Summary
To metastasize, a tumor cell must acquire abilities such as the capacity to colonize new tissue and evade immune surveillance. Recent evidence suggests that microRNAs can promote the evolution of malignant behaviors by regulating multiple targets. We performed a microRNA analysis of human melanoma, a highly invasive cancer, and found that miR-30b/30d upregulation correlates with stage, metastatic potential, shorter time to recurrence and reduced overall survival. Ectopic expression of miR-30b/30d promoted the metastatic behavior of melanoma cells by directly targeting the GalNAc transferase GALNT7, resulted in increased synthesis of the immunosuppressive cytokine IL-10, and reduced immune cell activation and recruitment. These data support a key role of miR-30b/30d and GalNAc transferases in metastasis, by simultaneously promoting cellular invasion and immunosuppression.
INTRODUCTION
Far more attention has been given to the process of malignant transformation than to metastasis, yet it is the spread of transformed cells that accounts for 90% of deaths from solid tumors (Gupta and Massague, 2006). The capacity of a tumor cell to metastasize depends upon its ability to escape the primary tumor, intravasate into circulation, survive transit, extravasate into distant tissue, and colonize it, while evading immune surveillance and promoting various changes to the local tissue environment (Gupta and Massague, 2006). The conventional view of tumor progression assumed that malignant cells evolve these aggressive functions over time, but we are beginning to appreciate that metastatic traits may be acquired earlier rather than later in oncogenesis (Gupta et al., 2005; Scheel et al., 2007; Talmadge, 2007).
Accumulating evidence suggests that alterations in microRNA (miRNA) expression might prove crucial in promoting metastasis (Croce and Calin, 2005; Ma et al., 2007; Ma et al.; Tavazoie et al., 2008). This is an intuitively compelling idea because miRNAs have been found to serve important regulatory functions during numerous developmental and pathological processes by altering multiple target genes, and therefore multiple cellular activities, simultaneously (Gupta and Massague, 2006). Expression profiling has identified miRNA signatures for a number of tumors that correlate with disease stage and clinical outcome (Calin and Croce, 2006). The extent to which these alterations in miRNA expression actually influence metastasis is difficult to decipher since in many cases the miRNAs exert confounding effects on cell growth and proliferation within the primary tumor (Tavazoie et al., 2008). Given both the importance of metastasis to cancer-associated lethality and our relatively tenuous grasp of how it is executed by tumor cells, we sought to investigate the role of miRNAs in one of the most invasive tumor types, melanoma.
RESULTS
Expression of miR-30b and 30d in human melanoma marks the progression from primary to metastatic tumors
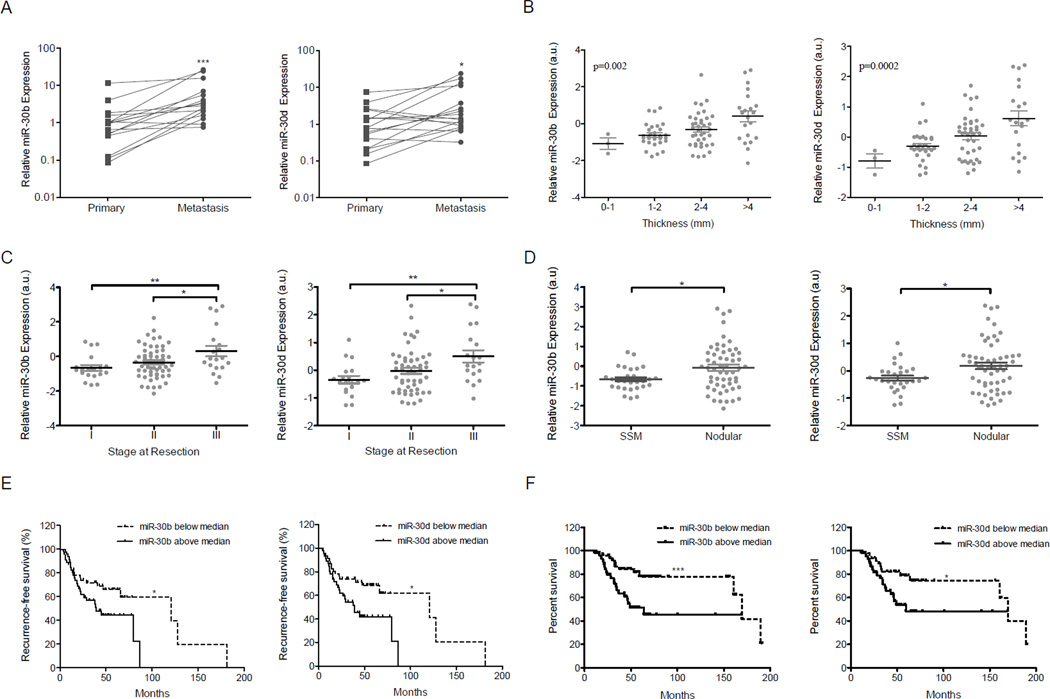
MiRNA array analysis of 59 metastatic melanoma tumor samples (Segura et al., 2010), followed by quantitative RT-PCR validation, revealed high expression levels of miR-30b and -30d. These two miRNAs form a cluster on 8q24, a common amplicon in melanoma (Ehlers et al., 2005). No statistically significant upregulation of miR-30b/30d was observed from congenital nevi to primary melanomas (Fig. S1A). However, in a subset of 17 paired samples (primary tumor and a metastasis from the same patient), we found a statistically significant increase in expression of these miRNAs from the primary to the metastatic stage (p=0.0007 for miR-30b, p=0.026 for miR-30d) (Figure 1A). A miRNA profile of primary melanomas (n=92; Table S1) revealed that higher levels of miR-30b and -30d corresponded with increased tumor thickness (p=0.002 for miR-30b, p=0.0002 for 30d; Fig. 1B) and advancing stage (I to III) (p=0.004 for miR-30b, p=0.001 for 30d; Fig. 1C), suggesting an association between miR-30b/d expression and tumor progression. By histological subtype, the more invasive nodular melanomas (NM) had higher miR-30b/30d levels than superficial spreading melanomas (SSM) (p=0.015 for miR-30b, p=0.0189 for 30d Fig. 1D). Furthermore, the subgroup of primary melanomas that had metastasized (n=44) showed higher levels of miR-30b and -30d expression than those that had not spread (n=48) during a period of 24 months or more of follow-up (p=0.048 for miR-30b, p=0.037 for miR-30d; data not shown). Accordingly, miR-30b and miR-30d levels above the median correlated with shorter time to recurrence (p=0.04 for miR-30b and p=0.01 for miR-30d Fig. 1E) and lower overall survival of melanoma patients (with p=0.0004 for miR-30b and p=0.02 for miR-30d; Fig. 1F). Multivariate analysis using COX PH models indicated that the expression level of miR-30d is a statistically significant independent predictor for melanoma mortality (p= 0.004) when adjusted for primary tumor thickness and ulceration status. The expression level of miR-30b is only marginally significant as an independent predictor for death with melanoma when adjusted for primary tumor thickness and ulceration (p= 0.054). These data support an association between miR-30b/30d upregulation and increased melanoma aggressiveness, and suggest a potential use of these miRNAs as prognostic biomarkers.
Fig. 1. miR-30b and miR-30d overexpression is associated with metastatic behavior in melanoma, shorter time to recurrence, and lower overall survival.
A. Increased relative levels of miR-30b and miR-30d in 17 metastatic cases compared to the levels in their matched primary tumors, as measured by quantitative RT-PCR. B–C. MiR-30b and miR-30d normalized array levels in 92 primary cases with (B) increased thickness and (C) increased stage. ANOVA test was applied in B. D. MiR-30b and miR-30d normalized array levels in superficial spreading melanomas (SSM; n=28) vs. nodular melanoma (NM; n=56). E–F. Graphs show shorter time to recurrence (E) and lower overall survival (F; n=92) in patients with high (above median value) as opposed to low (below median value) miR-30b/30d levels. (*p<0.05; ** p<0.01; ***p<0.001). See also Figure S1 and Table S1.
MiR-30b/30d overexpression correlates with genomic amplification in a subset of human melanoma samples
The miR-30b/30d cluster (8q24.22–8q24.23) is located in the vicinity of a genomic region containing the oncogene c-MYC (8q24.21), which is frequently amplified in multiple cancer types, including medulloblastoma (Lu et al., 2009); uveal melanoma (Ehlers et al., 2005); head, neck and cervical squamous cell carcinomas; bladder (Visapaa et al., 2003), lung and prostate cancer (Van Den Berg et al., 1995). c-MYC amplification is usually associated with tumor progression.
We found the miR-30b/30d genomic region amplified in 12 out of 33 metastatic melanoma tissues (36.4% of cases, Fig. S1B), of which approximately half harbored concomitant c-MYC gene copy gains (Fig. S1C), suggesting that the miR-30b/30d gains are generally independent of c-MYC amplification. Interestingly, we noted a higher fraction of patients carrying the miR-30b/30d amplification died within the study period (Fig. S1B), suggesting this genetic trait is associated with more aggressive disease.
MiR-30b or miR-30d modulation alters the invasive potential of melanoma cells without affecting cell proliferation
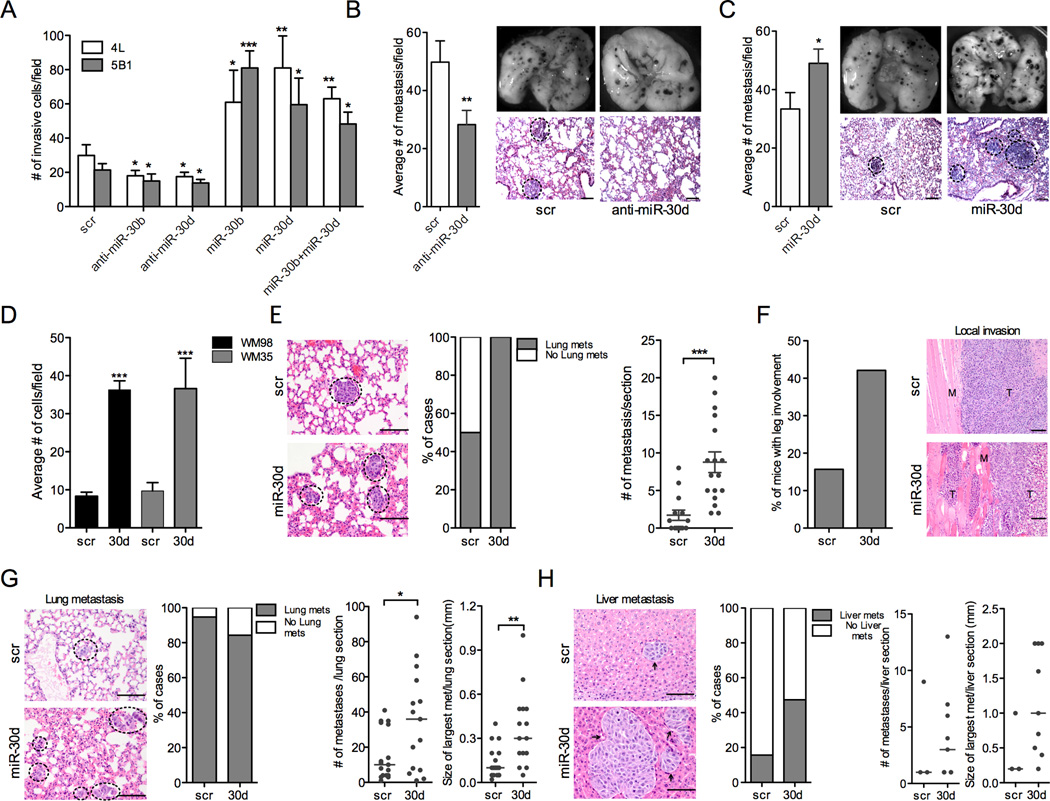
Since upregulation of miR-30b and 30d is associated with progression from primary to metastatic melanoma, we asked whether these miRNAs enhance the invasive behavior of melanoma cells. Using a fibronectin transwell invasion assay, we found that ectopic expression of miR-30b and 30d (Fig. S2A) strongly stimulated the invasive capacity of two metastatic melanoma cell lines, 113/6-4L (hereafter, 4L) and 131/4-5B1 (hereafter 5B1) (Cruz-Munoz et al., 2008) (Fig. 2A; p=0.037 and p=0.0002 for miR-30b and p=0.009 and p=0.011 for miR-30d in 4L and 5B1, respectively). In contrast, silencing of miR-30b or miR-30d by antisense oligonucleotide (anti-miR) transfection (Fig. S2A) suppressed cell migration (p=0.026 and p=0.032 for miR-30b; p=0.041 and p=0.044 for miR-30d in 4L and 5B1 respectively; Fig. 2A).
Fig. 2. MiR-30b and miR-30d promote melanoma invasion and metastasis in vitro and in vivo.
A. Transwell invasion assay of indicated cell lines with miR-30b, -30d, or both, either silenced or overexpressed (mean ±SEM). scr = scrambled control. B–C. In vivo metastasis assay with B16F10 mouse melanoma cells transfected with scr, anti-miR-30d or miR-30d mimics injected through the lateral tail vein of C57BL/6J mice. Histogram in (B) shows that anti-miR-30d suppressed metastasis, while miR-30d increased metastatic behavior (C); (mean ±SEM). Right: macroscopic pictures of mouse lungs and H&E-stained sections of lung metastases at termination of the experiment. Black dotted circles mark metastatic foci. D. Transwell invasion assay with primary melanoma cell lines WM98 and WM35 transduced with scrambled control or miR-30d (mean ±SEM). E. In vivo metastasis assay with WM98 melanoma cells stably transduced with GIPZ-scr or GIPZ-miR-30d injected through the lateral tail vein of NOG/SCID mice. Histogram shows the percentage of mice that developed lung metastases in each cohort. Whisker plots show the distribution of the number of metastases per section. Representative H&E-stained sections of lungs are shown. Scale bars represent 100µm. Black dotted circles mark metastatic foci. F–H. Pre-clinical model of human melanoma metastasis. 5B1 cells stably transduced with either scr or miR-30d vectors, were injected sub-cutaneously into the flanks of NOG/SCID mice. F. H&E-stained sections show increased local invasion of miR-30d-transduced tumors. Histogram represents the percentage of mice in each cohort with primary tumors that invaded grossly into the leg. G–H. Representative micrographs of H&E-stained sections of lungs (G) or livers (H). Histograms show the percentage of mice that developed lung or liver metastases in each cohort. Whisker plots show the distribution of the number or size (largest in each section) of metastasis per section. Bars represent the median value (*p<0.05; ** p<0.01; ***p<0.001). For all micrographs scale bars represent100µm. See also Fig. S2.
To determine whether the increase in invasive behavior could be explained, at least in part, by increased cell proliferation, we compared the growth rates of cells transduced with miR-30b or miR-30d or scrambled control. We found no statistically significant differences by means of trypan-blue exclusion or crystal violet staining (data not shown). Therefore, we conclude that miR-30b/30d increase melanoma cells’ capacity to migrate through the extracellular matrix, an essential ability for metastasis.
Next, since miR-30b and miR-30d are co-expressed from the same cluster, we tested the effect of inducing both simultaneously. Neither additive nor synergistic effects were detected in the Boyden chamber assay (Fig. 2A), indicating that the two miRNAs have redundant pro-invasive functions. This is not surprising, since they share the same seed region and thus likely operate through common targets.
MiR-/30b/30d overexpression enhances metastasis, whereas its silencing represses metastasis in vivo
Our in vitro results led us to study the impact of miR-30d downregulation in a classic in vivo model of lung metastasis: we transiently transduced B16F10 mouse melanoma cells in vitro with scrambled or anti-miR-30d oligonucleotides (Fig. S2B) and injected them into the tail veins of immunocompetent mice 8 to12 weeks of age. Eleven days post-injection we sacrificed the mice and dissected the lungs for macro- and microscopic histology. Lungs of B16F10/anti-miR-30d injected mice harbored significantly fewer micro- and macroscopic metastases than scramble control (p=0.0085; Fig. 2B). Conversely, mice injected with B16F10 cells transiently transduced with miR-30d mimic oligonucleotides generated more metastatic foci than control cells transfected with scrambled oligonucleotide (p=0.0218; Fig. 2C). Then, we compared the metastatic potential of B16 transiently transfected with miR-30b, miR-30d or combinations of miR-30b and 30d mimic oligonucleotides, injected through the tail vein. MiR-30d and miR-30b had similar pro-metastatic effects and the combination of the two showed only a slight increase over miR-30d alone (Fig. S3A–C). Therefore, both our in vitro and in vivo results indicate that miR-30b and miR-30d have redundant effects on invasion and metastasis. Given this functional redundancy, we focused primarily on miR-30d in the following experiments.
We asked whether miR-30d could confer metastatic potential to melanoma cells devoid of such ability, such as the primary melanoma cells WM35 and WM98. In vitro invasion assays revealed the ability of miR-30d to significantly enhance the invasive capacity of WM35 and WM98 primary human melanoma cells (Fig. 2D). In vivo, WM98 cells display very poor seeding and colonization of mouse lungs upon tail vein injection, but miR-30d upregulation dramatically increased both the incidence of lung metastasis and the total number of metastasis per lung section (Fig. 2E). These in vitro and in vivo results evidence the strong pro-metastatic potential of this miRNA.
Given the limitations of tail vein injection models at recapitulating all the steps of metastasis, we decided to test the effects of miR-30d in a more preclinical system, in which human melanoma cells are injected in the flanks of immunocompromised mice. These mice form a tumor mass within about 2 weeks, from which cells migrate and reach the lungs in 8–10 weeks with occasional spread to the liver and other organs (Cruz-Munoz et al., 2008). 5B1 cells stably transduced with lentiviruses carrying pre-miR-30d (GIPZ/miR-30d) or a scrambled sequence (GIPZ/scr) were inoculated in the flanks of NOD/Shi-scid/IL-2Rγnul (NOG/SCID) mice. Local muscle invasion involving the proximal leg was more commonly found among the GIPZ/miR-30d injected mice than in the GIPZ/scr group (Fig. 2F). Moreover, the proportion of mice that developed liver metastases at completion of the experiment was higher in the miR-30d cohort (9/19) than in the scramble (3/19) (Fig. 2H; p=0.038). Both the number and size of lung and liver micrometastases found 11 weeks after the initial injection were elevated in mice of the GIPZ/miR-30d cohort (Figure 2G–H). These data demonstrate that miR-30d augments the ability of melanoma cells to either intravasate, extravasate, seed, and/or colonize a distant site.
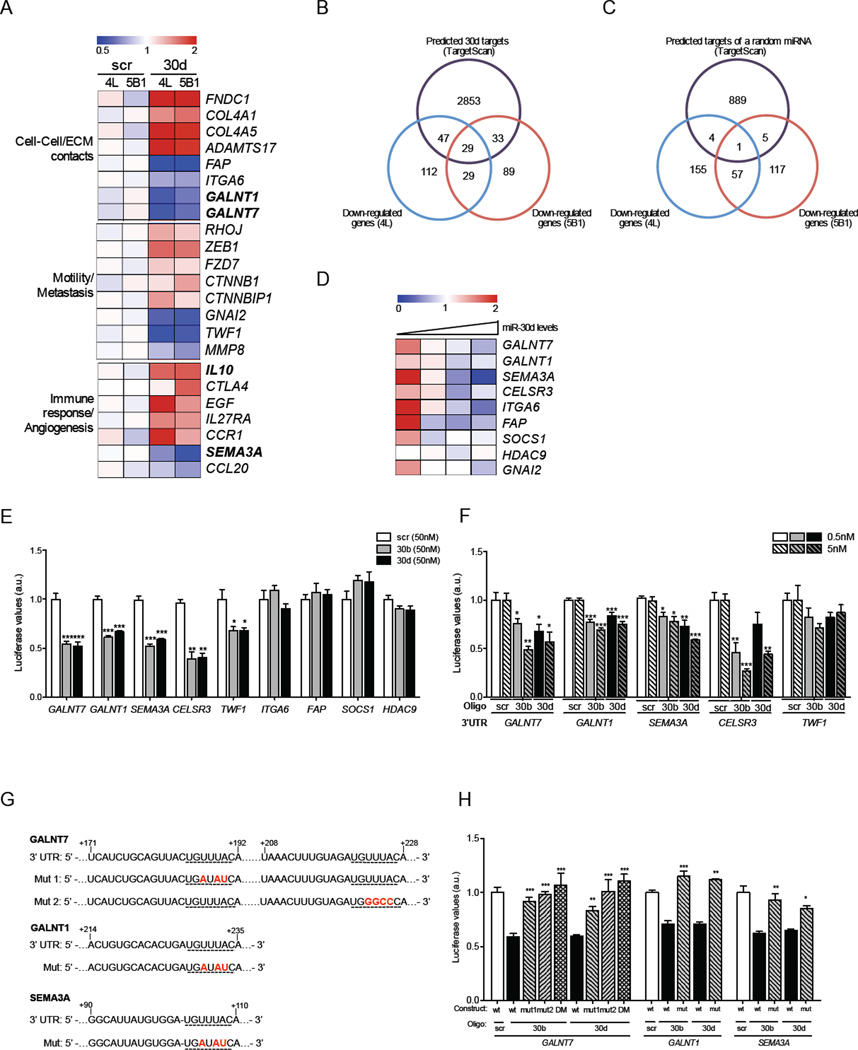
GALNT7, GALNT1, SEMA3, CELSR3 and TWF1 are miR-30b/30d targets in melanoma cells
To identify cellular pathways modulated by miR-30d upregulation and to define specific gene targets that might mediate its pro-metastatic effects, we conducted a global transcriptome analysis of 4L and 5B1 cells transduced with miR-30d or scrambled oligonucleotides using Affymetrix arrays. Using thresholds of a minimum fold change of 1.33 and a p value of <0.05, we found 784 genes to be differentially expressed by the two cell lines. Gene ontology analysis revealed candidate genes whose altered expression could contribute to the invasive phenotype induced by miR-30d (Fig. 3A). Of the 784 altered genes, we found 217 genes down-regulated in 4L, 180 downregulated in 5B1, and 58 downregulated in both lines. Nearly one-third of the downregulated genes were direct miR-30d targets predicted by public algorithms (TargetScan) (Lewis et al., 2005) (Fig. 3B), and included GNAI2, a validated miR-30d target (Yao et al., 2009). Meanwhile, the overlap with targets of a randomly selected miRNA was minimal (Fig. 3C). Interestingly, data mining of our previously published mRNA profile of human metastatic melanoma tissues (Bogunovic et al., 2009) revealed that miR-30d levels inversely correlate with expression of several targets identified in our array analysis, including SEMA3A, GALNT1, and GALNT7 (Fig. 3D), further supporting the physiological relevance of this regulatory mechanism. The expression levels of other GALNT family members, many of which carry recognition sites for miR-30d in their 3’-untranslated regions (3’-UTR), also inversely correlated with miR-30d levels in those tissues (Fig. S4). Using 3’ UTR luciferase reporter assays and quantitative RT-PCR, we confirmed that GALNT7, GALNT1, SEMA3, CESLR3 and TWF1, are direct targets of miR-30b/30d (Fig. 3E, F). Mutations in the miRNA recognition sites (Fig. 3G) rendered the constructs unresponsive to miR-30b or miR-30d induction (Fig. 3H), further confirming that these are direct miR-30b/30d targets.
Fig. 3. MiR-30b/30d directly targets SEMA3A, TWF1, CESLR3, GALNT7 and GALNT1.
A–C Microarray analysis performed in independent biological duplicates for each indicated cell line. A. Heatmap showing the average normalized relative expression levels of genes involved in cell-cell/ECM contacts, motility, metastasis, immune response, or angiogenesis in the indicated cell lines transfected with either scr or miR-30d mimics. B. Venn diagram depicting the overlap between predicted miR-30d targets (TargetScan) and probes significantly downregulated in response to miR-30d overexpression in two cell lines. C. Venn diagram illustrating the overlap between predicted targets of an unrelated miRNA (miR-199a-3p) and genes significantly downregulated in response to miR-30d overexpression in two cell lines. D. Heatmap depicting the expression levels of selected predicted targets in 18 human metastatic melanoma tissues with increasing levels of miR-30d. Each column represents an average expression of 4–8 samples with similar miR-30d levels. See also Figure S4. E. Reporter assay in 293T cells transfected with miR-30b or miR-30d and constructs carrying the luciferase cDNA fused to the 3’UTR of selected predicted targets (mean ±SEM). F. Reporter assay in 293T cells transfected with luciferase constructs fused to the 3’UTR of GALNT7, GALNT1, SEMA3A, CESLR3 and TWF1 and significantly lower concentrations of miR-30b/30d. G–H. Reporter assay in 293T cells transfected with luciferase constructs carrying GALNT7, GALNT1 and SEMA3A 3’UTRs mutated in miR-30b/30d binding sites. DM= double mutant (mean ±SEM). (*p<0.05; ** p<0.01; ***p<0.001).
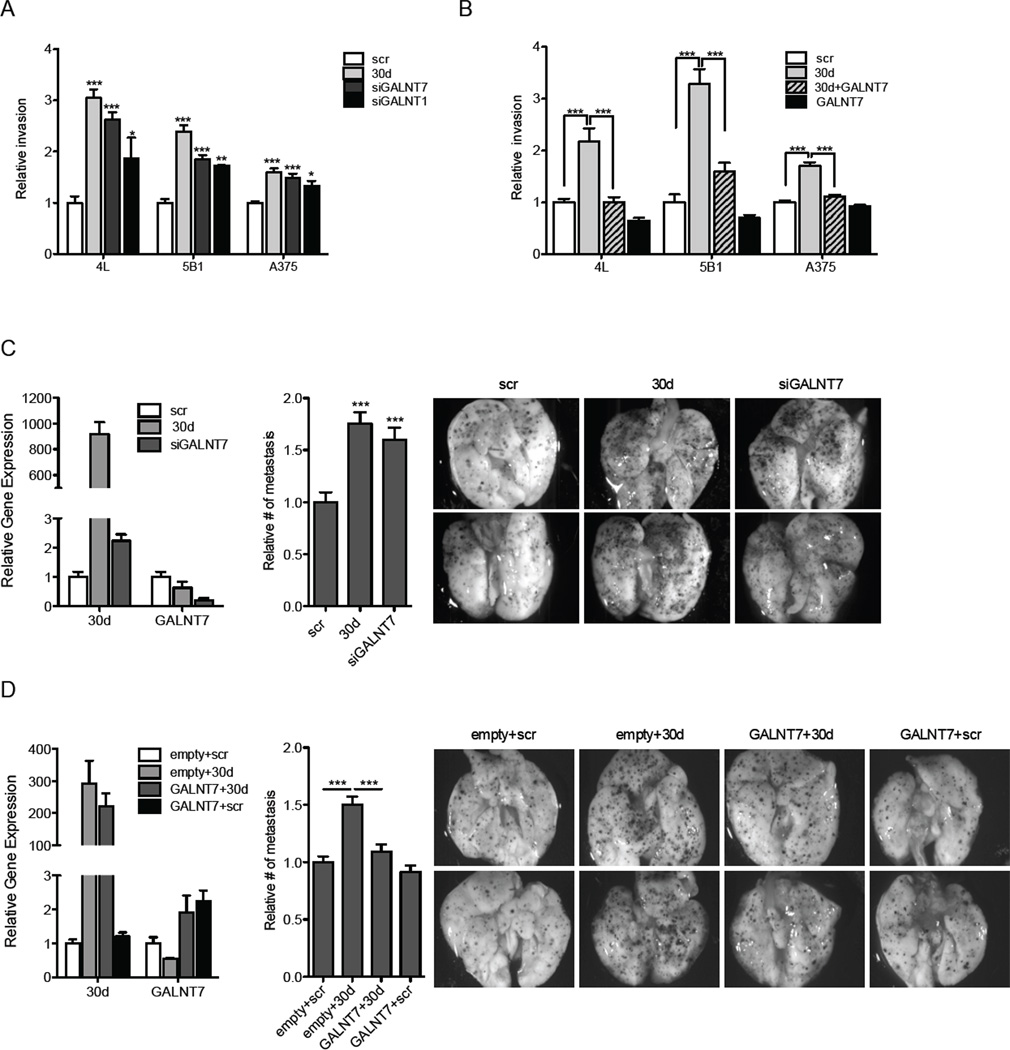
GALNT7 is a critical mediator of miR-30d pro-invasive effects in vitro and pro-metastatic in vivo
Next, we investigated which, if any, miR-30d direct targets mediate the capacity for cellular invasion. Several candidates seemed appealing: CELSR3 is involved in contact-mediated cell-to-cell communication (Wu and Maniatis, 1999), TWF1 encodes for twinfilin (Palmgren et al., 2002) which regulates cell motility and Semaphorin 3A (SEMA3A) exerts antiangiogenic properties (Maione et al., 2009). GalNAc transferases (GalNAc-Ts) initiate mucin-type O-linked glycosylation in the Golgi apparatus by catalyzing the transfer of N-Acetylgalactosamine (GalNAc) to serine and threonine residues on target proteins. These post-translational modifications affect the structure of numerous transmembranal substrates, determining their functional interaction with the extracellular environment (Ten Hagen et al., 2003).
Given their described roles, all these molecules seemed plausible candidates to shape the pro-metastatic influence of miR-30b/30d. To determine whether any of them were critical mediators of miR-30d’s role in cellular invasion, we silenced each of them using RNA interference (RNAi) in melanoma cell lines. While downregulating SEMA3A, CELSR3 and TWF1 did not enhance invasion, repression of GALNT7, and to a lesser extent GALNT1, recapitulated the pro-invasive effects of miR-30d (p<0.0001 for GALNT7; p=0.004 for 5B1 and 0.01 for A375 for GALNT1; Fig. 4A and Fig. S2C). Moreover, co-expression of a GALNT7 cDNA lacking the 3’UTR was able to suppress miR-30d promotion of cell invasion, indicating that GALNT7 silencing critically mediates miR-30d’s pro-migratory effects in melanoma (p<0.0001; Fig. 4B and Fig. S2D).
Fig. 4. GALNT modulation accounts for miR-30d pro-invasive effects in vitro and in vivo.
A–B. Transwell invasion assay of indicated cell lines transfected with scr, miR-30d, siGALNT7, siGALNT1, GALNT7 cDNA, or co-transfected with miR-30d and GALNT7 cDNA (mean ±SEM). C–D. In vivo metastasis assay with B16F10 mouse melanoma cells. C. Cells were transfected with scr, miR-30d mimics or siGALNT7 oligos and injected through the lateral tail vein of C57BL/6J mice. Levels of knockdown or over expression are shown on the left. Histogram and macroscopic pictures are shown. D. Cells were stably transduced with either pEIGW-Empty or pEIGW-mmuGALNT7. 24 hours prior to injection, cells were transfected with either scr control or miR-30d oligonucleotides. Cells were injected through the lateral tail vein of NOG/SCID mice. Levels of overexpression are shown on the left. Histogram and representative macroscopic pictures are depicted (mean ±SEM; *p<0.05; ** p<0.01; ***p<0.001). See also Fig. S3.
GALNT7 silencing by siRNA oligonucleotides was able to mirror miR-30d’s promotion of B16F10 metastatic capacity upon tail vein injection (p=0.0006; Fig. 4C). Finally, we compared the metastatic potential of B16F10 cells transduced with miR-30d oligonucleotides and either an empty lentiviral vector or one expressing the murine GALNT7 cDNA. Concomitant ectopic expression of GALNT7 and miR-30d interfered with miR-30d’ pro-metastatic effect (p=0.0002; Fig. 4D). Overall, our in vitro and in vivo data support GALNT7 inhibition as a key contributor of miR-30d’s pro-metastatic effects in melanoma cells.
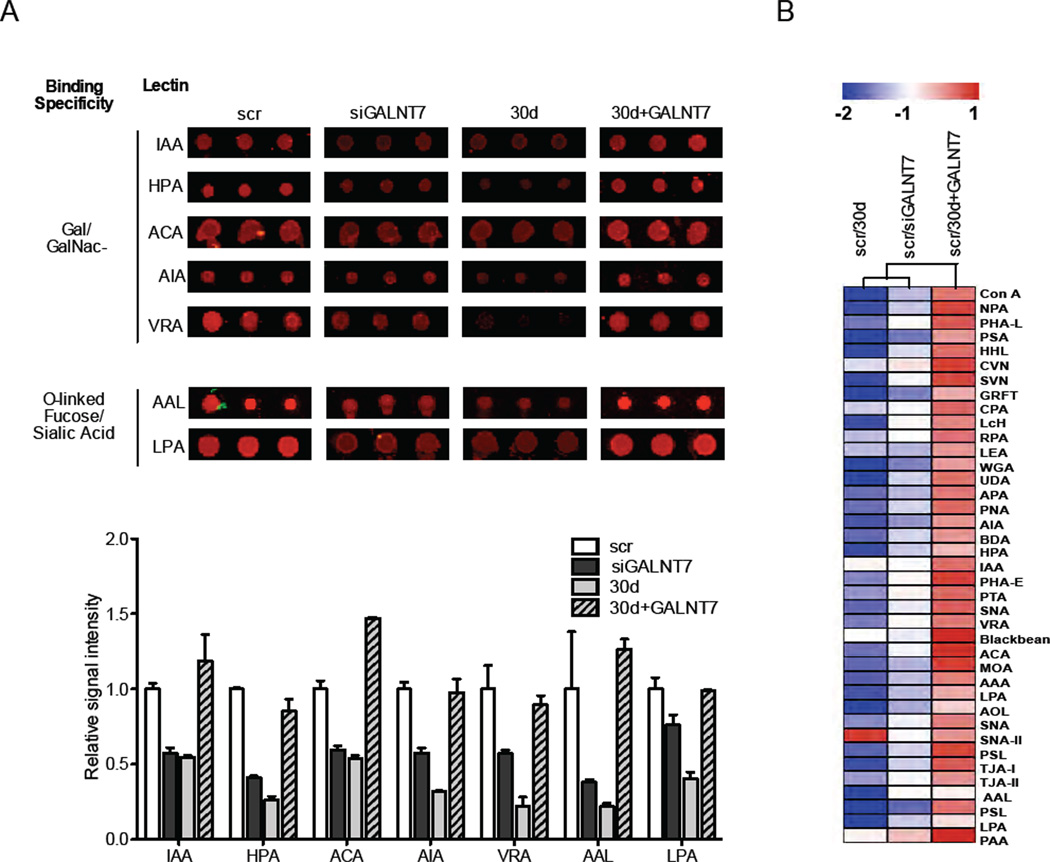
MiR-30d overexpression and GALNT7 inhibition produce similar glycomic changes, which are rescued by GALNT7 ectopic expression
We hypothesized that inhibiting GALNT7 could promote cell invasion by modifying the O-glycosylation patterns of membrane proteins that interact with the extracellular matrix and cells of the tumor microenvironment. To test this hypothesis, we obtained glycomic profiles of melanoma cells transfected with miR-30d only, siGALNT7 only, miR-30d together with GALNT7 cDNA, and scrambled-control miRNA, using lectin microarrays consisting of 84 discrete carbohydrate-binding proteins (Krishnamoorthy et al., 2009). We analyzed cellular micellae from isolated cell membranes, which previous work has shown to contain both glycoproteins and glycolipids representative of the cell surface (Pilobello et al., 2007). Our lectin microarray analysis revealed an overall decrease in glycosylation in both siGALNT7 and miR30d-transduced cells relative to scrambled control, affecting both N- and O-linked glycosylation (Fig. S5). The most pronounced conserved effects across both miR-30d and siGALNT7, based on single color array data, were on secondary modifications such as fucose and sialic acid as well as terminal GalNAc, confirming a predominant effect on O-linked glycosylation (Fig. 5A). Importantly, co-transfection of GALNT7 cDNA rescued these glycosylation defects for both N- and O-linked glycans (Fig. 5A and Fig. S5A). To facilitate direct comparisons among samples, we utilized a more sensitive ratiometric two-color approach (Krishnamoorthy et al., 2009) in which cell membrane micellae from scrambled-transduced cells served as a common biological reference. These data confirmed the observed general reduction in both N-and O-linked glycosylation (Fig. 5B and Fig. S5B). Similar but not completely overlapping changes were induced by siGALNT1 (data not shown). That similar glycosylation changes are induced by both miR-30d upregulation and siGALNT7, and are restored by re-expressing GALNT7, supports the key contribution of GALNT7 repression to miR-30d-associated phenotypes. It is likely that those modified glycosylation patterns act as direct or indirect mediators of miR-30d’s pro-metastatic role.
Fig. 5. GALNT modulation accounts for miR-30d-mediated alterations in membranous O-linked glycans.
A–B. Lectin microarray analysis of 5B1 cells transiently transfected with scrambled control, miR-30d, siGALNT7, or co-transfected with both miR-30d and GALNT7 cDNA. C. Array spots (in triplicates) showing raw signal intensities of galactosamine- or N-Acetylgalactosamine (Gal/GalNAc)-, fucose- or sialic acid-binding lectins in the four treatment groups. The similarities and differences are quantitated in the histogram that represents the average relative binding signal intensity of the lectins depicted above (mean±SEM). D. Heatmap representing normalized signal intensity of a dual color lectin array. Each treatment sample (labeled with Cy3) was hybridized in a 1:1 molar ratio with a scr sample (labeled with Cy5) as internal control. See also Fig. S5.
In order to determine the contribution of chemokine receptors signaling to the pro-metastatic role of miR-30d, we investigated the effects of blocking intracellular signaling by incubating the melanoma cells with Pertussis toxin (PTX), which is known to catalyze the ADP-ribosylation of the α subunits of the heterotrimeric G protein, and prevents Gi proteins from interacting with G-protein coupled receptors on the cell membrane. Pretreatment of melanoma cells with PTX (100ng/ml; 24h) had little to no effect on miR-30d’s pro-metastatic potential in a tail vein injection experiment (Fig. S6). This suggests that Gi-dependent chemokine signaling does not contribute significantly to the effects of miR-30d or GALNT7 on extravasation, seeding or colonization, but we cannot rule out a Gi-independent chemokine signaling.
MiR-30d stimulates the expression of the immunosuppressive cytokine IL-10 by repressing GALNT7
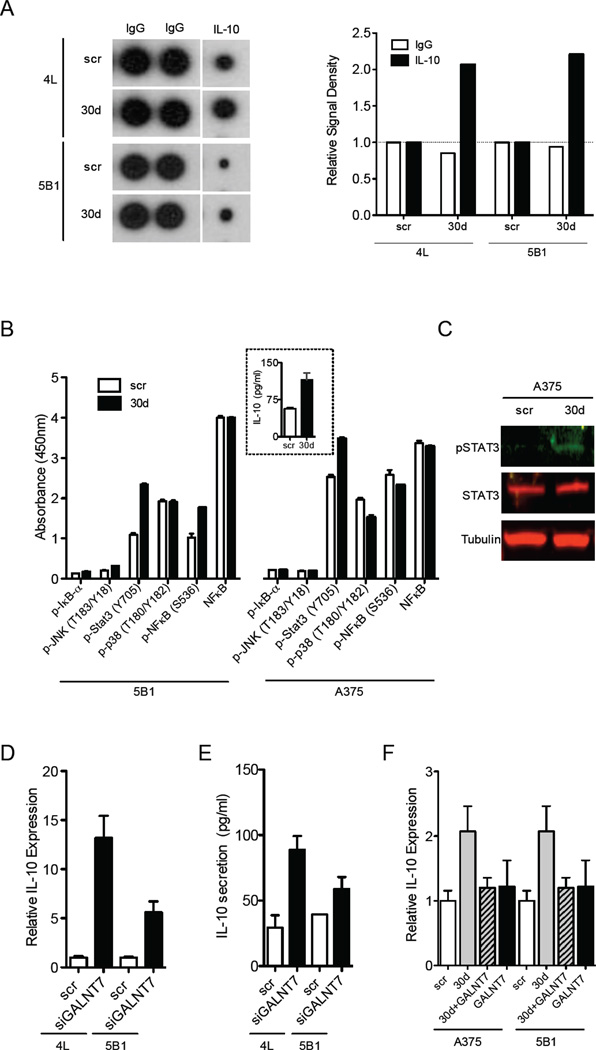
Our microarray analyses in two independent cell lines revealed that miR-30d ectopic expression results in mRNA upregulation of some immune modulators, among them the immunosuppressive immunoglobulin CTLA4 and the immunosuppressive cytokine interleukin-10 (IL-10) (Fig. 3A). Using human cytokine antibody arrays and ELISA, we confirmed that melanoma cells transfected with miR-30d and 30b mimics secrete significantly more IL-10 than scrambled controls (Fig. 6A and Fig. S3D). In search of a mechanism accounting for miR-30d-mediated induction of IL-10, we tested the effect of miR- 30d on the major signaling pathways known to modulate IL-10 levels (i.e., PI3K, STAT3, NF-κB, p38MAPK, JNK). Melanoma cells that overexpress miR-30d displayed increased levels of phospho-Tyr705-STAT3 (Fig. 6B–C), which is known to transcriptionally activate IL-10 as well as numerous pro-metastatic genes (Yu et al., 2009). Although STAT3 activation could partially explain the elevated IL-10 expression, we asked whether any of our identified miR-30d direct targets could contribute to it. Surprisingly, we found that GALNT7 silencing is sufficient to induce IL-10 synthesis and secretion to levels comparable to those induced by miR-30d (Fig. 6D–E) or miR-30b (Fig. S3D), and that GALNT7 overexpression counteracts miR-30d (Fig. 6F) or miR-30b-mediated (Fig. S3D) IL-10 upregulation. These results suggest that miR-30d/30b induce IL-10 at least in part by repressing GALNT7, revealing an unexpected role for a single GalNAc transferase in linking tumor cell invasion and immune modulation.
Fig. 6. MiR-30d promotes IL-10 secretion by suppressing GALNT7.
A. Levels of IL-10 in melanoma cells transduced with miR-30d relative to scr control measured by cytokine array. Quantification of signal density is presented on the right. B. Levels of phosphorylation of proteins that might explain the increase in IL-10 secretion from miR-30d-transfected cells as measured by solid phase ELISA in indicated cell lines. Inset: levels of IL-10 secreted from the cells, quantified by ELISA (mean ±SEM). C. Western blot of phospho-STAT3 levels in scr or miR-30d-transfected cells. Tubulin served as loading control. D. IL-10 mRNA levels in siGALNT7-transfected melanoma cell lines 4L and 5B1 relative to scr control as measured by qRT-PCR (mean ±SEM). E. Secretion of IL-10 to the supernatant of siGALNT7-transfected melanoma cell lines 4L and 5B1 as compared to scr-transfected cells, measured by ELISA. F. IL-10 mRNA (measured by qRT-PCR) levels in indicated melanoma cells transfected with scr, GALNT7 cDNA, or co-transfected with both miR-30d and GALNT7. (mean ±SEM). *p<0.05). See also Fig. S6.
MiR-30d upregulation triggers immunosuppressive properties at the metastatic site
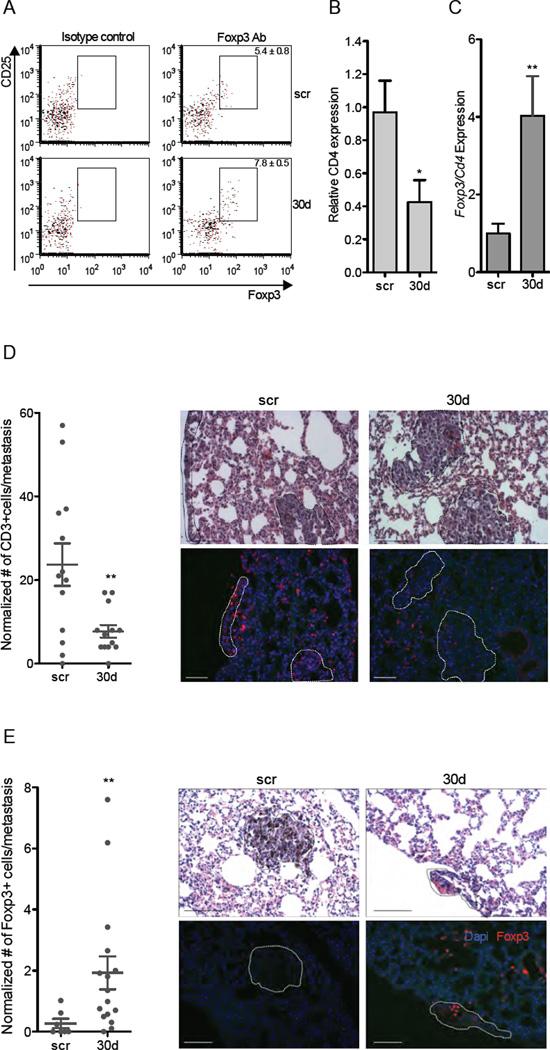
To determine whether aberrant miR-30d expression is able to promote an immunosuppressive environment in vivo, we compared the recruitment of T cells (CD3+), regulatory T-cells (Tregs, CD4+CD25+Foxp3+), activated dendritic cells (DCs, MHCII+F480−CD86+), and myeloid-derived suppressor cells (MDSCs, CD11b+Gr1+) in the lungs of immunocompetent mice injected with B16F10/scr or B16F10/miR-30d cells through the tail vein. FACS analysis showed that lungs of B16F10/miR-30d-injected mice contain significantly more Tregs (p=0.03; Fig. 7A) than the equivalent scrambled controls, with moderate changes in activated DCs and no significant changes in MDSCs (Table S2). Differences were more prominent when individually macro-dissected metastases were analyzed; we noted that metastases from mice injected with B16F10/miR-30d displayed lower local levels of CD4 mRNA (p=0.039; Fig. 7B) and higher levels of Foxp3 mRNA (normalized to CD4 levels in the tissue; p<0.01; Fig. 7C) than those from mice injected with B16F10/scr cells. Immunofluorescence stainings confirmed both the reduction in T-cell accrual (Fig. 7D) and the increased recruitment of Tregs to the metastases of B16F10/miR-30d cells (Fig. 7E). In accordance, immunohistochemistry analysis of human metastatic melanomas (n=32) revealed some association between miR-30d levels and FOXP3 expression in infiltrating lymphocytes (p=0.11; data not shown). Interestingly, we found a significant correlation between FOXP3 expression and miR-30d levels in the tumor cells themselves (n=45; p=0.02; Fig. S7A). To confirm that FOXP3 is indeed expressed by melanoma cells, we conducted HMBA-45 immunohistochemistry stainings in consecutive tissue sections (Fig. S7B). Overall, these results suggest that miR-30d might contribute to metastasis not only by promoting migration but also by suppressing immune surveillance.
Fig. 7. MiR-30d associates with enhanced immunosuppressive features at the metastatic site.
A. Representative flow cytometry of regulatory T cells (CD4+ CD25+ Foxp3+) isolated from whole lungs of mice injected with B16F10/scr or B16F10/miR-30d (mean ± SEM). Isotype controls are shown on the left for each treatment group. See also Table S2. B. CD4 mRNA levels in macrometastases dissected from B16F10/miR-30d relative to B16F10/scr injected mice (mean ±SEM). C. Foxp3 mRNA expression in macro-dissected metastases extracted from lungs of mice injected with B16F10/miR-30d relative to B16F10/scr injected mice (mean ±SEM). D. CD3 immunofluorescence staining of metastases of B16F10/mir-30d and B16F10/scr- injected mice shows recruitment of T cells to scr-transfected compared to B16F10/miR-30d transfected metastasis. Corresponding H&E stainings on consecutive sections are shown in upper panels and metastatic foci are circled. Scatter plot depicts the number of recruited CD3+ T cells to the metastasis in several mice per group. The number of cells was normalized to the area of metastasis. E. FoxP3 immunofluorescence staining shows recruitment of regulatory T lymphocytes to B16F10/miR-30d compared to B16F10/scr metastases. Corresponding H&E stainings on consecutive sections are shown in upper panels and metastatic foci are circled. Scatter plot depicts the number of recruited CD3+ T cells to the metastasis in several different mice for each group. Scale bars represent 100µm (*p<0.05; ** p<0.01). See also Fig. S7.
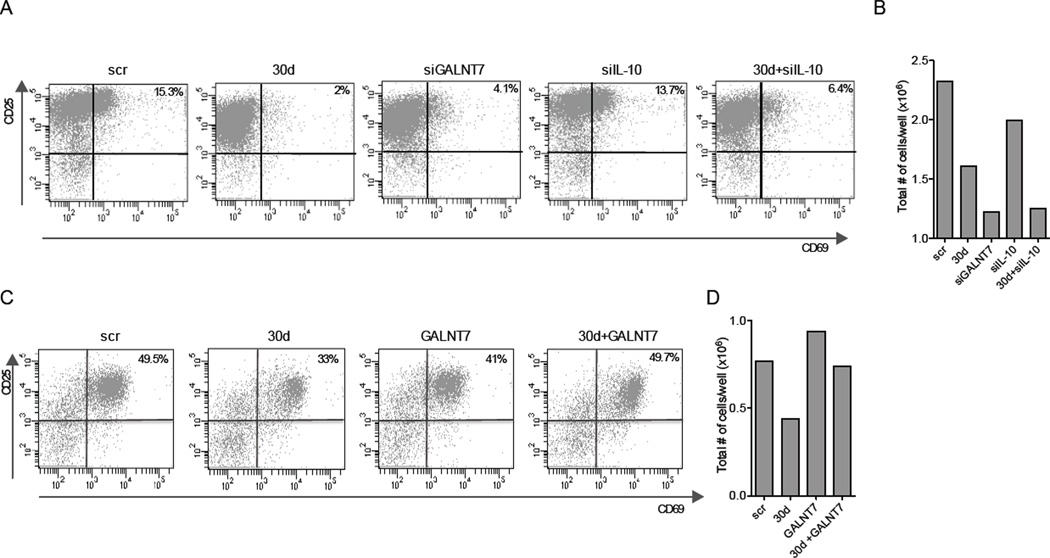
In order to explore the mechanism of immune modulation by miR-30d, we tested the ability of miR-30d upregulation or GALNT7 silencing to alter the secretion of immunomodulatory molecules by melanoma cells. For that, CD4+ splenocytes isolated from FoxP3-GFP mice (Bettelli et al., 2006) were activated ex vivo with CD28 and CD3 antibodies and then incubated in the presence of supernatants from scrambled, miR-30d, or siGALNT7-transfected A375 melanoma cells. We observed that supernatants from miR-30d or siGALNT7 display increased ability to suppress T cell activation (assessed by expression of surface markers CD25 and CD69) (Fig. 8A), and total T cell number (Fig. 8B) than those from scrambled-transfected cells. Importantly, these effects of miR-30d were reversed by co-expression of GALNT7 cDNA in 5B1 melanoma cells (Fig. 8C, D). In addition, supernatants from miR-30d and siGALNT7 promoted T cell differentiation into regulatory T cells (Tregs), as indicated by the number of CD25+ GFP+ (Foxp3+) cells (Fig. S7C). Moreover, we found that concomitant silencing of IL-10 by siRNA only partially counteracted miR-30d immunosuppressive activities (Fig. 8A, B and Fig. S7C). Therefore, our data suggest that IL-10 is one of multiple immunomodulatory molecules in miR-30d’s regulated cellular secretome.
Fig. 8. GALNT modulation accounts for miR-30d-mediated immunosupressive effects ex vivo.
A. FACS analysis of activated (CD25+CD69+ gated on CD4+) T lymphocyes isolated from spleens of Foxp3-GFP mice, stimulated by CD28 and CD3 antibodies and incubated for 72h in the presence of supernatants from A375 melanoma cells transfected with scr, miR-30d, siGALNT7, siIL10 and miR-30d+siIL-10. B. Total number of T cells at conclusion of the experiment (representative experiment, n=3). C. FACS analysis of activated (CD25+CD69+ CD4+) T lymphocytes isolated from spleens of Foxp3-GFP mice, stimulated by CD28 and CD3 antibodies and incubated for 72h in the presence of supernatants from 5B1 melanoma cells transfected with scr+pCDNA3-Empty (scr), miR-30d+pCDNA3-Empty (30d), scr+pCMV-GALNT7 (GALNT7) and miR-30d+pCMV-GALNT7 (30d+GALNT7). D. Total number of T cells at conclusion of the experiment (a representative experiment).
DISCUSSION
We have demonstrated that miR-30d/30b overexpression enhances the invasive capacity of melanoma cells in vitro and increases their metastatic potential in vivo, predominantly by suppressing GALNT7. Downregulation of miR-30d produced the opposite effects, while direct silencing of GALNT7 replicated most effects of miR-30d/b overexpression. Changes in glycosylation patterns have been associated with tumor progression for some time (Dennis et al., 1999), yet the specific molecular mechanisms underlying abnormal glycosylation and the downstream processes directly or indirectly contributing to metastasis remain poorly characterized. Strikingly, aberrant miRNA-mediated regulation of a GalNAc transferase promoted both cell motility and immunosuppressive mechanisms, which could synergize during metastasis.
Glycosylation in tumor progression
Our lectin arrays revealed that GALNT7 silencing or miR-30d upregulation have specific effects on O-glycans and, to a lesser extent, on N-glycosylated substrates. Alterations in O-glycans have many biological consequences in cancer, because potential ligands responsible for interactions between cancer cells and their microenvironment are changed. This influences the growth and survival of the cell and its interactions with lectins and cell-surface receptors on neighboring cells or immune cells, all of which are important for its ability to metastasize (Brockhausen, 2006). GalNAc transferases (GalNAc-Ts) initiate mucin-type O-linked glycosylation in the Golgi apparatus by catalyzing the transfer of GalNAc to serine and threonine residues on target proteins (Ten Hagen et al., 2003). GalNAc-Ts have different but overlapping substrate specificities and patterns of expression. Our glycomic analysis revealed that GALNT7 silencing has broad effects on the glycosylation of melanoma cells beyond its known transferase activity. These effects could be moderated through mislocalization of other enzymes in the pathway due to loss of the transferase or alterations in protein localization and stability that influence the general glycosylation phenotype. Regardless, the glycomic signature is clearly rescued by overexpression of GALNT7, indicating that the effects are specific to this enzyme. This modified glycocode might, at least partially, account for miR-30d/siGALNT7 phenotype, even though the direct effectors (i.e. glycans, signaling pathways) of their pro-invasive and immunomodulatory actions could not be elucidated at this time. Pre-incubation with pertussis toxin did not affect miR-30d’s pro-metastatic effects, suggesting that Gi-dependent chemokine signaling is not a key player, at least under those experimental conditions.
We have shown that expression of GALNT7 and GALNT1 is controlled by miR-30b/30d levels, which increase during melanoma tumor progression in parallel with advancing stage and metastatic potential at the time of diagnosis. Intriguingly, in addition to GALNT1 and GALNT7, other GalNAc-T family members carry miR-30d recognition sites. We found that the expression of many of them inversely correlated with miR-30d levels in human samples, suggesting that this miRNA might coordinately regulate the entire GALNT family. Not much is known about the regulation of the GALNT family, though another miRNA, miR-378, with a potential effect in osteoblast differentiation was shown modulate GALNT7 (Kahai et al., 2009).
It is interesting to note that a pro-metastatic role for miR-30d has also been recently shown in hepatocellular carcinoma (Yao et al., 2009), and that miR-30d levels in the sera of lung cancer patients correlate with poor prognosis (Hu et al., 2010). Together, these studies indicate that this miRNA cluster (and possibly GALNT suppression) might exert a common pro-metastatic effect in various cancers. The pleiotropic effects (immunosuppressive and pro-invasive) of miR-30d described here for melanoma—mostly mediated by GalNAc-T suppression—might thus be relevant in other tumor types. In fact, metastatic clones derived from colorectal cancer cells have altered expression of various GalNAc-Ts in comparison with their non-metastatic parental counterparts (Kato et al., 2010).
Curiously, miR-30e, which shares a seed region with miR-30b/d but is located in a separate genomic location, has shown an anti-metastatic role in breast cancer (Yu et al., 2010). A plausible explanation for this apparent paradox is that whereas O-glycosylation of specific substrates, particularly of mucins, promotes breast or colon cancer progression (Brockhausen, 2006), the expression of mucins and their contribution to metastasis in melanoma is known to be limited (Bhavanandan, 1991). In addition, the cell-type specific repertoire of GalNAc-Ts, which vary with cellular differentiation and malignant transformation (Mandel et al., 1999), could account for the opposing outcomes of miR-30e and miR-30b/30d induction in different tumors. These observations underscore the context-dependence of miRNA functions in cancer.
Although the pro-metastatic action of miR-30d is critically mediated by GALNT7 silencing, we observed that GALNT7 ectopic expression did not completely counteract miR-30d’s effects in vivo, suggesting a contribution of other miR-30d targets. In fact, in addition to the GALNT family, we validated other miR-30d targets such as SEMA3A, which exerts anti-angiogenic functions (Maione et al., 2009; Serini et al., 2003), and CESLR3 and TWF1, which are involved in cell-to-cell interactions and migration. Downregulation of these genes, however, failed to promote melanoma cell invasion through a fibronectin coat. They may nevertheless mediate other metastatic abilities not tested here, such as vascularization, adhesion or motility. The contributions of SEMA3A, CESLR3 and TWF1 to miR-30b/30d pro-metastatic function need to be further investigated.
Immune modulation in melanoma
Melanoma is a paradigmatically immunogenic tumor, with abundant inflammatory infiltrates in both cutaneous and metastatic lesions, yet it manages to evade this upregulated host immune response (Lee et al., 2005; Real et al., 2001; Redondo et al., 2003). We found that miR-30d overexpression correlates with reduced CD3+ T cells recruitment and accumulation of Tregs at the metastatic site in vivo. Consistently, we demonstrated that miR-30d upregulation alters melanoma cells’ secretome such that it suppresses T cell activation and favors Treg induction ex vivo. These effects can be partially mediated by increased IL-10 secretion, which results from GALNT7 suppression. This miR-30b/30d- GALNT7-IL-10 axis could provide a mechanistic explanation for the immunosuppressive behavior of some metastatic melanomas. We observed some association between higher miR-30d expression and more FOXP3-positive lymphocytes (putative Tregs) in human metastatic samples. Surprisingly, high 30d levels significantly correlated with FOXP3 expression in the tumor cells themselves. The expression of FOXP3 by tumor cells was already reported in several cancer types (Hinz et al., 2007; Merlo et al., 2009), including melanoma (Ebert et al., 2008). It has been proposed that tumor cells expressing FOXP3 share immunosuppressive effects with Tregs (Martin et al., 2010) which might represent a new mechanism of immune evasion in melanoma. Both cell- and non-cell autonomous mechanisms, potentially exerted by miR-30d, could cooperate to restrain the host antitumoral response. MiR-30d pro-metastatic effects, critically mediated by GALNT7 suppression, are prominent even in the absence of a functional host immune system, as indicated by our experiments conducted in NOG/SCID mice. However, our results in immunocompetent mice reveal notable immunosuppressive effects associated with miR-30d upregulation, which might synergize with its pro-invasive properties during metastasis.
The control of immuno-stimulant or immuno-suppressive molecules by miRNAs in the context of tumor formation and progression is largely unexplored. MiR-21, a miRNA with established tumorigenic role (Esquela-Kerscher and Slack, 2006), has been shown to negatively regulate TLR4 via targeting the proinflammatory tumor suppressor PDCD4 (Sheedy et al., 2010), but the contribution of this mechanism to the tumorigenic activities of miR-21 remains unknown. Evidence of another miRNA directly targeting IL-10, miR-106a, has been recently reported (Sharma et al., 2009). It is interesting to note in this context that we found reduced miR-106a levels in miR-30d transduced cells (data not shown), which suggests a possible miRNA network converging on the modulation of IL-10 levels.
A recent report showed that Snail-mediated induction of epithelial to mesenchymal transition induces an immunosuppressive response in melanoma cells mainly by inducing the cytokine TSP1 (Kudo-Saito et al., 2009). These data and our current results reveal that cell migration and immune evasion are intimately connected during metastasis, and our findings suggest that GalNAc transferases can serve as a link between the two.
In sum, this study shows that a single miRNA can exert both pro-invasive and immunomodulatory effects, and that both actions could be critically mediated by one target, GALNT7. Our data could have important prognostic implications: higher miR-30d expression correlates with advanced melanoma and aggressive biological behavior, and is a predictor of time to death with melanoma, independent of thickness. Moreover, miR-30d targeting represents a plausible therapeutic approach: targeting miR-30d in tumor cells with chemically modified oligonucleotides or artificial decoys [reviewed in (Tong and Nemunaitis, 2008) and (Valastyan and Weinberg, 2009)] could de-repress the endogenous GALNT7 levels, simultaneously counteracting both its pro-invasive and immunosuppressive effects. More research is needed to understand the effects of concurrently de-repressing other miR-30d targets, but the possibility remains that miRNA targeting could synergistically impede metastasis as much as miRNA upregulation promotes it.
EXPERIMENTAL PROCEDURES
Cell lines
Cell lines were cultured as previously described (Segura et al., 2009) (Cruz-Munoz et al., 2008). HEK293T and A375 cells were purchased from American Type Culture Collection (ATCC). The B16F10 mouse melanoma cell line and the human WM35 and WM98 cell lines were acquired from the Wistar Institute. Cell lines 113/6-4L (4L) and 131/4-5B1 (5B1) were isolated and cultured as previously described (Cruz-Munoz et al., 2008). 4L, 5B1, A375 and B16F10 are metastatic melanoma cell lines whereas WM35 and WM98 were derived from primary melanomas.
Luciferase assays
HEK293T were seeded into 96-well plates and co-transfected with 3’UTR vectors and indicated amounts of miR-30b or -30d mimics or miRIDIAN mimic negative control (Dharmacon). Luciferase activity was measured using the Dual-Glo™ Luciferase Assay System (Promega). Renilla luciferase activity was normalized to corresponding firefly luciferase activity and plotted as a percentage of the control.
Ex vivo T cell activation, followed by FACS analysis
CD4+ splenocytes were isolated from Foxp3-GFP mice using a CD4+ T Cell Isolation Kit (Miltenyi Biotec). 1×106 cells were then incubated in 24wells with conditioned media of melanoma cells supplemented with 25µl of CD3/CD28 T-activators beads (Invitrogen). FACS analysis was performed after 72h on cells stained for CD4 (APC-Cy7-conjugated; BioLegend), CD25 (APC-conjugated; e bioscience) and CD69 (PE-conjugated; BD Biosciences).
Clinical Specimens
Human melanoma specimens (primary, metastatic) were collected at the time of surgery. Approval to collect specimens was granted by NYU IRB protocol #10362, "Development of an NYU interdisciplinary melanoma cooperative group: A clinicopathological database". Informed consent was obtained from all subjects included.
Mouse experiments
were conducted following protocols approved by the NYU Institutional Animal Care Use Committee (IACUC) (protocol number #080109).
Statistical Methodologies
Statistical significance was determined by paired or unpaired t-test in cases of standardized expression data. One-way ANOVA was performed for multiple group comparisons (GraphPad Prism Software). Wilcoxon matched pairs test and Mann-Whitney tests for nonparametric analyses of non-Gaussian data. Chi-square test and McNemar’s test were used for testing association among unmatched and matched categorical variables. In particular, chi-square test and Fisher’s exact test were used to assess association of miR-30d with FOXP3 staining in T-cells and in melanoma cells (Fig. S7A). Multivariable COX PH models were used to analyze time-to-recurrence and overall survival when adjusted for primary tumor thickness and ulceration status. Log rank test was used for analyses presented in Figures 1E and 1F.
Supplementary Material
HIGHLIGHTS.
-
□
MiR-30b/d levels associate with advanced stage and poor outcome melanoma patients
-
□
MiR-30d promotes metastasis and immunosuppressive features in vivo
-
□
MiR-30d enhances invasion and IL-10 secretion by targeting GalNAc transferases
-
□
MiR-30d upregulation leads to GALNT7-dependent alterations in cellular glycosylation
Significance.
MicroRNAs are emerging as key contributors to tumor metastasis because of their ability to regulate multiple targets, and thereby alter several functions, simultaneously. We found a miRNA cluster that promotes metastasis by concurrently enhancing invasive capabilities of melanoma cells and suppressing immune surveillance mechanisms, allowing the tumor cells to migrate and invade foreign tissue. Both these effects of miR-30b/30d are mediated by direct suppression of GalNAc transferases. Aberrant glycosylation has previously been connected to tumor progression, but the underlying molecular mechanisms and their impact on specific cellular pathways are poorly understood. Our work places the control of glycosylation as a molecular link between tumor cell migration and immune evasion, two processes that act synergistically during metastasis.
ACKNOWLEDGMENTS
We thank members of the NYU Cancer Institute Genomics Facility for array analysis, and Dr. Cindy Loomis and members of the NYU Cancer Institute Histopathology and Immunohistochemistry Core Laboratories for tissue processing and histological stainings. We thank Drs. Dan Littman and Vijay Kuchroo for the FoxP3-GFP mice. We are grateful to Silvia Menendez and Lisa Koetz for technical assistance, Chin-Siean Tay for IF stainings, and to Dr. Michelle Krogsgaard and members of her lab for discussions and technical assistance. This work was funded by the ConCerN Foundation, the Melanoma Research Foundation, the Marc Jacobs Campaign, and funds from the NIH–NCI Cancer Center Support Grant P30CA016087. L.M. is supported by a NIH 7 DP2 OD004711-02 grant. M.F.S. is supported by a National Cancer Center fellowship, and R.D.M. by an EMBO post-doctoral fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBER
Accession number for microarray data. GSE27718
For additional methods, see Supplemental Experimental Procedures.
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST. The authors declare no conflict of interest.
REFERENCES
- Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- Bhavanandan VP. Cancer-associated mucins and mucin-type glycoproteins. Glycobiology. 1991;1:493–503. doi: 10.1093/glycob/1.5.493. [DOI] [PubMed] [Google Scholar]
- Bogunovic D, O'Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, et al. Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci U S A. 2009;106:20429–20434. doi: 10.1073/pnas.0905139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhausen I. Mucin-type O-glycans in human colon and breast cancer: glycodynamics and functions. EMBO Rep. 2006;7:599–604. doi: 10.1038/sj.embor.7400705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Cruz-Munoz W, Man S, Xu P, Kerbel RS. Development of a preclinical model of spontaneous human melanoma central nervous system metastasis. Cancer Res. 2008;68:4500–4505. doi: 10.1158/0008-5472.CAN-08-0041. [DOI] [PubMed] [Google Scholar]
- Dennis JW, Granovsky M, Warren CE. Glycoprotein glycosylation and cancer progression. Biochim Biophys Acta. 1999;1473:21–34. doi: 10.1016/s0304-4165(99)00167-1. [DOI] [PubMed] [Google Scholar]
- Ebert LM, Tan BS, Browning J, Svobodova S, Russell SE, Kirkpatrick N, Gedye C, Moss D, Ng SP, MacGregor D, et al. The regulatory T cell-associated transcription factor FoxP3 is expressed by tumor cells. Cancer Res. 2008;68:3001–3009. doi: 10.1158/0008-5472.CAN-07-5664. [DOI] [PubMed] [Google Scholar]
- Ehlers JP, Worley L, Onken MD, Harbour JW. DDEF1 is located in an amplified region of chromosome 8q and is overexpressed in uveal melanoma. Clin Cancer Res. 2005;11:3609–3613. doi: 10.1158/1078-0432.CCR-04-1941. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Gupta GP, Massague J. Cancer metastasis: building a framework. Cell. 2006;127:679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Mani S, Yang J, Hartwell K, Weinberg RA. The evolving portrait of cancer metastasis. Cold Spring Harb Symp Quant Biol. 2005;70:291–297. doi: 10.1101/sqb.2005.70.033. [DOI] [PubMed] [Google Scholar]
- Hinz S, Pagerols-Raluy L, Oberg HH, Ammerpohl O, Grussel S, Sipos B, Grutzmann R, Pilarsky C, Ungefroren H, Saeger HD, et al. Foxp3 expression in pancreatic carcinoma cells as a novel mechanism of immune evasion in cancer. Cancer Res. 2007;67:8344–8350. doi: 10.1158/0008-5472.CAN-06-3304. [DOI] [PubMed] [Google Scholar]
- Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, Chen Y, Xu L, Zen K, Zhang C, Shen H. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- Kahai S, Lee SC, Lee DY, Yang J, Li M, Wang CH, Jiang Z, Zhang Y, Peng C, Yang BB. MicroRNA miR-378 regulates nephronectin expression modulating osteoblast differentiation by targeting GalNT-7. PLoS ONE. 2009;4:e7535. doi: 10.1371/journal.pone.0007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Takeuchi H, Kanoh A, Miyahara N, Nemoto-Sasaki Y, Morimoto-Tomita M, Matsubara A, Ohashi Y, Waki M, Usami K, et al. Loss of UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase 3 and reduced O-glycosylation in colon carcinoma cells selected for hepatic metastasis. Glycoconj J. 2010;27:267–276. doi: 10.1007/s10719-009-9275-4. [DOI] [PubMed] [Google Scholar]
- Kudo-Saito C, Shirako H, Takeuchi T, Kawakami Y. Cancer metastasis is accelerated through immunosuppression during Snail-induced EMT of cancer cells. Cancer Cell. 2009;15:195–206. doi: 10.1016/j.ccr.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Lee JH, Torisu-Itakara H, Cochran AJ, Kadison A, Huynh Y, Morton DL, Essner R. Quantitative analysis of melanoma-induced cytokine-mediated immunosuppression in melanoma sentinel nodes. Clin Cancer Res. 2005;11:107–112. [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lu Y, Ryan SL, Elliott DJ, Bignell GR, Futreal PA, Ellison DW, Bailey S, Clifford SC. Amplification and overexpression of Hsa-miR-30b, Hsa-miR-30d and KHDRBS3 at 8q24.22–q24.23 in medulloblastoma. PLoS ONE. 2009;4:e6159. doi: 10.1371/journal.pone.0006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maione F, Molla F, Meda C, Latini R, Zentilin L, Giacca M, Seano G, Serini G, Bussolino F, Giraudo E. Semaphorin 3A is an endogenous angiogenesis inhibitor that blocks tumor growth and normalizes tumor vasculature in transgenic mouse models. J Clin Invest. 2009;119:3356–3372. doi: 10.1172/JCI36308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandel U, Hassan H, Therkildsen MH, Rygaard J, Jakobsen MH, Juhl BR, Dabelsteen E, Clausen H. Expression of polypeptide GalNAc-transferases in stratified epithelia and squamous cell carcinomas: immunohistological evaluation using monoclonal antibodies to three members of the GalNAc-transferase family. Glycobiology. 1999;9:43–52. doi: 10.1093/glycob/9.1.43. [DOI] [PubMed] [Google Scholar]
- Martin F, Ladoire S, Mignot G, Apetoh L, Ghiringhelli F. Human FOXP3 and cancer. Oncogene. 2010;29:4121–4129. doi: 10.1038/onc.2010.174. [DOI] [PubMed] [Google Scholar]
- Merlo A, Casalini P, Carcangiu ML, Malventano C, Triulzi T, Menard S, Tagliabue E, Balsari A. FOXP3 expression and overall survival in breast cancer. J Clin Oncol. 2009;27:1746–1752. doi: 10.1200/JCO.2008.17.9036. [DOI] [PubMed] [Google Scholar]
- Palmgren S, Vartiainen M, Lappalainen P. Twinfilin, a molecular mailman for actin monomers. J Cell Sci. 2002;115:881–886. doi: 10.1242/jcs.115.5.881. [DOI] [PubMed] [Google Scholar]
- Real LM, Jimenez P, Kirkin A, Serrano A, Garcia A, Canton J, Zeuthen J, Garrido F, Ruiz-Cabello F. Multiple mechanisms of immune evasion can coexist in melanoma tumor cell lines derived from the same patient. Cancer Immunol Immunother. 2001;49:621–628. doi: 10.1007/s002620000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redondo P, Sanchez-Carpintero I, Bauza A, Idoate M, Solano T, Mihm MC., Jr Immunologic escape and angiogenesis in human malignant melanoma. J Am Acad Dermatol. 2003;49:255–263. doi: 10.1067/s0190-9622(03)00921-6. [DOI] [PubMed] [Google Scholar]
- Scheel C, Onder T, Karnoub A, Weinberg RA. Adaptation versus selection: the origins of metastatic behavior. Cancer Res. 2007;67:11476–11479. doi: 10.1158/0008-5472.CAN-07-1653. discussion 11479–11480. [DOI] [PubMed] [Google Scholar]
- Segura MF, Belitskaya-Levy I, Rose AE, Zakrzewski J, Gaziel A, Hanniford D, Darvishian F, Berman RS, Shapiro RL, Pavlick AC, et al. Melanoma MicroRNA Signature Predicts Post-Recurrence Survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A. 2009;106:1814–1819. doi: 10.1073/pnas.0808263106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serini G, Valdembri D, Zanivan S, Morterra G, Burkhardt C, Caccavari F, Zammataro L, Primo L, Tamagnone L, Logan M, et al. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature. 2003;424:391–397. doi: 10.1038/nature01784. [DOI] [PubMed] [Google Scholar]
- Sharma A, Kumar M, Aich J, Hariharan M, Brahmachari SK, Agrawal A, Ghosh B. Posttranscriptional regulation of interleukin-10 expression by hsa-miR-106a. Proc Natl Acad Sci U S A. 2009;106:5761–5766. doi: 10.1073/pnas.0808743106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O'Leary JJ, Ruan Q, Johnson DS, Chen Y, O'Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- Talmadge JE. Clonal selection of metastasis within the life history of a tumor. Cancer Res. 2007;67:11471–11475. doi: 10.1158/0008-5472.CAN-07-2496. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Hagen KG, Fritz TA, Tabak LA. All in the family: the UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- Tong AW, Nemunaitis J. Modulation of miRNA activity in human cancer: a new paradigm for cancer gene therapy? Cancer Gene Ther. 2008;15:341–355. doi: 10.1038/cgt.2008.8. [DOI] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. Assaying microRNA loss-of-function phenotypes in mammalian cells: emerging tools and their potential therapeutic utility. RNA Biol. 2009;6:541–545. doi: 10.4161/rna.6.5.10081. [DOI] [PubMed] [Google Scholar]
- Van Den Berg C, Guan XY, Von Hoff D, Jenkins R, Bittner, Griffin C, Kallioniemi O, Visakorpi, McGill, Herath J, et al. DNA sequence amplification in human prostate cancer identified by chromosome microdissection: potential prognostic implications. Clin Cancer Res. 1995;1:11–18. [PubMed] [Google Scholar]
- Visapaa H, Seligson D, Eeva M, Gaber F, Rao J, Belldegrun A, Palotie A. 8q24 amplification in transitional cell carcinoma of bladder. Appl Immunohistochem Mol Morphol. 2003;11:33–36. doi: 10.1097/00129039-200303000-00006. [DOI] [PubMed] [Google Scholar]
- Wu Q, Maniatis T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell. 1999;97:779–790. doi: 10.1016/s0092-8674(00)80789-8. [DOI] [PubMed] [Google Scholar]
- Yao J, Liang L, Huang S, Ding J, Tan N, Zhao Y, Yan M, Ge C, Zhang Z, Chen T, et al. MicroRNA-30d promotes tumor invasion and metastasis by targeting Galphai2 in hepatocellular carcinoma. Hepatology. 2009;51:846–856. doi: 10.1002/hep.23443. [DOI] [PubMed] [Google Scholar]
- Yu F, Deng H, Yao H, Liu Q, Su F, Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.