The demand for parallel, multiplex analysis of protein biomarkers from ever smaller biospecimens is an increasing trend for both fundamental biology and clinical diagnostics.[1–3] The most highly multiplex protein assays rely on spatially encoded antibody microarrays,[4–6] and small biospecimens samples are now routinely manipulated using microfluidics approaches. The integration of antibody microarray techniques with microfluidics chips has only been explored relatively recently. One challenge arises from the relative instability of antibodies to microfluidics fabrication conditions. In recent years, several groups have devised methods to transform standard DNA microarrays in situ into protein microarrays and cell-capture platforms.[7–13] These approaches capitalize on the chemical robustness of DNA oligomers, and the reliable assembly of DNA-labeled structures via complementary hybridization. Recently, Fan et al. utilized a microfluidics-based flow patterning technique to generate DNA barcode-type arrays at 10 × higher density than standard, spotted microarrays.[14] The DNA barcodes were converted into antibody arrays using the DNA-encoded antibody library (DEAL) technique, and then applied towards the measurement of a highly multiplex panel of proteins from a pinprick of whole blood.
A second challenge involves scaling such miniaturized DNA microarrays so that a large surface area can be encoded. This problem is non-trivial, as it involves identifying chemistries for patterning 10−5 m wide, 1 m long strips of biomolecules with a uniformity that permits those patterns to be utilized in hundreds to thousands of quantitative protein assays per chip. Herein, we explore the surface chemistry associated with microfluidics-based flow patterning of DNA barcodes, with an eye towards producing highly reproducible and robust barcodes. We then apply the optimized chemistry towards assaying a panel of cytoplasmic proteins from single cells.
We explore three different flow patterning surface chemistries: two rely upon the electrostatic adsorption of DNA onto a poly-L-lysine (PLL) surface, and the third utilizes flow patterning of dendrimers onto aminated glass substrates, followed by covalent attachment of DNA oligomers onto the dendrimer scaffolds. For the electrostatic adsorption cases, we investigate, using both theory and experiment, the role that counterions play in flow patterning within the confined dimensions of a microfluidic channel, and we find that solvent mixtures which associate counterions more strongly to the negatively charged DNA oligomers yield more reproducible and robust barcodes. We then demonstrate the utility of the best flow patterning chemistry by combining it with DEAL to construct antibody barcodes for quantitatively assaying a panel of phosphorylated proteins, associated with oncogenic pathways, from single cells that are representative of the brain cancer glioblastoma multiforme (GBM).
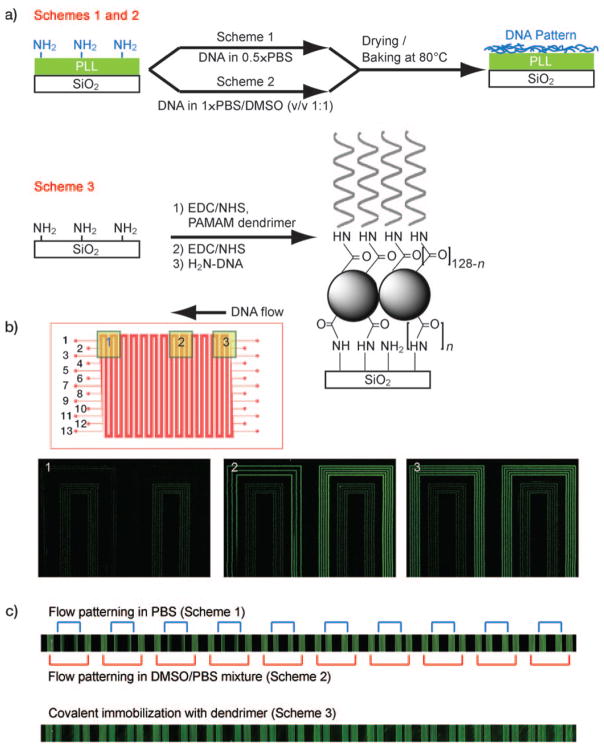
The microfluidics flow patterning chip is comprised of a patterned polydimethylsiloxane (PDMS) layer adhered to an aminated or PLL-coated glass substrate that provides the base surface for the microchannels. The microchannels are long (about 55 cm), meandering channels that span ca. 0.85 cm2 of our substrate, and are used to pattern a DNA barcode over most of the glass surface (Figure 1b). After the flow patterning is completed, the PDMS layer is replaced with a second micro-patterned PDMS layer that is designed to support a biological assay, such as the previously reported blood proteomics chip,[14] or the single-cell proteomics chips utilized herein. For the microfluidic patterning method to be useful, it must generate a DNA barcode that exhibits high and uniform DNA loading over the entire substrate. We evaluated the patterning chemistries illustrated in Figure 1a, Schemes 1–3. Schemes 1 and 2 are drawn from the conventional protocol for pin-spotted microarrays—a solution containing the DNA is introduced, the solvent is evaporated, and subsequent thermal or UV treatment is employed to cross-link the deposited DNA to the substrate. In Scheme 1 ssDNA oligomers dissolved in phosphate-buffered saline (PBS) are utilized, whereas in Scheme 2 ssDNAs in a 1:1 mixture of 1×PBS and dimethyl sulfoxide (DMSO) are employed. DMSO is used in conventional microarray preparation to improve feature consistency by reducing the rate of solvent evaporation and by denaturing the DNA[15] although, as described below, its role in this process is different. In Scheme 3 a covalent immobilization method based upon a dendrimer scaffold is utilized.[16] Poly(amidoamine) (PAMAM) dendrimers (generation 4.5, carboxylate surface) have previously shown promise as DNA and protein microarray substrates. Dendrimers do not form entangled chains[17] and because harsh crosslinking procedures are avoided, dendrimer-immobilized DNA retains high accessibility and activity in microarray applications. Moreover, the highly branched structure of the dendrimers provides a high density of reactive sites for surface attachment and for DNA coupling, thus leading to a high overall binding capacity. For all cases, a high level of DNA loading has been shown to decrease non-specific binding when compared to standard microarray substrates.[11, 18–20]
Figure 1.
a) Surface treatment schemes. b) Design of the DNA patterning device (top) and fluorescence image of DNAs filled into the channel (still in solution). Outer five channels are filled with DNAs in 1:1 mixture of PBS and water (Scheme 1). The five inner are filled with DNA in a 1:1 mixture of PBS and DMSO (Scheme 2). Three channels in between are left empty for visualization. c) Fluorescence images of patterned DNAs by three schemes.
Figure 1b (top) shows the PDMS chip design used for bar-code patterning. Thirteen discrete channels (for a thirteen-element barcode) allow for a multiplex microarray. We loaded five adjacent channels according to Scheme 1, skipped three channels, and then loaded the remaining five channels according to Scheme 2. The use of fluorescently-tagged DNA permitted measurements of the DNA distribution within each individual channel immediately after introducing the solutions. Figure 1b demonstrates a clear difference in aqueous DNA distribution across the chip: DNA loaded according to Scheme 1 (outer five channels) is notably lower in concentration near the middle of the chip (Figure 1b, Region 2) and is barely detectable near the channel exit (Figure 1b, Region 1). Conversely, DNA loaded according to Scheme 2 (inner five channels) presents an even, consistent distribution across the entire chip. Notably, Scheme 1 yields a relatively higher fluorescence intensity at the input side of the chip. These results clearly indicate that, for Scheme 1, the ssDNA oligomers are accumulating upstream during the early stages of flow, and so are depleted from the advancing solution by the time it reaches mid-chip. The actual patterning of the glass substrate occurs when solvent is evaporated (Figure S2, Supportiing Information). Indeed, the final patterning results after solvent evaporation and cross-linking (Figure 1c, top) reflect the trend established by the aqueous fluorescence images; Scheme 2 produces uniform DNA barcodes across the substrate, while Scheme 1 does not. Line profiles corresponding to Figure 1c can be found in Figure S1 (Supporting Information).
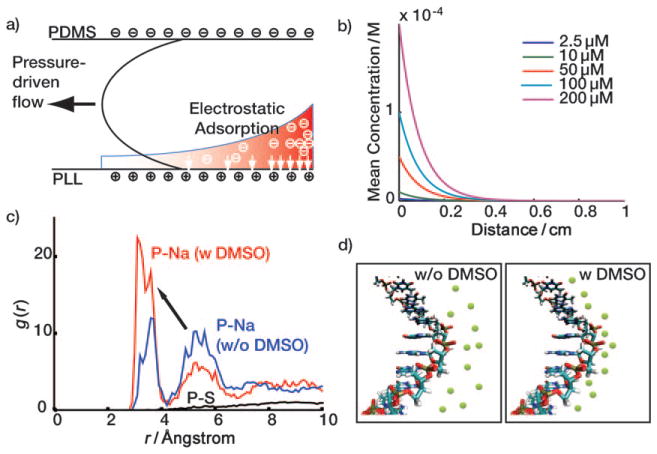
In order to understand the difference in patterning uniformity between Schemes 1 and 2, we considered the electrostatic environment for each case. As depicted in Figure 2a, the PDMS side walls carry a slightly negative zeta potential, whereas the PLL surface has a strong positive zeta potential.[21] When the ssDNA solution in Scheme 1 is introduced to the channel, ssDNA near the PLL matrix is electrostatically immobilized, thereby generating a concentration gradient.[22] As the solution flows towards the channel exit, the ssDNA oligomers are continually depleted via deposition onto the PLL surface. Figure 2b shows the results from a rough simulation designed to capture the mean concentration of aqueous ssDNA as the solution traverses a channel. The simulation implies that the effect of electrostatic adsorption proves dominant even at high DNA concentrations, a result that agrees well with the observed behavior for Scheme 1 in Figure 1b. A detailed description of the model and assumptions employed can be found in the Supporting Information. We tested this model via the strong negative charging of all four channel surfaces via O2 plasma treatment. Consistent with the model, both Schemes 1 and 2 exhibited equivalently uniform distribution of fluorescence intensity across the chip (Figure S3b, Supporting Information). We note that lack of the positive charges on the bottom surface failed to hold DNAs during the drying procedure and that the plasma treatment induces the irreversible bonding of PDMS and glass, which limits further use beyond this experimental test.
Figure 2.
Electrostatic adsorption of DNAs on PLL surface and DMSO effect. a) The filling step. b) Simulation result of electrostatic adsorption of DNAs to PLL surface. c) Molecular simulation of the DMSO effect: the radial distribution function of P atom of the phosphate group and the sodium ions. The presence of DMSO pumps sodium ions from the 2nd shell to the 1st shell (arrow). d) Schematics for DMSO effect. Green circles represent sodium ions.
The results from Schemes 1 and 2 imply that DMSO alleviates the electrostatic adsorption effect. In order to understand this more fully, we performed molecular dynamics (MD) simulations of DNA in PBS and PBS/DMSO solutions; 3 ns of NPT [NPT is a simulation in which number of moles (N), pressure (P) and temperature (T) are held constant]. The MD simulations were performed with the last 1 ns trajectory used for analysis. We examined the radial distribution function of phosphorous atoms in the DNA backbone with respect to various elements of the surrounding solvent. For example, the radial distribution function of P and the O atom of a water molecule is virtually unperturbed by the addition of DMSO (Figure S4, Supporting Information). Consequently, it is unsurprising that the radial distribution function of P and the S atom of DMSO (Figure 2c, black solid line) reveals that DMSO is not forming a solvation structure with the DNA backbone. However, Figure 2c demonstrates a clear interaction between P and Na+ ions, which delineates into two well-defined shell structures: the first is located at r <4.3 Å while the second is located at 4.3 Å <r <6.6 Å. These are similar to the locations of the first and the second water solvation structures. By integrating the radial distribution functions, we determined the number of molecules per phosphate in the first and second shells for both PBS and PBS/DMSO solutions. Although the number H2O molecules per shell is virtually independent of DMSO, DMSO does significantly increase the number of Na+ ions in the first shell (from 0.14 to 0.24), and it decreases the number of Na+ ions in the second shell (from 0.61 to 0.34). Conversely, the number of DMSO molecules is almost zero in the first shell (0.01) but becomes significant in the second shell (0.20). Thus, we conclude that DMSO, with a lower dielectric constant relative to water (47.2 vs 80), destabilizes the solvation energy of Na+ in the second shell. This thermodynamic change prompts the sodium ions to move to the first shell where they are stabilized by electrostatic interactions with the negatively charged phosphate groups. The increased number of sodium ions near the DNA backbone screens the negative charges of phosphate groups more efficiently, thereby reducing electrostatic interactions of the DNA with the PLL surface, resulting in uniform DNA distribution throughout the channels. Although the addition of DMSO to DNA patterning solutions yields the same ultimate effect for both traditional spotted arrays and microfluidics-patterned barcodes, the underlying mechanisms are completely different. We conclude that Scheme 2 is intrinsically superior relative to Scheme 1.
We now turn towards analyzing Scheme 3, and comparing it against Scheme 2. For this scheme, the PAMAM dendrimers are first covalently attached to the aminated glass surface, and then (aminated) ssDNA oligomers are covalently attached to the dendrimers. The lack of a solvent evaporation step makes Scheme 3 significantly more rapid than Scheme 2. We flowed activated PAMAM dendrimers, followed by aminated ssDNA, through ten microfluidic channels (Figure 1b). Note that the aqueous DNA distribution is expected to be uniform because the substrate surface is comprised of charge-neutral N-hydroxysuccinimide (NHS)-modified carboxylates which minimize electrostatic interactions. The resulting DNA microarray was assayed for uniformity with complementary DNAs labeled with Cy3-fluorophores. Visual analysis indicates good uniformity across the chip (Figure 1c, bottom). In order to quantify the patterning quality for all three schemes, we obtained signal intensities for each channel at sixteen locations within the patterning region and calculated the coefficient of variation (CV). The CV is defined as the standard deviation divided by the mean and expressed as a percentage. CVs for Schemes 1, 2, and 3 registered 69.8 %, 10.5 %, and 10.9 %, respectively. Thus, we conclude that Schemes 2 and 3 offer consistent DNA loading across the entire substrate.
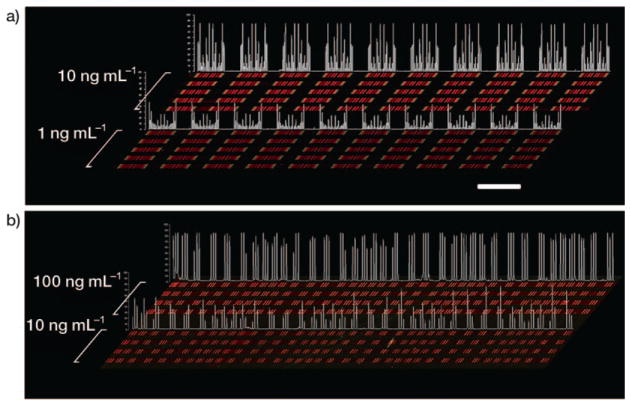
Having established that Schemes 2 and 3 produce consistent, large-scale DNA barcodes, we then extended our analysis of array consistency to protein measurements. We previously demonstrated that, when using the DEAL platform for multiplex protein sensing in microfluidics channels, the sensitivities of the assays directly correlate with the amount of immobilized DNA,[14] up to the point where the DNA coverage is saturated. We performed multiple protein assays along the length of our DNA stripes to ensure that the results described above would translate into stable and sensitive barcodes for protein sensing. All protein assays were performed in microfluidic channels which were oriented perpendicular to the patterned barcodes (five channels for Scheme 2 and four channels for Scheme 3). This allowed us to test distal microarray repeats with a single small analyte volume. For barcodes prepared using Scheme 2, we utilized the DEAL technique to convert them into antibody barcodes designed to assay the following proteins: phosphorylated (phospho)-steroid receptor coactivator (Src), phospho-mammalian target of rapamycin (mTOR), phospho-p70 S6 kinase (S6K), phospho-glycogen synthase kinase (GSK)-3α/β, phospho-p38α, phospho-extracellular signal-regulated kinase (ERK), and total epidermal growth factor receptor (EGFR) at 10 ngmL−1 and 1 ngmL−1 concentrations. This panel samples key nodes of the phosphoinositide 3-kinase (PI3K) signaling pathway within GBM, and are used below for single-cell assays.[23] For barcodes prepared using Scheme 3, we similarly converted the DNA barcodes into antibody barcodes designed to detect three proteins [interferon (INF)-γ, tumor necrosis factor (TNF)α, and interleukin (IL)-2] at 100 ngmL−1 and 10 ngmL−1. All the DNAs used were pre-validated for the orthogonality in order to avoid cross-hybridization and the sequences can be found in the Supporting Information, Table 1. The detection scheme is similar to a sandwich immunoassay. Captured proteins from primary antibodies were fluorescently visualized by biotin-labeled secondary antibodies and Cy5-labeled streptavidin. For both cases, data averaged from multiple DNA repeats across the chip yielded CVs that were commensurate with those of the underlying DNA barcodes (from 10 ngmL−1 concentration, 7% for scheme 2 and 17% for Scheme 3, respectively). Figure 3 shows line profiles of the signal intensities along with the raw data, and demonstrate a better uniformity for barcodes prepared according to Scheme 2. While we found that Scheme 3 could produce barcodes that were close in quality to those of Scheme 2, the absolute (chip-to-chip) consistency of Scheme 3 is hard to guarantee due to its use of the unstable coupling reagents 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and NHS.[24] Moreover, although Scheme 3 is faster, the detailed procedure itself is more labor-intensive. Scheme 2 can potentially be automated. Thus, we chose Scheme 2 as the preferred barcode patterning method. With Scheme 2, over 90 % of the patterned slides showed good quality for the test.
Figure 3.
Contrast-enhanced raw data extracted from multi-protein calibration experiments performed on a substrate prepared according to a) Scheme 2 and b) Scheme 3. Each red bar represents a unique protein measurement, and is clustered with up to ten additional proteins (for Scheme 2). The clusters become symmetrical due to the winding nature of the barcode pattern, so that each cluster actually contains two measurements of each protein. Clustering is less evident in (b) because lower-density barcode pattern was employed. Recombinant proteins were analyzed across five discrete channels per concentration for (a) and four discrete channels per concentration for (b); quantitative data for statistical analysis was extracted from all the repeats in each of the channels. By utilizing identical DEAL cocktails followed by identical standard protein cocktails, the reproducibility was also checked. The identical signal patterns within individual channels and between channels of similar concentrations demonstrate the good uniformity and quality of DNA barcodes. Signal intensity profiles sampled from one analysis channel per concentration are quantified in white. Scale bar: 2 mm.
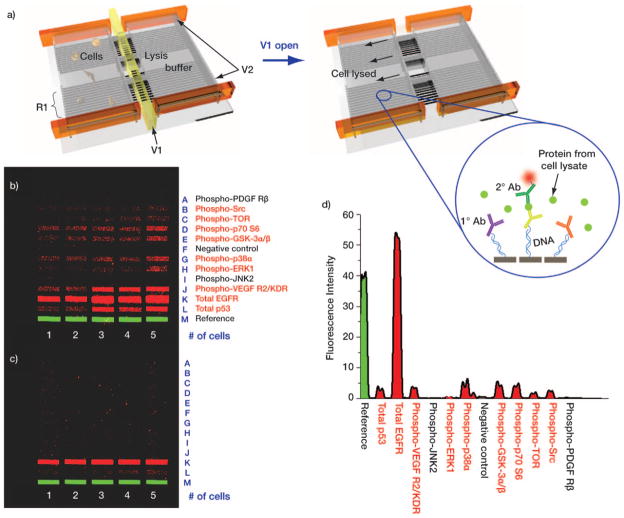
We validated the use of the antibody barcodes by applying them towards the multiplex assay of cytoplasmic proteins from single cells. There is a significant body of evidence that demonstrates that genetically identical cells can exhibit significant functional heterogeneity—behavior that cannot be captured by proteomics techniques that average data across a population.[25] We therefore designed a highly parallel microfluidic device capable of isolating single/few numbers of cells in chambers with a full complement of antibody barcodes designed to detect intracellular proteins (Figure S5, Supporting Information). Figure 4a shows a schematic of the device and the DEAL-based protein detection scheme. The small chamber size keeps the finite number of protein molecules concentrated, thereby enhancing sensitivity. Assaying such a panel of proteins would not be possible without a high density antibody array, such as the barcodes utilized herein, for the following reasons. First, all the barcodes should fit into such a small chamber for multiplexing. Second, since data averaging in such a spatially-constrained scheme is impractical, it is critical to have consistent DNA loading across the microrarray if data comparisons are to be meaningful.
Figure 4.
a) Schematic representation of the single-cell, intracellular protein analysis device. Single or few cells are incubated in an isolated chamber under varying stimuli. Intracellular proteins are assayed by introducing a pre-aliquoted lysis buffer, whereupon the released proteins bind to the DEAL (DNA-labeled antibody) barcode within the chamber. V1: valve for lysis buffer control, V2: valve for isolated chamber formation, and R1: DNA barcode array converted into DEAL antibody array. b), c) Contrast-enhanced images of developed barcode assays highlight the benefits of using Scheme 2 (b) versus Scheme 1 (c). Protein names listed in red font correspond to those which were detected using Scheme 2 barcodes. d) Representative fluorescence intensity profile from the single-cell lysate of (b).
We chose the U87 GBM cell line as a model system for our platform. GBM is the most common malignant brain tumor found in adults, and is the most lethal of all cancers. As the name implies, GBM exhibits extensive biological variability and heterogeneous clinical behavior.[26] EGFR is an important GBM oncogene and therapeutic target.[27] Thus, we assayed for eleven intracellular proteins associated with the EGFR-activated PI3 K signaling pathway. We provide representative sets of data for protein detection from the lysate of one to five cells (Figures 4b and c). Eight proteins were detected from single-cell lysate and up to nine proteins were detected from five cells when using barcodes patterned by Scheme 2 (Figures 4b, d), whereas only one protein could be detected from barcodes prepared by Scheme 1 (Figure 4c). All the separate protein assays were screened for cross-reactivity (Figure S6), and, for the cases where recombinant proteins were available, quantitation curves for each protein assay were measured (Figure S7). More detailed statistical analysis of these cells, as well as genetic variants thereof, is currently being investigated.
We identified a protocol for generating high-quality, high-density DNA barcode patterns by comparing three microfluidics-based patterning schemes. We find, through both experiment and theory, that the electrostatic attractions between positively-charged PLL substrates and the negatively-charged DNA backbone induces significant non-uniformity in the patterning process, but that those electrostatic interactions may be mediated by adding DMSO to the solution, resulting in uniform and highly reproducible barcodes patterned using ~55 cm long channels that template barcodes across an entire 2.5 cm wide glass slide. Dendrimer-based covalent immobilization also yields good ultimate uniformity, but is hampered by a relatively unstable chemistry that limits run-to-run reproducibility. DNA barcodes were coupled with the DEAL technique to generate antibody barcodes, and then integrated into specifically designed microfluidic chips for assaying cytoplasm proteins from single and few lysed U87 model cancer cells. Successful detection of a panel of such proteins represents the potential of our platform to be applied to various biological and, perhaps, clinical applications.
Experimental Section
Microfluidic Chip Fabrication for DNA Patterning
Microfluidic-patterning PDMS chips were fabricated by soft lithography. The master mold was prepared using either a negative photoresist, SU8 2010, with photolithography or an etched silicon mold generated by a deep reactive ion etching (DRIE) process. The mold has long meandering channels with a 200×20 μm cross section. The distance from channel to channel is also 20 μm, which generates 100 higher density than standard, spotted microarrays. Sylgard PDMS (Corning) prepolymer and curing agent were mixed in a 10:1 ratio (w/w), poured onto the mold, and cured (80°, 1 hour). The cured PDMS slab was released from the mold, inlet/outlet holes were punched, and the device was bonded onto a PLL coated (C40–5257 m20, Thermo scientific) or aminated glass slide (48382–220, VWR) to form enclosed channels. The number of microfluidic channels determines the size of the microarray; 13 parallel microchannels were used in this study.
Patterning of DNA Barcode Arrays
For the DNA filling test, a 30-mer DNA oligomer labeled with Cy3 fluorescence tag on the 5′ end (5′-/Cy3/-AAA AAA AAA ATA CGG ACT TAG CTC CAG GAT-3′) in a 1:1 mixture (v/v) of 1×PBS buffer and DMSO or a 1:1 mixture (v/v) of 1×PBS buffer and deionized (DI) water was used. The final DNA concentration was 2.5 μm. DNA solution was pushed into the channel under a constant pressure (2.5 psi). Immediately after the channels were fully filled, fluorescence images were obtained by confocal microscopy.
Dendrimer-based microarrays were prepared using aminated substrates. Generation 4.5 Poly(amidoamine) (PAMAM) dendrimers (470457–2.5G, Aldrich), 5 % wt in MeOH, were mixed 1:1 (v/v) with EDC/NHS (0.2 m) in MES buffer (0.1m, pH 6.0). After 5 min of incubation, the activated dendrimers were introduced to the microfluidic channels, and allowed to flow (2 h). Following a brief MeOH rinse to remove unbound dendrimers, the channels were filled with EDC/NHS (0.2 m) in MES (0.1 m, pH 5.3) with NaCl (0.5 m). After 0.5 h, 5′ aminated DNA sequences in 1×PBS (200 μm) were introduced to the channels and allowed to flow (2 h). Thereafter, the microfluidic device was removed from the substrate, and the latter was rinsed copiously with DI water. Prepared substrates that were not used immediately were stored in a desiccator.
To generate the DNA barcode array for multi-protein detection and single-cell lysis test, 13 orthogonal DNA oligomer solutions (sequences are provided in the Supporting Information, Table 1) in 1×PBS buffer (400 μm) were mixed with DMSO (in 1:2 ratio, v/v) and flowed into each of the microfluidic channels (Scheme 2). For Scheme 1, DNA solutions in 1×PBS buffer were used. The DNA-filled chip was placed in a desiccator until the solvent evaporated completely, leaving only DNA molecules behind. Finally, the PDMS elastomer was removed from the glass substrate and the microarray-patterned DNAs were cross-linked to the PLL by thermal treatment (80 °C, 4 h). The slide was gently rinsed with DI water prior to use in order to remove salt crystals remaining from the solution evaporation step.
Microfluidic Chip Fabrication for Multi-Protein Detection
The PDMS microfluidic chip for the cell experiment was fabricated by two-layer soft lithography.[28] A push-down valve configuration was utilized with a thick control layer bonded together with a thin flow layer. The molds for the control layer and the flow layer were fabricated with SU8 2010 negative photoresist (~20 μm thickness) and SPR 220 positive photoresist (~18 μm), respectively. The photoresist patterns for the flow layer were rounded via thermal treatment. The thick control layer was molded with a 5:1 mixture of GE RTV 615 PDMS prepolymer part A and part B (w/w) and the flow layer was formed by spin-coating a 20:1 mixture of GE RTV 615 part A and part B (w/w) on the flow layer mold (2000 rpm, 60 sec). Both layers were cured (80 °C, 1 hour), whereupon the control layer was cut from its mold and aligned to the flow layer. An additional thermal treatment (80 °C, 1 hour) ensured that the two layers bonded into a monolithic device, which was then peeled from its mold and punched to create appropriate access holes. Finally, the PDMS chip was thermally bonded to the DNA microbarcodes-patterned glass slide to form the working device.
Cell Culture
The human GBM cell line U87 was cultured in DMEM (American Type Culture Collection, ATCC) supplemented with 10 % fetal bovine serum (FBS, Sigma–Aldrich). U87 cells were serum-starved for 1 day and then stimulated by EGF (50 ngmL–1, 10 min) before they were introduced into the device.
Multi-Protein Detection
Protein detection assays were initiated by blocking the chip with 3% bovine serum albumin (BSA) in PBS to prevent non-specific binding. This 3 % BSA/PBS solution was used as a working buffer for most subsequent steps. After blocking, a cocktail containing all eleven (Scheme 2) or three (Scheme 3) DNA–antibody conjugates (~0.5 μg mL−1, 100 μL) in working buffer was flowed through the micro channels for 1 h. The unbound DNA–antibody conjugates were washed away with fresh buffer. Then, target proteins were flowed through the microfluidic channels for 1 hour. These were followed by a 200 μL cocktail containing biotin-labeled detection antibodies (~0.5 μgmL−1) in working buffer, and thereafter a 200 μL mixture of 1 μgmL−1 Cy5-labeled streptavidin and 25 nm Cy3-labeled M’ ssDNA in working buffer to complete the immune sandwich assay. DNA sequence M is used for a location reference. The microchannels were rinsed with working buffer once more before the PDMS chip was removed; the bare microarray slide was rinsed sequentially with 1×PBS, 0.5 × PBS, DI water, and was finally subjected to spin-drying.
On-Chip Cell Lysis and Multiplexed Intracellular Protein Profiling from Single Cells
The multi-protein detection procedure described above was slightly modified for intracellular protein profiling experiments. Again, the chip was initially blocked with a 3% BSA/PBS working buffer, followed by a 200 μL cocktail containing all eleven DNA–antibody conjugates (~0.5 μgmL−1, Supporting Information, Table 2) in working buffer (continuously flowed for 1 h). Unbound DNA-antibody conjugates were washed off with fresh buffer. The lysis buffer (Cell Signaling) was loaded into the lysis buffer channels while valve 1 (V1 in Figure 4a) was kept closed by applying 15–20 psi constant pressure. Then, cells were introduced to the cell loading channels and microfluidic valves (V2 in Figure 4a) were closed by applying 15–20 psi constant pressure; this converts the eight channels into 120 isolated microchamber sets. After cell numbers were counted under microscope, V1 valves were released to allow diffusion of lysis buffer to the neighboring microchamber containing different numbers of cells. The cell lysis was performed on ice for two hours. After that, the V2 valves were released and the unbound cell lysate was quickly removed by flowing the fresh buffer. Then, a cocktail containing biotin-labeled detection antibodies (~0.5 μgmL−1, 200 μL) in working buffer was flowed into the chip for 1 h on ice, followed by flowing a 200 μL mixture of Cy5-labeled straptavidin (1 μg mL−1) and Cy3-labeled M’ ssDNA (25 nm) in working buffer to complete the sandwich immunoassay. Finally, the microchannels were rinsed with working buffer, the PDMS chip was removed, and the bare microarray slide was rinsed sequentially with 1×PBS, 0.5×PBS, DI water, before spin-drying. The layout of the chip and used inlets for different solutions were described in Figure S5.
Data Analysis
The microarray slide was scanned with the GenePix 200B (Axon Instruments) to obtain a fluorescence image of both Cy3 and Cy5 channels. All scans were performed with the same setting of 50% (635 nm) and 15% (532 nm) laser power, 500 (635 nm) and 450 (532 nm) optical gain. The averaged fluorescence intensities for all barcodes in each chamber were obtained and matched to the cell number by custom-developed Excel or MATLAB codes.
Molecular Dynamic Simulations
The MD simulations were performed with the all-atom AMBER2003 force field[29–30] using the Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) code.[31] As an initial structure, a single strand of DNA (5′-ACCCATGGAGCATTCCGGG-3′) whose base pairs were randomly chosen was built using Namot2 program.[32] Near the DNA strand, 19 sodium counter ions were included to neutralize the negatively charged 19 phosphate groups on the DNA backbone. Then, this is immersed in a solvation box composed of either 1) 5206 water molecules+106 DMSO molecules or 2) only 5206 water molecules. We used TIP4P model to describe the water interactions.[33] We performed 3 ns NPT MD simulations using Nosé–Hoover thermostat with a damping relaxation time of 0.1 ps and Andersen–Hoover barostat with a dimensionless cell mass factor of 1.0. The last 1 ns trajectory is employed for the analysis. To compute the electrostatic interactions, the particle-particle particle-mesh method[34] was employed using an accuracy criterion of 10−4.
Supplementary Material
Acknowledgments
This work was supported by National Cancer Institute Grant No. 5U54 CA119347 (J.R.H., P.I.) and by a gift from the Jean Perkins Foundation. H. K, T. A. P and W. A. G also acknowledge support from the WCU programs through NRF of Korea funded by the MEST (R31–2008–000–10055–0).
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cphc.201000528.
References
- 1.Heath JR, Davis ME. Annu Rev Med. 2008;59:251–265. doi: 10.1146/annurev.med.59.061506.185523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heath JR, Phelps ME, Hood L. Mol Imaging Biol. 2003;5:312–325. doi: 10.1016/j.mibio.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 3.Khalil IG, Hill C. Curr Opin Oncol. 2005;17:44–48. doi: 10.1097/01.cco.0000150951.38222.16. [DOI] [PubMed] [Google Scholar]
- 4.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat Biotechnol. 2005;23:1294–1301. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 5.Niemeyer CM. Nano Today. 2007;2:42–52. [Google Scholar]
- 6.Boozer C, Ladd J, Chen S, Jiang S. Anal Chem. 2006;78:1515–1519. doi: 10.1021/ac051923l. [DOI] [PubMed] [Google Scholar]
- 7.Wacker R, Niemeyer CM. ChemBioChem. 2004;5:453–459. doi: 10.1002/cbic.200300788. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder H, Adler M, Gerigk K, Muller-Chorus B, Gotz F, Niemeyer CM. Anal Chem. 2009;81:1275–1279. doi: 10.1021/ac802228k. [DOI] [PubMed] [Google Scholar]
- 9.Wacker R, Schrçder H, Niemeyer CM. Anal Biochem. 2004;330:281–287. doi: 10.1016/j.ab.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 10.Douglas ES, Chandra RA, Bertozzi CR, Mathies RA, Francis MB. Lab on a Chip. 2007;7:1442–1448. doi: 10.1039/b708666k. [DOI] [PubMed] [Google Scholar]
- 11.Bailey RC, Kwong GA, Radu CG, Witte ON, Heath JR. J Am Chem Soc. 2007;129:1959–1967. doi: 10.1021/ja065930i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niemeyer CM, Sano T, Smith CL, Cantor CR. Nucleic Acids Res. 1994;22:5530–5539. doi: 10.1093/nar/22.25.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sano T, Smith C, Cantor C. Science. 1992;258:120–122. doi: 10.1126/science.1439758. [DOI] [PubMed] [Google Scholar]
- 14.Fan R, Vermesh O, Srivastava A, Yen BK, Qin L, Ahmad H, Kwong GA, Liu CC, Gould J, Hood L, Heath JR. Nat Biotechnol. 2008;26:1373–1378. doi: 10.1038/nbt.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dufva M. Biomol Eng. 2005;22:173–184. doi: 10.1016/j.bioeng.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 16.Le Berre V, Trevisiol E, Dagkessamanskaia A, Sokol S, Caminade AM, Majoral JP, Meunier B, Francois J. Nucleic Acids Res. 2003;31:88e. [Google Scholar]
- 17.Bosman AW, Janssen HM, Meijer EW. Chem Rev. 1999;99:1665–1688. doi: 10.1021/cr970069y. [DOI] [PubMed] [Google Scholar]
- 18.Benters R, Niemeyer CM, Drutschmann D, Blohm D, Wohrle D. Nucleic Acids Res. 2002;30:10e. doi: 10.1093/nar/30.2.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Angenendt P, Glokler J, Sobek J, Lehrach H, Cahill DJ. J Chromatogr A. 2003;1009:97–104. doi: 10.1016/s0021-9673(03)00769-6. [DOI] [PubMed] [Google Scholar]
- 20.Ajikumar PK, Ng JK, Tang YC, Lee JY, Stephanopoulos G, Too HP. Langmuir. 2007;23:5670–5677. doi: 10.1021/la063717u. [DOI] [PubMed] [Google Scholar]
- 21.Kuo AT, Chang CH, Wei HH. Appl Phys Lett. 2008;92:244102–244103. [Google Scholar]
- 22.Benn JA, Hu J, Hogan BJ, Fry RC, Samson LD, Thorsen T. Anal Biochem. 2006;348:284–293. doi: 10.1016/j.ab.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research Network. Nature. 2008;455:1061–1068. [Google Scholar]
- 24.Kausaite A, Dijk Mv, Castrop J, Ramanaviciene A, Baltrus JP, Acaite J, Ramanavicius A. Biochem Molec Biol Educ. 2007;35:57–63. doi: 10.1002/bmb.22. [DOI] [PubMed] [Google Scholar]
- 25.Krutzik PO, Irish JM, Nolan GP, Perez OD. Clin Immunol. 2004;110:206–221. doi: 10.1016/j.clim.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Proc Natl Acad Sci USA. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee JC, Vivanco I, Beroukhim R, Huang JHY, Feng WL, DeBiasi RM, Yoshimoto K, King JC, Nghiemphu P, Yuza Y, Xu Q, Greulich H, Thomas RK, Paez JG, Peck TC, Linhart DJ, Glatt KA, Getz G, Onofrio R, Ziaugra L, Levine RL, Gabriel S, Kawaguchi T, O’Neill K, Khan H, Liau LM, Nelson SF, Rao PN, Mischel P, Pieper RO, Cloughesy T, Leahy DJ, Sellers WR, Sawyers CL, Meyerson M, Mellinghoff IK. PLoS Med. 2006;3:e485. doi: 10.1371/journal.pmed.0030485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580–584. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 29.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. J Comput Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 30.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- 31.Plimpton S. J Comput Phys. 1995;117:1–19. [Google Scholar]
- 32.Tung CS, Carter ES., II Comput Appl Biosci. 1994;10:427–433. doi: 10.1093/bioinformatics/10.4.427. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 34.Hockney RW, Eastwood JW. Computer Simulation Using Particles. McGraw-Hill; New York: 1981. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.