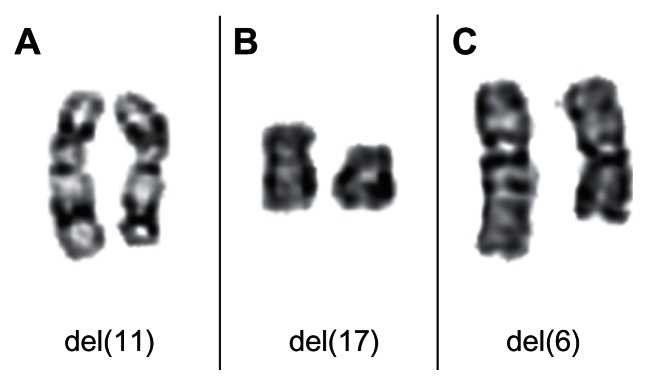
Partial karyotypes showing recurrent chromosomal deletions in CLL. (A) Chromosomal pair 11 from a CLL showing a del(11)(q22). (B) Chromosomal pair 17 from a CLL showing a del(17)(p12). (C) Chromosomal pair 6 from a CLL showing a del(6)(q13q22).
Abbreviation: CCL, chronic lymphocytic leukemia.
Deletions of the short arm of chromosome 17 involving 17p13: another p53 story
It appears that the same pathways are disturbed in CLL with deletions of chromosomal band 17p13 (Figure 5B), which is seen in about 5% of cases. These deletions always cause a loss of the TP53 tumor suppressor gene. Interestingly, in the majority of cases with cytogenetically visible monoallelic deletion, mutations have been detected in the remaining TP53 allele as well.32 In rare cases, biallelic mutations of TP53 have been found without 17p deletions, leading to clinical, pathological, and prognostic features identical to 17p deletions. However, these genetic aberrations carry the same devastating prognostic and predictive impact as 17p deletions.33 Additional techniques such as sequencing are now being applied to analyze mutations affecting TP53 that escape detection by classical cytogenetics.
Rare, but recurrent cytogenetic abnormalities in CLL
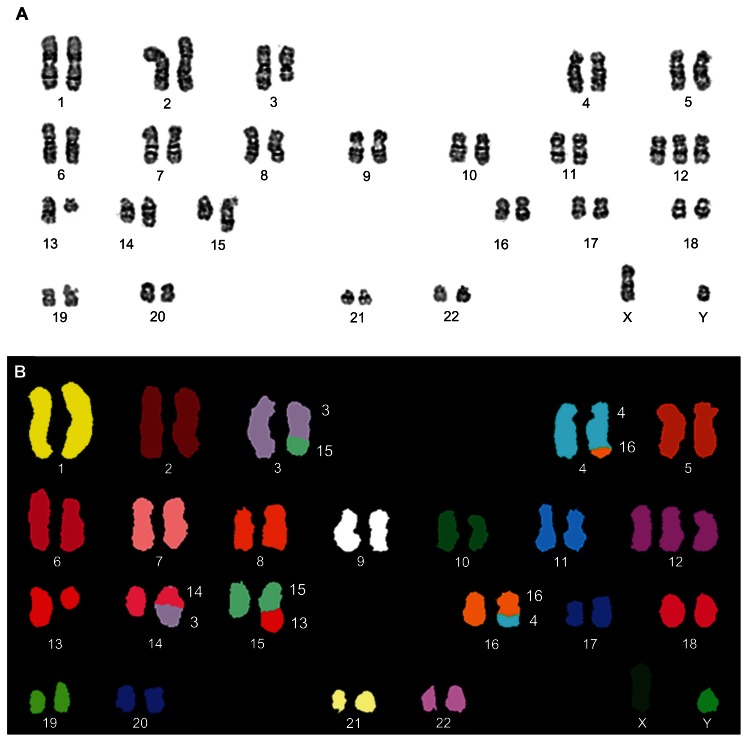
Other recurrent but rare karyotypic abnormalities in CLL are deletion of the long arm of chromosome 6 (Figure 5C), translocations involving 14q32 chromosome 3, and trisomies of the long arm of chromosome 8.34–40 The uncovering of complex karyotype aberrations in CLL is sometimes impossible without the application of molecular cytogenetic methods (Figure 6). Complex aberrant karyotypes (defined as the existence of three or more clonal abnormalities identified by chromosomal band analysis changes) have generally been described to characterize an adverse prognostic group of CLL.41 Only a few cases of Richter’s cytogenetic transformation of CLL have been studied in detail.42
Figure 6.
Complex aberrant karyotypes in CLL are associated with an adverse prognosis. Example of a CLL with a complex karyotypic rearrangement involving chromosomes 3, 4, 13, 14, 15, and 16. Karyotype: 47,XY,der(3)t(3;15)(q21;q22),t(4;16)(q31;q24),+12,del(13)(q14),der(14)t(3;14)(q21;q24),der(15)t(13;15)(q14;q22). (A) Representative G-banded karyotype. (B) SKY-FISH analysis.
Abbreviations: CCL, chronic lymphocytic leukemia; SKY-FISH, spectral karyotyping and fluorescent in situ hybridization.
Prognostic and predictive implications of chromosomal abnormalities and miRNA deregulation in CLL
Numerous studies have provided clear evidence that chromosomal alterations in CLL have an impact on the patient’s prognosis. Nevertheless, the significance of predictive markers may change with emerging new therapies, and it will not be easy to clearly distinguish between the prognostic and predictive significance of changes. Those alterations that are detectable at the level of classical cytogenetics (ie, the analyses of chromosomal bands, or by FISH with commonly used DNA probes) and deletions of the Tp53 locus are highly relevant indicators of poor prognosis.
Recent studies indicate that the impact of these deletions and equivalent mutations in TP53 not only has prognostic impacts but also affects the response to therapy. Standard CLL-therapy is currently based on the use of fludarabine, but this treatment is generally ineffective in patients showing impairment of p53.43 However, not all restrictions of p53 functions are detectable through the application of these techniques, and thus sequencing and functional assays are also used to identify genetic subgroups of CLL. For example, in vitro exposure to etoposide plus nutlin-3a allows the detection and identification of ATM and TP53 mutations.44 Deletions of the ATM locus that are usually detectable by means of classical cytogenetics are generally associated with a poor prognosis as well, but recent studies suggest that chemoimmunotherapy may overcome the prognostic impact of 11q deletion and point to 11q deletion as a potential predictive factor for increased benefit by fludarabine– cyclophosphamide–rituximab (FCR) treatment.45,46
Trisomy 12 as the sole cytogenetic deviation is associated with a intermediate prognostic impact (median survival = 114 months)47 but recent studies suggest that CLL patients with trisomy 12 and NOTCH1 (Notch homolog 1) mutations have a prognostic course worse than that with trisomy 12 alone. NOTCH1 mutations contribute to a significantly worse survival in trisomy 12 CLL patients compared to that of trisomy 12 alone.48
CLL with 13q deletion as the sole cytogenetic abnormality usually has a good prognosis. However, it has become obvious from recent data that both the percentage of deleted nuclei and presence of larger deletions involving the RB1 locus can help to further refine the prognosis of del 13q-only cases. In particular, CLL carrying <70% of 13q deleted nuclei with deletions not including the RB1 locus were characterized by particularly long time-to-treatment. On the other hand, 13q deletions seen in <70% of nuclei but involving the RB1 locus, or 13q deletion in ≥70% of nuclei with or without RB1 deletions, are apparently associated with a shorter time-to-treatment.49
The expression of microRNAs is a rapidly emerging field in cancer diagnosis and monitoring. In CLL, the expression of miR-150, miR-223, and miR-29b/c correlates with mutated IgHV (immunoglobulin heavy variable) and a favorable clinical course. Similarly, low expression of miR-223 and miR-29c was found to correlate with unmutated IgHV, ZAP-70 (zeta-chain-associated protein kinase 70) expression, and disease progression.47 However, while miRNA expression patterns present a possible diagnostic and predictive tool, more data will be required before they can be integrated into daily clinical diagnostic practice.
Future directions in genetic analyses of CLL
The type and frequency of aberrations among the tumor cell population are major characteristics of CLL, so genetic analyses offer valuable tools for a prognostic and predictive stratification of patient groups. Currently, a combination of classical G-band analysis and FISH seems to be the best method of collecting relevant data. Despite its much higher resolution, FISH on interphase nuclei using a panel of probes covering the most frequent genomic inbalances seen in CLL still cannot replace classical cytogenetics, which is superior in detecting complex karyotypic alterations that are of prognostic impact. As adjuncts, the application of molecular cytogenetic methods for the interpretation of complex karyotypes (Figure 6B) has considerable significance for selected cases following classical cytogenetics and interphase FISH. Array-based comparative genomic hybridization will become another important tool for the analysis of selected cases of CLL.50–52 Perhaps even more important is the combination of genetic analysis with functional or gene expression studies. As an example of this type of study, Decker et al were able to demonstrate that smoothened-inhibitor responsiveness correlated with elevated GLI1 and PTCH1 (protein Patched homolog 1) transcript levels and the presence of trisomy 12, while no other karyotype correlated with responsiveness.53
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumors of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- 2.Lanasa MC. Novel insights into the biology of CLL. Hematology Am Soc Hematol Educ Program. 2010;2010:70–76. doi: 10.1182/asheducation-2010.1.70. [DOI] [PubMed] [Google Scholar]
- 3.Cramer P, Hallek M. Prognostic factors in chronic lymphocytic leukemia – what do we need to know? Nat Rev Clin Oncol. 2011;8:38–47. doi: 10.1038/nrclinonc.2010.167. [DOI] [PubMed] [Google Scholar]
- 4.Klein U, Dalla-Favera R. New insights into the pathogenesis of chronic lymphocytic leukemia. Semin Cancer Biol. 2010;20:377–383. doi: 10.1016/j.semcancer.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 5.Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide-induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: A study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108:3152–3160. doi: 10.1182/blood-2006-02-005322. [DOI] [PubMed] [Google Scholar]
- 6.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21:2442–2451. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 7.Ho AK, Hill S, Preobrazhensky SN, Miller ME, Chen Z, Bahler DW. Small B-cell neoplasms with typical mantle cell lymphoma immuno-phenotypes often include chronic lymphocytic leukemias. Am J Clin Pathol. 2009;131:27–32. doi: 10.1309/AJCPPAG4VR4IPGHZ. [DOI] [PubMed] [Google Scholar]
- 8.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 9.Döhner H, Stilgenbauer S, Döhner K, Bentz M, Lichter P. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J Mol Med (Berl) 1999;77:266–281. doi: 10.1007/s001090050350. [DOI] [PubMed] [Google Scholar]
- 10.Kalachikov S, Migliazza A, Cayanis E, et al. Cloning and gene mapping of the chromosome 13q14 region deleted in chronic lymphocytic leukemia. Genomics. 1997;42:369–377. doi: 10.1006/geno.1997.4747. [DOI] [PubMed] [Google Scholar]
- 11.Hernández JA, Rodríguez AE, González M, et al. A high number of losses in 13q14 chromosome band is associated with a worse outcome and biological differences in patients with B-cell chronic lymphoid leukemia. Haematologica. 2009;94:364–371. doi: 10.3324/haematol.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rawstron AC, Bennett FL, O’Connor SJ, et al. Monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. N Engl J Med. 2008;359:575–583. doi: 10.1056/NEJMoa075290. [DOI] [PubMed] [Google Scholar]
- 13.Belge G, Roque L, Soares J, et al. Cytogenetic investigations of 340 thyroid hyperplasias and adenomas revealing correlations between cytogenetic findings and histology. Cancer Genet Cytogenet. 1998;101:42–48. doi: 10.1016/s0165-4608(97)00057-5. [DOI] [PubMed] [Google Scholar]
- 14.Dahlén A, Debiec-Rychter M, Pedeutour F, et al. Clustering of deletions on chromosome 13 in benign and low-malignant lipomatous tumors. Int J Cancer. 2003;103:616–623. doi: 10.1002/ijc.10864. [DOI] [PubMed] [Google Scholar]
- 15.Cigudosa JC, Rao PH, Calasanz MJ, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91:3007–3010. [PubMed] [Google Scholar]
- 16.Liu Y, Hermanson M, Grandér D, et al. 13q deletions in lymphoid malignancies. Blood. 1995;86:1911–1915. [PubMed] [Google Scholar]
- 17.Stilgenbauer S, Nickolenko J, Wilhelm J, et al. Expressed sequences as candidates for a novel tumor suppressor gene at band 13q14 in B-cell chronic lymphocytic leukemia and mantle cell lymphoma. Oncogene. 1998;16:1891–1897. doi: 10.1038/sj.onc.1201764. [DOI] [PubMed] [Google Scholar]
- 18.Rosenwald A, Ott G, Krumdiek AK, et al. A biological role for deletions in chromosomal band 13q14 in mantle cell and peripheral t-cell lymphomas? Genes Chromosomes Cancer. 1999;26:210–214. doi: 10.1002/(sici)1098-2264(199911)26:3<210::aid-gcc4>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Corcoran M, Rasool O, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15:2463–2473. doi: 10.1038/sj.onc.1201643. [DOI] [PubMed] [Google Scholar]
- 20.Migliazza A, Bosch F, Komatsu H, et al. Nucleotide sequence, transcription map, and mutation analysis of the 13q14 chromosomal region deleted in B-cell chronic lymphocytic leukemia. Blood. 2001;97:2098–2104. doi: 10.1182/blood.v97.7.2098. [DOI] [PubMed] [Google Scholar]
- 21.Fabbri M, Bottoni A, Shimizu M, et al. Association of a microRNA/ TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mertens D, Wolf S, Schroeter P, et al. Down-regulation of candidate tumor suppressor genes within chromosome band 13q14.3 is independent of the DNA methylation pattern in B-cell chronic lymphocytic leukemia. Blood. 2002;99:4116–4121. doi: 10.1182/blood.v99.11.4116. [DOI] [PubMed] [Google Scholar]
- 24.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Linsley PS, Schelter J, Burchard J, et al. Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Mol Cell Biol. 2007;27:2240–2252. doi: 10.1128/MCB.02005-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fulci V, Chiaretti S, Goldoni M, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 27.Van Dyke DL, Shanafelt TD, Call TG, et al. A comprehensive evaluation of the prognostic significance of 13q deletions in patients with B-chronic lymphocytic leukaemia. Br J Haematol. 2010;148:544–550. doi: 10.1111/j.1365-2141.2009.07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fangazio M, De Paoli L, Rossi D, Gaidano G. Predictive markers and driving factors behind Richter syndrome development. Expert Rev Anticancer Ther. 2011;11:433–442. doi: 10.1586/era.10.237. [DOI] [PubMed] [Google Scholar]
- 29.Ouillette P, Erba H, Kujawski L, et al. Integrated genomic profiling of chronic lymphocytic leukemia identifies subtypes of deletion 13q14. Cancer Res. 2008;68:1012–1021. doi: 10.1158/0008-5472.CAN-07-3105. [DOI] [PubMed] [Google Scholar]
- 30.Vanni R, Van Roy N, Lecca U, Speleman F. Uterine leiomyoma cytogenetics. III. Interphase cytogenetic analysis of karyotypically normal uterine leiomyoma excludes possibility of undetected trisomy 12. Cancer Genet Cytogenet. 1992;62:40–42. doi: 10.1016/0165-4608(92)90035-7. [DOI] [PubMed] [Google Scholar]
- 31.Schoenmakers EF, Wanschura S, Mols R, Bullerdiek J, Van den Berghe H, Van de Ven WJ. Recurrent rearrangements in the high mobility group protein gene, HMGI-C, in benign mesenchymal tumours. Nat Genet. 1995;10:436–444. doi: 10.1038/ng0895-436. [DOI] [PubMed] [Google Scholar]
- 32.Gronowitz JS, Hagberg H, Källander CF, Simonsson B. The use of serum deoxythymidine kinase as a prognostic marker, and in the monitoring of patients with non-Hodgkin’s lymphoma. Br J Cancer. 1983;47:487–495. doi: 10.1038/bjc.1983.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 34.Döhner H, Stilgenbauer S, James MR, et al. 11q deletions identify a new subset of B-cell chronic lymphocytic leukemia characterized by extensive nodal involvement and inferior prognosis. Blood. 1997;89:2516–2522. [PubMed] [Google Scholar]
- 35.Kröber A, Seiler T, Benner A, et al. V(H) mutation status, CD38 expression level, genomic aberrations, and survival in chronic lymphocytic leukemia. Blood. 2002;100:1410–1416. [PubMed] [Google Scholar]
- 36.Dewald GW, Brockman SR, Paternoster SF, et al. Chromosome anomalies detected by interphase fluorescence in situ hybridization: correlation with significant biological features of B-cell chronic lymphocytic leukaemia. Br J Haematol. 2003;121:287–295. doi: 10.1046/j.1365-2141.2003.04265.x. [DOI] [PubMed] [Google Scholar]
- 37.Shanafelt TD, Witzig TE, Fink SR, et al. Prospective evaluation of clonal evolution during long-term follow-up of patients with untreated early-stage chronic lymphocytic leukemia. J Clin Oncol. 2006;24:4634–4641. doi: 10.1200/JCO.2006.06.9492. [DOI] [PubMed] [Google Scholar]
- 38.Stilgenbauer S, Bullinger L, Benner A, et al. Incidence and clinical significance of 6q deletions in B cell chronic lymphocytic leukemia. Leukemia. 1999;13:1331–1334. doi: 10.1038/sj.leu.2401499. [DOI] [PubMed] [Google Scholar]
- 39.Stilgenbauer S, Sander S, Bullinger L, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92:1242–1245. doi: 10.3324/haematol.10720. [DOI] [PubMed] [Google Scholar]
- 40.Oscier DG, Gardiner AC, Mould SJ, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100:1177–1184. [PubMed] [Google Scholar]
- 41.Haferlach C, Dicker F, Weiss T, et al. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49:851–859. doi: 10.1002/gcc.20794. [DOI] [PubMed] [Google Scholar]
- 42.Bockorny B, Codreanu I, Dasanu CA. Hodgkin lymphoma as Richter transformation in chronic lymphocytic leukaemia: a retrospective analysis of world literature. Br J Haematol. 2012;156:50–66. doi: 10.1111/j.1365-2141.2011.08907.x. [DOI] [PubMed] [Google Scholar]
- 43.Hallek M, Fischer K, Fingerle-Rowson G, et al. Addition of rituximab to fludarabine and cyclophosphamide in patients with chronic lymphocytic leukaemia: a randomised, open-label, phase 3 trial. Lancet. 2010;376:1164–1174. doi: 10.1016/S0140-6736(10)61381-5. [DOI] [PubMed] [Google Scholar]
- 44.Best OG, Gardiner AC, Majid A, et al. A novel functional assay using etoposide plus nutlin-3a detects and distinguishes between ATM and TP53 mutations in CLL. Leukemia. 2008;22:1456–1459. doi: 10.1038/sj.leu.2405092. [DOI] [PubMed] [Google Scholar]
- 45.Tsimberidou AM, Tam C, Abruzzo LV, et al. Chemoimmunotherapy may overcome the adverse prognostic significance of 11q deletion in previously untreated patients with chronic lymphocytic leukemia. Cancer. 2009;115:373–380. doi: 10.1002/cncr.23993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zenz T, Fröhling S, Mertens D, Döhner H, Stilgenbauer S. Moving from prognostic to predictive factors in chronic lymphocytic leukaemia. Best Pract Res Clin Haematol. 2010;23:71–84. doi: 10.1016/j.beha.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 47.Parker TL, Strout MP. Chronic lymphocytic leukemia: prognostic factors and impact on treatment. Discov Med. 2011;11:115–123. [PubMed] [Google Scholar]
- 48.Del Giudice I, Rossi D, Chiaretti S, et al. NOTCH1 mutations in +12 chronic lymphocytic leukemia (CLL) confer an unfavorable prognosis, induce a distinctive transcriptional profiling and refine the intermediate prognosis of +12 CLL. Haematologica. 2011 Dec 29; doi: 10.3324/haematol.2011.060129. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dal Bo M, Rossi FM, Rossi D, et al. 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes Chromosomes Cancer. 2011;50:633–643. doi: 10.1002/gcc.20885. [DOI] [PubMed] [Google Scholar]
- 50.Gunn SR, Bolla AR, Barron LL, et al. Array CGH analysis of chronic lymphocytic leukemia reveals frequent cryptic monoallelic and biallelic deletions of chromosome 22q11 that include the PRAME gene. Leuk Res. 2009;33:1276–1281. doi: 10.1016/j.leukres.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 51.Kay NE, Eckel-Passow JE, Braggio E, et al. Progressive but previously untreated CLL patients with greater array CGH complexity exhibit a less durable response to chemoimmunotherapy. Cancer Genet Cytogenet. 2010;203:161–168. doi: 10.1016/j.cancergencyto.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rinaldi A, Mian M, Kwee I, et al. Genome-wide DNA profiling better defines the prognosis of chronic lymphocytic leukaemia. Br J Haematol. 2011;154:590–599. doi: 10.1111/j.1365-2141.2011.08789.x. [DOI] [PubMed] [Google Scholar]
- 53.Decker S, Zirlik K, Djebatchie L, et al. Trisomy 12 and elevated GLI1 and PTCH1 transcript levels are biomarkers for Hedgehog-inhibitor responsiveness in CLL. Blood. 2012;119:997–1007. doi: 10.1182/blood-2011-06-359075. [DOI] [PubMed] [Google Scholar]