Abstract
microRNAs (miRNAs) encode a novel class of small, non-coding RNAs that regulate gene expression post-trancriptionally. miRNAs comprise one of the major non-coding RNA families, whose diverse biological functions and unusual capacity for gene regulation have attracted enormous interests in the RNA world. Over the past 16 years, genetic, biochemical and computational approaches have greatly shaped the growth of the field, leading to the identification of thousands of miRNA genes in nearly all metazoans. The key molecular machinery for miRNA biogenesis and silencing has been identified, yet the precise biochemical and regulatory mechanisms still remain elusive. However, recent findings have shed new light on how miRNAs are generated and how they function to repress gene expression. miRNAs provide a paradigm for endogenous small RNAs that mediate gene silencing at a genome-wide level. The gene silencing mediated by these small RNAs constitutes a major component of gene regulation during various developmental and physiological processes. The accumulating knowledge about their biogenesis and gene silencing mechanism will add a new dimension to our understanding about the complex gene regulatory networks.
Introduction
The founding member of the miRNA family, lin-4, was identified in 1993 through a forward genetic screen for defects in temporal control of larval development in C. elegans 1, 2. lin-4 is a small non-coding RNA 21 nucleotide in length, generated by two sequential processing of its nascent precursor transcript (pri-miRNA) that formed a hairpin secondary structure 1. Its major target lin-14 was discovered through the same genetic screen, whose gain-of-function mutation phenocopied the loss-of-function mutation of lin-4 2. Besides the genetic evidence, the identification of seven imperfect lin-4 binding sites within the lin-14 3′UTR further indicated the ability of lin-4 to directly repress lin-14 expression. Both in vivo data and in vitro data suggested that these lin-4 binding elements are both necessary and sufficient for lin-4 mediated gene repression 1, 2. This first set of observations has established a paradigm in which a miRNA is derived from a structurally distinct precursor RNA to mediate the target repression at the post-transcriptional level.
Unfortunately, this exciting finding did not generate the profound impacts it deserved in the following years, as researchers failed to discover lin-4 counterparts in organisms other than the worm. lin-4, despite of its potent biological effects and its unique gene regulatory mechanisms, was considered an isolated story in C. elegans. It was only through the identification of the second worm miRNA, let-7, that the evolutionary conservation of miRNAs was uncovered, and the biological significance of these small RNAs started to be appreciated 3.
Almost coincident with the discovery of miRNAs, a highly related biological process, RNA interference (RNAi), gained enormous popularity as a functional genetic tool 4. RNAi was initially identified as a molecular mechanism for exogenous RNA molecules to mediate the cleavage of specific host mRNAs through perfect sequence complementarity 5. siRNAs, small non-coding RNAs similar to miRNAs in structure, were recognized as the key components of the RNAi pathway 6. Fundamentally, siRNAs and miRNAs are very similar in terms of their molecular characteristics, biogenesis pathways and effector pathways. But they differ in the mode of target recognition and the precise mechanisms for gene silencing 4, 7–9. Given the extensive evolutionary conservation of the RNAi pathway, miRNAs were hypothesized as a source of endogenous small RNAs that can harness the RNAi or RNAi-like mechanisms to mediate post-transcriptional gene silencing. Thus, it is conceivable that miRNAs exist in almost all metazoans that harbor the RNAi machinery. Not surprisingly, three different groups simultaneously carried out the initial cloning efforts, and identified abundant species of novel miRNAs in multiple model organisms, including worms, flies and mammals 10–12. To date, hundreds of miRNAs have been identified in various model organisms, yet the number of the miRNA species is still rising due to the combined efforts of molecular cloning, bioinformatic prediction and pyrosequencing. miRNAs exhibit tissue specific and developmentally regulated expression patterns, and novel miRNAs continue to be found whenever a new cell/tissue type is examined.
The biogenesis of miRNAs
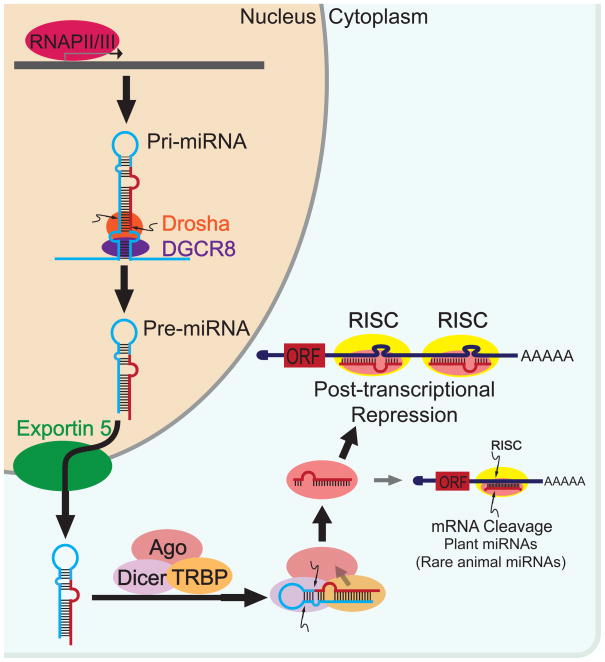
Primary miRNA transcripts (pri-miRNAs), which contain one or multiple stem-loop hairpin structures, are mostly derived from PolII-mediated transcription (Fig 1). The sequence encoding the mature miRNA is mostly embedded in one arm of the stem-loop structure, and eventually liberated after two sequential processing events during miRNA biogenesis 13. In the first step towards the canonical miRNA maturation pathway, pri-miRNA is cleaved by the microprocessor complex to yield the pre-miRNA, a hairpin-shaped intermediate precursor ~65 nucleotides in length 14 (Fig1). The microprocessor complex consists of the RNaseIII enzyme Drosha that mediates the pri-miRNA cleavage, and a double stranded RNA (dsRNA) binding protein DGCR8/Parsha 15, 16. DGCR8/Parsha binds to the base of the hairpin stem to position Drosha cleavage at a site about one helical RNA turn away 17 (Fig1). Although most miRNA biogenesis is initiated with Drosha processing, an alternative Drosha-independent pathway exists, in which debranched introns mimic the structural features of pre-miRNAs 18–20. Pre-miRNAs are then exported from the nucleus to the cytoplasm by Explortin 5 21, where another RNaseIII enzyme Dicer, assisted by its dsRNA binding partner TRBP/loquacious, catalyzes the second processing event for miRNA biogenesis and liberates the mature miRNA duplexes 22 (Fig1).
Fig 1. The current model for miRNA biogenesis.
RNA polymerase (RNAP) II/III transcribes a nascent pri-miRNA, which is processed by the microprocessor complex, Drosha and DGCR8, to produce a hairpin precursor, pre-miRNA. Pre-miRNA is exported into the cytoplasm by Exportin5, and subsequently processed by Dicer to yield mature miRNA duplex. The RISC loading complex contains Dicer, Ago and TRBP, which catalyzes pre-miRNA cleavage, mediates miRNA strand selection, and preferentially load mature miRNA strand onto the Ago protein. The Ago:miRNA complex then dissociates from the RISC loading complex, and become the core of the RISC complex to regulate post-transcriptional gene repression of specific target mRNAs.
The biogenesis of miRNAs is not a static process; instead, several lines of evidence suggest the existence of a molecular network that tightly regulates the biogenesis and the stability of specific miRNAs in a cell-type dependent and context dependent manner. One of the best studied examples is the repression of let-7 biogenesis in undifferentiated embryonic stem cells and neuronal stem cells 23, where the RNA-binding protein, Lin28, specifically binds to conserved nucleotides in the hairpin loop within the pri-let-7 precursors to prevent the Drosha processing 23–25. In addition, Lin28 also induces the uridylation of pre-let-7 at its 3′ end to block its processing by Dicer, and to mediate its degradation 24. Given the molecular parallel between the Drosha complex and the Dicer complex, it would not be surprising if more enhancers and/or repressors are identified to regulate the biogenesis of specific miRNAs on both processing steps.
The mature miRNA duplexes consist of the mature miRNA strand and the miRNA* strand, which are derived from two separate arms of the hairpin stem within the miRNA precursors. The imperfect base-pairing between miRNA and miRNA*, as predetermined by the stem-loop structure of the miRNA precursor, gives rise to a thermodynamically less stable 5′ end for the miRNA strand to facilitate its unwinding 26 (Fig1). This unique feature underlies the structural basis for the preferential loading of the miRNA strand into the effector complex, the RNA-induced silencing complex (RISC). The remaining miRNA* strand then undergoes subsequent degradation 26. The strand selection and the RISC loading of miRNAs are tightly coupled to the miRNA biogenesis described above. This is achieved by the RISC loading complex that contains Dicer, TRBP and the key component of the RISC complex, the Argonaut family proteins (Ago) 27 (Fig1). Pre-miRNAs get incorporated into the RISC loading complex, which catalyzes the pre-miRNA cleavage, mediates the miRNA strand selection, and promotes the miRNA loading into the Ago proteins 27. Finally, the Ago:miRNA component dissociates from the rest of the complex, and forms the core of the RISC complex that harbors the gene silencing activity (Fig1).
Target recognition by miRNAs
The key function of miRNAs within the RISC complex is to mediate target recognition, as the artificial tethering of the Ago proteins to an mRNA is sufficient to render gene silencing 28. Plant miRNAs and rare animal miRNAs can recognize their targets with nearly perfect sequence complementarity, thus triggering mRNA cleavage at the site of complementarity 8. However, most animal miRNAs bind their target mRNAs through multiple imperfect base-pairings within the 3′UTR, thus mediating post-transcriptional gene silencing through a cleavage-independent manner 8. The 2–8 nucleotides at the 5′ end of a miRNA are designated as the “seed” sequence. Its complementarity to the target mRNA, combined with a favorable sequence context flanking the seed-matching motif, largely determines the specificity for target recognition. In addition, the base-paring at the miRNA 3′ end and cooperative miRNA binding also have an impact on miRNA target recognition 29. The small size of miRNAs, combined with the imperfect base-pairing nature of their target recognition, allows miRNAs to regulate a large number of mRNAs with varying degrees of affinities 29. Conversely, a single mRNA target could be subjected to the combined regulation of multiple miRNAs.
The sequence complementarity between a miRNA and its targets has made it possible for computational prediction of miRNA targets. Classic examples of miRNA:target interactions were combined with extensive reporter analysis in cell culture studies to allow bioinformatic prediction of miRNA targets 28, 30, 31. Such prediction programs yield hundreds of mRNAs as targets of a given miRNA. This scenario has been recently validated for two specific miRNAs through quantitative mass spectrometry in over-expression studies 32, 33. Although miRNAs have the capacity to regulate a large number of mRNAs, the specific biological context, through a yet unknown mechanism, is also likely to play a role in determining the identity of miRNA targets and the affinity of miRNA:mRNA interaction. In addition, synergy among different miRNAs and the fine regulation of miRNA expression levels further adds to the varying degrees of target repression. It is this characteristic of miRNA regulation, perhaps, that allows miRNAs to exhibit substantial capacity and complexity in gene regulation, making them a key component of the gene regulatory networks underlying various developmental and physiological processes.
Mechanisms of miRNA-mediated gene silencing
The exact mechanisms for miRNA-mediated post-transcriptional repression are still being heavily investigated, yet studies using various miRNA systems have suggested different possibilities. Post-transcriptional repression mediated by miRNAs can occur through the inhibition of protein synthesis, the degradation of target mRNAs, and the translocation of target mRNAs into cytoplasmic processing bodies (P-bodies). Although seemingly different, these scenarios may not be mutually exclusive. For example, translation inhibition and mRNA degradation can occur concurrently as a result of miRNA-mediated gene silencing; the accumulation of miRNA targets in the P-body can be a consequence of inhibition of translational initiation; and the P-body may well be the site for mRNA degradation as it is enriched for mRNA degradation machinery. It is conceivable that the miRNA pathway mediates gene silencing through a variety of mechanisms; yet for a specific miRNA, the exact repression mechanisms may vary depending on the targets and the given biological context. This hypothesis may explain the diverse observations made by different groups that investigated the miRNA repression mechanisms using different biological systems.
miRNA-mediated translational repression
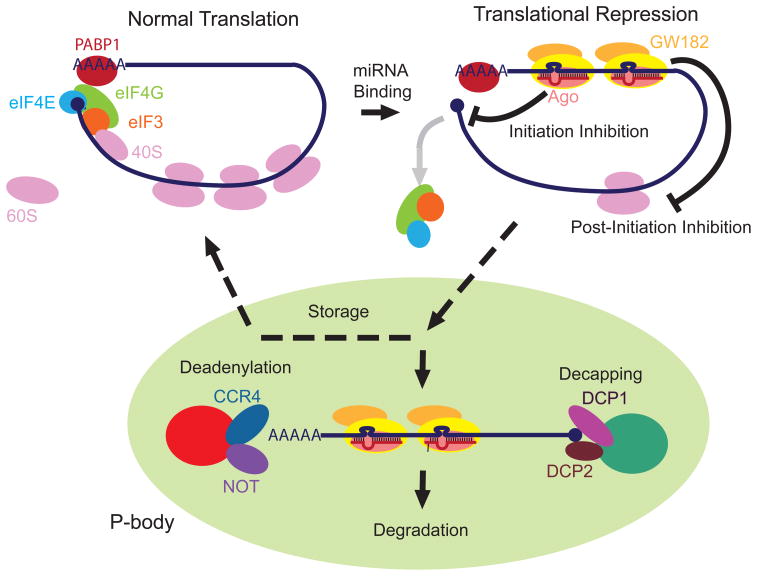
Translational repression is the first mechanism proposed for miRNA-mediated gene silencing, as some of the first identified miRNA targets exhibit significant down-regulation at the protein level without affecting mRNA stability 34. Translation occurs in three general steps: initiation, elongation, and termination. In cap-dependent eukaryotic translation, the translation initiation complex recognizes the mRNA 5′ cap through the cap binding component eIF4E, which interacts with the scaffolding core of the initiation complex, eIF4G. eIF4G associates with another initiation factor eIF3 to recruit the 40S ribosome subunit to the mRNA. The 40S ribosome subunit moves along the 5′ UTR of the mRNA until being assembled with the 60S subunit at the start codon for translational elongation. eIF4G also interacts with polyadenylate-binding protein 1 (PABP1) that binds the mRNA poly(A) tail. As a result, the initiation complex ‘circularizes’ the mRNA to strengthen the cap binding ability of eIF4e and facilitate the ribosome recycling 35.
Emerging evidence has indicated the indispensable role of the mRNA 5′ cap structure in miRNA-mediated gene silencing (Fig2). In HeLa cells, otherwise perfect miRNA targets were immune to miRNA-mediated repression when undergoing cap-independent translation, such as that initiated at the internal ribosome entry site (IRES) 36, 37. Experiments in cell-free systems also support these findings, as an in vitro reconstituted miRNA repression system requires a functional m7G cap in the target mRNA 38, 39. Consistently, in some polysome gradient analyses, miRNA-mediated repression was accompanied by reduced ribosome loading on the target mRNAs, while derepression of miRNA silencing restores the polysome association with the target mRNAs 37. These findings suggest that miRNAs act to inhibit ribosome assembly onto the target mRNA in a cap-dependent manner. A model for the underlying mechanistic basis comes from a recent study that compared the sequence homology between human Ago2 and eIF4E, which centered around the Ago mid domain that directly binds the m7G cap to repress translation, possibly by precluding the eIF4E recruitment 40 (Fig2). When mutations were introduced into the two conserved phenylalanine residues within the mid domain, human AGO2 lost its cap binding ability, and subsequently failed to mediate gene silencing induced by miRNAs 40. However, the competitive binding by Ago may not be a universal mechanism for miRNA-mediated silencing in all organisms. In contrast to the human data, the same mutation in the mid domain of Drosophila Ago1 abolished its ability to catalyze miRNA silencing without affecting m7G cap binding, suggesting the importance of a cap-independent mechanism under specific biological context 41. It is worth noting that both studies described above were carried out using a reporter over-expression studies in tissue culture cells. Thus, the physiological roles of Ago proteins in repressing cap-dependent translation need to be addressed in future studies.
Fig 2. The current model for miRNA-mediated gene repression.
Upon being bound by miRNAs, mRNAs become translationally repressed via a mechanism involving the proteins Ago and GW182, the latter of which is a known component of P-bodies. The inhibition can be either at the level of initiation of translation or post-initiation. Then, through a mechanism that is still not completely known, the silencing complexes containing the mRNAs aggregate to form P-bodies. Within the P-bodies, the mRNA can either be stored and released for translation at a later time or be marked for degradation via deadenylation or decapping.
Interestingly, a number of studies challenged the hypothesis described above, and proposed a model for post-initiation repression by miRNAs 42; Petersen, 2006 #206} (Fig2). Most evidence comes from the observation that miRNAs, repressed miRNA target mRNAs or RISC components can be associated with translationally competent polysomes 42, 43, and that IRES-dependent translation can be repressed by miRNAs in one particular study 44. These findings are in disagreement with the cap-dependent repression model. Despite the number of studies supporting a post-initiation model, there are debates about the reliability in differentiating between miRNPs and translationally competent polysomes in polysome gradient assays. As suggested by a recent study, formation of pseudo-polysomes, dense miRNA protein complexes that resemble polysomes in their sedimentation characteristics, can be observed even when ribosome assembly and polyribosome formation are blocked during initiation 45. It is likely that the differences in experimental approaches and biological systems contributed to this landscape of varying results. Despite the lack of consensus in the miRNA field, one can not exclude the possibility that post-initiation repression may work with cap-dependent repression under certain physiological context to achieve miRNA-mediated gene silencing.
miRNA-mediated mRNA degradation
Although miRNA-mediated mRNA degradation was once dismissed as a repression mechanism in some of the early studies, increasing evidence suggests that miRNA play an important role in regulating the stability of its target mRNAs. Both genome-wide mRNA profiling experiments 46 and detailed characterizations of specific miRNA targets 47 indicate that many, but not all, of the target mRNAs are subjected to partial degradation as induced by miRNAs. Studies in zebrafish, flies and mammalian systems suggest that miRNAs could lead to the deadenylation and/or decapping of their target mRNAs, followed by 3′->5′ and 5′->3′ decay, respectively 48. In some demonstrated cases, deadenylation and decapping were achieved through the recruitment of GW182 to the target mRNAs via the Ago-mediated protein interaction 48. GW182, a key component of the processing body (P-body), further recruits the deadenylating complex CCR4-NOT and/or the decapping complex DCP1/2 to the repressed mRNA target 49 (Fig2). It is worth noting that most repressed mRNAs only undergo partial degradation, the significance of which is not well understood. It is conceivable that partial degradation of mRNAs is desirable under specific physiological conditions, as a portion of the remaining mRNAs may eventually return to productive translation (Fig2).
The relative contribution of mRNA degradation and translational repression to miRNA-mediated gene silencing is still not clear. There are clear examples where miRNA-mediated gene silencing occurs predominantly through one of the two mechanisms. However, there are also plenty of examples where both mechanisms play a role in silencing. mRNA degradation results in permanent removal of miRNA targets, while translational repression leads to temporary inhibition of miRNA targets, which may resume translation upon derepression. Under physiological conditions, miRNAs mostly act as developmental switches or mediators for stress responses, and the relative contribution of these two modes of miRNA repression may well depend on the specific miRNA functions.
miRNAs and P-bodies
Although there is still debate about the exact mechanism of miRNA-mediated repression, evidence has been mounting in favor of the segregation of miRNA function in discrete cytoplasmic foci, known as P-bodies or GW-bodies 35. P-bodies have previously been shown to be dynamic cellular assemblies for mRNA degradation and storage 50. The first functional link between miRNAs and the P-bodies came from studies that demonstrated the accumulation of Ago proteins in P-bodies via a miRNA-dependent and target mRNA- dependent manner 51. The translocation of target mRNAs into the P-bodies is mediated through the physical interaction between Ago and GW182, which results in decreased mRNA association with polysomes, and the subsequent degradation of the mRNAs 41, 51 (Fig2). Knock-down of GW182 disrupted P-body formation and maintenance, and abrogated miRNA-mediated silencing 51. It is hypothesized that the P-body translocation insulates the miRNA-targeted mRNAs from active translation machinery and facilitates their turn-over. P-bodies can also serve as storage sites for repressed mRNAs, which, under certain physiological conditions, can be recycled back into the cytoplasm and reassemble with translation machinery 52. P-bodies contain a number of molecular machineries that mediate the two major mRNA degradation pathways through deadenylation and decapping, respectively. Among these, CCR4-NOT are the key apparatus for mRNA degradation through deadenylation and 3′->5′ decay, and DCP1/2 and XRN1 are the key players for mRNA degradation through decapping and 5′->3′ decay 49, 53 (Fig2). Depletion of these P-body components invariably impairs the target mRNA degradation induced by miRNAs 38, 41, 53.
Although it is tempting to hypothesize that P-bodies provide one of the sites for miRNA-mediated gene silencing, the formation of microscopically visible P-bodies does not always precede miRNA silencing. When the P-body proteins Lsm1 and Lsm3 were depleted, microscopically visible P-bodies vanished, but miRNA function was not compromised 41. Such findings indicate that microscopically visible P-bodies are not a prerequisite for miRNA silencing. It is proposed that submicroscopic miRNP complexes are sufficient to mediate silencing before the emergence of the visible P-body loci. Currently, P-bodies are loosely defined by molecular markers as a dynamic assembly of microscopically visible foci, but this definition may give rise to a great deal of heterogeneity within the P-bodies themselves. A less ambiguous characterization of P-bodies will greatly contribute to our understanding of the roles P-bodies play in miRNA-mediated gene repression.
Finally, evidence has emerged that along with P-bodies, stress granules may also play a role in miRNA-mediated repression 54. In fact, the association of Ago proteins to stress granules requires the presence of miRNAs, while this association is not necessary for P-bodies 54. However, recent findings have blurred the distinction between P-bodies and stress granules, as a subset of them share common molecular markers 55. The extent to which stress granules contribute to miRNA-mediated repression, and how they interact with P-body-mediated repression are still largely unknown.
Conclusions
We are still at the beginning to understand the realm of non-coding RNAs, whose functional importance is appreciated only recently. miRNAs represent one of the best studied non-coding RNA families. The diverse biological functions and unique gene repression mechanisms of miRNAs place them as integral components in the gene regulatory network for development and physiology. Although most miRNA genes share similar molecular structures, extensive complexity exists in the molecular basis underlying their biogenesis pathway and their silencing effects. To date, the key biochemical components for the major miRNA biogenesis pathway have been identified, and the regulatory network that promotes or inhibits miRNA biogenesis under specific physiological conditions has begun to emerge. More controversies exist for the precise mechanisms underlying miRNA-mediated gene silencing, as multiple models are established based on various biochemical studies using different biological systems. Since these studies often employ chimeric reporter mRNAs as miRNA target mRNAs, it is not clear if the different mechanistic models proposed reflect the different biological properties of these reporter constructs, or they truly recapitulate the range of diverse miRNA regulation under differing developmental or physiological contexts. It is conceivable that multiple mechanisms regulate miRNA mediated gene silencing. Yet, the key question is whether there exists specificity in this process. Under a given biological context, would a single mechanism or a combination of mechanisms be employed for gene silencing by a specific miRNA? In addition, are different models of miRNA function, if they co-exist, regulated differently? And are they each correspond to unique biological outcome? As we are witnessing the unparalleled discovery efforts aiming for small RNAs discovery, particularly miRNA identification, the understanding of these questions will undoubtedly provide important insights into the core of small RNA biology.
Acknowledgments
We thank Mona Foth, Pengcheng Bu and Virginie Olive for stimulating discussions and helpful input. L.H. is a Searle Scholar, and is supported by a K99/R00 grant from NCI.
References
- 1.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Slack FJ, et al. The lin-41 RBCC gene acts in the C. elegans heterochronic pathway between the let-7 regulatory RNA and the LIN-29 transcription factor. Mol Cell. 2000;5:659–69. doi: 10.1016/s1097-2765(00)80245-2. [DOI] [PubMed] [Google Scholar]
- 4.Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 5.Fire A, et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–11. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 6.Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–2. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- 7.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 10.Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–8. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- 11.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–4. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 12.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–62. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- 13.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. Embo J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee Y, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 15.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 16.Han J, et al. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Han J, et al. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 18.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–6. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutvagner G, et al. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–8. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- 23.Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heo I, et al. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 25.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. Rna. 2008;14:1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 27.MacRae IJ, Ma E, Zhou M, Robinson CV, Doudna JA. In vitro reconstitution of the human RISC-loading complex. Proc Natl Acad Sci U S A. 2008;105:512–7. doi: 10.1073/pnas.0710869105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 29.Grimson A, et al. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 31.Rajewsky N. microRNA target predictions in animals. Nat Genet. 2006;38 (Suppl):S8–13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- 32.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 34.Olsen PH, Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216:671–80. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 35.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 36.Humphreys DT, Westman BJ, Martin DI, Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc Natl Acad Sci U S A. 2005;102:16961–6. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pillai RS, et al. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–6. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 38.Wakiyama M, Takimoto K, Ohara O, Yokoyama S. Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system. Genes Dev. 2007;21:1857–62. doi: 10.1101/gad.1566707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–60. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- 40.Kiriakidou M, et al. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 2007;129:1141–51. doi: 10.1016/j.cell.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 41.Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346–53. doi: 10.1038/nsmb.1405. [DOI] [PubMed] [Google Scholar]
- 42.Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–7. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- 43.Nottrott S, Simard MJ, Richter JD. Human let-7a miRNA blocks protein production on actively translating polyribosomes. Nat Struct Mol Biol. 2006;13:1108–14. doi: 10.1038/nsmb1173. [DOI] [PubMed] [Google Scholar]
- 44.Petersen CP, Bordeleau ME, Pelletier J, Sharp PA. Short RNAs repress translation after initiation in mammalian cells. Mol Cell. 2006;21:533–42. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 45.Thermann R, Hentze MW. Drosophila miR2 induces pseudo-polysomes and inhibits translation initiation. Nature. 2007;447:875–8. doi: 10.1038/nature05878. [DOI] [PubMed] [Google Scholar]
- 46.Lim LP, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 47.Bagga S, et al. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–63. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 48.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–21. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 49.Behm-Ansmant I, et al. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes Dev. 2006;20:1885–98. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–46. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, et al. A role for the P-body component GW182 in microRNA function. Nat Cell Biol. 2005;7:1261–6. doi: 10.1038/ncb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–21. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 53.Sheth U, Parker R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–8. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc Natl Acad Sci U S A. 2006;103:18125–30. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kedersha N, et al. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J Cell Biol. 2005;169:871–84. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]