Abstract
Introduction
Cancer of the testes is currently the most frequent neoplasm and a leading cause of morbidity in men 15–35 years of age. Its incidence is increasing. Embryonal carcinoma is its most malignant form, which either may be resistant or may develop resistance to therapies, which results in relapses. Cancer stem cells are hypothesized to be drivers of these phenomena.
Specific aim
The specific aim of this work was identification and isolation of spectra of single, living cancer stem cells, which were acquired directly from the patients’ biopsies, followed by testing of their pluripotency.
Patients. Methods
Biopsies were obtained from the patients with the clinical and histological diagnoses of the primary, pure embryonal carcinomas of the testes. The magnetic and fluorescent antibodies were genetically engineered. The SSEA-4 and TRA-1–60 cell surface display was analyzed by multiphoton fluorescence spectroscopy (MPFS), flow cytometry (FCM), immunoblotting (IB), nuclear magnetic resonance spectroscopy (NMRS), energy dispersive x-ray spectroscopy (EDXS), and total reflection x-ray spectroscopy (TRXFS). The single, living cells were isolated by magnetic or fluorescent sorting followed by their clonal expansion. The OCT4A, SOX2, and NANOG genes’ transcripts were analyzed by qRTPCR and the products by IB and MPFS.
Results
The clones of cells, with the strong surface display of TRA-1–60 and SSEA-4, were identified and isolated directly from the biopsies acquired from the patients diagnosed with the pure embryonal carcinomas of the testes. These cells demonstrated high levels of transcription and translation of the pluripotency genes: OCT4A, SOX2, and NANOG. They formed embryoid bodies, which differentiated into ectoderm, mesoderm, and endoderm.
Conclusion
In the pure embryonal carcinomas of the testes, acquired directly from the patients, we identified, isolated with high viability and selectivity, and profiled the clones of the pluripotent stem cells. These results may help in explaining therapy-resistance and relapses of these neoplasms, as well as, in designing targeted, personalized therapy.
Keywords: Cancer of the testis, testicular carcinoma, germ cell tumor, embryonal carcinoma, pluripotency, stem cell, Fv, TRA-1–60, SSEA-4, OCT4A, SOX2, NANOG
Introduction
Cancer of the testis, also known as testicular cancer or testis’ cancer, is the most frequent neoplasm in men of 15–35 years of age [1–7]. Significant increases in incidence trends have been recorded with the rate factor of 2.3 in the USA between 1995–1989 and 0.8 between 1989–2009. Its incidence nearly doubled globally in the years 1989–2009. Men with Northern Europe heritage are at the highest risk [8–10]. In utero exposure of male fetuses, to endocrine disruptors and other environmental pollutants, is suggested as one of the causes [11–15]. In histological classification, germ cell tumors (GCTs) constitute more than 95% of these tumors. Embryonal carcinomas of the testes (ECT) are the most malignant forms of GCTs [4–6].
The average 5-Year Relative Survival Rates (5Y-RSR) have improved significantly from 83% to 96% in the United States between 1975 to 2007 [2]. Nevertheless early diagnosis plays a critical role as 5Y-RSR reached 99% for distribution of 69% of patients, who were diagnosed with the localized cancer of the testis, but dropped down to 95.8% for those 18%, who were diagnosed with the cancer already spreading to the regional lymph nodes, and fell further down to 72.5% for those 12%, who were diagnosed with the metastasized cancer. However, the major concerns are raised by the increased mortality delayed beyond 5 years, the increased numbers of secondary neoplasms, and late relapses [16–25].
Initial history taking may help to provide warning signs [26–32]. In particular, prior history of the cancer in the contra-lateral testis increases the risks and may suggest survey biopsies, which may reveal pre-invasive carcinoma in situ. Infertility, oligospermia, or gynecomastia may also be warning signs. Reported symptoms may include pain in the testicle, scrotum, or abdomen, scrotal heaviness or firmness. During physical examination, swelling, redness, and often sensitivity to touch may be determined. Only 5% of patients may report gastrointestinal, endocrinological, neurological, or respiratory problems, which may point to metastases. Ultrasonography (USG) is the first choice of non-ionizing radiation based imaging [33]. Non-conclusive USG has to be refined by magnetic resonance imaging (MRI) [34]. Genetic testing may detect aneuploidy and multiple copies of the chromosome 12 [35]. Blood tests may reveal increased levels of alpha-fetoprotein, beta human chorionic gonadotropin, and TRA-1–60 [35–39]. In pathomorphology, signs of hemorrhaging and necrosis are manifestations of rapidly growing malignant tumors [40–43]. Moreover, presence of high percentage of embryonal carcinoma cells in mixed tumors, which appear as polygonal and undifferentiated, but with washed-out nuclei and prominent nucleoli, prompts poor prognosis. Furthermore, mixed GCTs create not only serious diagnostic issues – due to often non-representative sampling of heterogeneous populations of cancer cells within tumors, but also therapeutic challenges – due to differences in responsiveness to systemic therapies augmenting orchiectomy, in nearly all patients, to suppress dissemination [44–49]. Therefore, finding unique molecular biomarkers, which not only facilitate histological classification, but also define malignant potential, is critical for planning personalized, targeted therapies.
SSEA-4 and SSEA-3 - globo-series glycolipids, which were displayed on surfaces of the majority of undifferentiated cells from human, cultured, testicular embryonal carcinoma lines, became the hallmark of undifferentiated stem cells [50–51]. SSEA-1 was displayed instead on these cells upon their differentiation [52]. TRA-1–60 and TRA- 1–81- mucin-like antigens, which were displayed on and shed from the embryonal carcinomas, also became biomarkers of pluripotent cells [53]. Pluripotency of these cells was demonstrated by their ability to form embryoid bodies in vitro and to differentiate into three, morphologically distinct, germ layers: ectoderm, endoderm, mesoderm. Moreover, their ability to differentiate was demonstrated by induction with dimethyl-sulfoxide (DMSO), retinoic acid (RA), or hexamethylene-bisacetamide (HMBA), into all tissue types including muscular, neuronal, and epithelial [54–63]. Dynamics of these biomarkers’ cell surface display showed similarities with the cultured pluripotent embryonic stem cells from inner masses of human blastocysts, thus possessing totipotential [64–73]. They were detected on stem cells of the human fetal testes [74]. These biomarkers were also displayed on pluripotent stem cells derived from bone marrow [75]. CD30, CD117, CD44, CD133, CD29, SSEA-5, and MHC, were identified as biomarkers of stem cells’ fractions or differentiation stages, but were not uniquely present on all human, pluripotent stem cells. Moreover, they were reported on cells in some studies, but undetected in the others, also by immunocytochemistry on paraffin sections from cancerous and healthy testes [76–91].
Transcription factors: NANOG, OCT4, and SOX2, determined as capable of inducing pluripotency of differentiated cells, constituted the group of the unique biomarkers of pluripotent stem cells [92–94]. The genes’ expression profiles were very similar in the cultured pluripotent cells of the lines of embryonal carcinomas of the testes and of the embryonic inner mass, as well as, in the pluripotent stem cells identified directly in the embryonal carcinomas of the ovaries [94]. These factors were also tested in formaldehyde-fixed, paraffin-embedded (FFPE) and snap-frozen (SF) tissues, which were acquired from the patients tumors’ and healthy testes’, but often with conflicting results. Gene transcripts of these factors were also quantified by RTPCR and microarrays on homogenized tissues recovered from FFPE, SF, and fresh cancerous and healthy tissue samples, but often with varying results [97–103]. Randomness of selection and small sizes of samples not reflecting completeness of human cancer cell heterogeneity, variability in the samples’ preparation methods, differences in cultures’ environments, lack of the direct correlations between pathomorphology and functionality of the living cancer cells, heterogeneity of clones in cell lines, difficulties in extrapolating cell culture data onto in vivo phenomena, incompatibility between biomarkers for humans versus other species, all created the problems, to mention only these few, with streamlining of the acquired data into the clinics. Meanwhile, the clones of the living, pluripotent human stem cells with the SSEA-4 and TRA-1–60 displayd were not isolated directly from the testicular tumors, the transcription factors were not quantified in these clones, and the clones of pluripotent stem cells were not imaged in these cancers in vivo. To address these problems, the novel biomarkers and approaches are being developed to study spectra of single, living cells, ex vivo - acquired directly from tumors or in vivo, in situ - in their natural environment [104–114].
Our work herein is the first attempt to identify and isolate, directly from the biopsies of the patients diagnosed with germ cell tumors of the testes, spectra of single, living cells with the cell surface display of SSEA-4 and TRA-1–60, and to follow up with the single cells’ cloning and evaluating their pluripotency.
Materials and Methods
Patients. Samples
All the samples were obtained in accordance with the Declaration of Helsinki with the Patients’ Informed Consent and with the Institutional Review Boards’ approval. The samples from the patients, who were being clinically and histopathologically diagnosed with the germ cell tumors (GCT): men with testicular GCTs (n=103); women with ovarian GCTs (n=43); women (n=3) or men (n=3) with the extragonadal GCTs were included into this study. All the samples were encoded to protect the patients’ identity. The cases of the primary, pure, advanced embryonal carcinomas of the testes (ECT) were selected for this study. Collection of the samples from the tumor, ascites, metastases, healthy tissue, bone marrow, and blood was performed according to the standard surgical procedures. The batches of the samples were either immediately labeled with the single or dual chain variable fragment (Fv) antibodies, or incubated for cultures / clonal expansion, or rapidly cryoimmobilized or chemically preserved for bio-banking [96]. Healthy brain, heart, ovary, and testis tissues, which were dissected as the safety margins or prophylaxis, and bone marrow mononuclear and peripheral blood mononuclear cells obtained prior to cancer therapy, were the controls.
Genetically engineered single or dual chain variable fragment (Fv) antibodies
The genetically engineered, variable fragment (Fv) antibodies were prepared as described [94], thus only briefly outlined below. Antibodies for TRA-1–60, SSEA-4, CD-45, CD-34, CD-19, CD-20 were genetically engineered from the B cells of patients suffering cancers. Anti-DOTA, anti-DTPA, and anti-TETA were genetically engineered from the B cells of patients undergoing multiple rounds of imaging and therapy involving chelates as the contrast agents. The pooled B cells from these patients were used to isolate mRNA, reverse transcribe, and create the cDNA libraries of complementarity determining regions (CDR) and framework regions (FWR) for anti-cancer-antibodies (ACA) coding sequences. The cds, after insertion into the plasmids containing chelates’ harboring coding sequences under the CMV promoters, were propagated and expressed in human myelomas as described (Fv clones TRA-1–6024, SSEA-437 were used in this project). The native TRA-1–60, SSEA-4, CD-45, CD-34, CD-19, CD-20 were purified, which followed by modification with biotin, digoxigenin, or fluorescein. The modified TRA-1–60, SSEA-4, CD-45, CD-34, CD-19, CD-20 were anchored onto anti-biotin, anti-dig, or anti-FITC saturated pans and served as baits for selection of the Fv clones from the ACA libraries. The chelates were saturated with Gd, Tb, Eu, Mn, or Ru. The elemental compositions were validated with EDXS (Noran, Middleton, WI, USA) or TRXFS (Bruker AXS, Fitchburg, WI, USA). The fluorescent properties were measured with the RF-5301PC spectrofluorometer (Shimadzu, Tokyo, Japan). The specificity and sensitivity of the Fvs were tested with the EELS and EDXS [115]. The magnetic relaxivities were measured on the DMX 400 WB or AVANCE II NMR spectrometers (Bruker Optics, Dallas, TX, USA). The monoclonal antibodies targeting TRA-1–60, SSEA-4, CD45, CD34, CD-19, CD-20 served as the positive controls, and antibodies towards 6His, FLAG, DOTA, TETA, and DTPA were the negative controls. To target human OCT-4, SOX2, NANOG, the genetically engineered, variable fragment (Fv) antibodies were prepared as described [94].
Flow cytometry (FCM). Fluorescently activated cell sorting (FACS). Multiphoton Fluorescence Spectroscopy (MPFS)
The cell clusters were thoroughly disintegrated into single cell suspension by short treatment with the PIPES buffered DNase, RNase, trypsin, and collagenase as described [94]. The negative selection involved depletion of hematopoietic progenitor cells with the Fvs anti-CD34; differentiated hematopoietic cells with the Fvs anti-CD45, apoptotic cells with the Fvs anti-PS, the dead cells with the Fvs anti-DNA to reach above 99.5% of purity. The remaining samples were further enriched by the positive selection with the Fvs for TRA-1–60 or SSEA-4. The enriched populations of the cells labeled with the fluorescent Fvs targeting TRA-1–60 and SSEA-4 were measured with the Calibur, Vantage SE, or Aria (Becton-Dickinson, Franklin Lakes, NJ, USA) or the FC500 (Beckman-Coulter, Brea, CA, USA). The fluorescently labeled cells were imaged with the Axiovert (Zeiss, Oberkochen, D, EU) equipped with the Enterprise argon ion (457 nm, 488 nm, 529 nm lines) and ultraviolet (UV) (364 nm line) lasers; Odyssey XL digital video-rate confocal laser scanning imaging system operated up to 240 frames/s under control of Intervision software (Noran, Madison, WI, USA), and the Diaphot (Nikon, Tokyo, Japan) equipped with the Microlase diode-pumped Nd:YLF solid state laser (1048 nm line), the pulse compressor with the pulses’ rate 300 fs at 120 MHz and the MRC600 scanning system under control of Comos software (the multi-photon fluorescence station built based upon the NIH funds – PI: Dr J. White). Deconvolution of images was done on the Indy workstation (Silicon Graphics, Fremont, CA, USA).
Total Reflection X-ray Fluorescence Spectroscopy (TRXFS)
In this study, the ICP standard of 1000 mg/l of mono-element Gallium (CPI International, Denver, CO, USA) was added to 500 microL of each sample to the final concentration of 10 mg/l. The data were generated from the S2 Picofox TXRF spectrometer equipped with a molybdenum (Mo) X-ray target and the Peltier cooled Xflash Silicon Drift Detector (Bruker AXS, Fitchburg, WI, USA). Scan times ranged up to 1000 seconds. The automatic sample changer, which can hold up to 25 samples, was also used along with the SPECTRA 7 software for the instrument control, data collection, and analysis (Bruker AXS, Fitchburg, WI, USA).
Nuclear Magnetic Resonance (NMR). Magnetically Activated Cell Sorting (MACS)
The cells were labeled for positive selection with the superparamagnetic Fvs targeting TRA-1–60 and SSEA-4, and for the negative selection targeting CD45, CD34, dsDNA, and PS, while suspended in the physiological buffer supplemented with serum and glucose. The small aliquots were dispensed into the magnetism-free NMR tubes (Shigemi, Tokyo, Japan). The relaxation times T1 were measured in resonance to the applied FLAIR pulse sequences on the NMR spectrometers: DMX 400 WB or AVANCE II NMR (Bruker, Billerica, MA) or the Signa clinical scanners (GE, Milwaukee, WI, USA). The superparamagnetic Fvs were also used to isolate the labeled cells from the solution using the magnetic sorter to reach above 99.5% of purity (the sorter designed and built based upon the NSF funds – PI: Dr M. Malecki).
Electron Energy Loss Spectroscopy (EELS). Energy Dispersive X-Ray Spectroscopy (EDXS)
The samples, which were cryo-immobilized, presented the life-like supramolecular organization. Molecular imaging was pursued as described [112]. The field emission, scanning transmission, electron microscope FESTEM HB501 (Vacuum Generators, Kirkland, WA, USA) was equipped with the energy dispersive x-ray spectrometer (EDXS) (Noran, Middleton, WI, USA) and post-column electron energy loss spectrometer (EELS) (Gatan, Pleasanton, CA). The cryo-energy filtering transmission electron microscope 912 Omega was equipped with the in-column, electron energy loss spectrometer (EELS) (Zeiss, Oberkochen, D, EU). The cryo-energy filtering transmission electron microscopes 410 and 430 Phillips were equipped with the post-column, electron energy loss spectrometer (EELS) (Noran, Middleton, WI, USA). The field emission, scanning electron microscope SEM1530 (Zeiss, Oberkochen, D, EU) was equipped with the energy dispersive x-ray spectrometer (EDXS) (Noran, Middleton, WI, USA). The field emission, scanning electron microscope 3400 was equipped with the energy dispersive x-ray spectrometer (EDXS) (Hitachi, Tokyo, Japan). The images and spectra were acquired using the ccd camera operating under the image acquisition and processing software (SIS, Herzogenrath, D, EU or Emispec Systems, Tempe, AZ, USA).
Cultures. Embryoid bodies. Clonal Expansion. Differentiation
The cancer cells were grown as described [55,60,96]. Cell clusters were separated into single cell suspension by short treatment with the PIPES buffered DNase, RNase, hyaluronidase, trypsin, and collagenase. The cells were labeled with the fluorescent or magnetic Fvs and isolated by FACS or MACS [94, 104–105]. Cultures were established as described for metastatic testicular embryonal carcinoma line [54] or human embryonic stem cells [66]. The cultures of the human, male embryonic stem cells - hESC H1, H13, H14 with normal XY karyotypes [66], human line of embryonal carcinoma of the testis cells derived from metastasis of teratoma-hECT NT2D1, muscle cells - RD, brain neuronal cells - HCN-2 were the controls (ATCC, Manassas, VA). Moreover, healthy tissue of ovaries (HTO) from prophylactic oopherectomy, healthy tissue from testes (HTT) dissected during orchiectomy - testosterone depleting measure during therapy of prostate cancer, healthy neural tissue (HTN) – from healthy margins removed during surgery of brain tumors, healthy margins of cardiac muscle excised during cardiac surgery (HTC) were the controls. Finally, bone marrow and peripheral blood mononuclear cells were the controls. Differentiation of the embryoid bodies into the three germ layers was conducted and validated based upon testing transcription of the specific genes with qRTPCR, whereas amplicons for neurofilament 68kDa, α-fetoprotein, and ζ-globin identified formation of ectoderm, endoderm, and mesoderm respectively as described [70]. Differentiation towards specific lineages was induced on plated cells by 10−5 M retinoic acid, 3 mM hexamethylene bisacetamide (HMBA), 1% dimethyl sulfoxide (DMSO), 250 ng/ml nerve growth factor, epidermal growth factor, or vascular endothelial growth factor added to the media (Sigma-Aldrich, Saint Louis, MO, USA). The evaluation was based upon labeling with the Fvs anti-neurofilaments and bungarotoxin – to validate differentiation towards neurons, anti-GFAP – into glia, anti-desmin and cardiac myosin - into muscle, anti-cytokeratins – into epithelium.
Immunoblotting (IB)
The cells were either frozen in liquid nitrogen, crushed, and thawed or/and disintegrated with ultrasonicator (Branson Ultrasonic, Danbury, CT, USA) within the sample buffers for native protein analysis. They were stored in liquid nitrogen or electrophoresed in the native buffer (Invitrogen, Carlsbad, CA, USA). They were vacuum or electro-transferred onto the PVDF membranes (Amersham, Buckinghamshire, UK, EU). The membranes carrying the transferred proteins were soaked within human serum and labeled with the Fvs. The samples of muscle myosin, neuronal filament protein, lamins, actin, and cytokeratins served as the controls. The images of the blots were acquired and quantified with Fluoroimager (Molecular Dynamics, Sunnyvale, CA, USA) or Storm 840 (Amersham, Buckinghamshire, UK, EU). The levels of the products were also calculated, as the ratio between the protein concentration in the examined patient’s cells and the controls.
Quantitative Reverse Transcription and Polymerase Chain Reaction (qRTPCR)
Total isolated mRNA served as the template to generate cDNA through reverse transcription using random hexamers and reverse transcriptase (ABI, Foster City, CA, USA) as described [96]. The cDNAs’ and amplicons’ qualities were tested by polymerase chain reaction of beta actin and GAPDH, as the reference genes (ABI, Foster City, CA, USA). For evaluation of the gene expression levels for OCT4A, OCT4B, OCT4B, the primers sets were designed using Primer Express (ABI, Foster City, CA, USA) based upon the sequences imported from the Public Domain GenBank (NCBI), and synthesized on the 380A DNA Synthesizer (ABI, Foster City, CA, USA). The PCR reactions were carried using the mix of the cDNA, the synthesized primers, dNTPs, and Taq DNA polymerase (Hoffmann–La Roche, Basel, H) on the Robocycler (Stratagene, San Diego, CA, USA), Mastercycler (Eppendorf, Hamburg, D, EU), or 7500, 7900 systems (ABI, Foster City, CA, USA). The images of the gels were acquired and quantified with Fluoroimager (Molecular Dynamics, Sunnyvale, CA, USA) or Storm 840 (Amersham, Buckinghamshire, UK, EU). The levels of the transcripts were all normalized against GAPDH or actin, and thereafter calculated as the ratios between the transcript concentration in the examined patient’s cells versus the cells from the healthy control tissues and cultures.
Statistical analysis
Fisher’s exact test was used to examine the association of the gene expression between the human ECT cells versus the controls: cultured human embryonal carcinoma cells of the testis (NT2D1), cultured human embryonic stem cells (H1, H13, H14), healthy tissue of the ovaries (HTO), healthy tissue of the testis (HTT), peripheral blood mononuclear cells (PBMC), or bone marrow mononuclear cells (BMMC). Average gene expression measurements were run in triplicates for each patient, which were used for gene expression statistical analysis. For the comparisons, Wilcoxon signed rank test was used, and Wilcoxon rank sum test for the comparison of the independent groups of the ECT versus the controls. A two-sided p-value was computed in each comparison. The APC was accepted as statistically significant with p < .001. The graphs were displayed using GraphPad software (GraphPad Software, Inc, La Jolla, CA).
Results
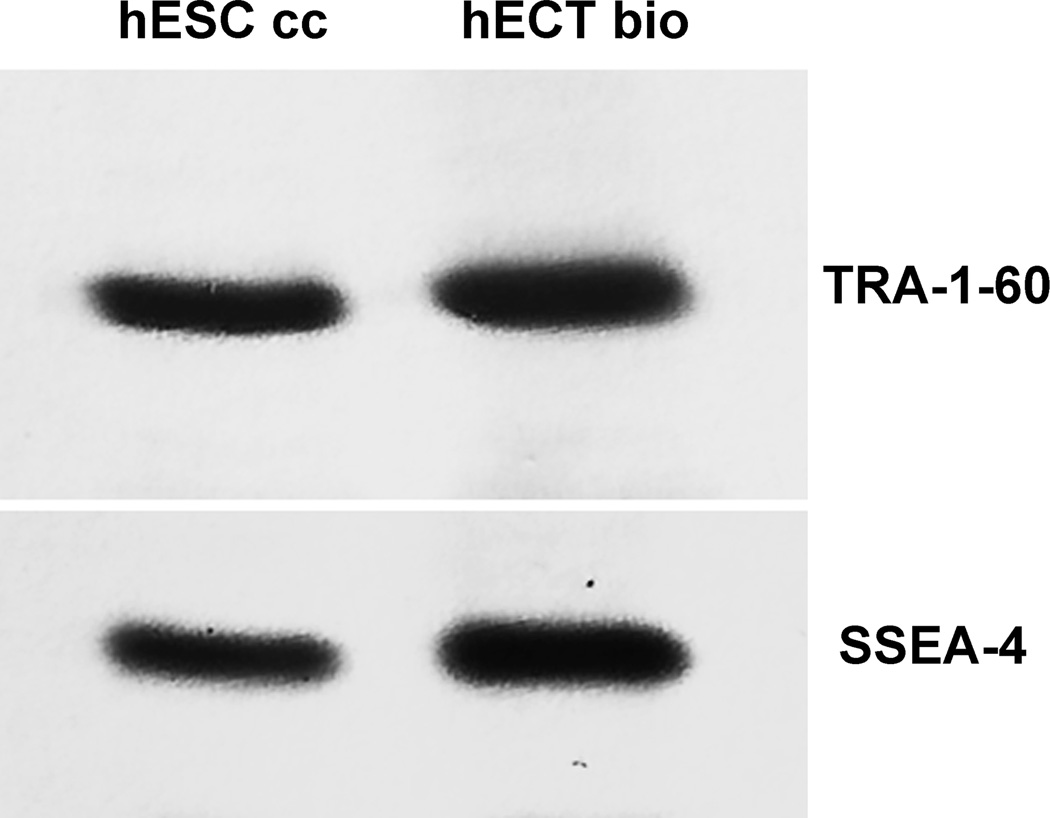
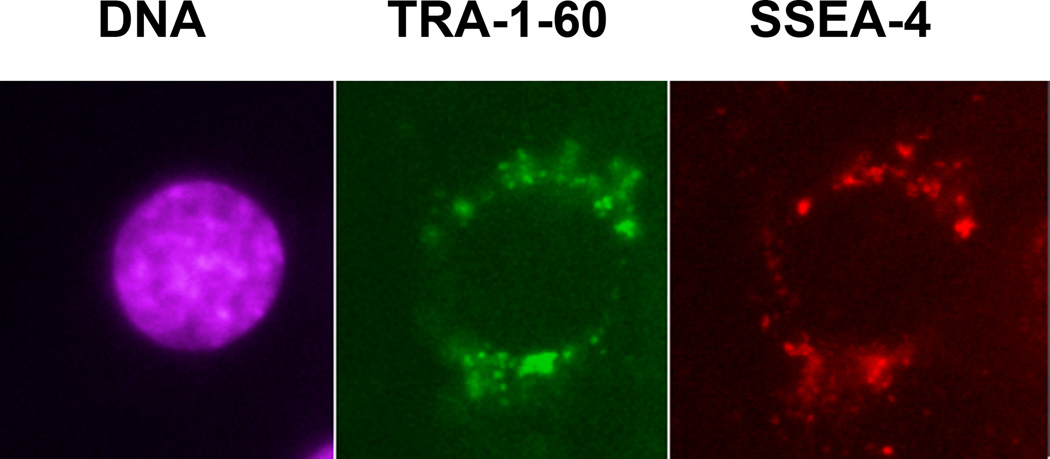
Displays of TRA-1–60 and SSEA-4 on surfaces of the cells biopsied directly from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes were presented in the figures 1–5. Specificity and sensitivity of Fvs were first determined on immunoblots as illustrated in the figure 1. The patients data (huECT bio) were validated on blots of the cultured human embryonic stem cells as positive controls (huESC cc) and mononuclear cells as the negative controls (huBMM bio). For all batches of the cells, acquired from each of the patients and from controls, the electrophoresis and blotting were run in triplicates. The blots in the figure 1 are representative for all the other samples. The labeling is highly sensitive and specific, as validated by the single bands against completely clean background. Both, TRA-1–60 and SSEA-4 showed the strong display. Their display was much higher on the ECT cells than on the ESC cells and absent on the BMM and PBM cells. Specificity and sensitivity of chelated, thus fluorescent, Fvs were highlighted by multiphoton fluorescence, as illustrated in the figure 2. For all batches of the cells, acquired from each of the patients and from the controls, the immunolabeling was run in triplicates, followed by acquisition of images from ten randomly selected cells. The images presented are representative for all the samples studied.
Figure 1.
TRA-1–60 and SSEA-4 displayed on the cells biopsied directly from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (in the fig. 1 for the patient encoded 001) were compared to those on the cultured human embryonic stem cells from the line H1 (huESC cc). The cells were homogenized, electrophoresed, transferred on membranes, and labeled with the Fvs. All samples were run in triplicates. The blots were representative for all the remaining patients’ biopsies and controls. The blots were quantified with the statistical significance accepted at p < 0.001.
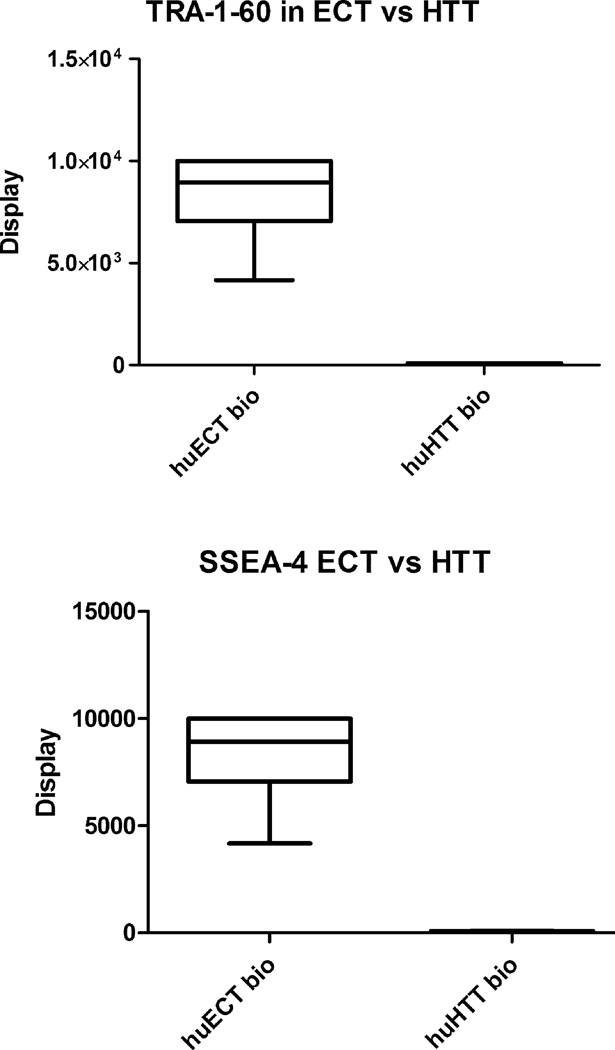
Figure 5.
Display of TRA-1–60 and SSEA-4 in the enriched samples of the cells acquired from the patients with embryonal carcinomas of the testes (ECT), relative to the cells biopsied from the healthy tissue of the testes (huHTT bio), was determined by EDXS. Since the Fvs were chelating metal ions, then they were generating the energies uniquely specific to the elements. Each sample was run in triplicates. The data presented are representative to all the samples and the controls. The relative display was quantified by comparing the values of the x-rays induced scintillation saturation set at the 10,000 counts’ ceiling (y-axis). The statistical significance was accepted at p < 0.001.
Figure 2.
The stem cells biopsied directly from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (in the fig. 1 for the patient encoded 001) were triple-labeled with the Fvs targeting DNA (violet), TRA-1–60 (green), and SSEA-4 (red), which were tested for specificity and sensitivity on blots as shown in the figure 1. All the batches for every patient were labeled in triplicates. The images presented in the figure are representative to all the patients and the controls studied. From each batch, the images of ten randomly selected cells were acquired at the three acquisition channels without correcting the wavelength induced shift. The images were acquired with multiphoton excitation fluorescence. The HFW: 30µm.
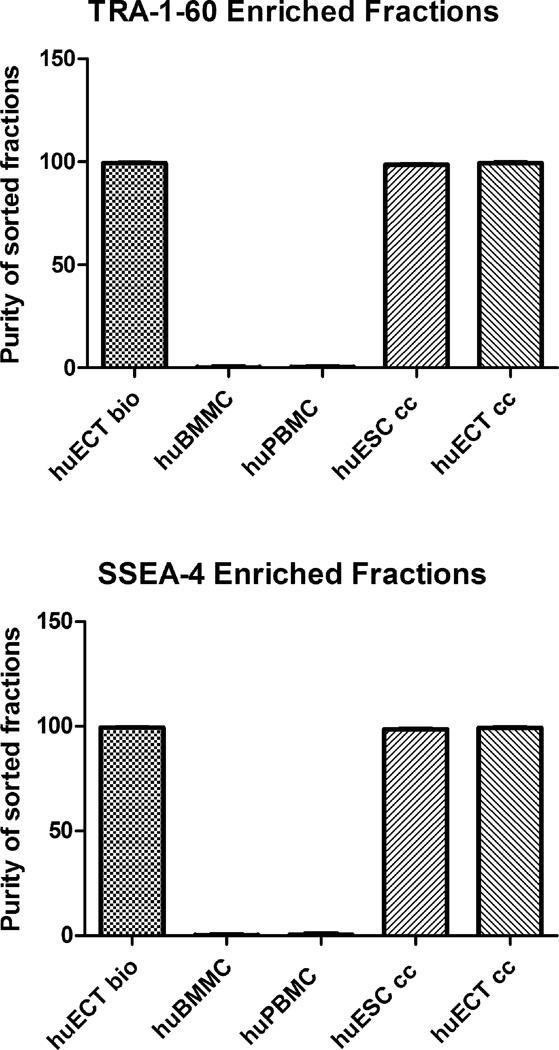
Considering heterogeneity of the tumor cells’ populations, fractions of cells displaying TRA-1–60 and SSEA-4, as labeled with superparamagnetic Fvs, were enriched by magnetic sorting (MACS). These enrichment processes included the embryonal carcinoma of the testes (Patients encoded 001–009) (huECT bio), the mononuclear cells from bone marrow (huBMMC) and from peripheral blood (huPBMC), the cultured human embryonic stem cell lines (H1, H13, H14) (huESC cc), and the cultured cells from metastasis to lungs of the testicular embryonal carcinoma (NT2D1) (huECT cc). Sorting was performed in triplicates for each patient and controls. The presented graphs are representative to all the runs. The outcomes of sorting were quantified by flow cytometry as documented in the figure 3. These fractions’ purities were above 99.5%. The signals from the BMM and PBM cells were not exceeding background levels. The statistical significance of differences between these fractions was accepted at p < 0.001.
Figure 3.
The cells were biopsied directly from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (in the fig. 3, the patients encoded 001–009), the mononuclear cells from bone marrow (huBMMC) and from peripheral blood (huPBMC), the cultured human embryonic stem cell lines (H1, H13, H14) (huESC cc), and the cultured cells from metastasis to lungs of the testicular embryonal carcinoma (NT2D1) (huECT cc), labeled with the superparamagnetic Fvs targeting TRA-1–6- and SSEA-4, and isolated with magnetic sorter to enrich the samples’ purity better than 99.5% with the statistical significance accepted at p < 0.001.
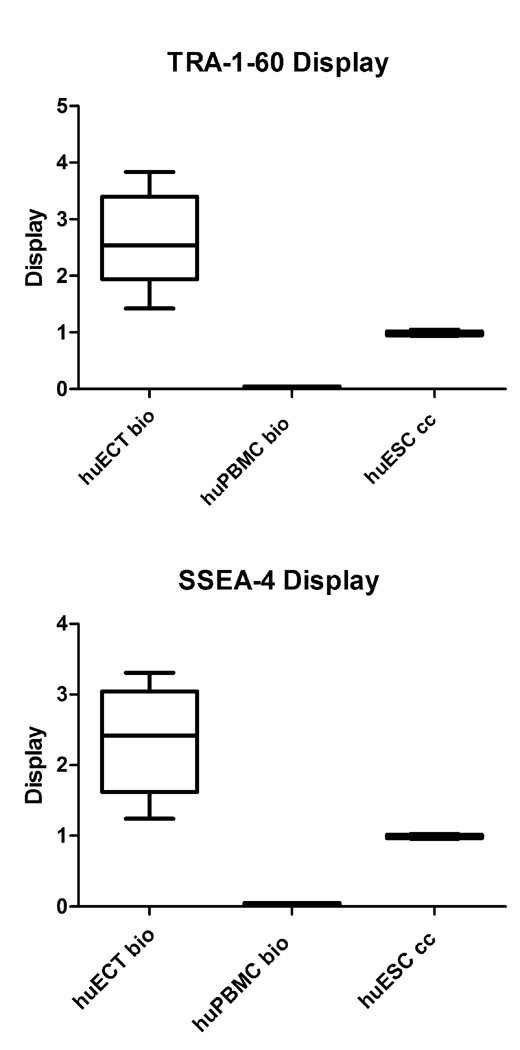
Differences between the aforementioned pools of the cells were quantified on blots and on the labeled cells with x-ray fluorescence as determined by energy dispersive x-ray spectroscopy on field emission scanning electron microscopes (EDXS), total reflection x-ray fluorescence spectroscopy (TRXRFS), and nuclear magnetic resonance spectroscopy (NMRS). The data from NMRS are presented in the figure 4. The samples were acquired from each patient, labeled, and measured in triplicates. The data presented are representative for all the samples studied. The relative display was determined based upon calculating the ratio between the relaxation times of the patients’ samples versus the relaxivity times of the control having the assigned value as 1. Displays of TRA-1–60 and SSEA-4 varied on the ECT cells acquired directly from the tumors of the patients. These displays were always much higher, than on the cultured embryonic stem cells (huESC cc) and on the cultured testicular embryonal carcinoma cells (huECT cc). Displays on the PBM and BMM cells were not detected above the threshold. The statistical significance was accepted at p < 0.001.
Figure 4.
Display of TRA-1–60 and SSEA-4 in the enriched samples of the cells acquired from the patients with embryonal carcinomas of the testes (ECT), relative to the cultured human embryonic stem cell lines (huESC cc) and the peripheral blood (huPBMC bio), was quantified by NMRS. Since the Fvs were chelating superparamagnetic metals, then they were causing shortening relaxation times, thus increased relaxivities, of the labeled samples. Each sample was run in triplicates. The data presented in the figure are representative for all the samples and controls. The relative display was quantified by calculating the ratios (y-axis), between the relaxivity of the patients’ samples in relation to the averaged value for the sample of the labeled huESC cells, which was considered as 1. The differences between the recordings are presented as the standard deviation. Labels as in the figure 2: the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (the patients encoded 001–009), the peripheral blood (huPBMC bio), the cultured human embryonic stem cell lines (H1, H13, H14) (huESC cc). The statistical significance was accepted at p < 0.001.
From a potential diagnostic point of view, the TRA-1–60 and SSEA-4 displays’ differences, between the cells within the embryonal carcinoma of the testes’ tumors (huECT bio) and the healthy tissues of the testes acquired by orchiectomy as surgical therapy for prostate cancers (huHTT bio), were essential. After the cells’ labeling with chelated Fvs, TRA-1–60 and SSEA-4 were quantified by EDXS FESEM, TRXRF, and NMRS. The samples acquired from each patient were run in triplicates. The data presented are representative for all the samples studied. The data acquired by EDXS are illustrated in the figures 5. The scintillation counter was set for 10,000 ceiling. Although spermatogonia in the HTT displayed these biomarkers, the displays’ intensities were quantitatively different. The solid statistical significance between them was accepted for p < 0.001.
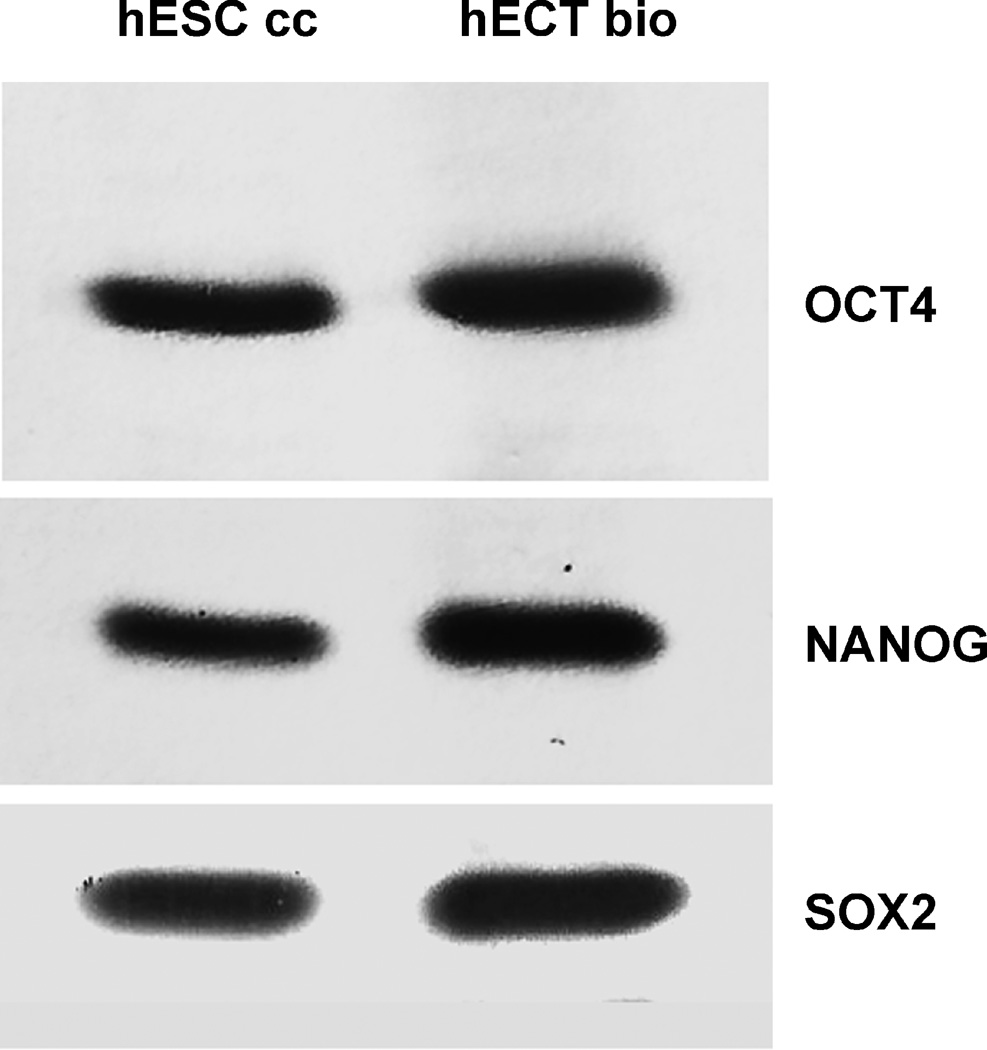
Transcription and translation of the genes OCT4A, NANOG, and SOX2 were demonstrated in the figures 6–11. Transcription factors’ labeling was validated in the figure 6. This labeling is very specific, as validated by the single bands and clean backgrounds. The intensity of labeling indicates much higher concentrations of the transcripts in the ECT biopsies, than in the cultured ESC and ECT cells.
Figure 6.
The cells were homogenized, electrophoresed, transferred on membranes, and labeled with the Fvs targeting the transcription factors: OCT4A, NANOG, SOX2 in the cells acquired from each patient diagnosed with the embryonal carcinoma of the testes (huECT bio) (the patients encoded 001–009) versus those from the cultured human embryonic stem cell lines (H1, H13, 14) (huESC cc). Each sample was run in triplicate. The blots presented in this figure are representative for all the samples and the controls run. The blots were quantified with the statistical significance accepted at p < 0.001.
Figure 11.
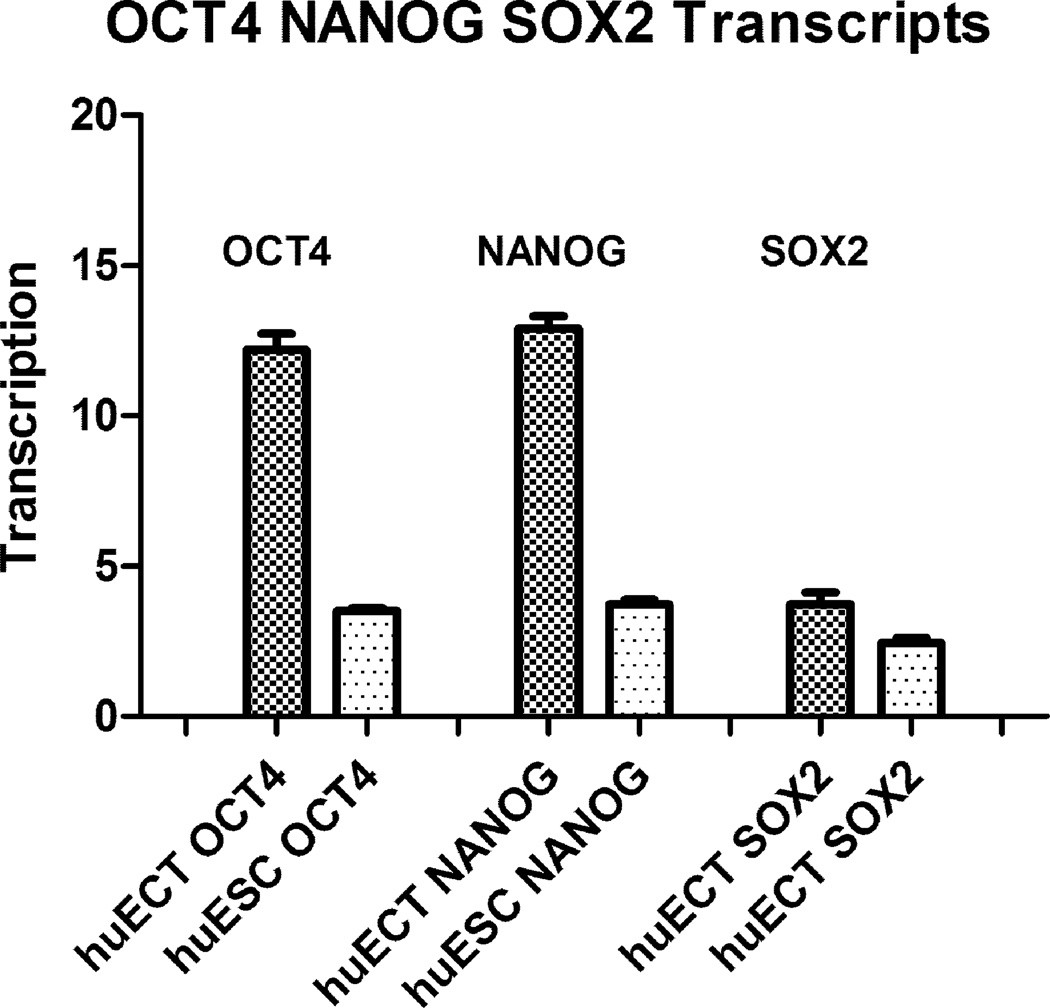
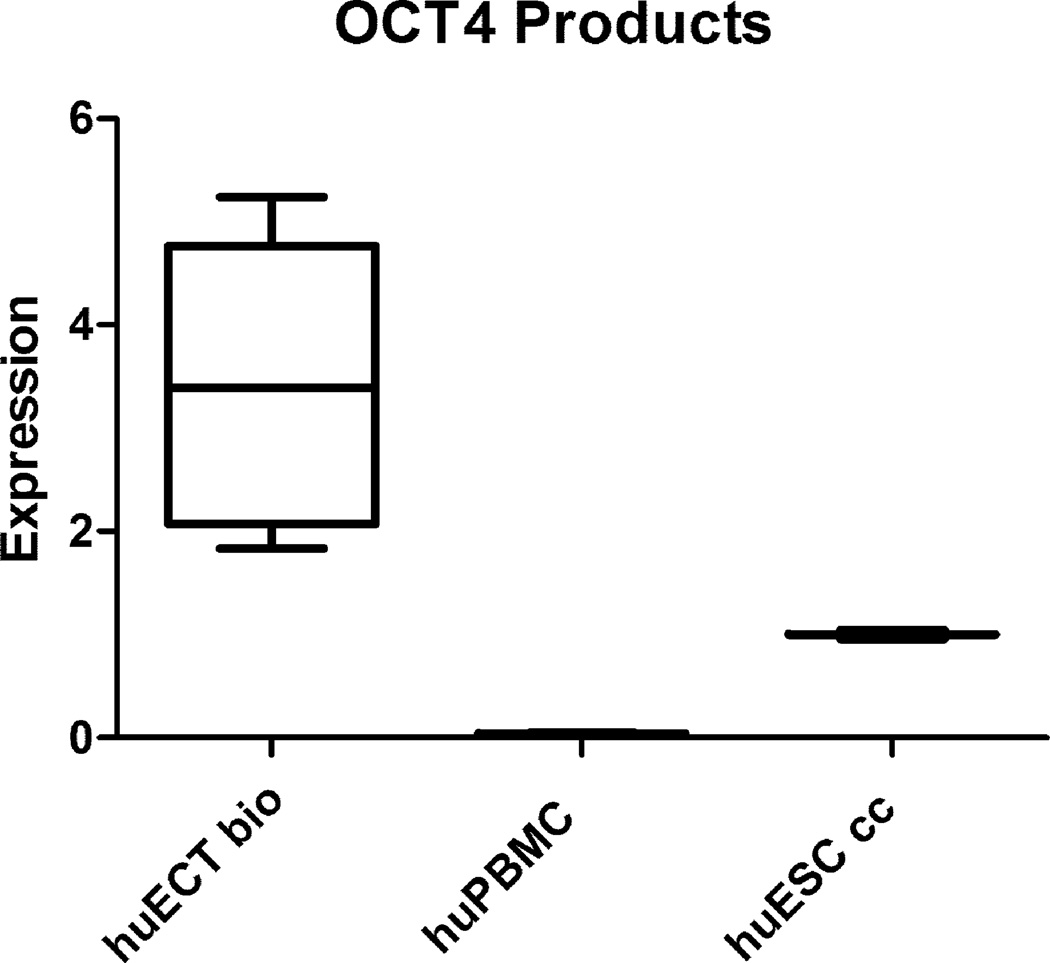
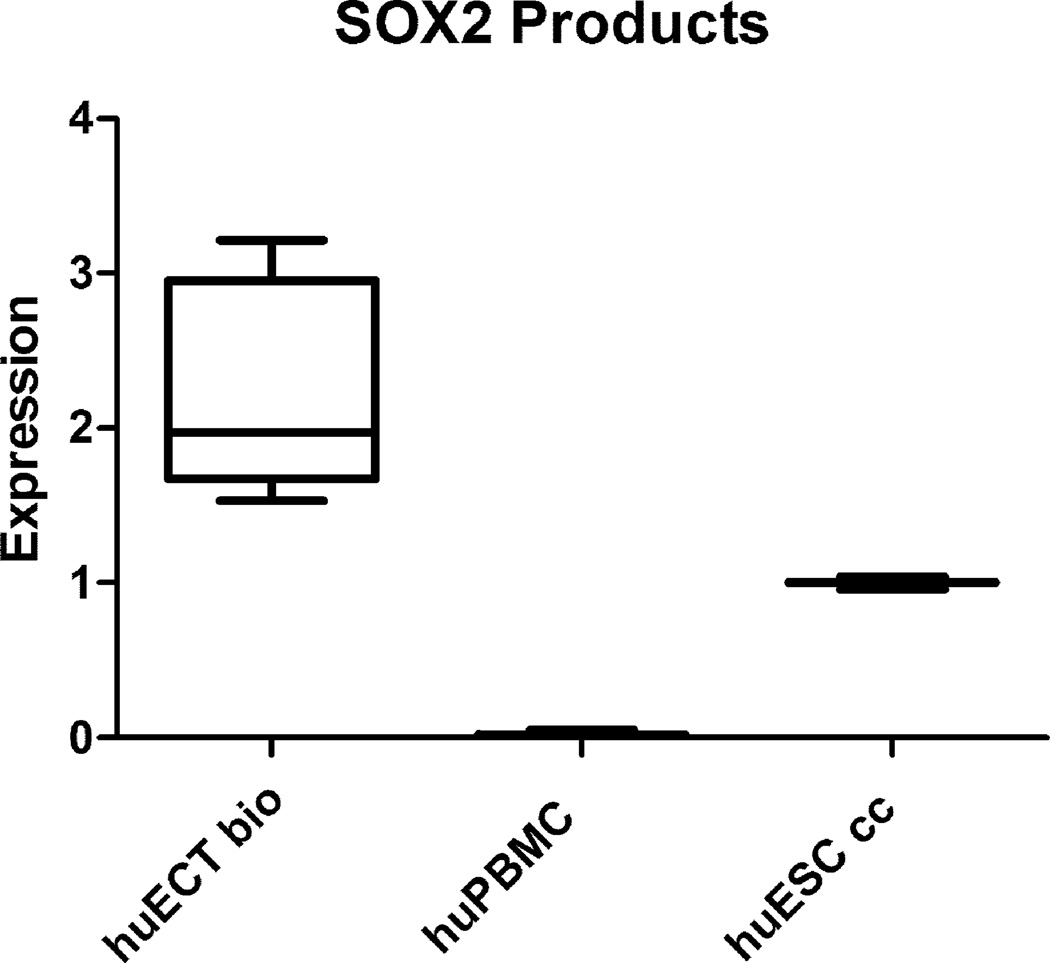
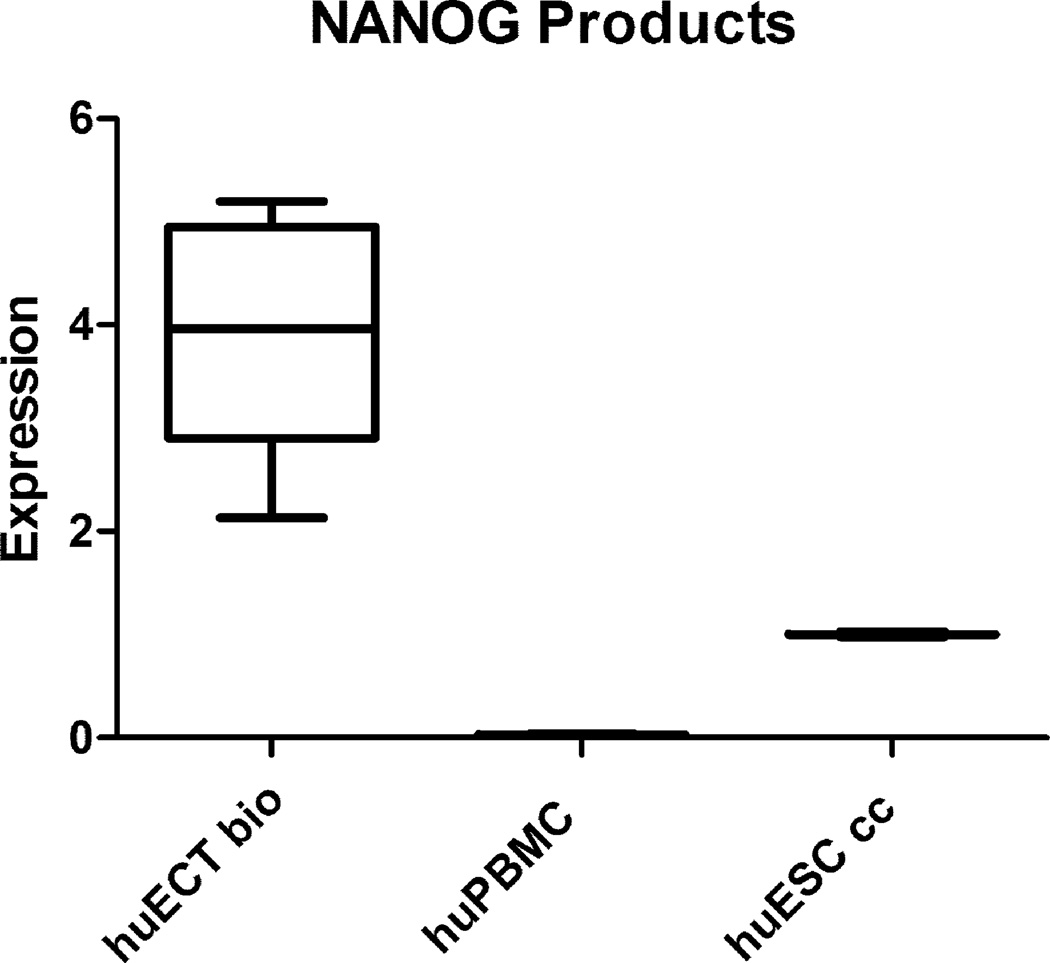
OCT4A, NANOG, and SOX2 genes transcripts’ concentrations were determined by qRTPCR in the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (the patients encoded 001–009), the cultured human embryonic stem cell lines (H1, H13, H14) (huESC cc), and the mononuclear cells from bone marrow (huBMMC) (below the detection threshold – not shown). The read-outs were calculated in relation to actin (y-axis). The statistical significance was accepted at p < 0.001.
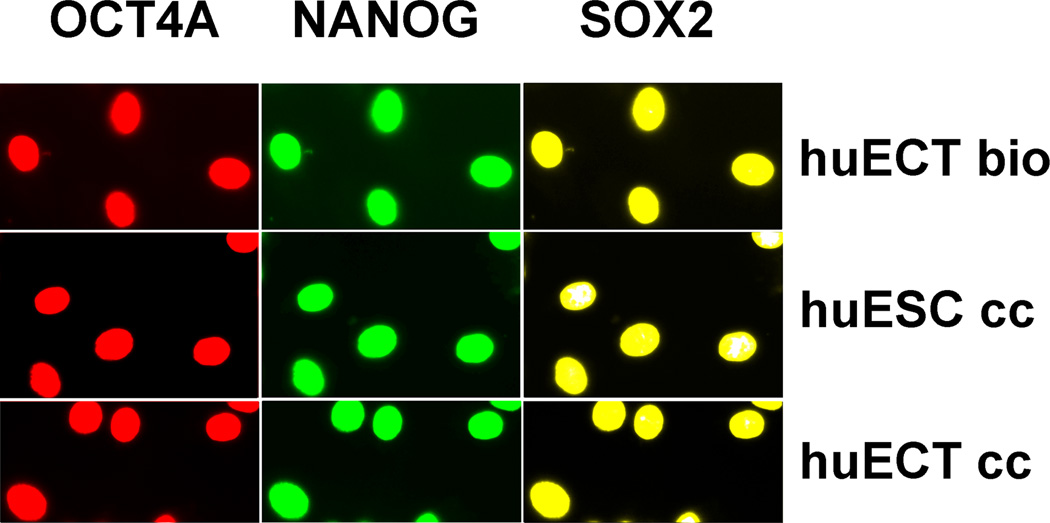
Nuclear localization of the transcription factors expressed from these genes was highlighted in the figure 7. All the samples were labeled in triplicates. The images presented in the figure are representative to all the samples studied. The cells acquired from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (Patient encoded 001), the cultured human embryonic stem cell line (cell line H1) (huESC cc), and the cultured cells from metastasis to lungs of the testicular embryonal carcinoma (cell line NT2D1) (huECT cc) were labeled with the Fvs. Metal binding domains of these Fvs were chelating metals, which rendered them fluorescent, in addition to changing their magnetism and mass. The nuclear localization of these transcription factors in the human ECT directly from biopsies is identical to that of the cultured huECT and huESC cells. Levels of expression, for each of the transcription factors, were quantified by scanning of the blots, EDXRS, TRXRFS, and NMRS. The data acquired are documented in the figures 8–10. For each modality, every sample was run in triplicates. The data presented are representative to all the samples studied. The results are very consistently validating much higher levels of gene expression for OCT4A, NANOG, SOX2 in the ECT cells biopsied from the patients (huECT bio) than in the cells from the cultures of embryonic stem cells (huESC cc).
Figure 7.
The stem cells biopsied directly from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (the patients encoded 001–009), the cultured human embryonic stem cell lines (H1, H13, H14) (huESC cc), and the cultured cells from metastasis to lungs of the testicular embryonal carcinoma (NT2D1) (huECT cc) were heavily saponin-permeabilized and triple-labeled with the Fvs targeting OCT4A - red, NANOG - green, and SOX2 – yellow. Specificity and sensitivity of the Fvs were validated on the blots as shown in the figure 6. All the batches for every patient were labeled in triplicates. From each batch, the images of ten randomly selected fields of view were acquired via the three acquisition channels without correcting the wavelength induced shift. The images were acquired with multiphoton excitation fluorescence. The images presented in the figure were acquired from the primary tumors of the patient (huECT bio) (Patient encoded 001), the cultured human embryonic stem cell line – cell line H1 (huESC cc), and the cultured cells from metastasis to lungs of the testicular embryonal carcinoma – cell line NT2D1 (huECT cc). The images presented in the figure are representative to all the patients and the controls studied. The HFW: 200µm.
Figure 8.
OCT4A gene expression products’ concentrations in the cells biopsied directly from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (the patients encoded 001–009), from peripheral blood (huPBMC), the cultured human embryonic stem cell lines (H1, H13, H14) (huESC cc) were quantified on blots described in the figure 6, normalized against actin, compared to the averaged values of huESC cc, which were considered as 1, as displayed as the ratio (y-axis). Each batch was run in triplicates. The data presented in this figure are representative to all the patients’ samples and the controls. The differences between the samples are shown as the standard deviations. The statistical significance was accepted at p < 0.001.
Figure 10.
SOX2 gene expression products’ concentrations were determined as described in the figure 10.
Transcription intensity for OCT4, NANOG, and SOX2 was also determined by quantitative reverse transcribed real time polymerase chain reaction (qRTPCR) as documented in the figure 11. The high concentrations of these transcripts indicated the high pluripotency of these cells. The levels of transcripts for these genes were much higher in the biopsied embryonal carcinoma cells of the testes, than in the cultured embryonic stem cells.
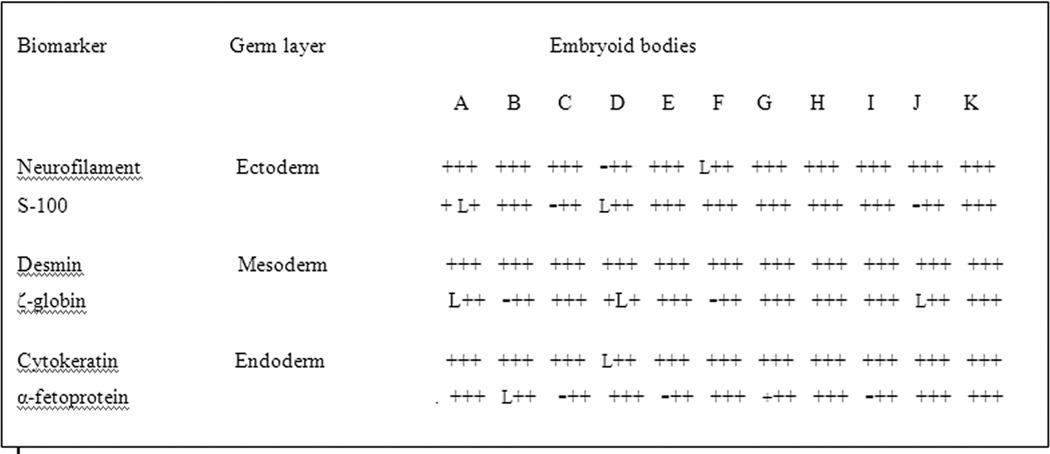
Formation of the embryoid bodies and differentiation into the three main germ layers were the direct demonstrations of the pluripotency of the cells in the ECT tumors as summarized in the figure 12. As the means to determine pluripotency was conducted for the cells acquired from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (Patients encoded 001–009 as A–I respectively) and the cultured human embryonic stem cell lines H1, H13 (huESC cc – J–K).
Figure 12.
Differentiation of embryoid bodies, as the means to determine pluripotency, was conducted for the cells acquired from the primary tumors of the patients diagnosed with the embryonal carcinoma of the testes (huECT bio) (the patients encoded 001–009, which are labeled in this figure as A–I respectively) and the cultured human embryonic stem cell lines (H1, H13 huESC cc, as J–K). Expression detected: +; no expression detected: −; lost sample: L.
Discussion
Novelty of this work relies on identification in the pure, primary embryonal carcinomas of the testes, the clones of living, pluripotent stem cells with the surface display of SSEA-4 and TRA-1–60. Pluripotency of these cells was confirmed by quantifying transcription and translation of the genes for the transcription factors controlling pluripotency, growing embryoid bodies from these cells, and their differentiation into three main germ layers. The main accomplishment of this work was revealing these pluripotency profiles in the cells acquired directly from the primary tumors. Before this work, the data concerned with the cell surface displayed biomarkers of pluripotent cells were studied on: cell cultures and fixed tissues. However, the biomarkers’ display is critically dependent on the environment, in which these cells are growing. For example, it was determined early on, that the cells in cultures change their molecular display profiles depending on the conditions including cell density, levels of oxygen, feeder layers, etc. Moreover, these cells radically change their surface display profiles in vitro and in vivo. Furthermore, the human stem cells have entirely different cell surface profiles than other species. For these reasons, all cultures of pluripotent stem cell lines need validation through revealing pluripotent cells’ gene products and transcripts, formation of embryoid bodies, differentiating in vitro into three germ layers. On the other hand, processing of tissues, for histopathology and immunohistochemistry, changes their antigenicity. Therefore, the results of the works on fixed tissues may not be applicable for streamlining in personalized diagnosis and therapy, but rather require validation on the directly biopsied living cells, as we have done herein.
Meanwhile, targeting contrast agents for molecular imaging in vivo, in situ of specific cancer cells and cancer stem cells are not yet available. In this project, we determined the display intensity profiles on the living, pluripotent cancer cells, using superparamagnetic and fluorescent, genetically engineered Fvs, which allowed us to determine the gene expression products on living cells, while using modern technologies of EDXS, TRXFS, and NMRFS. Our protocols, which utilize genetically bioengineered biotags targeting cancer specific biomarkers on living cells, are applicable for newest technologies of non-invasive in vitro diagnosis. These include TRXRFS, EDXS, and NMRS. However, the superparamagnetic and fluorescent properties of our Fvs tested in vitro, open the routes for minimally invasive diagnoses in vivo, while using these Fvs as the targeted contrast agents. Therefore, from the point of view of diagnostic accuracy and expedience of the diagnostic processes, the approach, which we promote in this study, probing living cells acquired directly from the tumor, is far more reliable for streamlining into clinical trials. We did not show in vivo images yet, but we did show specificity and sensitivity of these genetically engineered Fvs. Specifically, we demonstrated significant increase in relaxivity induced by the superparamagnetic Fvs in NMRS. For imaging purposes in vivo, it translates into an effective targeted contrast agent for MRI. We also demonstrated their ability to fluoresce. For imaging purposes, it translates into the in vivo imaging fluorescence opportunities, while considering options for deconvolution of the signal due to scattering and absorption by tissues. Specifically, we also demonstrated emission of the element specific energy. For imaging, it is applicable for the x-ray based imaging modalities. These novel molecular biology approaches promoted quantification of the data without radioactive or toxic compounds. Moreover, by bioengineering the probes specific for pluripotent stem cells, we created the foundation for molecular imaging of cancer and pluripotent stem cells in vivo.
The cells studied herein were enriched by selection for the pluripotency biomarkers displayed on surfaces of the living cells. Thereafter, these selected cells were characterized. The tumors consist of very heterogeneous populations of cells. It is also the case with the germ cells tumors of the testes. While embryonal carcinomas in their pure forms constitute only 20% of the tumors, almost 80% of all the tumors of the testes contain admixes of embryonal carcinoma cells. In those populations, only approximately 2% are SSEA-5+, which are particularly capable for developing teratomas and their separation reduces that risk. In general, the aforementioned clones of cells have very different characteristics. This is reflected in the diagnostic pathology, when the percentage of the embryonal carcinoma cells, contributing to the total tumor mass, serves for diagnosis of malignancy and prognosis of progression. An attempt to address this problem is laser assisted tissue dissection. It facilitates conducting molecular analysis on a more selective manner, i.e., on the tumor sections’ selected areas of the fixed and embedded or frozen tissues. Although due to processing, molecules, in those fixed tissues, come in an altered form anyway. To address these problems, we and others promote diagnostic works on the spectra on single, isolated living cells. This approach allows us to create a composite image of the clones propelling the tumor expansion. This approach serves as the foundation for crafting personalized therapy.
Similarities of the gene expression profiles of the cultured cells in the testicular embryonal carcinoma lines derived from the cultured metastases of the testicular teratomas to the lungs, the cultured cells from the human embryonic stem cell lines derived from the inner mass of blastocysts, the spermatogonia of the normal testes, the cells acquired directly from the primary tumor of the human embryonal carcinoma of the ovaries, with the profiles of the primary embryonal carcinomas of the testes revealed herein, indicate that these neoplasms originate from the cells corresponding to those, which are at the early stages of differentiation. They are also known as tumor initiating cells (TICs) [56, 66–71, 90, 91]. This has important clinical consequences. Stem cells exhibited resistance to radiotherapy [114–115]. Therefore, these cells present in the mixed tumors may survive therapy and become the sources of relapses. Moreover, radiation therapy may induce reprogramming of the cells, while leading to generation of more malignant cancers [116–117]. Nonseminomatous testicular germ cell tumors (NSGCTs), including embryonal carcinomas, were sensitive to cisplatin based chemotherapy [42–46,118]. However, in some cases, they develop resistance [119–121]. One of the mechanisms is loss of expression of OCT4 transcription factor [121]. This change may drive the relapses. Therefore, from the clinical point of view, identification and isolation of pluripotent stem cells, followed by characterization of their functional properties may help in improving effectiveness of therapy.
Conclusion
In the pure embryonal carcinomas of the testes, acquired directly from the patients, we identified, isolated with high viability and selectivity, and profiled the clones of the pluripotent stem cells. These results may help in explaining therapy-resistance and relapses of these neoplasms, as well as, in designing targeted, personalized therapy.
Figure 9.
NANOG gene expression products’ concentrations were determined as described in the figure 10.
Acknowledgments
First of all, we thank the patients for their consent. We thank for providing primers, hexamers, monoclonal antibodies, and cultures of the embryonic stem cells and the testicular embryonal carcinoma cells by Dr. J. Antosiewicz, Dr M. L. Greaser, Dr T. Kunicki, Dr J.V. Small, Dr. J. Swiergiel, Dr. W. Szybalski, and Dr. J. Thomson. We acknowledge with thanks access to the NIH National Nuclear Magnetic Resonance Facility, the SDSU Functional Genomics Center, Bruker AXS Laboratories, Bruker Optics Laboratories, and the NIH IMR National Biotechnology Resource. We also thank Dr I. Damjanov, Dr S. Jeffrey, Dr. J. Langmore, Dr. E. Lianidou, Dr J. Markley, Dr. P. Paterlini, Dr. E. Rajpert-DeMeyts, Dr D. Solter, and Dr W. Szybalski for the comments.
This work was supported by the funds from the NIH, NCRR, GM103399, RR000570, from the NSF 9420056, 9522771, 9902020, and 0094016, and from the PBMEF. The work was pursued at the National Biotechnology Resource, NIH, the Molecular Imaging Laboratory, UCSD, the National Biomedical NMR Resource, NIH, McArdle Cancer Research Laboratories, UW, the PBMEF, the BioSpin, the Bruker Optics, and the Genomics Center, SDSU; therefore the access to the instrumentation at those laboratories is gratefully acknowledged.
Abbreviations
- GCT
germ cell tumor
- ECT
embryonal carcinoma of the testis
- ECO
embryonal carcinoma of the ovary
- OCT4A (aka POU5F1)
Octamer-binding transcription factor 4A (aka POU-type homeodomain – containing DNA binding protein (on ch6)
- SOX2
SRY (sex determining region Y)-box 2 (on ch3)
- NANOG
homeobox transcription factor Nanog (on ch12)
- scFv
single chain variable fragment antibody
- dcFv
dual chain variable fragment antibody
- FCM
flow cytometry
- IB
immunoblotting
- MACS
magnetically activated cell sorting
- FACS
fluorescently activated cell sorting
- NMRS
nuclear magnetic resonance spectroscopy
- TRXFS
total reflection x-ray fluorescence spectroscopy
- MPFS
multiphoton fluorescence spectroscopy
- EELS
electron energy loss spectroscopy
- EDXS
energy dispersive x-ray spectroscopy
Footnotes
Preliminary results of this work were presented at the 62nd Annual Meeting of the American Society of Human Genetics, November 6–10, 2012, San Francisco, California.
Conflict of interest
The authors state no conflict of interest. Dr Marek Malecki owns the IP for the Fv gene coding sequences, gene transcripts, and gene expression products used in this work, as well as their streamlining to diagnosis and therapy, all protected at the USPTO and the WIPO.
Bibliography
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics. 2012. CA 1 Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M, et al., editors. SEER Cancer Statistics Review, 1975–2009. Bethesda, MD: National Cancer Institute; 2009. http://seer.cancer.gov/csr/1975_2009_pops09. [Google Scholar]
- 3.Garner MJ, Turner MC, Ghadirian P, et al. Epidemiology of testicular cancer: an overview. Int J Cancer. 2005;116(3):331–339. doi: 10.1002/ijc.21032. [DOI] [PubMed] [Google Scholar]
- 4.Pierce GB, Abell MR. Embryonal carcinoma of the testis. Pathol Annu. 1970;5:27–60. [PubMed] [Google Scholar]
- 5.Bosl GJ, Motzer RJ. Testicular germ-cell cancer. N Engl J Med. 1997;337(4):242–253. doi: 10.1056/NEJM199707243370406. [DOI] [PubMed] [Google Scholar]
- 6.Horwich A, Shipley J, Huddart R. Testicular germ-cell cancer. Lancet. 2006;367(9512):754–765. doi: 10.1016/S0140-6736(06)68305-0. [DOI] [PubMed] [Google Scholar]
- 7.McGlynn KA, Cook MB. Globally, testicular cancer incidence is highest among men of northern European ancestry and lowest among men of Asian and African descent. Future Oncol. 2009;5(9):1389–1402. doi: 10.2217/fon.09.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fosså SD, Kravdal O. Fertility in Norwegian testicular cancer patients. Br J Cancer. 2000;82(3):737–741. doi: 10.1054/bjoc.1999.0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giwercman A, Carlsen E, Keiding N, et al. Evidence for increasing incidence of abnormalities of the human testis: a review. Environ Health Perspect. 1993;(suppl 2)(101):65–71. doi: 10.1289/ehp.93101s265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobsen R, Bostofte E, Engholm G, et al. Risk of testicular cancer in men with abnormal semen characteristics. BMJ. 2000;321(7264):789–792. doi: 10.1136/bmj.321.7264.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strohsnitter WC, Noller KL, Hoover RN, et al. Cancer risk in men exposed in utero to diethylstilbestrol. J NatlCancer Inst. 2001;93(7):545–551. doi: 10.1093/jnci/93.7.545. [DOI] [PubMed] [Google Scholar]
- 12.Coupland CA, Forman D, Chilvers CE, et al. Maternal risk factors for testicular cancer: a population-based case-control study (UK) Cancer Causes Control. 2004;15(3):277–283. doi: 10.1023/B:CACO.0000024257.49409.1f. [DOI] [PubMed] [Google Scholar]
- 13.Weir HK, Marrett LD, Kreiger N, et al. Pre-natal and peri-natal exposures and risk of testicular germ-cell cancer. Int J Cancer. 2000;87(3):438–443. doi: 10.1002/1097-0215(20000801)87:3<438::aid-ijc20>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 14.Jensen TK, Jørgensen N, Punab M, et al. Association of in utero exposure to maternal smoking with reduced semen quality and testis size in adulthood. Am J Epidemiol. 2004;159(1):49–58. doi: 10.1093/aje/kwh002. [DOI] [PubMed] [Google Scholar]
- 15.Srivastava A, Kreiger N. Cigarette smoking and testicular cancer. Cancer Epidemiol Biomarkers Prev. 2004;13(1):49–54. doi: 10.1158/1055-9965.epi-03-0133. [DOI] [PubMed] [Google Scholar]
- 16.Travis LB, Beard C, Allan JM, et al. Testicular cancer survivorship: research strategies and recommendations. J Natl Cancer Inst. 2010;102(15):1114–1130. doi: 10.1093/jnci/djq216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haugnes HS, Bosl GJ, Boer H, et al. Long-term and late effects of germ cell testicular cancer treatment and implications for follow-up. J Clin Oncol. 2012;30(30):3752–3763. doi: 10.1200/JCO.2012.43.4431. [DOI] [PubMed] [Google Scholar]
- 18.Jakob A, Kollmannsberger C, Kanz L, et al. Late toxicity after chemotherapy of malignant testicular tumors. Urologe A. 1998;37(6):635–647. doi: 10.1007/s001200050227. [DOI] [PubMed] [Google Scholar]
- 19.Lampe H, Horwich A, Norman A, et al. Fertility after chemotherapy for testicular germ cell cancers. J Clin Oncol. 1997;15(1):239–245. doi: 10.1200/JCO.1997.15.1.239. [DOI] [PubMed] [Google Scholar]
- 20.Huddart RA, Norman A, Shahidi M, et al. Cardiovascular disease as a long-term complication of treatment for testicular cancer. J Clin Oncol. 2003;21(8):1513–1523. doi: 10.1200/JCO.2003.04.173. [DOI] [PubMed] [Google Scholar]
- 21.Meinardi MT, Gietema JA, van der Graaf WT, et al. Cardiovascular morbidity in long-term survivors of metastatic testicular cancer. J Clin Oncol. 2000;18(8):1725–1732. doi: 10.1200/JCO.2000.18.8.1725. [DOI] [PubMed] [Google Scholar]
- 22.Fosså SD, Aass N, Harvei S, et al. Increased mortality rates in young and middle-aged patients with malignant germ cell tumours. Br J Cancer. 2004;90(3):607–612. doi: 10.1038/sj.bjc.6601558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huddart RA, Norman A, Moynihan C, et al. Fertility, gonadal and sexual function in survivors of testicular cancer. Br J Cancer. 2005;93(2):200–207. doi: 10.1038/sj.bjc.6602677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Travis LB, Curtis RE, Storm H, et al. Risk of second malignant neoplasms among long-term survivors of testicular cancer. J Natl Cancer Inst. 1997;89(19):1429–1439. doi: 10.1093/jnci/89.19.1429. [DOI] [PubMed] [Google Scholar]
- 25.American Cancer Society. Second cancers caused by cancer treatment. http://www.cancer.org/acs/groups/cid/documents/webcontent/002043-pdf.
- 26.Skakkebaek NE. Possible carcinoma-in-situ of the testis. Lancet. 1972;2:516–517. doi: 10.1016/s0140-6736(72)91909-5. [DOI] [PubMed] [Google Scholar]
- 27.Tseng A, Homing SJ, Freiha FS, et al. Gynecomastia in testicular cancer patients. Prognostic and therapeutic implications. Cancer. 1985;56(10):2534–2538. doi: 10.1002/1097-0142(19851115)56:10<2534::aid-cncr2820561036>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 28.Kanto S, Hiramatsu M, Takeuchi A, et al. Carcinoma in situ detected by contralateral testicular biopsy of 55 germ cell tumor patients. Nihon Hinyokika Gakkai Zasshi. 2004;95(1):35–41. doi: 10.5980/jpnjurol1989.95.35. [DOI] [PubMed] [Google Scholar]
- 29.Fosså SD, Chen J, Schonfeld SJ, et al. Risk of contralateral testicular cancer: a population-based study of 29,515 U.S. men. J Natl Cancer Inst. 2005;97(14):1056–1066. doi: 10.1093/jnci/dji185. [DOI] [PubMed] [Google Scholar]
- 30.Harland SJ, Cook PA, Fossa SD, et al. Intratubular germ cell neoplasia of the contralateral testis in testicular cancer: defining a high risk group. J Urol. 1998;160(4):1353–1357. [PubMed] [Google Scholar]
- 31.Rodriguez PN, Hafez GR, Messing EM. Nonseminomatous germ cell tumor of the testicle: does extensive staging of the primary tumor predict the likelihood of metastatic disease? J Urol. 1986;136(3):604–608. doi: 10.1016/s0022-5347(17)44988-3. [DOI] [PubMed] [Google Scholar]
- 32.Gels ME, Hoekstra HJ, Sleijfer DT, et al. Detection of recurrence in patients with clinical stage I nonseminomatous testicular germ cell tumors and consequences for further follow-up. J Clin Oncol. 1995;13(5):1188–1194. doi: 10.1200/JCO.1995.13.5.1188. [DOI] [PubMed] [Google Scholar]
- 33.Aganovic L, Cassidy F. Imaging of the scrotum. Radiol Clin North Am. 2012;50(6):1145–1165. doi: 10.1016/j.rcl.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 34.Tsili AC, Argyropoulou MI, Astrakas LG, et al. Dynamic contrast-enhanced subtraction MRI for characterizing intratesticular mass lesions. AJR Am J Roentgenol. 2013;200(3):578–585. doi: 10.2214/AJR.12.9064. [DOI] [PubMed] [Google Scholar]
- 35.von Eyben FE, de Graaff WE, Marrink J, et al. Serum lactate dehydrogenase isoenzyme 1 activity in patients with testicular germ cell tumors correlates with the total number of copies of the short arm of ch 12 in the tumor. Mol Gen Genet. 1992;235(1):140–146. doi: 10.1007/BF00286191. [DOI] [PubMed] [Google Scholar]
- 36.Looijenga LH, Gillis AJ, Stoop HJ, et al. Chromosomes and expression in human testicular germ-cell tumors: insight into their cell of origin and pathogenesis. Ann N Y Acad Sci. 2007;1120:187–214. doi: 10.1196/annals.1411.000. [DOI] [PubMed] [Google Scholar]
- 37.Gels ME, Marrink J, Visser P, et al. Importance of a new tumor marker TRA-1–60 in the follow-up of patients with clinical stage I nonseminomatous testicular germ cell tumors. Ann Surg Oncol. 1997;4(4):321–327. doi: 10.1007/BF02303582. [DOI] [PubMed] [Google Scholar]
- 38.Ulbright TM. Germ cell tumors of the gonads: a selective review emphasizing problems in differential diagnosis, newly appreciated, and controversial issues. Mod Pathol. 2005;18S2:S61–S79. doi: 10.1038/modpathol.3800310. [DOI] [PubMed] [Google Scholar]
- 39.Ye H, Ulbright TM. Difficult differential diagnoses in testicular pathology. Arch Pathol Lab Med. 2012;136(4):435–446. doi: 10.5858/arpa.2011-0475-RA. [DOI] [PubMed] [Google Scholar]
- 40.Karellas ME, Damjanov I, Holzbeierlein JM. ITGCN of the testis, contralateral testicular biopsy and bilateral testicular cancer. Urol Clin North Am. 2007;34(2):119–125. doi: 10.1016/j.ucl.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 41.Giwercman A, Andrews PW, Jørgensen N, et al. Immunohistochemical expression of embryonal marker TRA-1–60 in carcinoma in situ and germ cell tumors of the testis. Cancer. 1993;72(4):1308–1314. doi: 10.1002/1097-0142(19930815)72:4<1308::aid-cncr2820720426>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 42.Dieckmann KP, Wilken S, Loy V, et al. Treatment of testicular intraepithelial neoplasia with local radiotherapy or with platinum-based chemotherapy. Ann Oncol. 2013 doi: 10.1093/annonc/mds628. {ahead of press) [DOI] [PubMed] [Google Scholar]
- 43.Fléchon A, Culine S, Droz JP. Intensive and timely chemotherapy, the key of success in testicular cancer. Crit Rev Oncol Hematol. 2001;37(1):35–46. doi: 10.1016/s1040-8428(00)00074-3. [DOI] [PubMed] [Google Scholar]
- 44.Voss MH, Feldman DR, Motzer RJ. High-dose chemotherapy and stem cell transplantation for advanced testicular cancer. Expert Rev Anticancer Ther. 2011;11(7):1091–1103. doi: 10.1586/era.10.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Voss MH, Feldman DR, Bosl GJ, et al. A review of second-line chemotherapy and prognostic models for disseminated germ cell tumors. Hematol Oncol Clin North Am. 2011;25(3):557–576. doi: 10.1016/j.hoc.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shin YS, Kim HJ. Current management of testicular cancer. Korean J Urol. 2013;54(1):2–10. doi: 10.4111/kju.2013.54.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pajonk F, Vlashi E, McBride WH. Radiation resistance of cancer stem cells: the 4 R's of radiobiology revisited. Stem Cells. 2010;28(4):639–648. doi: 10.1002/stem.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Giwercman A, von der Maase H, Berthelsen J, G, et al. Localized irradiation of testes with carcinoma in situ. J Clin Endocrinol Metab. 1991;73:596–603. doi: 10.1210/jcem-73-3-596. [DOI] [PubMed] [Google Scholar]
- 49.Oosterhuis JW, Andrews PW, Knowles BB, et al. Effects of cis-platinum on embryonal carcinoma cell lines in vitro. Int J Cancer. 1984;34(1):133–139. doi: 10.1002/ijc.2910340123. [DOI] [PubMed] [Google Scholar]
- 50.Kannagi R, Cochran NA, Ishigami F, et al. Stage-specific embryonic antigens (SSEA-3 and -4) are epitopes of a unique globo-series ganglioside isolated from human teratocarcinoma cells. EMBO J. 1983;2(12):2355–2361. doi: 10.1002/j.1460-2075.1983.tb01746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shevinsky LH, Knowles BB, Damjanov I, Solter D. A stagespecific embryonic antigen defined by monoclonal antibody to murine embryos, expressed on mouse embryos and human teratocarcinoma cells. Cell. 1982;30:697–705. doi: 10.1016/0092-8674(82)90274-4. [DOI] [PubMed] [Google Scholar]
- 52.Knowles BB, Aden DP, Solter D. Monoclonal antibody detecting a stage-specific embryonic antigen (SSEA-1) on preimplantation mouse embryos and teratocarcinoma cells. Curr Top Microbiol Immunol. 1978;8:51–53. doi: 10.1007/978-3-642-67448-8_8. [DOI] [PubMed] [Google Scholar]
- 53.Andrews PW, Banting G, Damjanov I, et al. Three monoclonal antibodies defining distinct differentiation antigens associated with different high molecular weight polypeptides on the surface of human embryonal carcinoma cells. Hybridoma. 1984;3(4):347–361. doi: 10.1089/hyb.1984.3.347. [DOI] [PubMed] [Google Scholar]
- 54.Andrews PW, Fenderson B, Hakomori S. Human embryonal carcinoma cells and their differentiation in culture. Int J Androl. 1987;10(1):95–104. doi: 10.1111/j.1365-2605.1987.tb00170.x. [DOI] [PubMed] [Google Scholar]
- 55.Andrews PW, Goodfellow PN, Shevinsky LH, et al. Cell-surface antigens of a clonal human embryonal carcinoma cell line: morphological and antigenic differentiation in culture. Int J Cancer. 1982;29(5):523–531. doi: 10.1002/ijc.2910290507. [DOI] [PubMed] [Google Scholar]
- 56.Andrews PW, Casper J, Damjanov I, et al. Comparative analysis of cell surface antigens expressed by cell lines derived from human germ cell tumours. Int J Cancer. 1996;66(6):806–816. doi: 10.1002/(SICI)1097-0215(19960611)66:6<806::AID-IJC17>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 57.Andrews PW, Damjanov I, Simon D, et al. A pluripotent human stem-cell clone isolated from the TERA-2 teratocarcinoma line lacks antigens SSEA-3 and SSEA-4 in vitro, but expresses these antigens when grown as a xenograft tumor. Differentiation. 1985;29(2):127–135. doi: 10.1111/j.1432-0436.1985.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 58.Andrews PW, Damjanov I, Simon D, et al. Pluripotent embryonal carcinoma clones derived from the human teratocarcinoma cell line Tera-2. Differentiation in vivo and in vitro. Lab Invest. 1984;50(2):147–162. [PubMed] [Google Scholar]
- 59.Matthaei KI, Andrews PW, Bronson DL. Retinoic acid fails to induce differentiation in human teratocarcinoma cell lines that express high levels of a cellular receptor protein. Exp Cell Res. 1983;143(2):471–474. doi: 10.1016/0014-4827(83)90076-9. [DOI] [PubMed] [Google Scholar]
- 60.Simeone A, Acampora D, Arcioni L, et al. Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature. 1990;346(6286):763–766. doi: 10.1038/346763a0. [DOI] [PubMed] [Google Scholar]
- 61.Lee VM, Andrews PW. Differentiation of NTERA-2 clonal human embryonal carcinoma cells into neurons involves the induction of all three neurofilament proteins. J Neurosci. 1986;6(2):514–521. doi: 10.1523/JNEUROSCI.06-02-00514.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Andrews PW, G ncz l E, Plotkin SA, et al. Differentiation of TERA-2 human embryonal carcinoma cells into neurons and HCMV permissive cells. Induction by agents other than retinoic acid. Differentiation. 1986;31(2):119–126. doi: 10.1111/j.1432-0436.1986.tb00392.x. [DOI] [PubMed] [Google Scholar]
- 63.Damjanov I, Horvat B, Gibas Z. Retinoic acid-induced differentiation of the developmentally pluripotent human germ cell tumor-derived cell line, NCCIT. Lab Invest. 1993;68(2):220–232. [PubMed] [Google Scholar]
- 64.Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282(5391):1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 65.Draper JS, Pigott C, Thomson JA, et al. Surface antigens of human embryonic stem cells: changes upon differentiation in culture. J Anat. 2002;200(Pt 3):249–258. doi: 10.1046/j.1469-7580.2002.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sperger JM, Chen X, Draper JS, et al. Gene expression patterns in human embryonic stem cells and human pluripotent germ cell tumors. Proc Natl Acad Sci U S A. 2003;100(23):13350–13355. doi: 10.1073/pnas.2235735100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yu J, Thomson JA. Pluripotent stem cell lines. Genes Dev. 2008;22(15):1987–1997. doi: 10.1101/gad.1689808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chaerkady R, Kerr CL, Kandasamy K, et al. Comparative proteomics of human embryonic stem cells and embryonal carcinoma cells. Proteomics. 2010;10(7):1359–1373. doi: 10.1002/pmic.200900483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pashai N, Hao H, All A, et al. Genome-wide profiling of pluripotent cells reveals a unique molecular signature of human embryonic germ cells. PLoS One. 2012;7(6):e39088. doi: 10.1371/journal.pone.0039088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Itskovitz-Eldor J, Schuldiner M, Karsenti D. Differentiation of human embryonic stem cells into embryoid bodies comprising the three embryonic germ layers. Mol Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 71.Amit M, Itskovitz-Eldor J. Derivation and spontaneous differentiation of human embryonic stem cells. J Anat. 2002;200:225–232. doi: 10.1046/j.1469-7580.2002.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kerr CL, Hill CM, Blumenthal PD, Gearhart JD. Expression of pluripotent stem cell markers in the human fetal testis. Stem Cells. 2008;26(2):412–421. doi: 10.1634/stemcells.2007-0605. [DOI] [PubMed] [Google Scholar]
- 73.Malecki M, Pietruszkiewicz J, Ratajczak M, et al. Nanoengineered antibodies against embryonal carcinoma's SSEA-3, SSEA-4, TRA-1–60, and TRA-1–81 demonstrate presence of pluripotent cells in situ, in healthy human bone marrow. Eur J Surg Onc. 2006;32:1. [Google Scholar]
- 74.Pallesen G, Hamilton-Dutoit SJ. Ki-1 (CD30) antigen is regularly expressed by tumor cells of embryonal carcinoma. Am J Pathol. 1988;133(3):446–450. [PMC free article] [PubMed] [Google Scholar]
- 75.Leroy X, Augusto D, Leteurtre E, et al. CD30 and CD117 (c-kit) used in combination are useful for distinguishing embryonal carcinoma from seminoma. J Histochem Cytochem. 2002;50(2):283–285. doi: 10.1177/002215540205000216. [DOI] [PubMed] [Google Scholar]
- 76.Rajpert-De Meyts E, Skakkebaek NE. The expression of CD44 adhesion molecules on seminoma cells. A new marker for early detection of the tumor. Cancer. 1996;78(11):2443–2445. [PubMed] [Google Scholar]
- 77.Hadziselimovic F, Herzog B, Emmons LR. The expression of CD44 adhesion molecules on seminoma cells. A new marker for early detection of the tumor. Cancer. 1996;77(3):429–430. doi: 10.1002/(SICI)1097-0142(19960201)77:3<429::AID-CNCR1>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 78.Miyake H, Hara I, Yamanaka K, et al. Expression patterns of CD44 adhesion molecule in testicular germ cell tumors and normal testes. Am J Pathol. 1998;152(5):1157–1160. [PMC free article] [PubMed] [Google Scholar]
- 79.Looijenga LH, Hersmus R, Gillis AJ, et al. Genomic and expression profiling of human spermatocytic seminomas: primary spermatocyte as tumorigenic precursor and DMRT1 as candidate chromosome 9 gene. Cancer Res. 2006;66(1):290–302. doi: 10.1158/0008-5472.CAN-05-2936. [DOI] [PubMed] [Google Scholar]
- 80.Gashaw I, Dushaj O, Behr R, et al. Novel germ cell markers characterize testicular seminoma and fetal testis. Mol Hum Reprod. 2007;13(10):721–727. doi: 10.1093/molehr/gam059. [DOI] [PubMed] [Google Scholar]
- 81.Tang C, Lee AS, Volkmer JP, et al. An antibody against SSEA-5 glycan on human pluripotent stem cells enables removal of teratoma-forming cells. Nat Biotechnol. 2011;29(9):829–834. doi: 10.1038/nbt.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andrews PW, Bronson DL, Wiles MV, et al. The expression of MHC antigens by human teratocarcinoma derived cell lines. Tissue Antigens. 1981;17(5):493–500. doi: 10.1111/j.1399-0039.1981.tb00736.x. [DOI] [PubMed] [Google Scholar]
- 83.Rajpert-De Meyts E, Kvist M, Skakkebaek NE. Heterogeneity of expression of immunohistochemical tumour markers in testicular carcinoma in situ: pathogenetic relevance. Virchows Arch. 1996;428(3):133–139. doi: 10.1007/BF00200655. [DOI] [PubMed] [Google Scholar]
- 84.Jørgensen N, Rajpert-De Meyts E, Graem N, et al. Expression of immunohistochemical markers for testicular carcinoma in situ by normal human fetal germ cells. Lab Invest. 1995;72(2):223–231. [PubMed] [Google Scholar]
- 85.J rgensen N, Giwercman A, M ller J, et al. Immunohistochemical markers of carcinoma in situ of the testis also expressed in normal infantile germ cells. Histopathology. 1993;22(4):373–378. doi: 10.1111/j.1365-2559.1993.tb00138.x. [DOI] [PubMed] [Google Scholar]
- 86.Jørgensen N, Müller J, Jaubert F, et al. Heterogeneity of gonadoblastoma germ cells: similarities with immature germ cells, spermatogonia and testicular carcinoma in situ cells. Histopathology. 1997;30(2):177–186. doi: 10.1046/j.1365-2559.1997.d01-580.x. [DOI] [PubMed] [Google Scholar]
- 87.Tokuyama S, Saito S, Takahashi T, et al. Immunostaining of stage-specific embryonic antigen-4 in intratubular germ cell neoplasia unclassified and in testicular germ-cell tumors. Oncol Rep. 2003;10(5):1097–1104. [PubMed] [Google Scholar]
- 88.Liu S, Tang Z, Xiong T, et al. Isolation and characterization of human spermatogonial stem cells. Reprod Biol Endocrinol. 2011;9:141. doi: 10.1186/1477-7827-9-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Izadyar F, Wong J, Maki C, et al. Identification and characterization of repopulating spermatogonial stem cells from the adult human testis. Hum Reprod. 2011;26(6):1296–1306. doi: 10.1093/humrep/der026. [DOI] [PubMed] [Google Scholar]
- 90.Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;30;131(5):861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 91.Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;21;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 92.Nakagawa M, Koyanagi M, Tanabe K, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26(1):101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 93.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17(1):42–49. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 94.Malecki M, Anderson M, Beauchaine M, et al. TRA-1–60(+), SSEA-4(+), Oct4A(+), Nanog(+) Clones of Pluripotent Stem Cells in the Embryonal Carcinomas of the Ovaries. J Stem Cell Res Ther. 2012;2(5):130–140. [PMC free article] [PubMed] [Google Scholar]
- 95.Ezeh UI, Turek PJ, Reijo RA, et al. Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are expressed in both seminoma and breast carcinoma. Cancer. 2005;104(10):2255–2265. doi: 10.1002/cncr.21432. [DOI] [PubMed] [Google Scholar]
- 96.Gillis AJ, Stoop H, Biermann K, et al. Expression and interdependencies of pluripotency factors LIN28, OCT3/4, NANOG and SOX2 in human testicular germ cells and tumours of the testis. Int J Androl. 2011;34(4Pt2):e160–e174. doi: 10.1111/j.1365-2605.2011.01148.x. [DOI] [PubMed] [Google Scholar]
- 97.Almstrup K, Hoei-Hansen CE, Wirkner U, et al. Embryonic stem cell-like features of testicular carcinoma in situ revealed by genome-wide gene expression profiling. Cancer Res. 2004;64(14):4736–4743. doi: 10.1158/0008-5472.CAN-04-0679. [DOI] [PubMed] [Google Scholar]
- 98.Bosl GJ, Lange PH, Nochomovitz LE, et al. Tumor markers in advanced nonseminomatous testicular cancer. Cancer. 1981;47(3):572–576. doi: 10.1002/1097-0142(19810201)47:3<572::aid-cncr2820470324>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 99.De Jong J, Stoop H, Dohle GR, et al. Diagnostic value of OCT3/4 for preinvasive and invasive testicular germ cell tumours. J Pathol. 2005;206:242–249. doi: 10.1002/path.1766. [DOI] [PubMed] [Google Scholar]
- 100.Rajpert-De Meyts E, Hanstein R, Jorgensen N, et al. Developmental expression of POU5F1 (OCT- 3/4) in normal and dysgenetic human gonads. Hum Reprod. 2004;19:1338–1344. doi: 10.1093/humrep/deh265. [DOI] [PubMed] [Google Scholar]
- 101.Jones TD, Ulbright TM, Eble JN, et al. OCT4 staining in testicular tumors: a sensitive and specific marker for seminoma and embryonal carcinoma. Am J Surg Pathol. 2004;28:935–940. doi: 10.1097/00000478-200407000-00014. [DOI] [PubMed] [Google Scholar]
- 102.Pruszak J, Sonntag KC, Aung MH, et al. Markers and methods for cell sorting of human embryonic stem cell-derived neural cell populations. Stem Cells. 2007;25(9):2257–2268. doi: 10.1634/stemcells.2006-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fong CY, Peh GS, Gauthaman K, et al. Separation of SSEA-4 and TRA-1–60 labelled undifferentiated human embryonic stem cells from a heterogeneous cell population. Stem Cell Rev. 2009;5(1):72–80. doi: 10.1007/s12015-009-9054-4. [DOI] [PubMed] [Google Scholar]
- 104.Mantzaris NV. From single-cell genetic architecture to cell population dynamics: quantitatively decomposing the effects of different population heterogeneity sources for a genetic network with positive feedback architecture. Biophys J. 2007;92(12):4271–4288. doi: 10.1529/biophysj.106.100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Erdmann J. Sequencing cells one at a time offers singular insight into cancer. Nat Med. 2012;18(6):842. doi: 10.1038/nm0612-842a. [DOI] [PubMed] [Google Scholar]
- 106.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kroneis T, Geigl JB, El-Heliebi A, et al. Combined molecular genetic and cytogenetic analysis from single cells after isothermal whole-genome amplification. Clin Chem. 2011;57(7):1032–1041. doi: 10.1373/clinchem.2011.162131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu X, Hou Y, Yin X, et al. Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell. 2012;148(5):886–895. doi: 10.1016/j.cell.2012.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Malecki M, Szybalski W. Isolation of single, intact chromosomes from single, selected ovarian cancer cells for in situ hybridization and sequencing. Gene. 2012;493(1):132–139. doi: 10.1016/j.gene.2011.11.044. [DOI] [PubMed] [Google Scholar]
- 110.Alymani NA, Smith MD, Williams DJ, et al. Predictive biomarkers for personalised anti-cancer drug use: discovery to clinical implementation. Eur J Cancer. 2010;46(5):869–879. doi: 10.1016/j.ejca.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 111.David L, Morse DL, Gillies RJ. Molecular Imaging and Targeted Therapies. Biochem Pharmacol. 2010;80(5):731–738. doi: 10.1016/j.bcp.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Malecki M, Hsu A, Truong L, et al. Molecular immunolabeling with recombinant single-chain variable fragment (scFv) antibodies designed with metal-binding domains. Proc Natl Acad Sci U S A. 2002;99(1):213–218. doi: 10.1073/pnas.261567298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Malecki M, Malecki B. Methods for Manufacturing and Using of Molecular Death Tags. WIPO. 2011:1–123. WO2011162904. [Google Scholar]
- 114.Bao S, Wu Q, McLendon RE, Hao Y, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 115.Phillips TM, McBride WH, Pajonk F. The response of CD24 −/low /CD44 + breast cancer–initiating cells to radiation. J Natl Cancer Inst. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 116.Lagadec C, Vlashi E, Della Donna L, et al. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012;30(5):833–844. doi: 10.1002/stem.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Printz C. Radiation treatment generates therapy-resistant cancer stem cells from less aggressive breast cancer cells. Cancer. 2012;118(13):3225. doi: 10.1002/cncr.27701. [DOI] [PubMed] [Google Scholar]
- 118.Heinzelbecker J, Katzmarzik M, Weiss C, et al. During twenty years of Cisplatin-based therapy the face of nonseminomatous testicular germ cell tumors is still changing: an evaluation of presentation, management, predictive factors and survival. Int Braz J Urol. 2013;39(1):10–21. doi: 10.1590/S1677-5538.IBJU.2013.01.03. [DOI] [PubMed] [Google Scholar]
- 119.Steg AD, Bevis KS, Katre AA, et al. Stem cell pathways contribute to clinical chemoresistance in ovarian cancer. Clin Cancer Res. 2012;18(3):869–881. doi: 10.1158/1078-0432.CCR-11-2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Abdullah LN, Chow EKH. Mechanisms of chemoresistance in cancer stem cells. Clinical and Translational Medicine. 2013;2:3–12. doi: 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mueller T, Mueller LP, Luetzkendorf J, et al. Loss of Oct-3/4 expression in embryonal carcinoma cells is associated with induction of cisplatin resistance. Tumour Biol. 2006;27(2):71–83. doi: 10.1159/000092324. [DOI] [PubMed] [Google Scholar]