Abstract
After birth, contact to environmental antigens induces the production of IgA, which represents a first line of defense for the neonate. We sought to characterize the maturation of the repertoire of IgA H chain transcripts in circulating blood B cells during human ontogeny. We found that IgA H chain transcripts were present in cord blood as early as 27 weeks of gestation and that the restrictions of the primary antibody repertoire (IgM) persisted in the IgA repertoire. Thus, B cells harboring more “mature” VH regions were not preferred for class switch to IgA. Preterm and term neonates expressed a unique IgA repertoire, which was characterized by short CDR-H3 regions, preference of the JH proximal DH7-27 gene segment, and very few somatic mutations. During the first postnatal months, these restrictions were slowly released. Preterm birth did not measurably accelerate the maturation of the IgA repertoire. At a postconceptional age of 60 weeks, somatic mutation frequency of IgA H chain transcripts reached 25% of the adult values but still showed little evidence of antigen-driven selection. These results indicate that similar to IgG, the IgA repertoire expands in a controlled manner after birth. Thus, the IgA repertoire of the newborn has distinctive characteristics that differ from the adult IgA repertoire. These observations might explain the lower affinity and specificity of neonatal IgA antibodies, which could contribute to a higher susceptibility to infections and altered responses to vaccinations, but might also prevent the development of autoimmune and allergic diseases.
Keywords: Human, B-cells, antibodies, gene rearrangement, repertoire development
Introduction
IgA serves as the forward defense of the mucosal adaptive immune system where it can protect the organism by neutralizing toxins and by blocking the adherence and penetration of microorganisms. By means of its ability to penetrate the mucosa in conjunction with antigen and to consecutively induce effector immune responses, IgA also plays a key role in the maintenance of intestinal microflora and immune homeostasis (1, 2).
IgA production in mice is very low in the uninfected fetus and is stimulated after birth by exposure to commensal microorganisms and food antigens in the gut (3). Interestingly, in mice IgA production is particularly up-regulated during weaning (4, 5). Whereas in mice isolated lymphoid follicles, as inductive sites for B cell activation and expansion, develop after birth in response to the microflora, they are already present in humans at birth (6). In humans serum-IgA concentration increases during childhood and reaches adult levels during the second decade of life (7). Large amounts of IgA are secreted onto mucosal surfaces and by exocrine glands, including the mammary gland. Breast fed neonates take up high amounts of IgA through their mother’s milk, allowing a passive protection of the intestinal mucosa while the infant gradually establishes its own IgA production.
Hitherto IgA production during human ontogeny has only been examined quantitatively (serum levels), but not qualitatively (characteristics of antigen-binding sites) (7). Previous analyses of VH, DH and JH gene utilization, N-nucleotides and somatic mutations of immunoglobulin heavy chain gene transcripts have shown that the diversification of the primary (IgM) and the secondary (IgG) antibody repertoires are strictly regulated during ontogeny (8-12). Several observations in mice suggest that, in contrast to IgG, the IgA repertoire might not predominantly reflect a focused antigen driven selection but rather a diffuse, less selected production that might be directed against redundant epitopes of commensal microorganisms (13, 14). Moreover, normal serum IgA levels in mice can even be produced in the absence of organized secondary lymphoid structures such as Peyer’s patches and mesenteric lymph nodes (15). Studying the ontogeny of IgA production in human is important since although sharing many similarities, the regulation of IgA production differs between mouse and humans in several important aspects (6). We postulated that a systematic analysis of the human IgA repertoire during ontogeny might clarify if the circulating IgA repertoire underlies differing selective pressures than the other isotypes. In this study we have analyzed IgA transcripts from cord blood and from peripheral blood of preterm and term neonates during the first 6 months of life, using adult blood samples as a comparison. We found that the IgA repertoire diversifies slowly after birth. Due to short CDR-H3 regions and very low numbers of somatic mutations, the immature IgA repertoire distinctively differs from the adult IgA repertoire. These characteristics may explain the low antigen affinity and poly-reactivity of neonatal IgA antibodies (16) and contribute to the altered pattern of antigen reactivity that characterizes the very young (17).
Materials and Methods
Patient samples
Blood samples were collected from both extremely preterm neonates (25-30 weeks of gestation, n= 15) and from term neonates (39-42 weeks of gestation, n=14) at birth (cord blood) or at a postnatal age ranging from 1 to 35 weeks (venous blood); and from healthy adults (n=7). Postnatal samples were collected during routine blood tests which were most frequently performed for the control of blood gases, serum-electrolytes, or bilirubin. In each case, the blood for required clinical tests was collected first. Subsequently, 0.2 ml of EDTA blood was collected in a separate tube for this research project. The analysis of the IgA repertoires shown here is part of a project to describe IgM, IgG, and IgA repertoires during ontogeny. Each blood sample originates from a different individual. Cord blood was collected from umbilical cord arteries after thorough cleaning to avoid cross contamination with maternal blood. The numbers of blood samples and sequences of IgA transcripts from each sample are given in Table I. All subjects were caucasians. Gestational age was calculated from the first day of the last menstrual period and confirmed by early ultrasound and by clinical examination. Postconceptional age was calculated as gestational age plus postnatal age. The study was conducted in accordance with the guidelines of the World Medical Association’s Declaration of Helsinki. The Institutional Review Boards of the Free University of Berlin and of the Philipps-University Marburg approved the study protocol. The written consent of parents and adult donors was obtained.
Table I.
Somatic diversity and mutational frequency of IgA transcripts
| identifiera | gestational age (wk) |
postnatal age (wk) |
postcon ceptional age (wk) |
unique sequencesb |
clonotypes (% of total) |
somatic mutation frequencyc (CDR1 - FR3) |
antigen selection |
GenBank acc nod | |
|---|---|---|---|---|---|---|---|---|---|
| Preterm infants | |||||||||
| 1010 | 27 | 0 | 27 | 15 | 13 | (87) | 11.47 | 0.00% | DQ454820-DQ454835e |
| 1050 | 27 | 0 | 27 | 14 | 12 | (86) | 3.91 | 0.00% | DQ454848-DQ454861e |
| 2010 | 27 | 2 | 29 | 17 | 15 | (88) | 5.91 | 0.00% | DQ454879-DQ454895e |
| 2030 | 28 | 2 | 30 | 12 | 9 | (75) | 1.23 | 0.00% | DQ454896-DQ454908e |
| 2046 | 26 | 6 | 32 | 14 | 10 | (71) | 2.34 | 0.00% | JN376223-JN376236 |
| 2045 | 26 | 6 | 32 | 15 | 11 | (73) | 3.69 | 0.00% | JN376208-JN376222 |
| 2059 | 27 | 12 | 39 | 18 | 16 | (89) | 9.35 | 0.00% | JN376259-JN376276 |
| 2150 | 27 | 14 | 41 | 15 | 15 | (100) | 3.51 | 0.00% | JN376356-JN376369 |
| 2065 | 27 | 15 | 42 | 16 | 16 | (100) | 17.47 | 18.80% | JN376277-JN376292 |
| 2044 | 27 | 19 | 46 | 19 | 16 | (84) | 10.7 | 0.00% | JN376189-JN376207 |
| 2160 | 28 | 19 | 47 | 13 | 13 | (100) | 13.83 | 0.00% | DQ454916-DQ454929e |
| 2180 | 27 | 23 | 50 | 13 | 11 | (85) | 6.74 | 0.00% | JN376162-JN376174 |
| 2190 | 27 | 28 | 55 | 14 | 12 | (86) | 10.86 | 14.30% | JN376175-JN376188 |
| 2072 | 30 | 30 | 60 | 18 | 17 | (94) | 12.55 | 0.00% | JN376293-JN376310 |
| 2057 | 25 | 35 | 60 | 22 | 16 | (73) | 21.85 | 0.00% | JN376237-JN376258 |
| Term infants | |||||||||
| 3064 | 40 | 0 | 40 | 14 | 8 | (57) | 6.27 | 0.00% | JN376493-JN376506 |
| 3303 | 39 | 0 | 39 | 16 | 8 | (50) | 4.66 | 0.00% | JX173304-JX173319 |
| 3304 | 40 | 0 | 40 | 28 | 16 | (57) | 3.22 | 0.00% | JX173320-JX173347 |
| 3312 | 40 | 0 | 40 | 19 | 10 | (53) | 4.25 | 0.00% | JX173348-JX173366 |
| 4020 | 39 | 2 | 41 | 18 | 14 | (78) | 8.9 | 5.60% | JN376507-JN376524 |
| 4060 | 40 | 2 | 42 | 25 | 25 | (100) | 4.91 | 0.00% | JN376525-JN376549 |
| 4100 | 40 | 7 | 47 | 26 | 23 | (88) | 10.83 | 3.80% | DQ454943-DQ454970e |
| 4130 | 40 | 9 | 49 | 21 | 13 | (62) | 6.6 | 0.00% | JN376395-JN376415 |
| 4150 | 40 | 12 | 52 | 11 | 11 | (100) | 7.99 | 0.00% | DQ454971-DQ454984e |
| 4190 | 40 | 16 | 56 | 19 | 17 | (89) | 11.13 | 0.00% | JN376450-JN376468 |
| 4180 | 40 | 16 | 56 | 21 | 20 | (95) | 13.12 | 0.00% | DQ454986-DQ455007e |
| 4160 | 41 | 16 | 57 | 24 | 12 | (50) | 8.35 | 0.00% | JN376469-JN376492 |
| 4200 | 39 | 17 | 56 | 23 | 21 | (91) | 21.12 | 8.70% | JN376416-JN376438 |
| 4210 | 42 | 20 | 62 | 11 | 11 | (100) | 13.5 | 0.00% | JN376439-JN376449 |
| Adults | Age (years) |
||||||||
| 5020 | 22 | 14 | 10 | (71) | 68.20 | 21.4% | JN376608-JN376621 | ||
| 5030 | 36 | 14 | 10 | (71) | 67.81 | 28.6% | JN376622-JN376636 | ||
| 5040 | 39 | 15 | 15 | (100) | 54.37 | 26.7% | JN376637-JN376651 | ||
| 5050 | 37 | 13 | 13 | (100) | 76.57 | 46.2% | JN376652-JN376664 | ||
| 5060 | 33 | 35 | 33 | (94) | 70.79 | 28.6% | JN376687-JN376723 | ||
| 5070 | 28 | 37 | 27 | (73) | 71.93 | 32.4% | JN376724-JN376760 | ||
| 5080 | 29 | 22 | 20 | (91) | 87.52 | 54.6% | JN376665-JN376686 | ||
| Total sum (33 samples) | 661 | 539 | (82) | ||||||
Four digit number identifies the patient.
Unique sequences were defined as sequences with at least one nucleotide difference.
Somatic mutation frequency was calculated for all unique sequences within one blood sample
Website address: www.ncbi.nlm.nih.gov/genbank/
from Bauer et al. (18)
wk = weeks
Preparation of RNA and RT-polymerase chain reaction
Erythrocytes were lysed and leukocytes were recovered by centrifugation. Total RNA was isolated using the QIAamp RNA Blood Mini-Kit (Qiagen; Hilden, Germany) according to the manufacturer’s protocol. A combination of primers for the framework region 1 (FR1) of all human VH gene families (12) was used together with a consensus antisense primer specific for the first exon of the α1/2 constant region (Table II) (18). RT-PCR amplifications were carried out in a total volume of 50 μl containing 5 μl of RNA eluate, 2.5 mM MgCl2, 7.5 U recombinant RNase inhibitor, and 0.66 μM of each forward and reverse primer using a OneStep RT-PCR kit (Qiagen; Hilden, Germany). The following program was performed on a thermocycler (Sensoquest; Göttingen, Germany): 30 min at 50°C, 15 min at 94°C followed by 40 cycles using a cycle profile of 1 min at 94°C, 1 min at 66°C, and 1 min at 72°C, followed by a final extension of 10 min at 72°C. As a control for RNA quality GAPDH transcripts were amplified from each sample using humGAPDH-1 and humGAPDH-2 primers (Table II). PCR products were gel purified and DNA was extracted with QIAquick gel extraction kit (Qiagen).
Table II.
PCR primers used VH sense primer mixa
| IGHV1,3,5 | 5′-GTG CAG CTG GTG SAG TCT GG-3′ |
| IGHV2 | 5′-AGA TCA CCT TGA AGG AGT CTG G-3′ |
| IGHV4 | 5′-AGG TGC AGC TRC AGS AGT SG-3′ |
| IGHV6 | 5′-CAG CTG CAG CAG TCA GGT CC-3′ |
| CH antisense primer | |
| Cα1/2 reverse | 5′-GAG GCT CAG CGG GAA GAC CTT G-3′ |
| GAPDH-control | |
| humGAPDH 1 | 5′-AAT GCC TCC TGC ACC ACC AAC-3′ |
| humGAPDH 2 | 5′-GAC GGC AGG TCA GGT CCA CCA-3′ |
(S = G or C; R = A or G)
The VH sense primers are specific for the FR1 of all IgH VH families.
Cloning of PCR products
Ligation and transfection were performed using standard protocols according to the manufacturer’s instructions (TOPO-TA cloning kit, Invitrogen; Karlsruhe, Germany).
Sequence analysis
After the transformed cells had grown on agar plates, 20-35 clones from each subject were randomly selected. Plasmid DNA was extracted, linearized, and sequenced using an ABI capillary sequencer. Gene segments were aligned to germline gene segments using the ImMunoGeneTics (IMGT) database with V-QUEST (19). A minimum of six non-mutated nucleotides with at least two non-mutated nucleotides at each end were required to identify a diversity (D) gene (20). The CDR-H3 was defined to include those residues between the conserved cysteine (C104) of FR-H3 and the conserved tryptophan (W118) of FR-H4. To analyze the patterns of somatic mutations for signs of antigen-selection, we used the algorithms of Lossos et al. (21) and of Chang and Casali (22) as described previously (23). According to this method, the probability of antigen selection increases with the number of replacement mutations within the CDR regions. Sequences with a ratio of somatic mutation frequency in FR2 to FR3 > 3 or < 0.33 and a difference in somatic mutation frequency between FR2 and FR3 > 100‰ were excluded from the analysis to minimize the chance of including hybrid genes formed during the PCR amplification.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 5.0 (La Jolla, USA) and SPSS 17.0 (Chicago, USA). Normality distribution was assessed with Kolmogorov-Smirnov test. Differences between populations were assessed by a two-tailed Student’s t test for normally distributed data or a Mann-Whitney U test for non-normally distributed data, respectively. For categorical data, a chi-square test with post hoc-analysis was applied as described by Collis et al. (24). A p ≤ 0.05 was accepted significant. Means are given with standard error.
Results
To compare the age-related and environmental influences on the postnatal development of the IgA repertoire in preterm and term neonates, we analyzed a total of 752 functional IgA transcripts. Of these transcripts, 663 (88%) were unique, including 235 from 15 preterm neonates, 276 from 14 term neonates, and 150 from 7 adult venous blood samples. This analysis includes 129 sequences from 8 blood samples that had previously been studied for the presence of homology directed recombination (Table 1) (18).
The IgA H chain repertoire of preterm neonates retained the characteristics of fetal IgM H chain variable regions, including short N(D)N regions and overrepresentation of DH7–27
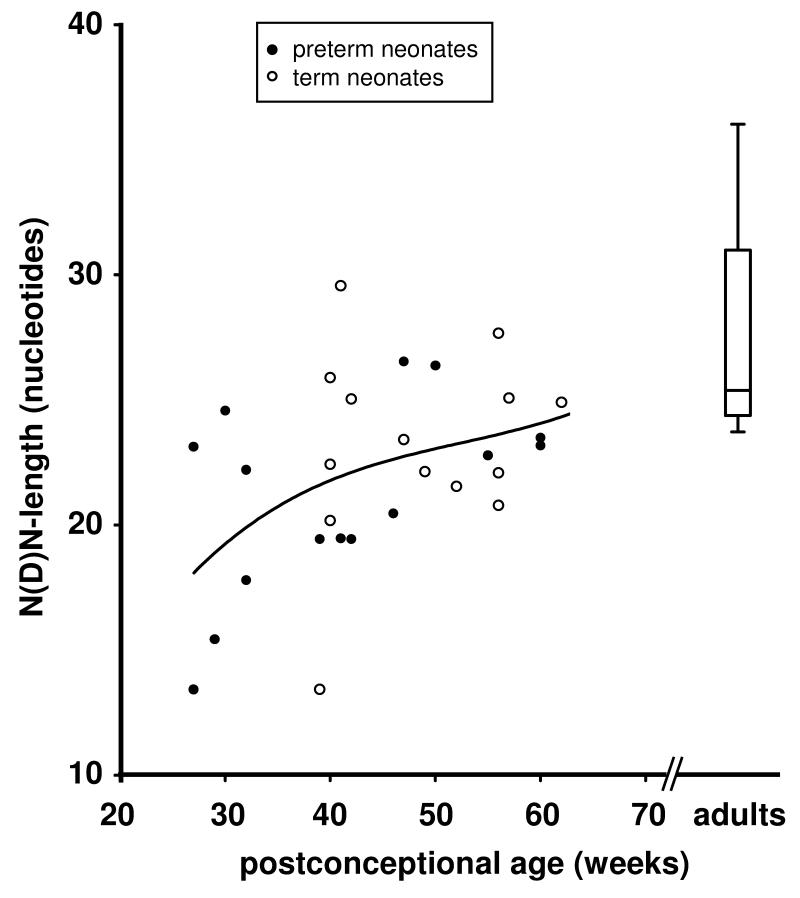
In preterm neonates, the N(D)N length increased during the period corresponding to the third trimester of gestation by 0.17 nucleotides per week (r=0.538, p<0.0001) (Fig. 1). At a postconceptional age of >50 weeks, the N(D)N length was similar in preterm and term neonates and had reached adult N(D)N length. Thus, N(D)N length increase was similar after premature birth and during intrauterine development. The increase in N(D)N length during ontogeny was mainly due to increasing numbers of N nucleotides that were added at the DH-JH-junction by 0.11 nucleotides per week (r=0.756, p<0.001).
FIGURE 1. N(D)N length of IgA transcripts.
Each data point represents the mean of one blood sample (see Table I). N(D)N length in preterm infants increased with postconceptional age (r=0.538, p<0.0001). Near term, the N(D)N length in preterm and term neonates reached adult levels. The polynominal non-linear best fit curve is shown for IgA sequences. Adult data are shown as mean, quartiles, and tukey whiskers.
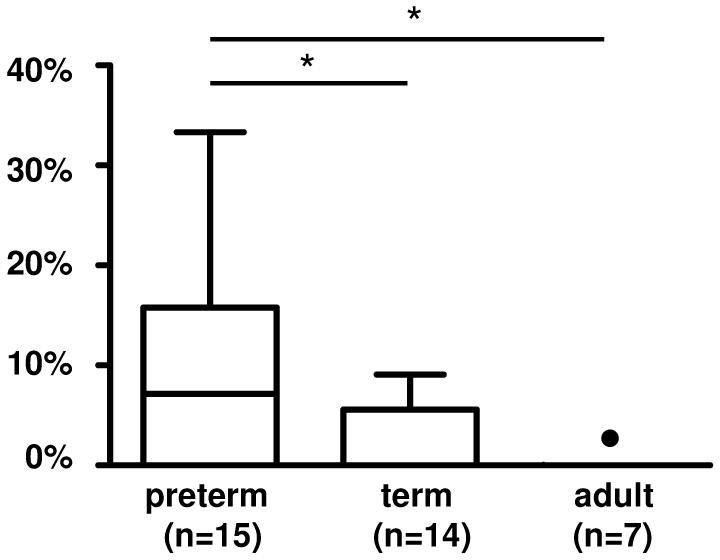
The frequency of the DH7-27, the most JH proximal DH gene segment, undergoes great changes during ontogeny in humans and in mice (reviewed in (25)). We found that DH7-27 was more frequently used in IgA transcripts from preterm (9.8±2.3%) than in term neonate blood B cells (2.5±1.0%, p<0.05) or adults (0.01±0.01%, p<0.001, Fisher exact test) (Fig 2). Use of the other VH, DH, and JH genes in IgA transcripts was similar. Briefly, in comparison to the frequency expected from the number of germline genes, the VH4 and VH6 families, as well as the JH4 genes were overrepresented, whereas the VH3 family was underrepresented in all groups of IgA transcripts studied (not shown). In all three groups, the VH6-1 gene segment was used most frequently, followed by VH4-59 (not shown). In summary, the VH, DH, and JH gene utilization was similar in IgA transcripts as in previously published IgM and IgG transcripts.
FIGURE 2. Usage of DH gene families in IgA transcripts.
The single member of the DH7 gene family, DH7-27, was overrepresented in IgA transcripts from preterm neonates, and its frequency decreased during ontogeny.
The somatic mutation frequency rises slowly in preterm neonates
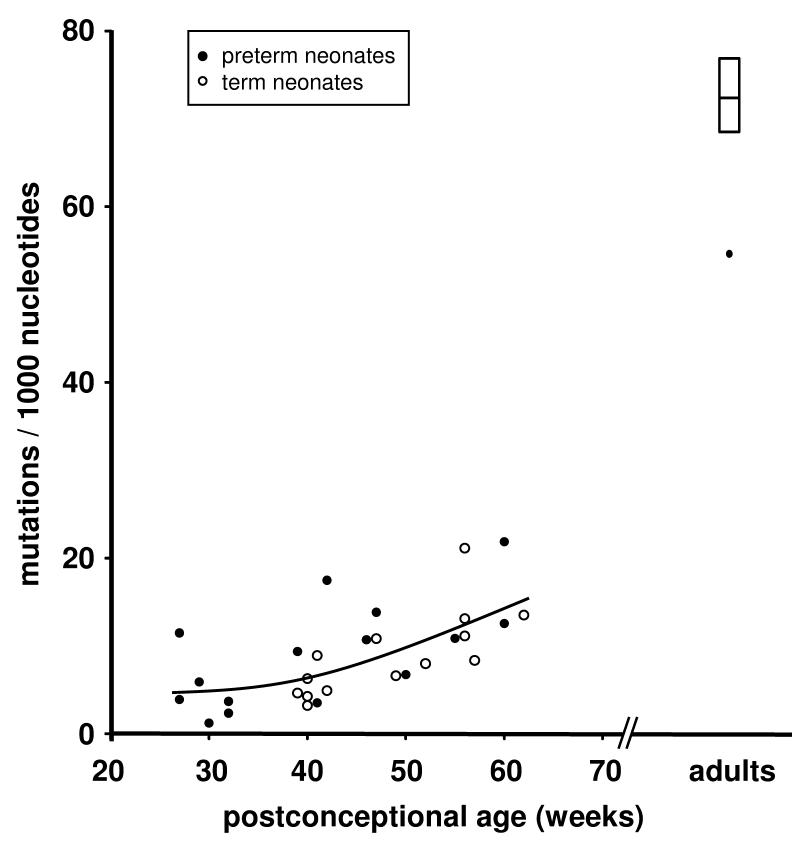
In neonates, the somatic mutation frequency within CDR-H1 to FR-H3 (number of mismatches to the most homologous VH gene segment per 1,000 nucleotides) increased during the time period studied by 0.35‰ (preterm, r=0.678, p<0.0001) and 0.44‰ (term, r=0.731, p<0.0001) per week respectively (Fig 3). At a postconceptional age of ~60 weeks the somatic mutation frequency was similar in preterm (17.7‰) and term neonates (15.9‰), but remained markedly below the somatic mutation frequency seen in adults (ranging from 54.4 to 97.5‰; median 71.9‰). The fraction of unmutated IgA transcripts was 42% in preterm neonates, 39% in term neonates (n.s.) and 3% in adults (p<0.0001 versus preterm and term neonates, two sided Chi2 test, respectively).
FIGURE 3. Average somatic mutation frequency of IgA transcripts.
Each data point represents the mean of one blood sample (see Table I). Somatic mutation frequency increased with postconceptional age (preterm, r=0.678, p<0.0001 / term, r=0.687, p<0.0001). At a postconceptional age of ~60 weeks the somatic mutation frequency was similar in preterm and term neonates. The polynominal non-linear best fit curve is shown for IgA sequences. Adult data are shown as mean, quartiles, and tukey whiskers.
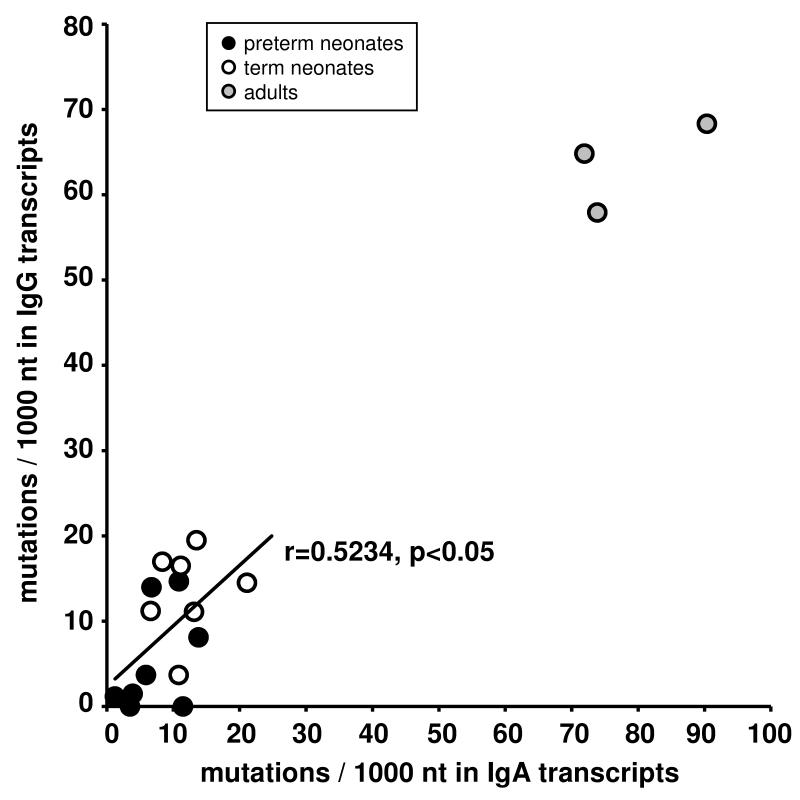
We found a close correlation between somatic mutation frequency of IgA transcripts and of previously published IgG transcripts in 15 blood samples from the same neonates that were studied for both isotypes (r= 0.5234, p<0.05) (Fig 4) (12), whereas the somatic mutation frequency remained very low in IgM transcripts throughout ontogeny.
FIGURE 4. Correlation between the somatic mutation frequencies of IgA and IgG transcripts from the same blood samples.
IgG transcripts had been obtained in a previous study (12) from the same 15 blood samples that were used to analyze IgA transcripts (r=0.5234, p< 0.05, excluding adult samples).
To exclude significant biasing by Taq polymerase error, we counted the mismatches within the 17 nucleotides of the C region that were included in the amplificates upstream of the reverse primer. The Taq error rate was estimated to be 0.61/1,000 nt in IgA transcripts, thus within the range observed for analogous methods for IgM and IgG transcripts. Therefore, the observed differences cannot be explained by Taq polymerase error.
IgA H chain transcripts of neonates display a very low degree of antigen selection
To evaluate whether IgA sequences demonstrated signs of antigen selection, we analyzed the distribution of replacement and silent mutations between framework regions (FR) and CDR as described previously (23). A previously described binomial distribution method (22) was used to determine the 95% confidence limits for the random enrichment of replacement mutations in the CDRs (26). This confidence limits are depicted as grey area in Figure 5. A data point falling outside this gray shaded area represents a sequence which has a high proportion of replacement mutations in the CDR. The probability that such a sequence has accumulated as many replacement mutations in the CDRs by mere random mutation is less than 0.05. Therefore, an allocation above the upper confidence limit was considered indicative of antigen-driven selection.
FIGURE 5. Antigen selection of IgA transcripts.
Inference of Ag selection in IgA transcripts from preterm neonates (A), term neonates (B) and adults (C). Shown is the ratio of replacement mutations in CDR-H1 and CDR-H2 (RCDR) to the total number of mutations in the V region (MV) plotted against MV. The shaded area represents the 95% confidence limits for the probability of random mutations. A data point falling outside these confidence limits represents a sequence that has a high proportion of replacement mutations in the CDR. The probability that these mutations occurred randomly is p < 0.05. 2 % of preterm and term neonate IgA transcripts exhibited statistical signs of antigen selection in comparison to 34 % of adult IgA transcripts (p<0.05, two-tailed Chi2 Test, respectively). Numbers of sequences are written above the dots; size of dots increases with the number of sequences with the same parameters.
According to this definition, 34% of the IgA transcripts of adults showed signs of antigen selection. Interestingly, the picture was strikingly different in neonates: The percentage of IgA sequences with signs of antigen selection was 2.1% (p<0.001 versus adults) in preterm neonates and 1.7% (p<0.001, two sided Chi2 test versus adults) in term neonates (Fig 5). Some neonatal sequences with few somatic mutations fulfill the mathematical criteria for antigen selection, although the biological significance of these findings is uncertain. Thus it is possible that our approach overestimates the number of antigen selected sequences in neonates.
The clonal diversity of each blood sample did not differ between the three groups, ranging from 71 to 100 percent in preterm neonates (median 87%), 50 to 100% in term neonates (median 91%) and 71 to 100% in adults (median 89%) (Table I).
Discussion
In this study we present the first ontogenetic analysis of the IgA repertoire in the perinatal period. We found that IgA H chain transcripts were present in cord blood as early as 27 weeks of gestation and that known restrictions of the primary antibody repertoire (IgM) persisted in the IgA repertoire. Preterm and term neonates possessed a unique IgA repertoire characterized by short CDR-H3 regions, biased DH gene usage, and very few somatic mutations. Thus, the IgA repertoire of the newborn is distinctly different from the adult IgA repertoire. During the first postnatal months, these restrictions were slowly released and the IgA H chain transcripts contained increasing evidence of somatic hypermutation that reflects antigen exposure. Preterm birth did not significantly accelerate the maturation of the IgA repertoire.
It has been suggested that the perinatal period is a window of opportunity for imprinting the B cell repertoire towards or against diseases of a dysbalanced immune system, such as allergies and autoimmune diseases (27, 28). These diseases represent misled antigen-driven secondary antibody reactions (26, 29). Thus studying the ontogeny of secondary antibody repertoires could lead to a better understanding of the pathogenesis of many immune diseases, and could help identifying key periods where manipulation of the repertoire might prevent diseases. We and others have demonstrated in previous studies that analyses of immunoglobulin transcripts can give valuable insights into the selective pressures acting on B cells during the recruitment into various B cell subpopulations and during physiologic and pathologic immune responses ((30, 31); review: (32)). Comparing the postnatal development of preterm neonates and term neonates represents a unique model that allows distinguishing between antigen-induced and maturity-induced mechanisms in the immune system.
Birth initiates the transition from the intrauterine germ free environment to the extrauterine confrontation with microbial and food antigens. This stimulus has a profound influence on the maturation of mucosa associated lymphatic tissues (e.g. Peyer’s patches) and other lymphoid tissues (7, 33, 34). Neonatal mice that were bred under germ-free conditions and fed an antigen-free diet failed to produce secondary antibody repertoires and were highly susceptible to infections when exposed to microbes later in life (35). The preterm neonates of our study were exposed to dietary antigens (formula milk or breast milk) from the first day of life and skin-to-skin contact was encouraged from the first week of life, enabling the colonization with commensals approximately three to four months earlier than in a term neonate. Despite this massive exposure to foreign antigens, the IgA heavy chain transcripts produced by the premature infants did not show more evidence of antigen-driven selection when they reached due date than term neonates of the same postconceptional age. This supports the view that many of the maturational steps involved in the ontogeny of the antibody repertoire are genetically regulated and are triggered independent of antigen exposure.
Most of the preterm neonates had at least one infection during the observed episode. Similar to previously published IgG sequences from the same blood samples, extensive statistical comparisons did not reveal differences between children with or without a history of infection (data not shown; (12)). This is not surprising, because compared to the continuous stimulation by non-pathogenic microbiota or dietary antigens, an episode of infection is a rare and time-limited antigenic exposure.
Our study provides molecular evidence that class switch recombination to IgA during intrauterine life is occurring without the high level of somatic mutations normally observed during adult antigen-driven selection. The diversity of IgA transcripts, which can be interpreted as a rough estimate for the clonal diversity, reached similar levels in extremely preterm neonates as in adults. Notably, both class switch recombination and somatic hypermutation are dependent on the enzymatic activity of AID, but the different processes are supported by different domains of the enzyme (36). This separation is in harmony with the observation that although specific antibody responses, including IgA, can be elicited by intrauterine infection or vaccination (37, 38), neonatal antibody responses are characterized by lower affinity and shorter half-life than in the mature organism (17).
After term, the somatic mutation frequency increased by approximately 0.7 ‰ per week, both in term and preterm neonates. Even at a postconceptional age of 60 weeks, equivalent to five months of age after term, the somatic mutation frequencies in preterm and term neonates were only approximately a quarter of the adult level, and only very few somatic mutations fulfilled the criteria of antigen-driven selection (21, 22). The lower number of somatic mutations in IgA transcripts from neonates could either reflect a lower activity of the enzyme AID, which normally introduces approximately 1 somatic mutation per 1,000 nucleotides and cell division into the immunoglobulin heavy chain gene (39), and/ or the cells in neonates could have undergone fewer cell divisions. Alternatively, the low number of somatic mutations might reflect a bias in neonates towards T cell independent IgA formation, as in mice, non-mutated IgA sequences appear to be generated through T cell independent mechanisms, whereas mutated IgA sequences require the presence of T cell mediated signals (40, 41). One can hypothesize that T cell help – and thus production of mutated IgA sequences – might be restricted during early ontogeny, e.g. due to the decreased expression of co-stimulatory receptors (CD40, CD80, CD86) (Reviewed in (17)).
In adults, IgA H chain transcripts from peripheral blood contained as many somatic mutations as IgA H chain transcripts from tonsils (42) or from intestinal plasma cells (43). Moreover, the somatic mutations within the IgA transcripts from adult peripheral blood showed all known characteristics of the antigen-driven selection that have been observed in both IgG and IgE repertoires as well (12, 26). The somatic mutation frequencies of IgA and IgG transcripts from the same blood samples correlated, indicating that comparable mechanisms may regulate the number of somatic mutations in IgG and IgA. This argues against the assumption that the IgA repertoire in circulating blood lymphocytes would reflect a rather diffuse, less selected antibody production. However, this hypothesis had been put forth for murine gut plasmablasts, that often arise from low affinity, polyreactive IgM-expressing B1 cells (14). It must be considered that important aspects of the human and murine IgA production differ. In example, phenotypic differences have recently been discovered between human and murine B1 cells (44), and, in contrast to mice, human peritoneal cells probably do not contribute significantly to the IgA secretion of the gut (45).
We found that, regarding DH utilization and CDR-H3 length, the IgA repertoire of neonates retained the same restrictions as the primary (IgM) repertoire (11, 12). Thus, class switch to IgA did not favor B cells with more “mature” VH regions. This could indicate that the main characteristics of the secondary immunoglobulin repertoire are already pre-determined during early B cell development prior to exogenous antigen contact. This finding is in line with observations in mouse models in which a bias had been introduced into the primary antibody repertoire by gene targeting the immunoglobulin heavy chain Diversity gene locus (32, 46-48). In agreement with our findings in the human neonatal IgM and IgA repertoires, somatic selection during class switch and affinity maturation was also insufficient to correct a bias within the primary IgM repertoire in these gene targeted mice. Interestingly, Kolar et al. have shown that the differences in CDR-H3 length and DH utilization were reduced in the IgM repertoires when NOD/SCID/β2m-/- mice were reconstituted with human fetal or adult lymphocyte progenitors. This indicates that the observed restrictions during early ontogeny are not exclusively caused by the B cell progenitors themselves but also by yet unknown factors of the environment (49).
Similar to IgM and IgG, the length of the CDR-H3 regions significantly increased by more than 5 nucleotides from extremely preterm neonates to adults. The paramount cause was the increased addition of non-template N-nucleotides between the rearranged Variable, Diversity and Joining gene segments. In theory, adding six random nucleotides to CDR-H3 would increase the potential diversity by 202 = 400 (two random amino acids out of twenty amino acids added). Similar to the fetus, TdT deficient mice produce very short CDR-H3 regions due to the absence of N-nucleotides. Interestingly, TdT deficient mice fill their peripheral lymphocyte pool more rapidly than wildtype mice (50, 51), but their secondary responses to NP19-CGG and to Lysozyme are weaker (52). In analogy, short CDR-H3 regions might enable human fetuses and neonates to rapidly establish their B cell populations in spite of the costs of lower antigen affinity of the antibody repertoire which appears to also persist in the secondary IgA and IgG repertoires.
A further constraint of neonatal IgA transcripts was the predominant use of the DH7-27 gene segment similar to IgM and IgG transcripts (11, 12). DH7-27 is the shortest DH gene segment and the only one of the 27 human DH gene segments that does not predominantly encode for neutral-hydrophilic amino acids. Based on these differences in length and amino acid composition of the CDR-H3 loops, it can be assumed that not only the diversity but also the structural repertoire of antigen binding sites must differ between preterm neonates, term neonates, and adults.
The structure of the antigen-binding site has strong influence on its function. It is highly probable that the differing characteristics of IgA transcripts during ontogeny are associated with differing preferences for antigen binding, since for example, anti-hapten antibodies usually have shorter CDR-H3 loops than anti-DNA-antibodies (24, 53). The antigen-binding site typically forms a groove when the CDR-H3 loop is shorter than 14 amino acids, whereas longer CDR-H3 loops protrude into the solvent (53, 54). According to this differentiation, the frequency of antigen binding grooves would decrease from 55.3% in preterm neonates to 38.1% in term neonates and would subsequently remain stable (40.5% in adults) (preterm vs term p<0.001, preterm vs adults p< 0.005). Taken together, since similar restrictions characterize the CDR-H3 repertoires of neonatal IgM, IgG, and IgA, it is possible that similar restrictions also apply to the antigen binding properties.
One limitation of our study is that due to low sample volumes, we have analyzed IgA transcripts from whole blood and not from sorted B cell subpopulations. Since longitudinal samples were unavailable for this study, each blood sample originates from a different individual, and we cannot measure the intraindividual development. It is possible that the observed differences during ontogeny partially arise from differing relations of B cell subpopulations (e.g. plasmablasts/plasma cells and memory B cells, and newly defined B1 cells (44, 55)) or differing routes of IgA formations (T cell dependent / independent) (41). Moreover, differing selective forces could act upon the membrane bound versus secretory IgA which cannot be distinguished with our experimental approach. The observed differences during ontogeny are probably not due to differing frequency of IgA1 and IgA2 producing cells, since in harmony with Schauer et al. we found similar IgA1 to IgA2 ratios of approximately 6.5:1 in preterm neonates, term neonates, and adult blood samples using quantitative PCR (data not shown, (56)). Feeding with breast milk versus formula milk has significant impact on the gut microbiota (57) and on the developing immune system (58), and may also influence the development of the IgA repertoire. However, due to the local feeding guidelines, all preterm neonates obtained formula milk from their first day and breast milk as soon as available. In consequence, most of our patients were not exclusively fed with one type of milk. Thus this cohort is unsuitable to study the potential influence of feeding type on the maturation of the IgA repertoire.
In summary, our study describes the development of the IgA repertoire during the critical time period of the stepwise acquisition of the ability to form antibodies against various antigens (59). We found that the IgA repertoires of preterm and term neonates are subject to the same restrictions as the IgM and IgG repertoires and that signs of antigen-driven selection accumulated very slowly. Thus, regarding DH utilization, CDR-H3 length, and somatic mutations, the diversification of the IgM, IgG, and IgA repertoires depends predominantly on maturation and less on antigen contact. The time of maturation for the IgA repertoire in the neonate obviously exceeds the time of passive IgA transfer through mother’s milk by far. From a teleological view it is highly unlikely that the strictly regulated diversification of the fetal and neonatal antibody repertoire would represent only immaturity per se. Instead the current view is that the altered immune status of the fetus and neonate, including antigen binding properties of the IgM, IgA and IgG repertoire, may be beneficial to establish tolerance to self antigens or maternal antigens and may contribute to the prevention of autoimmune and allergic diseases (59). On the other hand, the preterm neonate might be highly susceptible to infections and less responsive to vaccination because its immune system is still at the stage of tolerance induction instead of self-defense.
Acknowledgements
In memory of Karl Bauer † who supported this work during its initial stages. We gratefully thank Regina Stoehr and Sabine Jennemann for their excellent technical assistance. We thank Helge Prinz † from the Institut für Biometrie und Epidemiologie, Philipps University Marburg, for statistical help.
Footnotes
Disclosure The authors disclaim no conflict of interest.
Reference List
- 1.Corthesy B. Roundtrip ticket for secretory IgA: role in mucosal homeostasis? J. Immunol. 2007;178:27–32. doi: 10.4049/jimmunol.178.1.27. [DOI] [PubMed] [Google Scholar]
- 2.Suzuki K, Ha SA, Tsuji M, Fagarasan S. Intestinal IgA synthesis: a primitive form of adaptive immunity that regulates microbial communities in the gut. Semin. Immunol. 2007;19:127–135. doi: 10.1016/j.smim.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Kramer DR, Cebra JJ. Early appearance of “natural” mucosal IgA responses and germinal centers in suckling mice developing in the absence of maternal antibodies. J. Immunol. 1995;154:2051–2062. [PubMed] [Google Scholar]
- 4.Crabbe PA, Nash DR, Bazin H, Eyssen H, Heremans JF. Immunohistochemical observations on lymphoid tissues from conventional and germ-free mice. Lab. Invest. 1970;22:448–457. [PubMed] [Google Scholar]
- 5.Harris NL, Spoerri I, Schopfer JF, Nembrini C, Merky P, Massacand J, Urban JF, Jr., Lamarre A, Burki K, Odermatt B, Zinkernagel RM, Macpherson AJ. Mechanisms of neonatal mucosal antibody protection. J. Immunol. 2006;177:6256–6262. doi: 10.4049/jimmunol.177.9.6256. [DOI] [PubMed] [Google Scholar]
- 6.Gibbons DL, Spencer J. Mouse and human intestinal immunity: same ballpark, different players; different rules, same score. Mucosal Immunol. 2011;4:148–157. doi: 10.1038/mi.2010.85. [DOI] [PubMed] [Google Scholar]
- 7.Weemaes C, Klasen I, Goertz J, Beldhuis-Valkis M, Olafsson O, Haraldsson A. Development of immunoglobulin A in infancy and childhood. Scand. J. Immunol. 2003;58:642–648. doi: 10.1111/j.1365-3083.2003.01344.x. [DOI] [PubMed] [Google Scholar]
- 8.Mortari F, Wang JY, Schroeder HW., Jr. Human cord blood antibody repertoire. Mixed population of VH gene segments and CDR3 distribution in the expressed C alpha and C gamma repertoires. J. Immunol. 1993;150:1348–1357. [PubMed] [Google Scholar]
- 9.Cuisinier AM, Gauthier L, Boubli L, Fougereau M, Tonnelle C. Mechanisms that generate human immunoglobulin diversity operate from the 8th week of gestation in fetal liver. Eur. J. Immunol. 1993;23:110–118. doi: 10.1002/eji.1830230118. [DOI] [PubMed] [Google Scholar]
- 10.Schroeder HW, Jr., Zhang L, Philips JB., 3rd Slow, programmed maturation of the immunoglobulin HCDR3 repertoire during the third trimester of fetal life. Blood. 2001;98:2745–2751. doi: 10.1182/blood.v98.9.2745. [DOI] [PubMed] [Google Scholar]
- 11.Bauer K, Zemlin M, Hummel M, Pfeiffer S, Karstaedt J, Steinhauser G, Xiao X, Versmold H, Berek C. Diversification of Ig heavy chain genes in human preterm neonates prematurely exposed to environmental antigens. J. Immunol. 2002;169:1349–1356. doi: 10.4049/jimmunol.169.3.1349. [DOI] [PubMed] [Google Scholar]
- 12.Zemlin M, Hoersch G, Zemlin C, Pohl-Schickinger A, Hummel M, Berek C, Maier RF, Bauer K. The postnatal maturation of the immunoglobulin heavy chain IgG repertoire in human preterm neonates is slower than in term neonates. J. Immunol. 2007;178:1180–1188. doi: 10.4049/jimmunol.178.2.1180. [DOI] [PubMed] [Google Scholar]
- 13.Stoel M, Jiang HQ, van Diemen CC, Bun JC, Dammers PM, Thurnheer MC, Kroese FG, Cebra JJ, Bos NA. Restricted IgA repertoire in both B-1 and B-2 cell-derived gut plasmablasts. J. Immunol. 2005;174:1046–1054. doi: 10.4049/jimmunol.174.2.1046. [DOI] [PubMed] [Google Scholar]
- 14.Bao S, Beagley KW, Murray AM, Caristo V, Matthaei KI, Young IG, Husband AJ. Intestinal IgA plasma cells of the B1 lineage are IL-5 dependent. Immunology. 1998;94:181–188. doi: 10.1046/j.1365-2567.1998.00512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang HS, Chin RK, Wang Y, Yu P, Wang J, Newell KA, Fu YX. Signaling via LTbetaR on the lamina propria stromal cells of the gut is required for IgA production. Nat. Immunol. 2002;3:576–582. doi: 10.1038/ni795. [DOI] [PubMed] [Google Scholar]
- 16.Quan CP, Berneman A, Pires R, Avrameas S, Bouvet JP. Natural polyreactive secretory immunoglobulin A autoantibodies as a possible barrier to infection in humans. Infect. Immun. 1997;65:3997–4004. doi: 10.1128/iai.65.10.3997-4004.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Siegrist CA, Aspinall R. B-cell responses to vaccination at the extremes of age. Nat. Rev. Immunol. 2009;9:185–194. doi: 10.1038/nri2508. [DOI] [PubMed] [Google Scholar]
- 18.Bauer K, Hummel M, Berek C, Paar C, Rosenberger C, Kerzel S, Versmold H, Zemlin M. Homology-directed recombination in IgH variable region genes from human neonates, infants and adults: implications for junctional diversity. Mol. Immunol. 2007;44:2969–2977. doi: 10.1016/j.molimm.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiokawa S, Mortari F, Lima JO, Nunez C, Bertrand FE, 3rd, Kirkham PM, Zhu S, Dasanayake AP, Schroeder HW., Jr. IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J. Immunol. 1999;162:6060–6070. [PubMed] [Google Scholar]
- 21.Lossos IS, Tibshirani R, Narasimhan B, Levy R. The inference of antigen selection on Ig genes. J. Immunol. 2000;165:5122–5126. doi: 10.4049/jimmunol.165.9.5122. [DOI] [PubMed] [Google Scholar]
- 22.Chang B, Casali P. The CDR1 sequences of a major proportion of human germline Ig VH genes are inherently susceptible to amino acid replacement. Immunol. Today. 1994;15:367–373. doi: 10.1016/0167-5699(94)90175-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rogosch T, Kerzel S, Sikula L, Gentil K, Liebetruth M, Schlingmann KP, Maier RF, Zemlin M. Plasma Cells and Nonplasma B Cells Express Differing IgE Repertoires in Allergic Sensitization. J. Immunol. 2010;184:4947–4954. doi: 10.4049/jimmunol.0900859. [DOI] [PubMed] [Google Scholar]
- 24.Collis AV, Brouwer AP, Martin AC. Analysis of the antigen combining site: correlations between length and sequence composition of the hypervariable loops and the nature of the antigen. J. Mol. Biol. 2003;325:337–354. doi: 10.1016/s0022-2836(02)01222-6. [DOI] [PubMed] [Google Scholar]
- 25.Zemlin M, Schelonka RL, Bauer K, Schroeder HW., Jr. Regulation and chance in the ontogeny of B and T cell antigen receptor repertoires. Immunol. Res. 2002;26:265–278. doi: 10.1385/IR:26:1-3:265. [DOI] [PubMed] [Google Scholar]
- 26.Kerzel S, Rogosch T, Struecker B, Maier RF, Zemlin M. IgE transcripts in the circulation of allergic children reflect a classical antigen-driven B cell response and not a superantigen-like activation. J. Immunol. 2010;185:2253–2260. doi: 10.4049/jimmunol.0902942. [DOI] [PubMed] [Google Scholar]
- 27.Merbl Y, Zucker-Toledano M, Quintana FJ, Cohen IR. Newborn humans manifest autoantibodies to defined self molecules detected by antigen microarray informatics. J. Clin. Invest. 2007;117:712–718. doi: 10.1172/JCI29943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thornton CA, Macfarlane TV, Holt PG. The hygiene hypothesis revisited: role of materno-fetal interactions. Curr. Allergy Asthma Rep. 2010;10:444–452. doi: 10.1007/s11882-010-0148-5. [DOI] [PubMed] [Google Scholar]
- 29.Hiepe F, Dorner T, Hauser AE, Hoyer BF, Mei H, Radbruch A. Long-lived autoreactive plasma cells drive persistent autoimmune inflammation. Nat. Rev. Rheumatol. 2011;7:170–178. doi: 10.1038/nrrheum.2011.1. [DOI] [PubMed] [Google Scholar]
- 30.Brezinschek HP, Foster SJ, Brezinschek RI, Dorner T, Domiati-Saad R, Lipsky PE. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J. Clin. Invest. 1997;99:2488–2501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Souto-Carneiro MM, Sims GP, Girschik H, Lee J, Lipsky PE. Developmental changes in the human heavy chain CDR3. J. Immunol. 2005;175:7425–7436. doi: 10.4049/jimmunol.175.11.7425. [DOI] [PubMed] [Google Scholar]
- 32.Schroeder HW, Jr., Zemlin M, Khass M, Nguyen HH, Schelonka RL. Genetic control of DH reading frame and its effect on B-cell development and antigen-specifc antibody production. Crit. Rev. Immunol. 2010;30:327–344. doi: 10.1615/critrevimmunol.v30.i4.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnold LW, Pennell CA, McCray SK, Clarke SH. Development of B-1 cells: segregation of phosphatidyl choline-specific B cells to the B-1 population occurs after immunoglobulin gene expression. J. Exp. Med. 1994;179:1585–1595. doi: 10.1084/jem.179.5.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fagarasan S, Kinoshita K, Muramatsu M, Ikuta K, Honjo T. In situ class switching and differentiation to IgA-producing cells in the gut lamina propria. Nature. 2001;413:639–643. doi: 10.1038/35098100. [DOI] [PubMed] [Google Scholar]
- 35.Lanning D, Sethupathi P, Rhee KJ, Zhai SK, Knight KL. Intestinal microflora and diversification of the rabbit antibody repertoire. J. Immunol. 2000;165:2012–2019. doi: 10.4049/jimmunol.165.4.2012. [DOI] [PubMed] [Google Scholar]
- 36.Shinkura R, Ito S, Begum NA, Nagaoka H, Muramatsu M, Kinoshita K, Sakakibara Y, Hijikata H, Honjo T. Separate domains of AID are required for somatic hypermutation and class-switch recombination. Nat. Immunol. 2004;5:707–712. doi: 10.1038/ni1086. [DOI] [PubMed] [Google Scholar]
- 37.Silverstein AM. Congenital syphilis and the timing of immunogenesis in the human foetus. Nature. 1962;194:196–197. doi: 10.1038/194196a0. [DOI] [PubMed] [Google Scholar]
- 38.Ballow M, Cates KL, Rowe JC, Goetz C, Desbonnet C. Development of the immune system in very low birth weight (less than 1500 g) premature infants: concentrations of plasma immunoglobulins and patterns of infections. Pediatr. Res. 1986;20:899–904. doi: 10.1203/00006450-198609000-00019. [DOI] [PubMed] [Google Scholar]
- 39.Radu DL, Kodera T, Bona C. Expression of activation-induced cytidine deaminase decreases throughout the life. J. Cell. Mol. Med. 2003;7:141–145. doi: 10.1111/j.1582-4934.2003.tb00212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bemark M, Boysen P, Lycke NY. Induction of gut IgA production through T cell-dependent and T cell-independent pathways. Ann. N. Y. Acad. Sci. 2012;1247:97–116. doi: 10.1111/j.1749-6632.2011.06378.x. [DOI] [PubMed] [Google Scholar]
- 41.Bergqvist P, Stensson A, Lycke NY, Bemark M. T cell-independent IgA class switch recombination is restricted to the GALT and occurs prior to manifest germinal center formation. J. Immunol. 2010;184:3545–3553. doi: 10.4049/jimmunol.0901895. [DOI] [PubMed] [Google Scholar]
- 42.Yavuz S, Grammer AC, Yavuz AS, Nanki T, Lipsky PE. Comparative characteristics of mu chain and alpha chain transcripts expressed by individual tonsil plasma cells. Mol. Immunol. 2001;38:19–34. doi: 10.1016/s0161-5890(01)00036-0. [DOI] [PubMed] [Google Scholar]
- 43.Fischer M, Kuppers R. Human IgA- and IgM-secreting intestinal plasma cells carry heavily mutated VH region genes. Eur. J. Immunol. 1998;28:2971–2977. doi: 10.1002/(SICI)1521-4141(199809)28:09<2971::AID-IMMU2971>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 44.Griffin DO, Holodick NE, Rothstein TL. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70. J. Exp. Med. 2011;208:67–80. doi: 10.1084/jem.20101499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boursier L, Farstad IN, Mellembakken JR, Brandtzaeg P, Spencer J. IgVH gene analysis suggests that peritoneal B cells do not contribute to the gut immune system in man. Eur. J. Immunol. 2002;32:2427–2436. doi: 10.1002/1521-4141(200209)32:9<2427::AID-IMMU2427>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 46.Schelonka RL, Ivanov II, Jung DH, Ippolito GC, Nitschke L, Zhuang Y, Gartland GL, Pelkonen J, Alt FW, Rajewsky K, Schroeder HW., Jr. A single DH gene segment creates its own unique CDR-H3 repertoire and is sufficient for B cell development and immune function. J. Immunol. 2005;175:6624–6632. doi: 10.4049/jimmunol.175.10.6624. [DOI] [PubMed] [Google Scholar]
- 47.Ippolito GC, Schelonka RL, Zemlin M, Ivanov II, Kobayashi R, Zemlin C, Gartland GL, Nitschke L, Pelkonen J, Fujihashi K, Rajewsky K, Schroeder HW., Jr. Forced usage of positively charged amino acids in immunoglobulin CDR-H3 impairs B cell development and antibody production. J. Exp. Med. 2006;203:1567–1578. doi: 10.1084/jem.20052217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zemlin M, Schelonka RL, Ippolito GC, Zemlin C, Zhuang Y, Gartland GL, Nitschke L, Pelkonen J, Rajewsky K, Schroeder HW., Jr. Regulation of Repertoire Development through Genetic Control of DH Reading Frame Preference. J. Immunol. 2008;181:8416–8424. doi: 10.4049/jimmunol.181.12.8416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kolar GR, Yokota T, Rossi MI, Nath SK, Capra JD. Human fetal, cord blood, and adult lymphocyte progenitors have similar potential for generating B cells with a diverse immunoglobulin repertoire. Blood. 2004;104:2981–2987. doi: 10.1182/blood-2003-11-3961. [DOI] [PubMed] [Google Scholar]
- 50.Carey JB, Moffatt-Blue CS, Watson LC, Gavin AL, Feeney AJ. Repertoire-based selection into the marginal zone compartment during B cell development. J. Exp. Med. 2008;205:2043–2052. doi: 10.1084/jem.20080559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schelonka RL, Ivanov II, Vale AM, Dimmitt RA, Khaled M, Schroeder HW., Jr. Absence of N addition facilitates B cell development, but impairs immune responses. Immunogenetics. 2011;63:599–609. doi: 10.1007/s00251-011-0543-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weiss U, Rajewsky K. The repertoire of somatic antibody mutants accumulating in the memory compartment after primary immunization is restricted through affinity maturation and mirrors that expressed in the secondary response. J. Exp. Med. 1990;172:1681–1689. doi: 10.1084/jem.172.6.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ramsland PA, Kaushik A, Marchalonis JJ, Edmundson AB. Incorporation of long CDR3s into V domains: implications for the structural evolution of the antibody-combining site. Exp. Clin. Immunogenet. 2001;18:176–198. doi: 10.1159/000049197. [DOI] [PubMed] [Google Scholar]
- 54.Schroeder HW, Jr., Cavacini L. Structure and function of immunoglobulins. J. Allergy Clin. Immunol. 2010;125:S41–52. doi: 10.1016/j.jaci.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He B, Xu W, Santini PA, Polydorides AD, Chiu A, Estrella J, Shan M, Chadburn A, Villanacci V, Plebani A, Knowles DM, Rescigno M, Cerutti A. Intestinal bacteria trigger T cell-independent immunoglobulin A(2) class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 56.Schauer U, Stemberg F, Rieger CH, Borte M, Schubert S, Riedel F, Herz U, Renz H, Wick M, Herzog W. Establishment of age-dependent reference values for IgA subclasses. Clin. Chim. Acta. 2003;328:129–133. doi: 10.1016/s0009-8981(02)00418-7. [DOI] [PubMed] [Google Scholar]
- 57.Bezirtzoglou E, Tsiotsias A, Welling GW. Microbiota profile in feces of breast- and formula-fed newborns by using fluorescence in situ hybridization (FISH) Anaerobe. 2011;17:478–482. doi: 10.1016/j.anaerobe.2011.03.009. [DOI] [PubMed] [Google Scholar]
- 58.Amarri S, Benatti F, Callegari ML, Shahkhalili Y, Chauffard F, Rochat F, Acheson KJ, Hager C, Benyacoub J, Galli E, Rebecchi A, Morelli L. Changes of gut microbiota and immune markers during the complementary feeding period in healthy breast-fed infants. J. Pediatr. Gastroenterol. Nutr. 2006;42:488–495. doi: 10.1097/01.mpg.0000221907.14523.6d. [DOI] [PubMed] [Google Scholar]
- 59.Wood N, Siegrist CA. Neonatal immunization: where do we stand? Curr. Opin. Infect. Dis. 2011;24:190–195. doi: 10.1097/QCO.0b013e328345d563. [DOI] [PubMed] [Google Scholar]