Mitochondrial DNA nucleoids are distributed as many discrete foci in mitochondria. Nucleoid distribution is controlled by mitochondrial division and Fcj1 and Mos1, two evolutionarily conserved, mitochondrial proteins that maintain cristae junctions and tubular organelle morphology.
Abstract
Mitochondrial DNA (mtDNA) is packaged into DNA–protein complexes called nucleoids, which are distributed as many small foci in mitochondria. Nucleoids are crucial for the biogenesis and function of mtDNA. Here, using a yeast genetic screen for components that control nucleoid distribution and size, we identify Fcj1 and Mos1, two evolutionarily conserved mitochondrial proteins that maintain the connection between the cristae and boundary membranes. These two proteins are also important for establishing tubular morphology of mitochondria, as mitochondria lacking Fcj1 and Mos1 form lamellar sheets. We find that nucleoids aggregate, increase in size, and decrease in number in fcj1∆ and mos1∆ cells. In addition, Fcj1 form punctate structures and localized adjacent to nucleoids. Moreover, connecting mitochondria by deleting the DNM1 gene required for organelle division enhances aggregation of mtDNA nucleoids in fcj1∆ and mos1∆ cells, whereas single deletion of DNM1 does not affect nucleoids. Conversely, deleting F1Fo-ATP synthase dimerization factors generates concentric ring-like cristae, restores tubular mitochondrial morphology, and suppresses nucleoid aggregation in these mutants. Our findings suggest an unexpected role of Fcj1-Mos1 and organelle division in maintaining the distribution and size of mtDNA nucleoids.
INTRODUCTION
Mitochondria possess their own genome, called mitochondrial DNA (mtDNA), which encodes several essential components of oxidative phosphorylation. mtDNA is exposed to oxidative stress in mitochondria, and its mutations are associated with many human diseases (Wallace, 2010; Nunnari and Suomalainen, 2012). Like chromosomes for the nuclear genome, the mitochondrial genome is packaged into nucleoprotein complexes called nucleoids to protect from deleterious, oxidative damage (Chen and Butow, 2005; Spelbrink, 2010). Nucleoids are also important for the biogenesis of mtDNA, as they contain proteins that mediate DNA replication, repair, and recombination. Proteomic studies have identified >50 different proteins associated with nucleoids (Kaufman et al., 2000; Wang and Bogenhagen, 2006). Defects in these nucleoid proteins result in loss of mtDNA, demonstrating the importance of nucleoids for the maintenance of mtDNA (Gilkerson, 2009; Solieri, 2010).
Cells have multiple copies of mtDNA, and its copy number varies depending on the cell type. For example, yeast haploid cells contain ∼30 copies, whereas human cells have ∼300 copies (Chen and Butow, 2005; Spelbrink, 2010). mtDNA nucleoids are relatively uniform in shape and size. In yeast cells, one or two copies of mtDNA are packaged into each nucleoid (MacAlpine et al., 2000). Nucleoids are visualized as small discrete foci of ∼0.3 μm in diameter along mitochondrial tubules when stained by DNA-binding fluorescent dyes (MacAlpine et al., 2000). Such distribution of nucleoids ensures that each mitochondrion carries mtDNA (Chen and Butow, 2005). In addition, even distribution of nucleoids also facilitates efficient transmission of mtDNA into daughter cells during cell division. Because mtDNA cannot be synthesized de novo, faithful inheritance of mtDNA is essential for the maintenance of mitochondrial functions. Despite its importance, the molecular mechanisms that control the distribution of mtDNA nucleoids are largely unknown. Here we investigate how the size and distribution of mtDNA nucleoids are maintained.
RESULTS
Fcj1 and Mos1 are required for maintaining mtDNA nucleoid size
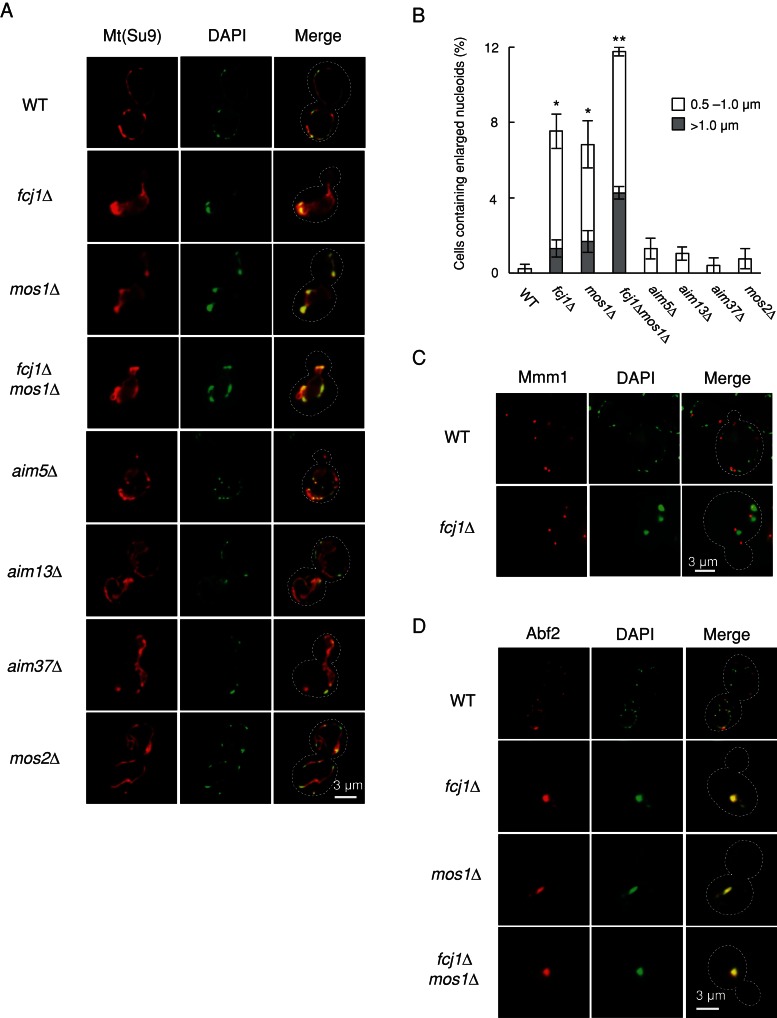
To identify mitochondrial components that control the size and distribution of mtDNA nucleoids, we selected 69 strains with reported alterations in mtDNA maintenance and/or inheritance from a yeast collection in which individual nonessential genes are deleted. These deletion strains include aim∆ (altered inheritance of mitochondria), rrg∆ (required for respiratory growth), and fmp∆ mutants (found in mitochondrial proteome; Supplemental Table S1; Sickmann et al., 2003; Hess et al., 2009; Merz and Westermann, 2009). To visualize mtDNA nucleoids, we incubated live yeast cells with 4′,6-diamidino-2-phenylindole (DAPI), which preferentially labels mtDNA in the absence of chemical fixation (Adams et al., 1997). Most strains (i.e., 63 strains) displayed normal-sized mtDNA nucleoids. However, 5 strains (aim10∆, aim15∆, rrg2∆, rrg4∆, and rrg8∆) exhibited decreased amounts of mtDNA, but their nucleoid size was unaffected (unpublished observations). More important, a strain lacking the FCJ1 gene (also called AIM28) possessed larger nucleoids. We generated independent fcj1∆ strains and examined their phenotypes. In wild-type (WT) cells, the average diameter of mtDNA nucleoids was ∼0.3 μm, with structures >0.5 μm not observed. However, 7% of fcj1∆ cells displayed nucleoids >0.5 μm in diameter (Figure 1, A and B). Among them, ∼15% of fcj1∆ cells contained a single or a few large mtDNA nucleoids with diameter of >1.0 μm. Total fluorescence intensity was greater in these large mtDNA nucleoids, suggesting that these larger structures may result from nucleoid aggregation or incomplete nucleoid division (Figure 1A). Overexpression of Fcj1 from the GAL1 promoter did not affect nucleoid size (Supplemental Figure S1). mtDNA is located near the mitochondria–endoplasmic reticulum (ER) contact sites between these organelles and observed next to the tethering complex containing Mmm1, Mmm2, Mdm10, and Mdm12 (Youngman et al., 2004; Kornmann et al., 2009; Nguyen et al., 2012). In cells lacking these proteins, mtDNA do not form nucleoids and diffuse in the matrix. We found that enlarged nucleoids are still located next to Mmm1-GFP in fcj1∆ cells (Figure 1C), suggesting that the mitochondria–ER contact site remained in fcj1∆ cells and enlarged nucleoids do not result from the loss of the contact site.
FIGURE 1:
Fcj1 and Mos1 are required for mtDNA nucleoid size. (A) WT, fcj1Δ, mos1Δ, fcj1Δmos1Δ, aim5∆, aim13∆, aim37∆, and mos2∆ cells expressing Su9-RFP (Mt) were grown in SGalSuc medium to early log phase and stained with DAPI. Cells were examined by DIC and fluorescence microscopy. Dotted lines outline cells based on DIC images. (B) Quantification of cells with larger mtDNA nucleoids. Cells that have nucleoids with diameter of 0.5–1.0 μm (white) and >1.0 μm (gray) were scored. At least 200 cells were examined in each experiment (n = 3). (C) WT and fcj1Δ cells expressing Mmm1-GFP were grown to log phase in SGalSuc medium and stained with DAPI for 15 min. Cells were viewed by fluorescence microscopy. (D) WT, fcj1Δ, mos1Δ, and fcj1Δmos1Δ cells expressing Abf2-GFP were stained with DAPI.
Fcj1 (called mitofilin in mammals) is a conserved mitochondrial inner membrane (IM) protein enriched at the cristae junction (CJ), which connects the cristae and boundary membranes. Fcj1 binds to other mitochondrial proteins, including Mos1/Mio10/Mcs10 (called MINOS1 in mammals), Mos2, Aim5, Aim13, and Aim37 (Rabl et al., 2009; Harner et al., 2011; Head et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011; Alkhaja et al., 2012; An et al., 2012). Loss of these proteins disconnects the cristae membrane from the boundary membrane, and their effects vary among different mutants, with the largest effect observed in fcj1∆ and mos1∆ cells (Rabl et al., 2009; Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011). In addition, in fcj1∆, mos1∆, mos2∆, aim5∆, aim13∆, and aim37∆ cells, mitochondria change their morphology from tubules to lamellar sheets (Figure 1A; Rabl et al., 2009; Hoppins et al., 2011). We found that mos1∆ cells showed enlarged mtDNA nucleoids similar to fcj1∆ cells (Figure 1, A and B). However, only ∼1% of aim5∆, aim13∆, aim37∆, and mos2∆ cells contain mtDNA nucleoids >0.5 μm in diameter (Figure 1, A and B). Thus Fcj1 and Mos1 are required for maintaining the size of mtDNA nucleoids. Although Fcj1 also interacts with Mia40, an intermembrane space (IMS) protein that mediates protein import (von der Malsburg et al., 2011), the depletion of Mia40 did not change nucleoid size (Supplemental Figure S2), showing that the nucleoid phenotype is not due to import defects in fcj1∆ cells.
When we generated fcj1∆mos1∆ double-deletion cells, the number of cells with larger (>0.5 μm) mtDNA nucleoids increased to ∼12%, with 40% of them exhibiting a single large mtDNA nucleoid with diameter of >1.0 μm (Figure 1, A and B). These results suggest partially overlapping functions for Fcj1 and Mos1 in the maintenance of mtDNA nucleoid size. In fcj1∆, mos1∆, and fcj1∆mos1∆ cells, mtDNA was still associated with Abf2, an HMG box–containing, DNA-binding protein required for packaging mtDNA into nucleoids (Miyakawa et al., 2010), as DAPI staining colocalized with Abf2–green fluorescent protein (GFP; Figure 1D). Therefore the enlarged morphology of mtDNA nucleoids was not due simply to dissociation of Abf2 from mtDNA. Finally, Southern blotting showed that WT, fcj1∆, mos1∆, and fcj1∆mos1∆ cells contained similar amounts of mtDNA (Supplemental Figure S3). Thus increased size of nucleoids appears not to affect the maintenance of mtDNA in these mutants.
Fcj1 is located adjacent to mtDNA nucleoids
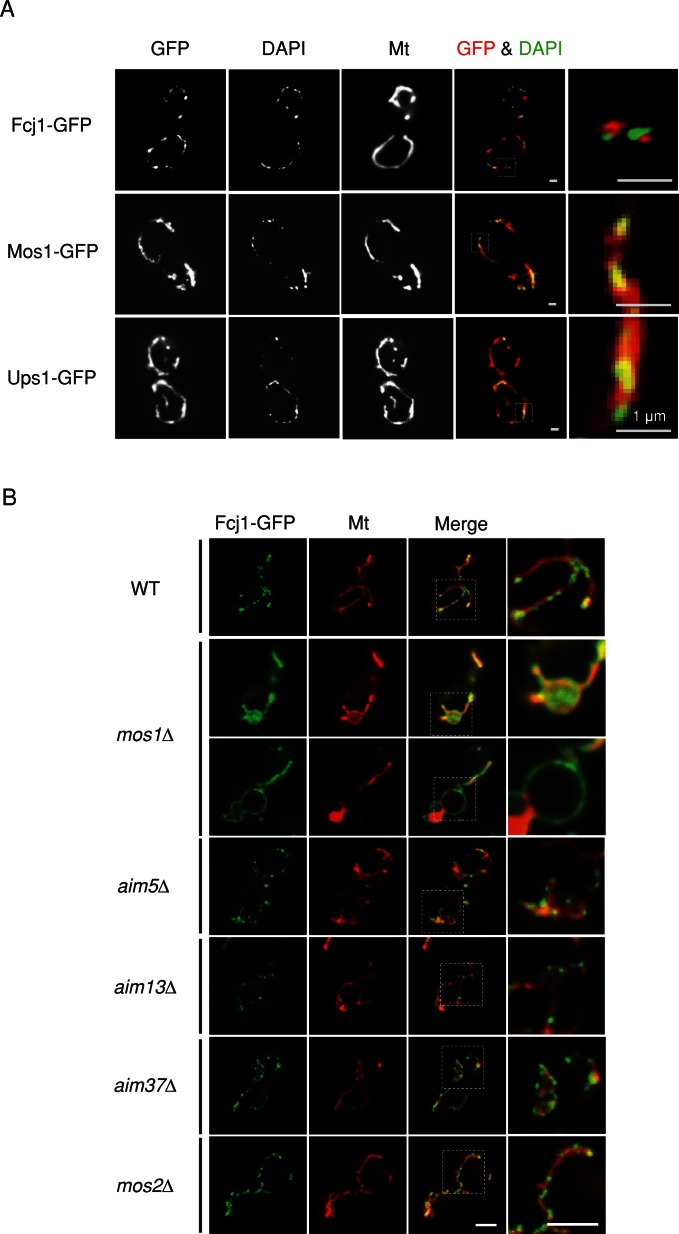
To examine the spatial relationship of mtDNA nucleoids with Fcj1 and Mos1, we replaced chromosomal FCJ1 and MOS1 by FCJ1-GFP and MOS1-GFP, respectively. GFP was fused to Fcj1 and Mos1 at their C-terminus, which faces the IMS. Both Fcj1-GFP and Mos1-GFP were functional, as cells expressing these GFP fusions showed normal morphology of mitochondria and nucleoids (Figure 2A) and grew normally (Supplemental Figure S4). A fraction of Fcj1-GFP formed punctate structures in mitochondria, with 80% located adjacent to mtDNA nucleoids (n = 120; Figure 2A). The formation of Fcj1-GFP puncta depended on Mos1, but not Aim5, Aim13, Aim37, or Mos2 (Figure 2B). In contrast, Mos1-GFP was uniformly distributed in mitochondria, similar to an IMS protein, Ups1-GFP (Figure 2A; Sesaki et al., 2006; Tamura et al., 2009). Because FCJ1 and MOS1 deletion has additional effects on nucleoid size, Mos1 likely has roles in addition to Fcj1-GFP puncta formation. Consistent with our finding that Fcj1-GFP puncta is closely positioned to nucleoids, a proteomic study showed that Fcj1 binds to the nucleoid protein Abf2 in yeast (von der Malsburg et al., 2011). Another study showed that mtDNA nucleoids purified from HeLa cells contained a mammalian homologue of Fcj1, mitofilin (Wang and Bogenhagen, 2006). However, we found that Fcj1 and Mos1 do not associate with nucleoids in sucrose density gradient centrifugation and that Abf2 does not interact with Fcj1 or Mos1 in coimmunoprecipitation studies (unpublished observations). Therefore either only a small fraction of Fcj1 or Mos1 bind to nucleoids or their interactions might be labile.
FIGURE 2:
Fcj1p-GFP forms puncate structures next mtDNA nucleoids. (A) GFP fusion proteins of Fcj1, Mos1, and Ups1 were expressed by integrating the GFP gene at their 3′ end in chromosomes. The GFP strains were transformed with Su9-RFP (Mt), grown to log phase, and stained with DAPI. Boxed regions show magnified images. (B) Fcj1-GFP was expressed in WT, mos1∆, aim5∆, aim13∆, aim37∆, and mos2∆ cells. The GFP strains were transformed with matrix-targeted Su9-RFP (Mt). Cells were grown to log phase, stained with DAPI, and observed by fluorescence microscopy. Boxed regions show magnified images.
Mitochondria form large, hollow spheres in fcj1∆ and mos1∆ cells that contain large mtDNA nucleoids
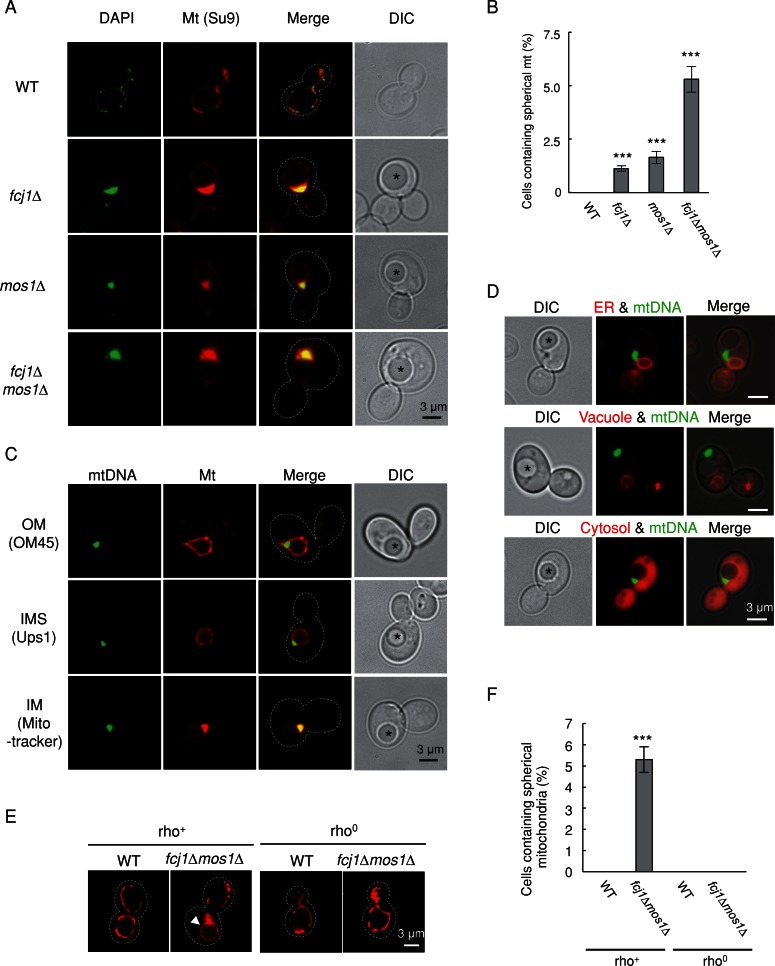
When we examined mitochondrial morphology in fcj1∆, mos1∆, and fcj1∆mos1∆ cells that contain large mtDNA nucleoids (>1.0 μm in diameter), essentially all of the cells displayed previously uncharacterized, enlarged spherical mitochondria (Figure 3, A and B). These mitochondria appeared to be hollow by matrix-targeted Su9–red fluorescent protein (RFP) and had mtDNA nucleoids around the rim. The spherical structures were clearly observed by differential interference contrast (DIC) microscopy (Figure 3A, asterisks). To further characterize the hollow, spherical mitochondria, we targeted GFP fusion proteins to different mitochondrial compartments, including the outer membrane (OM45-GFP) and the IMS (Ups1p-GFP) in fcj1∆mos1∆ cells (Figure 3C). We also used MitoTracker to stain the IM. OM45-GFP was observed along the rim of spherical mitochondria and mtDNA nucleoids. The IMS marker, Ups1-GFP, and MitoTracker also stained the contour of the spheres and showed some overlap with DAPI staining, similar to the matrix-targeted Su9-RFP. Remarkably, none of these markers labeled the interior of the spherical mitochondria. As expected, markers for vacuoles (FM4-64; Molecular Probes, Eugene, OR), the ER (Sec63-GFP), and the cytosol (GFP) did not label these spherical structures in fcj1∆mos1∆ cells (Figure 3D). This hollow mitochondrial sphere is, to the best of our knowledge, a novel morphology not reported in any yeast mutants examined previously.
FIGURE 3:
Round, hollow mitochondria are formed in fcj1Δ, mos1Δ, and fcj1Δmos1Δ cells that contain large mtDNA nucleoids. (A) WT, fcj1Δ, mos1Δ, and fcj1Δmos1Δ cells expressing Su9-RFP were grown in SGalSuc medium to early log phase and stained with DAPI. Cells were examined by DIC and fluorescence microscopy. Large spherical mitochondria are indicated by asterisks. (B) Quantification of cells that contain large, hollow mitochondria. At least 600 cells were examined in each experiment (n = 3). (C) Large, hollow mitochondria were analyzed by several mitochondrial markers, including OM45-GFP for the outer membrane (OM), Ups1-GFP for the IMS, and MitoTracker for the IM, in fcj1∆mos1∆ cells. mtDNA was labeled by either DAPI (with OM45p-GFP and Ups1p-GFP) or Abf2-GFP (with MitoTracker). (D) The ER, vacuoles, and the cytosol were marked by Sec63-GFP, FM4-64, and GFP, respectively. (E) WT and fcj1Δmos1Δ cells with (rho+) or without (rho0) mtDNA expressing Su9-RFP were examined by fluorescence microscopy. Dotted lines outline cells based on DIC images. Large, spherical mitochondria are indicated by an arrowhead. (F) Quantification of cells containing large, hollow mitochondria. At least 600 cells were examined in each experiment (n = 3).
The frequency of spherical mitochondria increased in fcj1∆mos1∆ cells (∼5%) compared with fcj1∆ and mos1∆ cells (∼1%; Figure 3B), correlating with the increased occurrence of mtDNA nucleoids with a diameter >1.0 μm (Figure 1B). In fcj1∆, mos1∆, and fcj1∆mos1∆ cells that contained normal nucleoids (<0.5 μm in diameter) or modestly enlarged nucleoids (0.5–1 μm), mitochondria showed normal tubular structures or large lamellar sheets (Figure 3A), as reported previously (Hoppins et al., 2011).
To understand whether the formation of hollow, spherical mitochondria requires mtDNA, we examined mitochondrial morphology in fcj1Δmos1Δ cells with (rho+) and without (rho0) mtDNA. As described, ∼5% of rho+ fcj1Δmos1Δ cells possessed enlarged, spherical mitochondria (Figure 3, E and F). However, these large, spherical mitochondria were missing in rho0 fcj1Δmos1Δ cells. Moreover, lamellar structures of mitochondria were not found in rho0 fcj1Δmos1Δ cells, consistent with previous observations showing that dimerization of F1Fo-ATP synthase mediated by Atp20 and Atp21 (therefore mtDNA that encodes subunits of this enzyme complex) is required for the formation of lamellar mitochondria (Hoppins et al., 2011). These results suggest that the generation of enlarged, spherical mitochondria requires mtDNA and that hollow, spherical mitochondria might derive from lamellar mitochondria.
Loss of Atp20 and Atp21 rescues nucleoid defects in cells lacking Fcj1 and Mos1
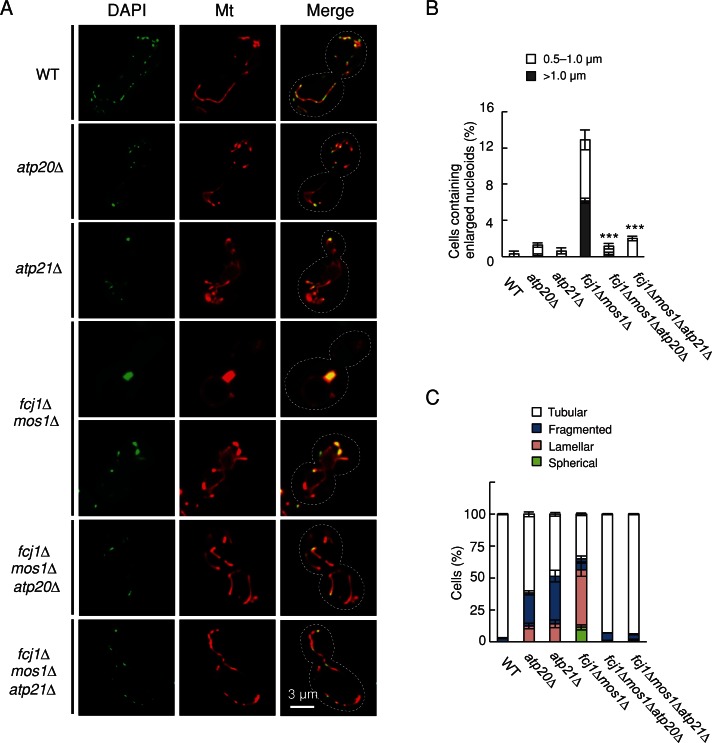
Because Atp20 and Atp21 control the structure of cristae membranes and loss of these proteins suppresses growth defects in fcj1∆ cells (Rabl et al., 2009), we examined nucleoid morphology in cells lacking these proteins. atp20∆ and atp21∆ cells exhibited normal-sized nucleoids (Figure 4, A and B). It is striking, however, that loss of Atp20 or Atp21 rescued nucleoid defects in fcj1∆mos1∆ cells (Figure 4, A and B). The inhibitor of F1Fo-ATP synthase, oligomycin, did not affect nucleoid size in fcj1∆mos1∆ cells, showing that dimerization of this enzyme, rather than its activity, is important (Supplemental Figure S5). In terms of mitochondrial morphology, atp20∆ and atp21∆ cells displayed lamellar or partially fragmented mitochondria (Figure 4C). As described earlier, fcj1∆mos1∆ mitochondria displayed lamellar sheets, hollow spheres, and small fragments (Figures 1A and 3A). These morphological defects were mutually suppressed in fcj1∆mos1∆atp20∆ and fcj1∆mos1∆atp21∆ cells (Figure 4, A and C). Taken together, these results suggest that Fcj1-Mos1 and Atp20-Atp21 play opposite roles in determining nucleoid size and organelle shape.
FIGURE 4:
Loss of Atp20 and Atp21 rescues nucleoid defects in fcj1∆mos1∆ cells. (A) WT, atp20∆, atp21∆, fcj1∆mos1∆, fcj1∆mos1∆atp20∆, and fcj1∆mos1∆atp21∆ cells expressing Su9-RFP were stained with DAPI. (B, C) Quantification of cells that contain increased size of mtDNA nucleoids (B) and mitochondrial morphology (C). At least 200 cells were examined in each experiment (n = 3).
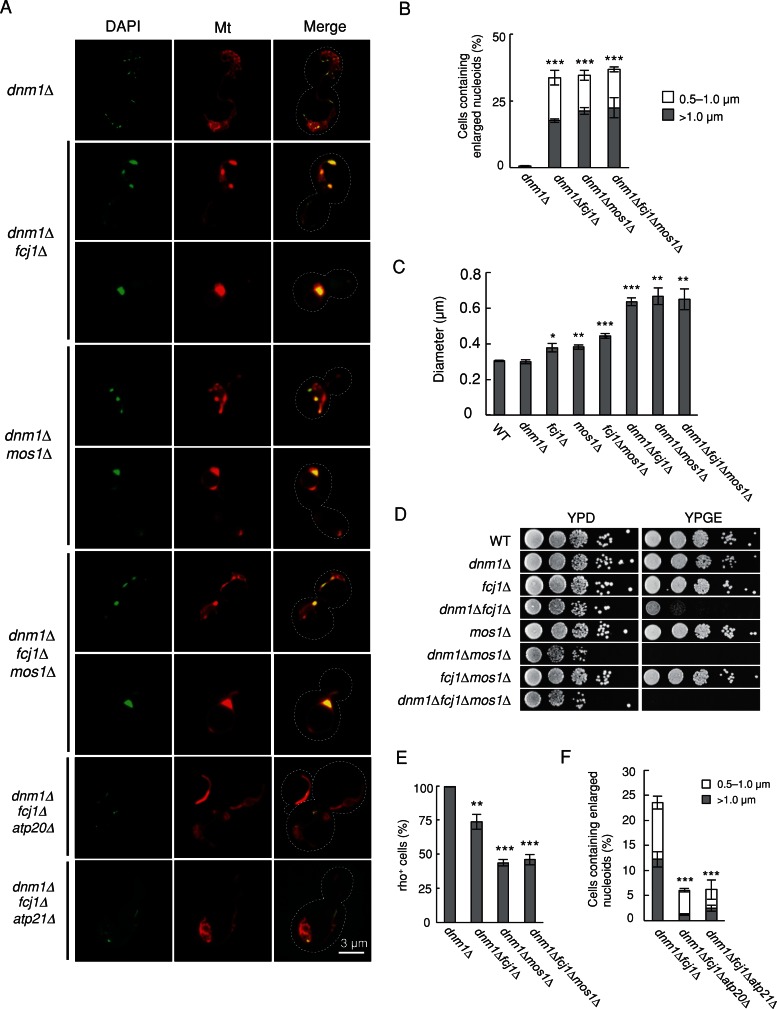
Blocking mitochondrial division exacerbates nucleoid defects in fcj1∆, mos1∆, and fcj1∆mos1∆ cells
Large nucleoids might arise from incomplete division or aggregation of nucleoids in cells lacking Fcj1 and Mos1. To distinguish these two possibilities, we examined nucleoids in cells defective in mitochondrial division. We reasoned that, if nucleoid division decreased, connecting mitochondria would not affect nucleoid size. In contrast, if nucleoids became aggregated, connecting mitochondria might accelerate nucleoid aggregation. We disrupted the DNM1 gene, which is required for mitochondrial division, in fcj1∆, mos1∆, and fcj1∆mos1∆ cells. dnm1∆ cells contained a single mitochondrion with interconnected tubules due to an imbalance that favors fusion over division and normal size of nucleoids, as reported (Bleazard et al., 1999; Sesaki and Jensen, 1999; Figure 5, A and B). We found that additional loss of Dnm1 increased the number of larger nucleoids to 35–40% in dnm1∆fcj1∆, dnm1∆mos1∆, and dnm1∆fcj1∆mos1∆ cells (Figure 5B), compared with 7–12% in fcj1∆, mos1∆, and fcj1∆mos1∆ cells (Figure 1B). In contrast, only slight increases in the number of enlarged nucleoids were observed in dnm1∆aim5∆, dnm1∆aim13∆, dnm1∆aim37∆, and dnm1∆mos2∆ cells (Supplemental Figure S6A). We also observed synergistic increases in diameter of nucleoids in cells lacking Fcj1, Mos1, and Dnm1 (Figure 5C). Thus Fcj1 and Mos1 are most likely important for the separation of individual nucleoids, and loss of these proteins might induce nucleoid aggregation. It appears that more nucleoids were gathered and formed large nucleoids when mitochondrial division is blocked in cells lacking Fcj1 and/or Mos1. Because a significant fraction of fcj1∆mos1∆dnm1∆ cells still has normal-sized nucleoids, there are likely additional mechanisms by which mtDNA nucleoids remain separate from each other to maintain their distribution within mitochondria.
FIGURE 5:
Connecting mitochondria enhances aggregation of nucleoids in fcj1∆, mos1∆, and fcj1∆mos1∆ cells. (A) dnm1∆, dnm1∆fcj1∆, dnm1∆mos1∆, and dnm1∆fcj1∆mos1∆ cells expressing Su9-RFP were stained with DAPI. (B) Quantification of cells that contain increased size of mtDNA nucleoids. At least 150 cells were examined in each experiment (n = 3). (C) Quantification of average nucleoid diameter in the indicated cells. At least 150 nucleoids were examined in each experiment (n = 3). (D) Serial dilutions of cells were spotted onto YPD and YPGE medium and incubated at 30°C for 2 and 5 d, respectively. (E) Quantification of cells that contain mtDNA. At least 600 cells were examined in each experiment (n = 3). (F) Quantification of cells that contain increased size of nucleoids. At least 150 cells were examined in each experiment (n = 3).
In addition to enlargement of nucleoids, we also noticed that dnm1∆fcj1∆, dnm1∆mos1∆, and dnm1∆fcj1∆mos1∆ cells more frequently lose mtDNA. When we spotted cells onto yeast/peptone/glycerol/ethanol (YPGE; a nonfermentable medium that requires mtDNA) and yeast/peptone/glucose (YPD; a fermentable medium that does not require mtDNA), fcj1∆, mos1∆, and fcj1∆mos1∆ cells grew very slowly on YPGE medium when DNM1 is additionally deleted, suggesting decreases in respiratory functions in dnm1∆fcj1∆, dnm1∆mos1∆, and dnm1∆fcj1∆mos1∆ cells (Figure 5D and Supplemental Figure S6B). To more directly examine mtDNA maintenance, we grew cells in the nonfermentable YPGE medium and then transferred these rho+ cells to a fermentable medium (SGalSuc). After culture for 2 d in SGalSuc medium, cells were stained with DAPI. Fcj1 or Mos1 deficiency increased the frequency of mtDNA loss in dnm1∆ cells (Figure 5E).
As observed for fcj1∆ and mos1∆ cells, increased nucleoid size in fcj1∆dnm1∆ cells was decreased with the additional loss of Atp20 or Atp21 (Figure 5, A and F). Moreover, the formation of hollow, spherical mitochondria was suppressed in dnm1∆fcj1∆atp20∆ and dnm1∆fcj1∆atp21∆ cells (Supplemental Figure S7). Lamellar mitochondria were still observed in the triple-knockout cells, although it was technically difficult to clearly distinguish net-like mitochondria from the sheet-like mitochondria.
Ultrastructural analysis of cristae
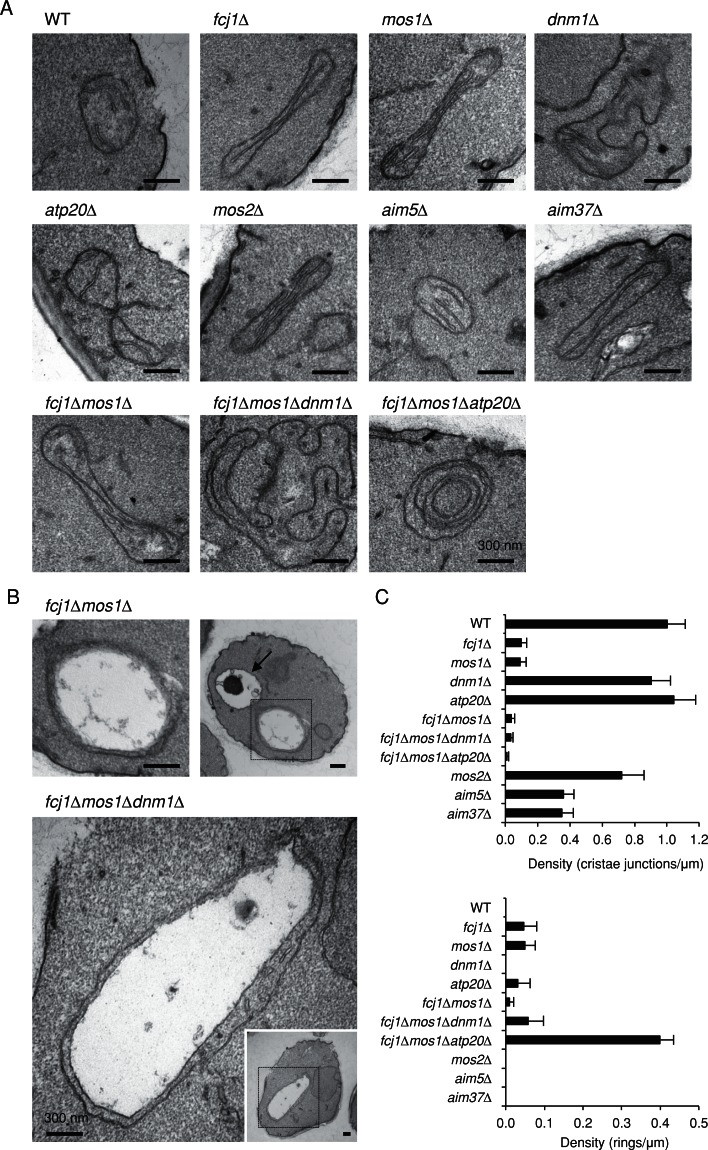
To analyze cristae organization, we performed electron microscopy. We observed lower densities of CJs in fcj1∆, mos1∆, mos2∆, aim5∆, and aim37∆ cells compared with WT cells, with the largest decreases seen in fcj1∆ and mos1∆ cells (Figure 6, A and C), consistent with previous studies (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011). Indistinguishable densities of CJs were observed in WT, atp20∆, and dnm1∆ cells; however, the number of cristae membrane tips was decreased in atp20∆ cells (Figure 6A), as reported (Rabl et al., 2009). fcj1∆mos1∆ and fcj1∆mos1∆dnm1∆ cells exhibit similar CJ densities, which is less than those of fcj1∆ and mos1∆ cells. We observed large, hollow mitochondria in fcj1∆mos1∆ and fcj1∆mos1∆dnm1∆ cells (Figure 6B), consistent with our findings made by light microscopy (Figures 2 and 5). In these large mitochondria, less-electron-dense compartments were surrounded by remnants of cristae. In fcj1∆mos1∆atp20∆ cells, we found dramatic increases in the frequency of cristae membrane that formed concentric ring-like structures (Figure 6, A and C).
FIGURE 6:
Ultrastructural analysis of mitochondria. (A, B) Electron micrographs of mitochondria in WT and the indicated mutants. Boxed regions show magnified images in B. An arrow indicates a vacuole in B. (C) Quantification of the density of crista junctions and concentric ring-like cristae relative to the length of mitochondrial perimeter. At least 22 mitochondria were analyzed for each genotype.
DISCUSSION
Fcj1 and Mos1 are involved in the maintenance of crista junctions, which may be critical for mtDNA nucleoid distribution. For example, in WT mitochondria, organized, periodic arrays of cristae membranes along the boundary membrane may function as partitions to make small compartments for mtDNA nucleoids in the matrix. When these partitions are greatly decreased in cells lacking Fcj1 and Mos1, a small but significant fraction of mtDNA nucleoids aggregate and increase in size. mos2∆, aim5∆, aim13∆, and aim37∆ cells modestly decrease the number of cristae junctions and maintain normal nucleoids. Therefore there may be a threshold for the density of crista junctions required for the maintenance of nucleoid distribution. Second, it is also possible that tubular morphology of mitochondria helps keep nucleoids from aggregation, since Fcj1 and Mos1 also function as tubule-forming components in mitochondria. In sheet mitochondria, nucleoids may collide more frequently due to increases in the direction of their movements in the matrix. Third, in fcj1∆ and mos1∆ cells, cristae membranes that are detached from the boundary membrane may push nucleoids toward the edge of lamellar sheets, leading to aggregation of nucleoids. Finally, nucleoids may be anchored to the IM to keep nucleoids in such small compartments formed by crista membranes, since mtDNA nucleoids are located adjacent to Fcj1. Whether Fcj1 and Mos1 mediate this potential anchoring mechanism awaits further studies. In the absence of Fcj1 and Mos1, mitochondrial division appears to help the separation of mtDNA nucleoids by creating physically separated organelles, maintaining nucleoids in separate mitochondria. This model is consistent with previous observations showing that nucleoids are dynamic and mobile in mammalian mitochondria (Garrido et al., 2003; Legros et al., 2004). Given that we found that cells only partially lose their ability to maintain normal mtDNA nucleoid size in the absence of Fcj1, Mos1, and Dnm1, there are likely additional mechanisms that control the size of mtDNA nucleoids that function alongside these proteins. It will be important to identify these mechanisms in future studies.
The nucleoid phenotypes of cells lacking Fcj1 and Mos1 appear to be different from those observed in several mutants with previously reported increased mtDNA nucleoid size. In yeast, Hsp60, a mitochondrial heat-shock protein associated with nucleoids, was proposed to participate in nucleoid division (Kaufman et al., 2003). In temperature-sensitive mutants of Hsp60, nucleoids are larger and exhibit a dumbbell-shaped morphology reminiscent of incomplete nucleoid division. In addition, cells lacking Abf2 showed defects in nucleoid packaging and therefore contained swollen nucleoids (Miyakawa et al., 2010). Mmm1, Mmm2, Mdm10, and Mdm12, which have been suggested to participate in ER–mitochondria tethering, mitochondrial shape, and phospholipid metabolism, also regulate mtDNA nucleoids (Kornmann et al., 2009; Nguyen et al., 2012). However, cells lacking Mmm1, Mmm2, Mdm10, and Mdm12 display diffusely distributed mtDNA and Abf2 within the matrix of their large mitochondria, suggesting that these four proteins may also be involved in the packaging of mtDNA into nucleoids (Youngman et al., 2004).
How the hollow mitochondrial sphere is formed in the absence of Fcj1 and Mos1 remains to be determined. We speculate that cristae membranes disconnected from the boundary membrane may fuse with each other and create additional compartments inside mitochondria. This additional compartment is not accessible by proteins imported from the cytosol, as indicated by the lack of staining on the interior of these spheres by the markers we tested. Note that the large, round mitochondria in fcj1∆ and mos1∆ cells are different from those found in mmm1∆, mmm2∆, mdm10∆, mdm12∆, and mdm33∆ cells, as the interiors could be stained with matrix markers (Berger et al., 1997; Hobbs et al., 2001; Messerschmitt et al., 2003). In addition, the formation of large, spherical mitochondria depends on the presence of mtDNA in fcj1∆ and mos1∆ cells, whereas mitochondrial shape in mmm1∆, mmm2∆, mdm10∆, and mdm12∆ cells is independent of mtDNA.
MATERIALS AND METHODS
Strains and media
Yeast strains, plasmids, and primers used in this study are listed in Supplemental Table S2. Disruption of complete open reading frame was performed using the HIS3, kanMX4, and URA3 genes as disruption markers as described (Sikorski and Hieter, 1989; Brachmann et al., 1998). Cells that carry kanMX4 were selected on YPD (1% yeast extract, 2% polypeptone, and 2% glucose) containing 500 μg/ml G418. rho0 cells were generated by incubating in YPD medium with 25 μg/ml ethidium bromide for 2 d. Cells were grown in YPD medium, SD medium (0.67% yeast nitrogen base without amino acids and 2% glucose), SCD medium (0.67% yeast nitrogen base without amino acids, 0.5% casamino acid, and 2% glucose), and SGalSuc medium (0.67% yeast nitrogen base without amino acids, 2% galactose, and 2% sucrose) with appropriate supplements (Adams et al., 1997).
Microscopy
Cells were viewed on an Olympus BX61 upright microscope (Olympus, Tokyo, Japan). Fluorescence and DIC images were captured with a CoolSnap HQ charge-coupled device (CCD) camera (Roper Photometrics, Tucson, AZ) using SlideBook software, version 5.0 (Intelligent Imaging Innovations, Denver, CO) and processed with Photoshop CS3 (Adobe, San Jose, CA; Tamura et al., 2010). For electron microscopy, cells were fixed, processed, and embedded in Epon resin, as described (Bauer et al., 2001). Ultrathin sections were obtained using an Ultracut E (Reichert-Jung, Germany), stained with 2% uranyl acetate and lead citrate, and viewed on a Hitachi H-7600 transmission electron microscope (Hitachi, Tokyo, Japan) equipped with the dual AMT CCD camera (Wakabayashi et al., 2009; Kageyama et al., 2012).
Southern blotting
DNA was extracted from yeast cells using the Yeast DNA Extraction Kit (Thermo Scientific, Waltham, MA). Then the DNA (5 μg) was digested with EcoRI, resolved by electrophoresis, and transferred to nitrocellulose membrane. Hybridization was performed using the ECL Direct Nucleic Acid Labelling and Detection System (Amersham, Piscataway, NJ) according to the manufacturer's instructions. The probe used for mtDNA corresponds to position 245–676 of the ATP6 gene, and the PCR primers corresponding to this probe were 5′-TGCTTAAAGGACAAATTGGAGG-3′ and 5′-TCATAGCTAAAGGTACAAAACCGA-3′. The probe used for nuclear DNA corresponds to position 399–779 of the 18S rRNA gene (RDN18), and the primers corresponding to this probe were 5′-AAACGGCTACCACATCCAAG-3′ and 5’-ACGCCTGCTTTGAACACTCT-3′.
Statistics
Values are mean ± SEM (n = 3). The p values were determined using the Student's t test: *p < 0.05; **p < 0.01; ***p < 0.001.
Supplementary Material
Acknowledgments
We thank members of the Sesaki and Iijima labs for helpful discussions. This work was supported by Japan Society for the Promotion of Science fellowships to K.I. and Y.T. and National Institutes of Health Grants to M.I. (GM084015) and H.S. (GM089853).
Abbreviations used:
- CJ
cristae junction
- IM
inner membrane
- IMS
intermembrane space
- mtDNA
mitochondrial DNA
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0125) on April 24, 2013.
*Present address: Research Center for Materials Science, Department of Chemistry, Graduate School of Science, Nagoya University, 464-8602, Chikusa-ku, Nagoya, Japan.
REFERENCES
- Adams A, Gottschling D, Kaiser C, Stearns T. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Alkhaja AK, et al. MINOS1 is a conserved component of mitofilin complexes and required for mitochondrial function and cristae organization. Mol Biol Cell. 2012;23:247–257. doi: 10.1091/mbc.E11-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J, Shi J, He Q, Lui K, Liu Y, Huang Y, Sheikh MS. CHCM1/CHCHD6, a novel mitochondrial protein linked to regulation of mitofilin and mitochondrial cristae morphology. J Biol Chem. 2012;287:7411–7426. doi: 10.1074/jbc.M111.277103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Herzog V, Bauer MF. Improved technique for electron microscope visualization of yeast membrane structure. Microsc Microanal. 2001;7:530–534. doi: 10.1017.S1431927601010522. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleazard W, McCaffery JM, King EJ, Bale S, Mozdy A, Tieu Q, Nunnari J, Shaw JM. The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol. 1999;1:298–304. doi: 10.1038/13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, GJ C, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains from Saccharomyces cerevisiae 288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Butow RA. The organization and inheritance of the mitochondrial genome. Nat Rev Genet. 2005;6:815–825. doi: 10.1038/nrg1708. [DOI] [PubMed] [Google Scholar]
- Garrido N, Griparic L, Jokitalo E, Wartiovaara J, van der Bliek AM, Spelbrink JN. Composition and dynamics of human mitochondrial nucleoids. Mol Biol Cell. 2003;14:1583–1596. doi: 10.1091/mbc.E02-07-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilkerson RW. Mitochondrial DNA nucleoids determine mitochondrial genetics and dysfunction. Int J Biochem Cell Biol. 2009;41:1899–1906. doi: 10.1016/j.biocel.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head BP, Zulaika M, Ryazantsev S, van der Bliek AM. A novel mitochondrial outer membrane protein, MOMA-1, that affects cristae morphology in Caenorhabditis elegans. Mol Biol Cell. 2011;22:831–841. doi: 10.1091/mbc.E10-07-0600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DC, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. J Cell Biol. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Zhang Z, Roda R, Fukaya M, Wakabayashi J, Wakabayashi N, Kensler TW, Reddy PH, Iijima M, Sesaki H. Mitochondrial division ensures the survival of postmitotic neurons by suppressing oxidative damage. J Cell Biol. 2012;197:535–551. doi: 10.1083/jcb.201110034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman BA, Kolesar JE, Perlman PS, Butow RA. A function for the mitochondrial chaperonin Hsp60 in the structure and transmission of mitochondrial DNA nucleoids in Saccharomyces cerevisiae. J Cell Biol. 2003;163:457–461. doi: 10.1083/jcb.200306132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman BA, Newman SM, Hallberg RL, Slaughter CA, Perlman PS, Butow RA. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc Natl Acad Sci USA. 2000;97:7772–7777. doi: 10.1073/pnas.140063197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legros F, Malka F, Frachon P, Lombes A, Rojo M. Organization and dynamics of human mitochondrial DNA. J Cell Sci. 2004;117:2653–2662. doi: 10.1242/jcs.01134. [DOI] [PubMed] [Google Scholar]
- MacAlpine DM, Perlman PS, Butow RA. The numbers of individual mitochondrial DNA molecules and mitochondrial DNA nucleoids in yeast are co-regulated by the general amino acid control pathway. EMBO J. 2000;19:767–775. doi: 10.1093/emboj/19.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz S, Westermann B. Genome-wide deletion mutant analysis reveals genes required for respiratory growth, mitochondrial genome maintenance and mitochondrial protein synthesis in Saccharomyces cerevisiae. Genome Biol. 2009;10:R95. doi: 10.1186/gb-2009-10-9-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messerschmitt M, Jakobs S, Vogel F, Fritz S, Dimmer KS, Neupert W, Westermann B. The inner membrane protein Mdm33 controls mitochondrial morphology in yeast. J Cell Biol. 2003;160:553–564. doi: 10.1083/jcb.200211113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa I, Kanayama M, Fujita Y, Sato H. Morphology and protein composition of the mitochondrial nucleoids in yeast cells lacking Abf2p, a high mobility group protein. J Gen Appl Microbiol. 2010;56:455–464. doi: 10.2323/jgam.56.455. [DOI] [PubMed] [Google Scholar]
- Nguyen T, Lewandowska A, Choi JY, Markgraf DF, Junker M, Bilgin M, Ejsing CS, Voelker DR, Rapoport TA, Shaw JM. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl R, et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol. 2009;185:1047–1063. doi: 10.1083/jcb.200811099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Dunn CD, Iijima M, Shepard KA, Yaffe MP, Machamer CE, Jensen RE. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p. J Cell Biol. 2006;173:651–658. doi: 10.1083/jcb.200603092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Jensen RE. Division versus fusion: Dnm1p and Fzo1p antagonistically regulate mitochondrial shape. J Cell Biol. 1999;147:699–706. doi: 10.1083/jcb.147.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, et al. The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–28. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solieri L. Mitochondrial inheritance in budding yeasts: towards an integrated understanding. Trends Microbiol. 2010;18:521–530. doi: 10.1016/j.tim.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Spelbrink JN. Functional organization of mammalian mitochondrial DNA in nucleoids: history, recent developments, and future challenges. IUBMB Life. 2010;62:19–32. doi: 10.1002/iub.282. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Endo T, Iijima M, Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. J Cell Biol. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Iijima M, Sesaki H. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 2010;29:2875–2887. doi: 10.1038/emboj.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, et al. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Wakabayashi J, Zhang Z, Wakabayashi N, Tamura Y, Fukaya M, Kensler TW, Iijima M, Sesaki H. The dynamin-related GTPase Drp1 is required for embryonic and brain development in mice. J Cell Biol. 2009;186:805–816. doi: 10.1083/jcb.200903065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- Wang Y, Bogenhagen DF. Human mitochondrial DNA nucleoids are linked to protein folding machinery and metabolic enzymes at the mitochondrial inner membrane. J Biol Chem. 2006;281:25791–25802. doi: 10.1074/jbc.M604501200. [DOI] [PubMed] [Google Scholar]
- Youngman MJ, Hobbs AE, Burgess SM, Srinivasan M, Jensen RE. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J Cell Biol. 2004;164:677–688. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.