Abstract
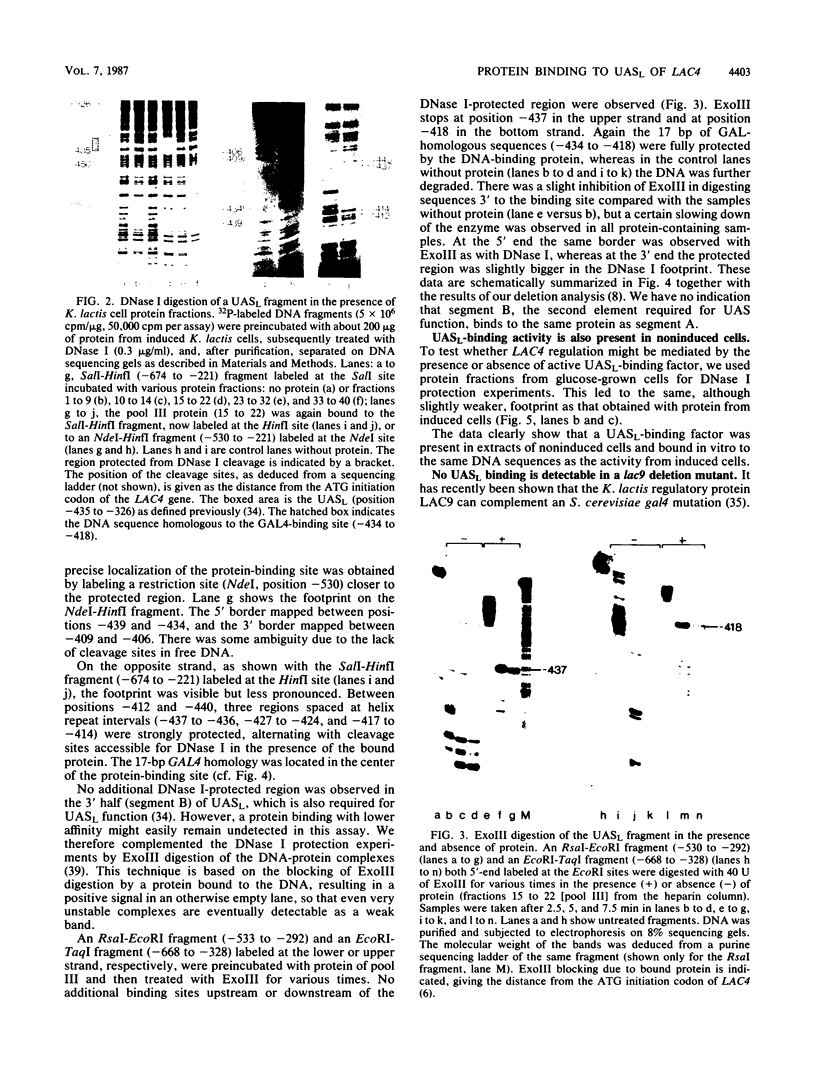
As shown previously, the beta-galactosidase gene of Kluyveromyces lactis is transcriptionally regulated via an upstream activation site (UASL) which contains a sequence homologous to the GAL4 protein-binding site in Saccharomyces cerevisiae (M. Ruzzi, K.D. Breunig, A.G. Ficca, and C.P. Hollenberg, Mol. Cell. Biol. 7:991-997, 1987). Here we demonstrate that the region of homology specifically binds a K. lactis regulatory protein. The binding activity was detectable in protein extracts from wild-type cells enriched for DNA-binding proteins by heparin affinity chromatography. These extracts could be used directly for DNase I and exonuclease III protection experiments. A lac9 deletion strain, which fails to induce the beta-galactosidase gene, did not contain the binding factor. The homology of LAC9 protein with GAL4 (J.M. Salmeron and S. A. Johnston, Nucleic Acids Res. 14:7767-7781, 1986) strongly suggests that LAC9 protein binds directly to UASL and plays a role similar to that of GAL4 in regulating transcription.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arcangioli B., Lescure B. Identification of proteins involved in the regulation of yeast iso- 1-cytochrome C expression by oxygen. EMBO J. 1985 Oct;4(10):2627–2633. doi: 10.1002/j.1460-2075.1985.tb03980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D. R., Sledziewski A., Young E. T. Deletion analysis identifies a region, upstream of the ADH2 gene of Saccharomyces cerevisiae, which is required for ADR1-mediated derepression. Mol Cell Biol. 1985 Jul;5(7):1743–1749. doi: 10.1128/mcb.5.7.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Kornberg R. D. Specific protein binding to far upstream activating sequences in polymerase II promoters. Proc Natl Acad Sci U S A. 1985 Jan;82(1):43–47. doi: 10.1073/pnas.82.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Lue N. F., Kornberg R. D. A GAL family of upstream activating sequences in yeast: roles in both induction and repression of transcription. EMBO J. 1986 Mar;5(3):603–608. doi: 10.1002/j.1460-2075.1986.tb04253.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A bacterial repressor protein or a yeast transcriptional terminator can block upstream activation of a yeast gene. Nature. 1984 Dec 13;312(5995):612–615. doi: 10.1038/312612a0. [DOI] [PubMed] [Google Scholar]
- Breunig K. D., Dahlems U., Das S., Hollenberg C. P. Analysis of a eukaryotic beta-galactosidase gene: the N-terminal end of the yeast Kluyveromyces lactis protein shows homology to the Escherichia coli lacZ gene product. Nucleic Acids Res. 1984 Mar 12;12(5):2327–2341. doi: 10.1093/nar/12.5.2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner A., de Cobos A. T., Griffiths D. E. The isolation and genetic characterization of extrachromosomal chloramphenicol and oligomycin-resistant mutants from the petite-negative yeast Kluyveromyces lactis. Mol Gen Genet. 1977 Apr 29;152(3):183–191. doi: 10.1007/BF00268816. [DOI] [PubMed] [Google Scholar]
- Das S., Breunig K. D., Hollenberg C. P. A positive regulatory element is involved in the induction of the beta-galactosidase gene from Kluyveromyces lactis. EMBO J. 1985 Mar;4(3):793–798. doi: 10.1002/j.1460-2075.1985.tb03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. C., Barr K. Characterization of lactose transport in Kluyveromyces lactis. J Bacteriol. 1983 Jun;154(3):1245–1251. doi: 10.1128/jb.154.3.1245-1251.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner M. M., Revzin A. A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 1981 Jul 10;9(13):3047–3060. doi: 10.1093/nar/9.13.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters: positive and negative elements. Cell. 1984 Apr;36(4):799–800. doi: 10.1016/0092-8674(84)90028-x. [DOI] [PubMed] [Google Scholar]
- Guarente L., Yocum R. R., Gifford P. A GAL10-CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc Natl Acad Sci U S A. 1982 Dec;79(23):7410–7414. doi: 10.1073/pnas.79.23.7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D. E., Hope I. A., Macke J. P., Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986 Oct 24;234(4775):451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lucchini G., Fink G. R. A synthetic HIS4 regulatory element confers general amino acid control on the cytochrome c gene (CYC1) of yeast. Proc Natl Acad Sci U S A. 1985 Jan;82(2):498–502. doi: 10.1073/pnas.82.2.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. GCN4 protein, synthesized in vitro, binds HIS3 regulatory sequences: implications for general control of amino acid biosynthetic genes in yeast. Cell. 1985 Nov;43(1):177–188. doi: 10.1016/0092-8674(85)90022-4. [DOI] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Klebe R. J., Harriss J. V., Sharp Z. D., Douglas M. G. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene. 1983 Nov;25(2-3):333–341. doi: 10.1016/0378-1119(83)90238-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Adachi Y., Toh-e A., Oshima Y. Function of positive regulatory gene gal4 in the synthesis of galactose pathway enzymes in Saccharomyces cerevisiae: evidence that the GAL81 region codes for part of the gal4 protein. J Bacteriol. 1980 Feb;141(2):508–527. doi: 10.1128/jb.141.2.508-527.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K., Toh-e A., Oshima Y. Genetic control of galactokinase synthesis in Saccharomyces cerevisiae: evidence for constitutive expression of the positive regulatory gene gal4. J Bacteriol. 1978 May;134(2):446–457. doi: 10.1128/jb.134.2.446-457.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Van Keuren M. L. Silver staining methods for polyacrylamide gel electrophoresis. Methods Enzymol. 1983;96:230–239. doi: 10.1016/s0076-6879(83)96021-4. [DOI] [PubMed] [Google Scholar]
- Perlman D., Hopper J. E. Constitutive synthesis of the GAL4 protein, a galactose pathway regulator in Saccharomyces cerevisiae. Cell. 1979 Jan;16(1):89–95. doi: 10.1016/0092-8674(79)90190-9. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Arcangioli B., Guarente L. Yeast HAP1 activator competes with the factor RC2 for binding to the upstream activation site UAS1 of the CYC1 gene. Cell. 1987 Apr 10;49(1):9–18. doi: 10.1016/0092-8674(87)90750-1. [DOI] [PubMed] [Google Scholar]
- Riley M. I., Hopper J. E., Johnston S. A., Dickson R. C. GAL4 of Saccharomyces cerevisiae activates the lactose-galactose regulon of Kluyveromyces lactis and creates a new phenotype: glucose repression of the regulon. Mol Cell Biol. 1987 Feb;7(2):780–786. doi: 10.1128/mcb.7.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruzzi M., Breunig K. D., Ficca A. G., Hollenberg C. P. Positive regulation of the beta-galactosidase gene from Kluyveromyces lactis is mediated by an upstream activation site that shows homology to the GAL upstream activation site of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):991–997. doi: 10.1128/mcb.7.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron J. M., Jr, Johnston S. A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986 Oct 10;14(19):7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheetz R. M., Dickson R. C. Mutations affecting synthesis of beta-galactosidase activity in the yeast Kluyveromyces lactis. Genetics. 1980 Aug;95(4):877–890. doi: 10.1093/genetics/95.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K., Chen W., Hill D. E., Hope I. A., Oettinger M. A. Constitutive and coordinately regulated transcription of yeast genes: promoter elements, positive and negative regulatory sites, and DNA binding proteins. Cold Spring Harb Symp Quant Biol. 1985;50:489–503. doi: 10.1101/sqb.1985.050.01.061. [DOI] [PubMed] [Google Scholar]
- Wray L. V., Jr, Witte M. M., Dickson R. C., Riley M. I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationships to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987 Mar;7(3):1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. An exonuclease protection assay reveals heat-shock element and TATA box DNA-binding proteins in crude nuclear extracts. Nature. 1985 Sep 5;317(6032):84–87. doi: 10.1038/317084a0. [DOI] [PubMed] [Google Scholar]