SUMMARY
The transcription factor HIF1A is a key mediator of the cellular response to hypoxia. Despite the importance of HIF1A in homeostasis and various pathologies, little is known about how it regulates RNA polymerase II (RNAPII). We report here that HIF1A employs a specific variant of the Mediator complex to stimulate RNAPII elongation. The Mediator-associated kinase CDK8, but not the paralog CDK19, is required for induction of many HIF1A target genes. HIF1A induces binding of CDK8-Mediator and the Super Elongation Complex (SEC), containing AFF4 and CDK9, to alleviate RNAPII pausing. CDK8 is dispensable for HIF1A chromatin binding and histone acetylation, but it is essential for binding of SEC and RNAPII elongation. Global analysis of active RNAPII reveals that hypoxia-inducible genes are paused and active prior to their induction. Our results provide a mechanistic link between HIF1A and CDK8, two potent oncogenes, in the cellular response to hypoxia.
INTRODUCTION
Hypoxia is a hallmark of rapidly growing tissues such as developing embryos and solid tumors. The cellular response to hypoxic stress involves the activation of Hypoxia Inducible Factors (HIFs), a group of DNA-binding proteins that induce transcription of numerous genes involved in angiogenesis, glycolysis, metabolic adaptation, erythropoiesis and cell survival (Semenza, 2012). Accordingly, the transcriptional response to hypoxia is essential for both proper embryonic development and growth of many solid tumors (Ryan et al., 1998). HIFs are heterodimeric transcription factors composed of α and β subunits (Wang et al., 1995). Whereas β subunits (ARNT1-3) are expressed constitutively, the expression of α subunits (HIF1A, HIF2A and HIF3A) is tightly regulated at the level of protein turnover by an oxygen-sensing pathway. In normoxia, enzymatic hydroxylation of key asparagine and proline residues in the α subunits inhibits their transactivation potential and generates a binding surface for the E3 ubiquitin ligase VHL, which triggers their degradation by the proteasome (Ivan et al., 2001; Jaakkola et al., 2001). Hydroxylation is impaired during hypoxia, leading to stabilization of α subunits, their nuclear accumulation, heterodimerization with β subunits, and consequent activation of target genes.
Although the biological importance of HIFs is undisputed, relatively little is known about the molecular mechanisms by which they control RNA polymerase II (RNAPII). Overexpression of HIF1A is known to drive the progression of various cancers, likely due to the adaptive advantage conferred by increased expression of target genes such as glycolytic enzymes (PGK, LDHA), pro-angiogenic factors (VEGF) and glucose transporters (GLUT1). Recently, HIF1A was implicated in the development of dietary obesity, atherosclerosis, pulmonary hypertension and abdominal aortic aneurysm formation (Semenza, 2012). Thus, a detailed understanding of HIF1A-dependent RNAPII transactivation will impact diverse fields of biomedical research. Much effort has been devoted to the characterization of the HIF proteins and the DNA elements they recognize (Mole et al., 2009), but little is known about HIF coactivators. The HIFα subunits contain an N-terminal activation domain (N-TAD) that overlaps the oxygen-dependent degradation domain and a C-terminal activation domain (C-TAD) that contains a conserved asparagine whose hydroxylation impairs binding to the coactivators CBP and p300 (Kallio et al., 1998). These two lysine acetyl-transferases are often depicted as the key HIF coactivators and, recently, PKM2 was identified as a cofactor enhancing HIF1A binding and p300 recruitment to hypoxia response elements (Luo et al., 2011). However, mutation of the CBP/p300 CH1 domain that is required for HIF interaction impairs expression of only a minority of HIF target genes, revealing the existence of additional coactivators (Kasper et al., 2005).
The Mediator coactivator complex is a widespread regulator of RNAPII activity conserved from yeast to humans (Bourbon, 2008; Conaway and Conaway, 2011). Since its early identification and characterization in yeast (Koleske et al., 1992; Thompson et al., 1993), Mediator has been shown to interact with RNAPII, general transcription factors, diverse DNA-binding proteins, transcription elongation factors and chromatin architecture proteins, thus serving as a signaling hub capable of integrating multiple inputs for precise control of RNAPII (Malik and Roeder, 2010). In mammals, Mediator is composed of 25–30 subunits assembled into distinct modules. The exact subunit composition of Mediator is variable, which may provide a mechanism for increased regulatory diversity and specificity (Sato et al., 2004). The CDK-module of Mediator carries the only known enzymatic activity in the complex and associates with the rest of Mediator in a facultative fashion (Sato et al., 2004). While the CDK-module was initially characterized as a repressor, recent work has shown that it is also required for gene activation across multiple transcriptional programs (for review see Galbraith et al. (2010)). Intriguingly, three subunits of this module have undergone gene duplications in the vertebrate lineage to generate the paralogous pairs CDK8/CDK19, MED12/MED12L and MED13/MED13L (Sato et al., 2004). Although very little is known about the functions of the different paralogs, it is expected that they each have specialized roles in mammals. For example, CDK8, but not CDK19, has been defined as an oncogene in colorectal cancer (Firestein et al., 2008).
In order to define specific functions for CDK8, we performed a comparative analysis of the two kinases in colorectal cancer cells. We found that they assemble into mutually exclusive Mediator complexes and regulate unique transcriptional programs. Strikingly, we found that CDK8, but not CDK19, is required for induction of many HIF1A target genes. Mechanistic studies revealed the existence of a novel HIF1A-dependent coactivation event acting in addition to HIF1A-induced histone acetylation. We found that HIF1A induces binding of CDK8 to its target genes during hypoxia and that its activation domain interacts with CDK8-Mediator. Prior to induction, hypoxia-inducible genes carry transcriptionally engaged, proximally paused RNAPII. Increased histone acetylation and recruitment of the BRD4 protein upon HIF1A activation does not suffice to activate paused RNAPII at these genes. Instead, we demonstrate that HIF1A employs CDK8-Mediator to recruit the Super Elongation Complex (SEC) along with CDK9. Global analysis of active RNA polymerases revealed that nearly all HIF1A targets are actively transcribed but display pausing during normoxia, suggesting that the transition to elongation is rate-limiting and fitting with the notion that CDK8 helps to stimulate pause-release. Thus, our studies constitute a significant advance in our understanding of how HIF1A and CDK8 regulate transcription and reveal a molecular mechanism by which these two oncogenes can collaborate.
RESULTS
CDK8 regulates gene expression programs distinct from CDK19
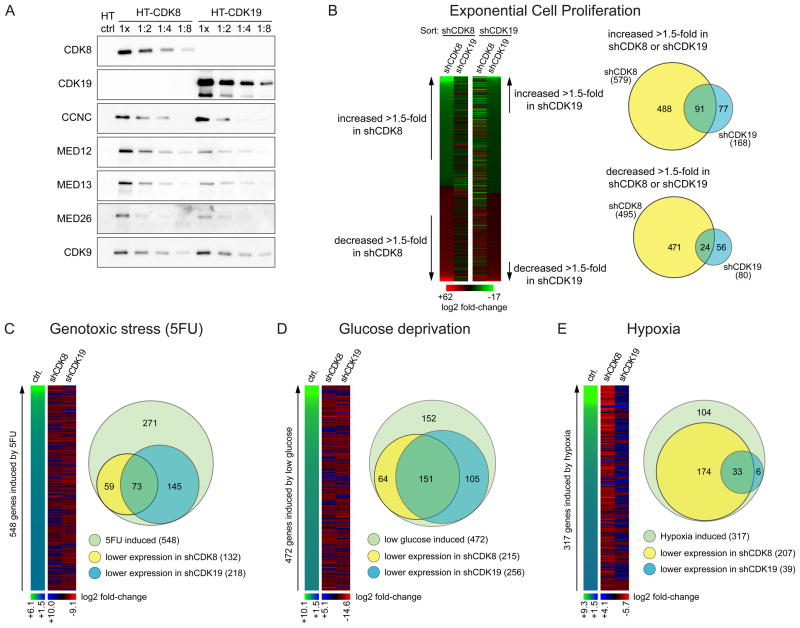
CDK8 and CDK19 are paralogous kinases previously found to associate with the Mediator complex (Figure S1A available online)(Sato et al., 2004). Using Q-RT-PCR, we observed wide co-expression of the two kinases across normal tissues, with a few sites showing higher expression for CDK8 (e.g. bladder, esophagus) or CDK19 (e.g. brain) (Figure S1B). Co-expression is also evident at the protein level in cell lines of diverse origins (Figure S1C). To investigate the possible functional specialization of CDK8 in colorectal cancer cells, we employed the well-characterized HCT116 cell line, which expresses significant amounts of both proteins (Figure S1D). Affinity complex purification of HaloTag versions of CDK8 and CDK19 revealed that the two kinases exist in mutually exclusive variants of Mediator, as CDK19 is undetectable in CDK8 complexes or vice versa (Figure 1A). Importantly, both kinases assemble with the rest of the CDK-module and core Mediator carrying MED26, a subunit shown to interact with the Super Elongation Complex (SEC) containing CDK9 (Takahashi et al., 2011), which is also detected in both complexes.
Figure 1. CDK8 and CDK19 regulate distinct gene expression programs.
(A) Western blots showing mutually exclusive interaction of CDK8 and CDK19 with the CDK-module of Mediator. CDK-module subunits (CDK8, CDK19, Cyclin C, MED12, MED13), MED26 and CDK9 were detected in pulldowns from protein extracts of HCT116 cells expressing HaloTag (HT) alone, HT-CDK8 or HT-CDK19. Increasing twofold dilutions are shown to indicate relative abundance of interacting subunits. (B) Microarray analysis showing the effects of CDK8 (shCDK8) or CDK19 (shCDK19) knockdown on the gene expression profiles of HCT116 cells under conditions leading to exponential cell proliferation, sorted by effect of shCDK8 or shCDK19 (>1.5 fold change from shControl, p<0.05). (C–E) Microarray analysis showing the effects of CDK8 or CDK19 knockdown on genes induced by (C) genotoxic stress upon 5-fluorouracil treatment (5FU, 12 h), (D) by glucose deprivation (24 h) or (E) by hypoxia (1% O2, 24 h). In all cases induction is defined as >1.5 fold, p<0.05. Heatmaps are color-coded by log2 fold-change, treated vs. untreated shControl cells for induction, shCDK8 or shCDK19 treated vs. shControl treated for knockdown effect. Venn diagrams compare the numbers of genes induced by each stimuli to those decreased by shCDK8 and shCDK19 under each condition (≥1.1 fold change, p<0.05). See also Figure S1 and Table S1.
With the goal of identifying gene expression networks controlled preferentially by CDK8, we performed a series of microarray analyses in HCT116 colorectal cancer cells where either kinase was stably knocked down (Figure S1D, Figure 1B-E, Table S1). First, we analyzed the effects on global gene expression under conditions that lead to exponential cell proliferation (Figure 1B). Importantly, the gene sets affected by each kinase are largely non-overlapping, revealing for the first time a specialization for each kinase in gene expression control. Of note, a greater number of genes are significantly induced or repressed by CDK8 knockdown than by CDK19 knockdown, indicating that CDK8 is a more widespread regulator of gene expression under these conditions in HCT116 cells. Next, we analyzed the impact of each kinase on stimulus-specific gene expression programs. Toward this end, we subjected HCT116 cells to three different stress stimuli: 5-fluorouracil (5FU), which provokes a genotoxic stress response involving p53 and other transcription factors; glucose deprivation, which affects gene expression programs downstream of AMPK and mTOR pathways; and hypoxia, which activates adaptive pathways, including those regulated by HIF1A and HIF2A (Figure 1C–E). In contrast to what is evident during exponential cell proliferation, CDK19 knockdown has a more pronounced effect than CDK8 depletion on the gene expression programs elicited by genotoxic stress and glucose deprivation. This indicates that the minor effects of CDK19 knockdown seen under standard culture conditions are not due to poor knockdown, but rather to a functional specialization of this kinase. Most strikingly, many more hypoxia-inducible genes show a requirement for CDK8 than for CDK19. Of 317 genes significantly induced by hypoxia, 207 (65%) showed reduced expression in CDK8-depleted cells, while only 39 (13%) showed reduced expression upon CDK19 knockdown.
These data demonstrate that the Mediator-associated kinases CDK8 and CDK19 have specialized to regulate different gene expression programs, with a predominant role for CDK8 in positive control of hypoxia-inducible genes.
CDK8 is widely required for activation of HIF1A target genes
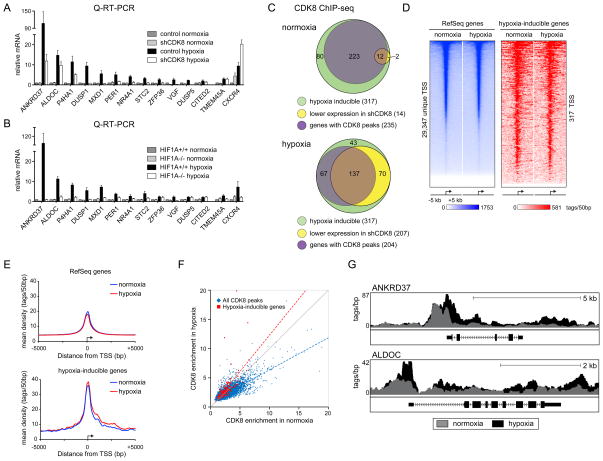
Since CDK8 knockdown leads to reduced expression of hypoxia-inducible genes, we hypothesized that CDK8 functions as a novel coactivator for HIF1A. To test this we used Q-RT-PCR to monitor the impact of CDK8 knockdown or HIF1A knockout on the expression of hypoxia-inducible genes that were affected by CDK8 knockdown according to our microarray analysis. Expectedly, Q-RT-PCR confirmed the requirement for CDK8 for full induction of all ten genes selected (ANKRD37 through CITED2, Figure 2A). For contrast purposes, we included TMEM45A and CXCR4, two genes that do not require CDK8. Importantly, HIF1A is also required for induction of most of this set of genes, with DUSP5 and CITED2 being the only exceptions (Figure 2B). Thus, CDK8 can co-regulate HIF1A target genes in HCT116 cells. Knockdown of CDK8 has no effect on the stabilization of HIF1A or HIF2A upon hypoxia (Figure S2A), suggesting that CDK8 acts at downstream steps in the hypoxic response. Furthermore, CDK8 depletion has minimal impact on the basal expression of most of these genes during normoxia (Figure S2B). CDK8 knockdown using a different shRNA also impaired induction of HIF1A target genes (Figures S2C–D).
Figure 2. CDK8 is a coactivator of many hypoxia-inducible genes.
(A) Relative expression of hypoxia-inducible genes as measured by Q-RT-PCR for control and CDK8 knockdown HCT116 cells in normoxia or after 24 h hypoxia (1% O2). Expression values were normalized to 18S rRNA and are expressed relative to the control normoxia value. Error bars represent SEM from three independent biological replicates. (B) Relative expression of hypoxia-inducible genes as measured by Q-RT-PCR for HIF1A+/+ and HIF1A−/− HCT116 cells in normoxia or after 24 h hypoxia (1% O2). (C) Venn diagrams displaying the proportion of hypoxia-inducible genes with CDK8 peaks and those negatively affected by CDK8 knockdown in normoxia and hypoxia. (D) CDK8 binding profiles around transcription start sites in normoxia and hypoxia as determined by ChIP-seq. Shown are 10 kb regions centered on all unique RefSeq TSS (left panel) and the TSS of hypoxia-inducible genes (right panel). Heatmaps are ranked by the CDK8 normoxia signal and the color scale represents tags per 50bp. (E) Metagenes centered on TSS showing CDK8 binding profiles for RefSeq genes (top) and hypoxia-inducible genes (bottom) in normoxia (blue) and hypoxia (red). (F) Dot plot comparing CDK8 enrichment in normoxia (x-axis) and hypoxia (y-axis) at all CDK8 peak regions (blue) and at peaks associated with hypoxia-inducible genes (red). The grey line represents a 1:1 relationship (no change) between normoxia and hypoxia. (G) Genome browser views of CDK8 binding at HIF1A target loci under both normoxic (grey) and hypoxic conditions (black). See also Figure S2 and Table S2.
To test if CDK8 could be acting directly at HIF1A target genes on a global scale, we performed CDK8 ChIP-seq experiments in HCT116 cells under normoxic and hypoxic conditions. This first global analysis of CDK8 chromatin binding revealed that ~43% of CDK8 peaks are found within the region spanning 3kb upstream of transcription start sites (TSSs) through to 5′ UTRs, whereas another ~50% are found in more distal intergenic and intragenic regions (Figure S2E). During normoxia, although 74% (235/317) of hypoxia-inducible genes are associated with significant CDK8 peaks, only 5% (12/235) of these display reduced basal expression in shCDK8 cells, suggesting that CDK8 is important mostly for their induced expression. Upon hypoxia, 64% (204/317) of hypoxia-inducible genes were associated with significant CDK8 peaks, and the expression of roughly 70% (137/204) of these genes is negatively affected by CDK8 knockdown (Figure 2C and Table S2).
When analyzed in a 10kb window around TSSs, a gradient of CDK8 occupancy becomes apparent both at all RefSeq genes and at hypoxia-inducible genes, with maximum signals centered over TSSs (Figure 2D–E). Further analysis reveals that hypoxia-inducible genes have a significantly higher level of CDK8 binding at their TSSs and also display more spreading of CDK8 into the gene body, as compared to the global average (Figure 2E and Figure S2F). Interestingly, hypoxia leads to a general decrease of CDK8 binding throughout the genome, concurrently with equal or increased CDK8 binding at peaks associated with hypoxia-inducible genes (Figure 2F). This is also apparent when looking at TSSs and 3′ ends of genes (Figure S2G–H). Thus, although hypoxia does not affect overall CDK8 protein expression (Figure S2A and S2C), it does produce a global change toward binding at the genes upregulated in this scenario. Representative examples of this pattern of CDK8 binding are shown for the HIF1A target genes ANKRD37 and ALDOC in Figure 2G.
CDK8 regulates RNAPII elongation without affecting HIF1A binding or histone acetylation
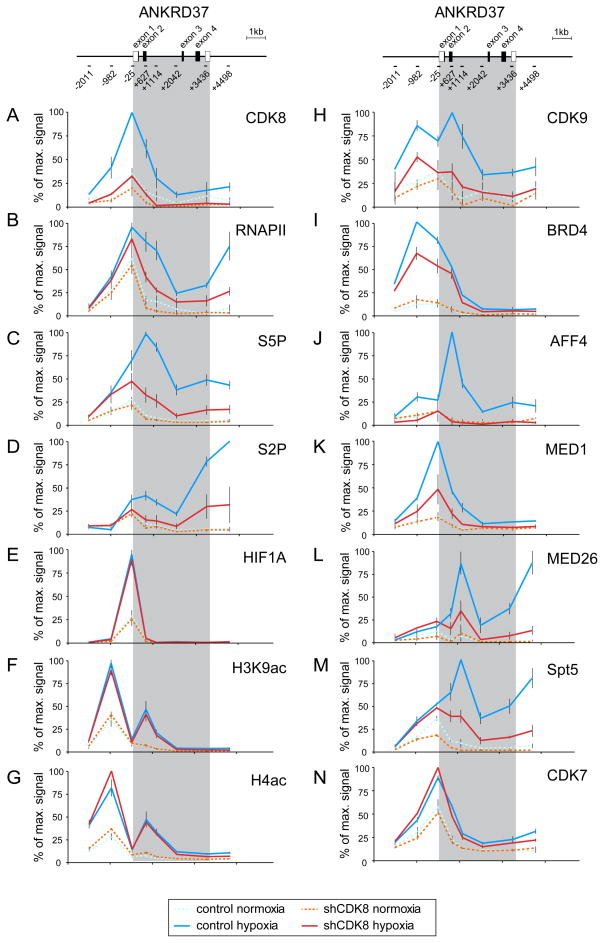
To investigate the mechanism by which CDK8 contributes to expression of HIF1A target genes, we performed a detailed quantitative ChIP analysis of the ANKRD37 and STC2 gene loci (Figures 3 and S3, respectively). Both genes undergo robust HIF1A- and CDK8-dependent induction upon hypoxia, but are not affected by CDK19 depletion (Figure S3A). As predicted from our ChIP-seq analysis, CDK8 is recruited to the ANKRD37 locus upon hypoxia, with peak signals around the TSS (Figure 3A). Expectedly, CDK8 ChIP signals are much reduced in shCDK8 cells. Interestingly, total RNAPII shows significant enrichment at the TSS under normoxia, when expression of ANKRD37 mRNA is 50–100 fold lower (Figure 3B). Upon hypoxia, a modest increase (<2 fold) in RNAPII is observed at the promoter, but a much more significant increase (up to 20 fold) in RNAPII occupancy is observed at the intragenic and 3′ regions. This RNAPII behavior is indicative of transcriptional pausing and is observed at many genes induced in response to specific stimuli (Boehm et al., 2003; Core and Lis, 2008; Rahl et al., 2010). Conversion of paused RNAPII into an elongation competent form involves the action of diverse elongation factors as well as various phosphorylation events, including phosphorylation of the C-terminal domain (CTD) heptad repeats of the RPB1 subunit of RNAPII. ChIP analysis of Ser5- and Ser2-phospho-CTD (S5P, S2P) revealed a significant increase in both phosphorylation events upon hypoxia (Figure 3C–D). In agreement with the typical patterns of these two phosphorylation marks, S5P peaks toward the 5′ end, whereas S2P is higher within the gene body and at the 3′ end. Importantly, CDK8 depletion leads to a substantial decrease in both total and phospho-RNAPII within the gene body of ANKRD37 (Figure 3B–D). Similar results were observed at the STC2 locus (Figure S3B). Taken together, this indicates that CDK8 is a positive regulator of RNAPII activity at these HIF1A target genes, likely acting to promote RNAPII elongation.
Figure 3. CDK8 regulates RNAPII elongation without affecting HIF1A binding or histone acetylation.
Quantitative ChIP analysis of (A) CDK8, (B) total RNAPII, (C) serine-5 and (D) serine-2-phosphorylated RNAPII CTD (S5P, S2P), (E) HIF1A, (F) H3K9ac and (G) H4ac histone acetylation, (H) the CDK9 subunit of P-TEFb, (I) the bromodomain protein BRD4, (J) the SEC subunit AFF4, the core mediator subunits (K) MED1 and (L) MED26, (M) the DSIF subunit Spt5 and (N) the TFIIH subunit CDK7 at the ANKRD37 locus in control and CDK8 knockdown HCT116 cells in normoxia or after 24 h hypoxia (1% O2). Values are plotted as a percentage of the maximum signal for that locus. Error bars represent SEM from three independent biological replicates. Grey shading indicates the transcribed region. See also Figure S3.
Upon hypoxia, HIF1A hydroxylation is impaired, which leads to increased HIF1A levels and enhanced association of the HIF1A C-TAD with p300/CBP (Kallio et al., 1998). Expectedly, ChIP analysis revealed a strong increase in HIF1A occupancy and acetylation of histones H3 and H4 upon hypoxia at the ANKRD37 and STC2 loci (Figure 3E–G, and S3B). Importantly, CDK8 depletion does not affect HIF1A chromatin binding or histone acetylation. In fact, CDK8 knockdown leads to a modest increase in HIF1A binding and histone H4 acetylation at the STC2 locus (Figure S3B). This demonstrates that HIF1A binding and the subsequent increase in histone acetylation are not sufficient to fully activate RNAPII, thus revealing the existence of an additional, previously unappreciated coactivation event involving CDK8 at these genes.
CDK8 is required for recruitment of the Super Elongation Complex
Given the fact that CDK8 depletion impairs RNAPII phosphorylation and elongation at HIF1A target loci, we next investigated its role in recruitment of P-TEFb (positive transcription elongation factor b), a critical regulator of RNAPII elongation. CDK9, the catalytic subunit of P-TEFb, overcomes RNAPII pausing in a kinase-dependent manner likely by phosphorylating various targets such as the negative elongation factor NELF, as well as RNAPII itself (Peterlin and Price, 2006). CDK9 recruitment to the ANKRD37 and STC2 loci is strongly stimulated by hypoxia but, importantly, is much reduced in CDK8 knockdown cells (Figure 3H and S3B). Numerous mechanisms have been described to explain P-TEFb binding to promoters during gene activation including recruitment by DNA-binding transactivators such as MYC (Rahl et al., 2010), recruitment by BRD4, a bromodomain protein that recognizes acetylated histones (Jang et al., 2005; Yang et al., 2005), and also by association with Mediator (Donner et al., 2010; Takahashi et al., 2011). Importantly, a fraction of P-TEFb exists within the so-called Super Elongation Complex (SEC) containing the scaffold protein AFF4, which interacts directly with MED26 (Lin et al., 2010; Takahashi et al., 2011). To investigate which of these P-TEFb recruitment mechanisms could be operating at HIF1A target genes, we performed ChIP analysis of BRD4, AFF4 and the core Mediator subunits MED1 and MED26 (Figure 3I–L). Hypoxia induces strong recruitment of BRD4, AFF4, MED1 and MED26 to ANKRD37, while CDK8 depletion modestly impairs BRD4 binding but completely abolishes AFF4 recruitment (Figure 3I,J). Interestingly, recruitment of the Mediator subunits MED1 and MED26 is also significantly reduced upon CDK8 knockdown, suggesting that the kinase subunit is required for full association of core Mediator (Figure 3K,L). Of note, CDK8 depletion also reduced the intragenic levels of the Spt5 subunit of DSIF, a factor that promotes RNAPII elongation after its phosphorylation by P-TEFb (Peterlin and Price, 2006) (Figure 3M). For contrast purposes, we show that induced recruitment of CDK7, the kinase subunit of the general transcription factor TFIIH, is not affected by CDK8 knockdown, indicating that preinitiation complex (PIC) formation is not impaired (Figure 3N). Overall, the data indicate that CDK8 co-activates HIF1A targets by mediating recruitment of P-TEFb and the SEC, perhaps via MED26, downstream or independently of histone acetylation and BRD4.
We next tested the effect of CDK8 knockdown on induction of the key HIF1A targets ANKRD37 and ALDOC in cancer cell lines of different origin and found a similar requirement for CDK8 (Figure S3C). We also carried out ChIP analysis of the ANKRD37 locus in U2OS osteosarcoma cells and found that, as for HCT116 cells, CDK8 knockdown impaired RNAPII elongation and CDK9 recruitment without affecting HIF1A binding or histone acetylation (Figure S3D-E).
HIF1A induces recruitment of CDK8-Mediator and P-TEFb to chromatin
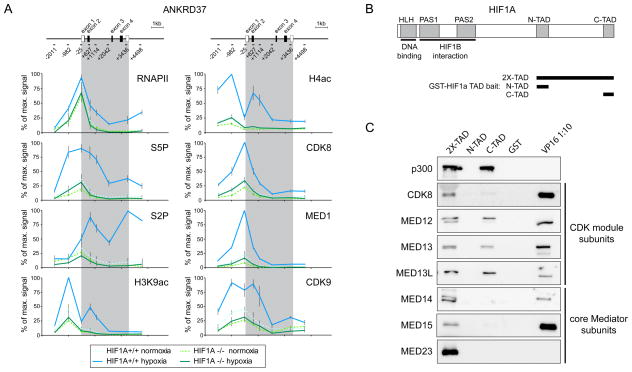
The data shown so far indicate that CDK8 is required at steps subsequent to HIF1A binding and histone acetylation and led us to hypothesize that HIF1A might recruit CDK8-Mediator to facilitate RNAPII elongation. To test this, we performed ChIP assays in HCT116 HIF1A+/+ versus HIF1A −/− cells (Figure 4A). Strikingly, large amounts of RNAPII are detected at the ANKRD37 promoter in HIF1A −/− cells, indicating that HIF1A is largely dispensable for RNAPII recruitment (Figure 4A). Importantly, hypoxia induces increased RNAPII occupancy in the gene body and 3′ end only in HIF1A+/+ cells. Thus, conversion of promoter-proximal into elongating RNAPII upon hypoxia requires HIF1A action. Accordingly, ChIP analysis of S5P and S2P shows that hypoxia-induced RNAPII phosphorylation at these key residues is dependent on HIF1A. As predicted from previous work, hypoxia-induced histone acetylation is strongest in HIF1A +/+ cells. In agreement with our hypothesis, HIF1A is also required for full recruitment of CDK8, the core Mediator subunit MED1, and CDK9 upon hypoxia.
Figure 4. HIF1A is required for recruitment of CDK8, Mediator and P-TEFb to chromatin.
(A) Quantitative ChIP analysis of total RNAPII, serine-5 and serine-2-phosphorylated RNAPII CTD (S5P, S2P), histone acetylation (H3K9ac and H4ac), CDK8, MED1 and CDK9 at the ANKRD37 locus in HIF1A+/+ and HIF1A−/− cells. Values are plotted as a percentage of the maximum signal. Error bars represent SEM from three independent biological replicates. Grey shading indicates the transcribed region. (B) Schematic of HIF1A protein domains cloned as GST-fusions for use as affinity bait proteins. (C) Western blot analysis of proteins purified by their affinity for GST-HIF1A or GST-VP16 transactivation domains. GST-alone served as a negative control. VP16-interacting proteins were diluted ten-fold lower to avoid overloading. See also Figure S4.
We next investigated if the transactivation domains (TADs) of HIF1A could interact with CDK8-Mediator. Toward this end, we used GST-fusion bait proteins corresponding to the N-TAD, the C-TAD or a larger region including both TADs and the intervening region (2X-TAD, Figure 4B) to capture interacting factors from HeLa cell nuclear extracts (Figure 4C). We employed a GST-VP16 transactivation domain and GST-only baits as positive and negative controls, respectively. Consistent with previous studies, the 2X-TAD and C-TAD baits interact with p300 (Kallio et al., 1998). Importantly, the 2X-TAD bait also effectively pulls down CDK8, additional CDK-module subunits (MED12, MED13 and MED13L) and core Mediator subunits (MED14, MED15 and MED23). None of these interactions were detected with the N-TAD and GST-only baits; however, MED12, MED13 and MED13L were detected using the C-TAD suggesting that a region comprising the C-TAD and the intervening region is required for optimal interaction with CDK8-Mediator. Overall, we conclude that the transactivation domain of HIF1A can associate with CDK8-Mediator, thus providing a physical basis for the observed HIF1A-dependent recruitment of this coactivator to hypoxia-inducible target gene loci.
Our gene expression analysis identified HIF1A targets that do not require CDK8 (e.g. CXCR4, Figure 2A and S4A), as well as non-hypoxia inducible genes that do require CDK8 (e.g. MAGI1, Figure S4A). ChIP analyses of these genes show that while CDK8 depletion does not significantly impair RNAPII activation or CDK9 recruitment at CXCR4, it does reduce RNAPII phosphorylation and CDK9 binding at MAGI1 (Figure S4B). Expectedly, analysis of UHMK1 whose expression is not induced by hypoxia or reduced by CDK8 knockdown, showed no impact of CDK8 depletion on RNAPII activity (Figure S4A–B). Of note, CXCR4 and UHMK1 carry very low levels of CDK8, CDK9 and RNAPII (see scales in Figure S4B).
Hypoxia-inducible genes are active and paused prior to HIF1A activation
Taken together, our ChIP data so far strongly suggest a requirement for CDK8 in stimulating transcription elongation at hypoxia-inducible HIF1A target genes. Our data are consistent with a number of genome-wide ChIP studies in which both active and inactive genes have high levels of RNAPII at their 5′ ends (Guenther et al., 2007; Rahl et al., 2010). However, ChIP analysis does not distinguish between a build-up of inactive promoter-bound RNAPII versus transcriptionally competent but paused RNAPII. In order to better define which step is rate limiting for induction of these genes, and that might be regulated by CDK8, we employed global nuclear run-on with deep sequencing (GRO-seq) to examine the genome-wide location of transcriptionally engaged RNA polymerases in HCT116 cells under normoxic conditions (Core et al., 2008).
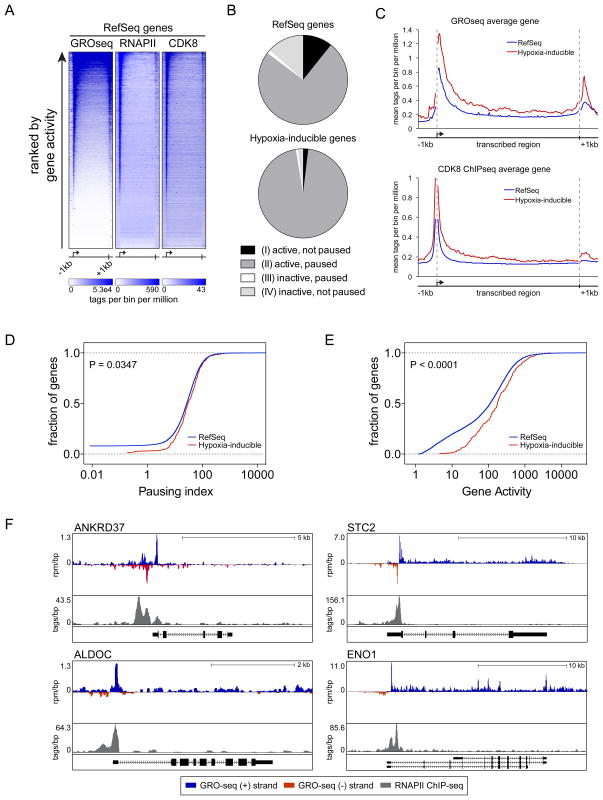
As reported previously for other cell types, we observed an accumulation of transcriptionally engaged RNA polymerases at the 5′ ends of a large fraction of RefSeq genes that corresponds with the location of RNAPII as defined by ChIP-seq (Figure 5A, (Core et al., 2008; Min et al., 2011; Rahl et al., 2010). Notably, when ranked by gene activity as measured by GRO-seq read density in gene bodies, it is clear that those genes with the highest levels of promoter-proximal active RNAPII also have substantial CDK8 enrichment at their 5′ end and intragenic regions. We next classified genes into four classes according to the GRO-seq read density in both the promoter proximal region and the gene body, as in Core et al. (2008): i) active, not paused; ii) active, paused, iii) inactive, paused; and iv) inactive, not paused. We then asked how hypoxia-inducible genes fit within these classes, which revealed that essentially all of them are active and paused during normoxia (96%, Figure 5B and Table S3). Hypergeometric tests show that there is a greater proportion of hypoxia-inducible genes that are active (p < 2.3 × 10−11), paused (p < 2.2 × 10−16) and paused, active (p < 2.2 × 10−16) compared to RefSeq genes. It is also apparent that on average hypoxia-inducible genes have higher densities of transcriptionally active RNAPII and CDK8 both at their promoter regions and within gene bodies than the RefSeq average, even before HIF1A is activated (Figure 5C). Furthermore, an increased fraction of active hypoxia-inducible genes display higher pausing indices and higher gene activity relative to all active RefSeq genes (Figure 5D–E). Examples of HIF1A targets that are both active and paused prior to their induction by hypoxia are shown in Figure 5F. Overall, these data indicate that pause-release is a rate-limiting step for induction of HIF1A targets.
Figure 5. Hypoxia-inducible genes are active and paused in normoxia.
(A) Gene profiles showing levels of transcriptionally engaged RNA polymerases (GRO-seq, sense strand) and RNAPII occupancy (ChIP-seq) in comparison to CDK8 occupancy (ChIP-seq) in normoxia for all RefSeq genes. Heatmap color scales represent the read density for 160 bins across the transcribed region of each gene with 1 kb upstream and downstream flanking regions (20 bins of 50 bp each). Heatmaps are ranked by gene activity as determined from GRO-seq. (B) Relative proportions of genes in each transcription class for RefSeq genes (top) and hypoxia-inducible genes (bottom). (C) Metagenes showing average GRO-seq (sense strand) and CDK8 ChIP-seq signals across RefSeq genes (blue) in comparison to the average signals across hypoxia-inducible genes (red). Units are mean tags per bin for 160 bins across the transcribed region of each gene with 1 kb upstream and downstream flanking regions (20 bins of 50 bp each) (D) Cumulative distribution plots of pausing index for active RefSeq genes (blue) and active hypoxia-inducible genes (red). Distributions are significantly different (Kolmogorov-Smirnov test, P = 0.0347). (E) Cumulative distribution plots of gene activity for active RefSeq genes (blue) and active hypoxia-inducible genes (red). Distributions are significantly different (Kolmogorov-Smirnov test, P < 0.0001). (F) Genome browser views showing GRO-seq reads in the sense- (blue) and anti-sense (red) direction relative to each gene, and RNAPII ChIP-seq signal (grey) at HIF1A target loci under normoxic conditions. See also Table S3.
DISCUSSION
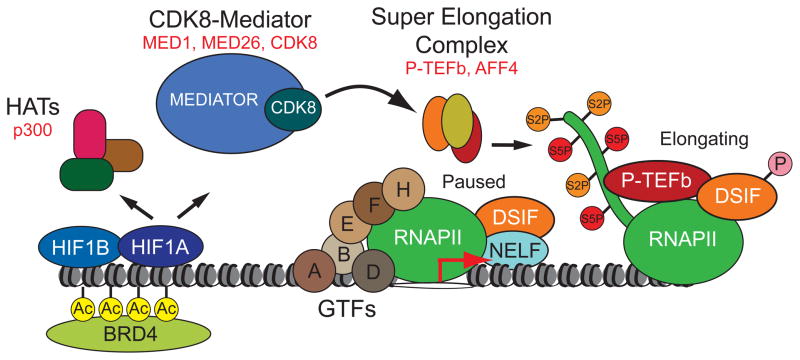
The cellular response to hypoxia is critical for normal physiology and underlies many disease processes. In particular, induction of HIF transcription factors due to intratumoral hypoxia promotes tumorigenesis across several cancer types, likely by upregulation of angiogenic factors, glucose transporters and glycolytic enzymes, as well as pro-survival and invasion factors. Accordingly, elevated HIF1A expression is associated with increased mortality in diverse cancers including those of the cervix, endometrium, breast, ovary and colon (Semenza, 2012). Although hundreds of genes across different cell types have been identified as direct targets of HIF1A and HIF2A, our knowledge of the mechanisms by which these factors regulate transcription remains incomplete. Our research described here has uncovered a novel HIF1A coactivation mechanism involving the Mediator subunit CDK8, an oncogene in colorectal cancer (Firestein et al., 2008). These findings constitute a significant advance in our understanding of how HIF1A regulates transcription, but also contribute to our view of Mediator in stimulus-specific control of gene expression and provide a link between HIF1A and CDK8, two potent oncogenes. Our mechanistic studies lead to a new model for HIF1A-dependent transactivation as depicted in Figure 6.
Figure 6. A new model of HIF1A transactivation.
See Discussion for details.
Prior to HIF1A activation, its target loci carry transcriptionally engaged, proximally paused RNAPII, indicating that HIF1A transactivation involves enhanced release from this paused state. In fact, our global studies of RNAPII behavior demonstrate that nearly all hypoxia-inducible genes are transcribed and paused in normoxic conditions. Upon hypoxia, HIF1A binds to chromatin and enhances histone acetylation and BRD4 recruitment. However, these events alone are not sufficient to stimulate RNAPII elongation. In addition, HIF1A recruits a variant of Mediator containing CDK8, MED1 and MED26. Of note, MED1 and MED26 recruitment is impaired upon CDK8 knockdown. More importantly, CDK8 is required for HIF1A-dependent recruitment of AFF4, the scaffold subunit of the SEC, and of CDK9, the catalytic subunit of P-TEFb. This could be due to the observed defect in MED26 binding, as MED26 has been shown to interact directly with AFF4 (Takahashi et al., 2011). Thus, release of paused RNAPII at hypoxia-inducible genes is driven by a HIF1A>CDK8-Mediator>P-TEFb axis.
While first reported in Drosophila, it has become apparent that RNAPII pausing is a conserved feature across many metazoan transcriptional programs (Core and Lis, 2008; Min et al., 2011; Muse et al., 2007; Rahl et al., 2010; Zeitlinger et al., 2007). Although our understanding of RNAPII pausing is still evolving, recent studies lead to a model where pausing is employed to create transcriptional competence at gene loci activated in response to developmental cues, cellular stress and various other signaling events. In this view, RNAPII pausing is not a repressive event, but rather a mechanism to protect stimulus-responsive promoters from the negative action of nucleosomes (Gilchrist et al., 2010). By preventing promoter occlusion, paused RNAPII ensures a basal level of transcription and readies gene loci for dynamic and synchronous responses to stimuli (Boettiger and Levine, 2009; Gilchrist et al., 2010). At these loci, the rate-limiting step is the recruitment of elongation factors, either directly or indirectly, by stimulus-activated transcription factors. Thus, RNAPII pausing would confer the hypoxia response network with a mechanism to induce gene expression in a robust and timely manner. Interestingly, previous studies have shown that activated HIF1A binds preferentially to ‘permissive’ loci, and it has been suggested that this may determine the observed cell-type specific expression of HIF1A target genes (Xia and Kung, 2009). Under normoxia, these ‘permissive’ loci display pre-loaded RNAPII, histone methylation marks associated with active genes and DNaseI hypersensitivity. Our data support and extend these findings by showing that RNAPII is transcriptionally engaged at virtually all HIF1A target loci in HCT116 cells prior to hypoxia and that HIF1A is ultimately required for recruitment of P-TEFb, whose activity is needed to stimulate RNAPII elongation. Furthermore, the observation that HIF1A-induced histone acetylation is not sufficient for full RNAPII activation reinforces the importance of elongation as a rate-limiting step at HIF1A targets. In fact, earlier work showed that the HIF1A-binding domain of p300/CBP is dispensable for transactivation of the majority of HIF1A targets tested, pointing to the existence of other primary coactivation events (Kasper et al., 2005). Our results predict that it might be possible to impair HIF1A function by targeting the biochemical steps leading to P-TEFb recruitment. Thus, a more detailed characterization of this pathway, which involves CDK8-Mediator, could reveal therapeutic strategies to attenuate HIF1A-driven pathological processes.
Since its early discovery as a key regulator of RNAPII activity in yeast (Koleske et al., 1992; Thompson et al., 1993), Mediator has proven to be a key player in a myriad of biological processes in metazoans, including stem cell pluripotency, hormone signaling, growth factor signaling, and lipid metabolism (Malik and Roeder, 2010). However, the precise molecular mechanism of action of Mediator remains elusive. Increasingly, Mediator has been involved in transcription elongation control. Early studies of Mediator in the serum response network revealed that ablation of the MED23 subunit abolishes serum-induced gene activation by a mechanism that could not be explained solely by effects on RNAPII recruitment (Wang et al., 2005). We later showed that CDK8 knockdown impairs elongation at serum response genes (Donner et al., 2010). More recently, the MED26 subunit of Mediator was found to interact directly with the SEC (Takahashi et al., 2011). Interestingly, biochemical purification experiments suggested that CDK8-containing Mediator is largely devoid of MED26 and vice-versa; however, this subunit segregation is not absolute and a fraction of Mediator contains both proteins (Figure 1A and Sato et al. (2004)).
Our finding that the HIF1A C-TAD interacts with CDK8-Mediator explains previous studies demonstrating that it has other transactivation functions beyond association with p300/CBP (Kasper et al., 2005). It will be interesting to further characterize the interaction between the HIF1A C-TAD and Mediator, and to define whether the interaction is sensitive to any of the post-translational modifications known to affect the transactivation potential of HIF1A, such as hydroxylation or phosphorylation.
In vertebrates, subunits of the CDK-module of Mediator have undergone gene duplication events to generate the paralog pairs CDK8/CDK19, MED12/MED12L and MED13/MED13L. These gene duplications are likely to create additional regulatory diversity in vertebrate cell types (Levine and Tjian, 2003), and there is evidence that these paralogs are not redundant. CDK8 knockout mice are embryonic lethal (Westerling et al., 2007), and germline mutations in CDK19 have been linked to a developmental syndrome (Mukhopadhyay et al., 2010). Our results reveal a functional specialization of CDK8 and CDK19 in gene expression control in colon cancer cells, with the hypoxic response being a striking example of their non-redundancy. Future studies will be aimed at defining the molecular basis of this specialization. While we have demonstrated here a differential requirement for the two kinases at hypoxia-inducible genes, it will be important to determine if this specialization arises from differential recruitment or by acting on different phosphorylation targets. Note that the two kinases differ mostly in their C-termini, which could differentially regulate their activity or recruitment to target gene loci.
Since CDK8, but not CDK19, is commonly amplified in colorectal cancers (Firestein et al., 2008), it is tempting to speculate that CDK8 oncogenicity is due to its effects on gene expression programs not regulated by CDK19. To what extent can the oncogenic effects of CDK8 overexpression be attributed to its role as a coactivator of hypoxia-inducible genes? Answering this question will require a careful genetic dissection of the hypoxic response versus the various oncogenic pathways that CDK8 has already been implicated in. CDK8 was first characterized as an oncogene during an unbiased screen for novel mediators of β-catenin-dependent transcription, and CDK8 is also a positive regulator of genes downstream of growth factor signaling (Donner et al., 2010; Firestein et al., 2008). More recently, CDK8 was found to promote the dedifferentiated state of both colon cancer cells and embryonic stem cells, acting partly via upregulation of MYC (Adler et al., 2012). Thus, overexpression of CDK8 may drive tumor survival and progression by affecting multiple interconnected pathways, and our work suggests that this may be in part by affecting the HIF1A transcriptional program.
EXPERIMENTAL PROCEDURES
Cell culture
Cells were cultured in McCoy’s 5A, DMEM, RPMI (Sigma) or DMEM/F12 (Life Technologies) supplemented with 10% FBS (Sigma) and antibiotic/antimycotic (Life Technologies) under 5% CO2 at 37°C. Cells were plated 24 h prior to experimental treatments and harvested in RIPA lysis buffer for protein or, either Tri-Reagent (Sigma) or RLT buffer (Qiagen), for total RNA. Hypoxia treatments were carried out in incubation chambers (Billups-Rothenberg) by flushing twice with 120 L of a mixture of 1% O2/5% CO2/94% N2 (Airgas) and incubated for 24 h at 37°C. 5-fluorouracil (Sigma) was used at 375 μM. Glucose deprivation was carried out by dropping glucose concentration from 4.5 g/L (high) to 1 g/L (low).
HaloTag Pull-Down Assay
Refer to Extended Experimental Procedures for a detailed protocol. Briefly, clarified lysates from HCT116 cells transfected with expression plasmids for HaloTag-CDK8 or -CDK19 were incubated with HaloLink Resin (Promega) for 15 min at 22°C, followed by extensive washing. Interacting proteins were eluted with SDS elution buffer and analyzed by Western blotting.
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was carried out as previously described (Donner et al., 2010; Donner et al., 2007). Antibodies and a detailed protocol are described in Extended Experimental Procedures. For ChIP-seq analysis, salmon-sperm DNA was omitted from the procedure and an Illumina sequencing library was prepared from the immunoprecipitated DNA as described in Extended Experimental Procedures. For quantitative ChIP analysis of the ANKRD37 and STC2 loci, real-time PCR was carried out on ChIP-enriched DNA against a standard curve of input DNA, with amplicons tiling across each locus. Enrichment values for each amplicon were calculated as percentage of the amplicon with maximum signal for each antibody at each locus. Primers are described in Extended Experimental Procedures.
HIF1A-TAD interaction assay
Briefly, bait proteins were prepared by expressing GST-HIF1A-TAD and -VP16 fusion proteins, or GST alone, in E. coli strain BL21, and bound to reduced glutathione sepharose resin, followed by high- and low-salt washes. Affinity resins containing each bound GST-bait protein were then incubated with HeLa nuclear extracts for 3 hr at 4°C. The beads were then washed extensively with buffer containing 0.5 M KCl, and finally purified proteins eluted with 30 mM reduced glutathione for analysis by Western blotting. Refer to Extended Experimental Procedures for a detailed protocol.
Microarray, ChIP-seq and GRO-seq analysis
See Supplemental Information for Extended Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
CDK8 is a positive regulator of hypoxia-inducible genes
HIF1A associates with and induces binding of CDK8-Mediator to its target loci
CDK8 is required for HIF1A-dependent recruitment of transcription elongation factors
Hypoxia-inducible genes are paused and active prior to HIF1A activation
Acknowledgments
We thank Dr. Shilatifard for AFF4 antibody, Dr. Taatjes and Dr. Lin for HeLa nuclear extracts and MED15 antibody, and Edward Head for helpful discussions. This work was supported primarily by NSF grant 0842974, and also by NIH RO1CA117907 grant (J.M.E.), a Butcher Seed Grant (J.M.E, R.D.D), the Boettcher Foundation’s Webb-Waring Biomedical Research program (R.D.D), DoD Breast Cancer Postdoctoral Fellowship W81XWH-10-1-0062 (X.W.), and NIH RO1CA140545 (W.C.H.). J.M.E. is an HHMI Early Career Scientist.
Footnotes
SUPPLEMENTAL INFORMATION. Supplemental information includes Extended Experimental Procedures, four figures, and three tables. Microarray, CDK8 ChIP-seq, and GRO-seq data are available under GEO accession numbers GSE38061, GSE38140, and GSE38258.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adler AS, McCleland ML, Truong T, Lau S, Modrusan Z, Soukup TM, Roose-Girma M, Blackwood EM, Firestein R. CDK8 maintains tumor de-differentiation and embryonic stem cell pluripotency. Cancer Res. 2012;72:2129–2139. doi: 10.1158/0008-5472.CAN-11-3886. [DOI] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettiger AN, Levine M. Synchronous and stochastic patterns of gene activation in the Drosophila embryo. Science. 2009;325:471–473. doi: 10.1126/science.1173976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon HM. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. Function and regulation of the Mediator complex. Curr Opin Genet Dev. 2011;21:225–230. doi: 10.1016/j.gde.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol. 2010;17:194–201. doi: 10.1038/nsmb.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 Is a Stimulus-Specific Positive Coregulator of p53 Target Genes. Mol Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, Freed E, Ligon AH, Vena N, Ogino S, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455:547–551. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith MD, Donner AJ, Espinosa JM. CDK8: A positive regulator of transcription. Transcription. 2010;1:4–12. doi: 10.4161/trns.1.1.12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist DA, Dos Santos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG. HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, von Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau Ubiquitylation Complex by O2-Regulated Prolyl Hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell. 2005;19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- Kallio PJ, Okamoto K, O’Brien S, Carrero P, Makino Y, Tanaka H, Poellinger L. Signal transduction in hypoxic cells: inducible nuclear translocation and recruitment of the CBP/p300 coactivator by the hypoxia-inducible factor-1alpha. EMBO J. 1998;17:6573–6586. doi: 10.1093/emboj/17.22.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Boussouar F, Boyd K, Xu W, Biesen M, Rehg J, Baudino TA, Cleveland JL, Brindle PK. Two transactivation mechanisms cooperate for the bulk of HIF-1-responsive gene expression. EMBO J. 2005;24:3846–3858. doi: 10.1038/sj.emboj.7600846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske AJ, Buratowski S, Nonet M, Young RA. A novel transcription factor reveals a functional link between the RNA polymerase II CTD and TFIID. Cell. 1992;69:883–894. doi: 10.1016/0092-8674(92)90298-q. [DOI] [PubMed] [Google Scholar]
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. doi: 10.1038/nature01763. [DOI] [PubMed] [Google Scholar]
- Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min IM, Waterfall JJ, Core LJ, Munroe RJ, Schimenti J, Lis JT. Regulating RNA polymerase pausing and transcription elongation in embryonic stem cells. Genes Dev. 2011;25:742–754. doi: 10.1101/gad.2005511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide association of hypoxia-inducible factor (HIF)-1alpha and HIF-2alpha DNA binding with expression profiling of hypoxia-inducible transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay A, Kramer JM, Merkx G, Lugtenberg D, Smeets DF, Oortveld MA, Blokland EA, Agrawal J, Schenck A, van Bokhoven H, et al. CDK19 is disrupted in a female patient with bilateral congenital retinal folds, microcephaly and mild mental retardation. Hum Genet. 2010;128:281–291. doi: 10.1007/s00439-010-0848-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. EMBO J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell. 2004;14:685–691. doi: 10.1016/j.molcel.2004.05.006. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CAS, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. Human Mediator Subunit MED26 Functions as a Docking Site for Transcription Elongation Factors. Cell. 2011;146:92–104. doi: 10.1016/j.cell.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Wang G, Balamotis MA, Stevens JL, Yamaguchi Y, Handa H, Berk AJ. Mediator requirement for both recruitment and postrecruitment steps in transcription initiation. Mol Cell. 2005;17:683–694. doi: 10.1016/j.molcel.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerling T, Kuuluvainen E, Mäkelä TP. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol. 2007;27:6177–6182. doi: 10.1128/MCB.01302-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Kung AL. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biology. 2009;10:R113. doi: 10.1186/gb-2009-10-10-r113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell. 2005;19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.