Spinal muscular atrophy is caused by deficiency of the survival motor neuron (SMN) protein. We show that the E3 ubiquitin ligase, mind bomb 1 (Mib1), ubiquitinates and targets SMN for degradation. Reducing Mib1 increases SMN levels, and decreasing the Caenorhabditis elegans orthologue of Mib1 mitigates a neuromuscular defect characteristic of SMN deficiency.
Abstract
Spinal muscular atrophy is an inherited motor neuron disease that results from a deficiency of the survival of motor neuron (SMN) protein. SMN is ubiquitinated and degraded through the ubiquitin proteasome system (UPS). We have previously shown that proteasome inhibition increases SMN protein levels, improves motor function, and reduces spinal cord, muscle, and neuromuscular junction pathology of spinal muscular atrophy (SMA) mice. Specific targets in the UPS may be more efficacious and less toxic. In this study, we show that the E3 ubiquitin ligase, mind bomb 1 (Mib1), interacts with and ubiquitinates SMN and facilitates its degradation. Knocking down Mib1 levels increases SMN protein levels in cultured cells. Also, knocking down the Mib1 orthologue improves neuromuscular function in Caenorhabditis elegans deficient in SMN. These findings demonstrate that Mib1 ubiquitinates and catalyzes the degradation of SMN, and thus represents a novel therapeutic target for SMA.
INTRODUCTION
Spinal muscular atrophy (SMA) is an autosomal recessive neurological disorder characterized by loss of lower motor neurons, leading to weakness and skeletal muscle atrophy, and it is one of the leading genetic causes of infant death. More than 90% of SMA results from deletion of the survival motor neuron (SMN1) gene, with all patients retaining at least one copy of the homologue SMN2 (Lefebvre et al., 1995). While SMN1 produces predominantly full-length SMN protein, SMN2 contains a translationally silent C-to-T transition within exon 7, causing this exon to be mostly skipped during mRNA splicing and producing a truncated protein (SMNΔ7) that is unstable and rapidly degraded (Coovert et al., 1997; Lorson et al., 1999; Lorson and Androphy, 2000; Monani et al., 1999; Burnett et al., 2009). Human SMN2 in transgenic mice mitigates the severity of the SMA disease phenotype on a mouse smn-null background, with a gene dosage effect (Hsieh-Li et al., 2000). Patients with mild SMA phenotypes often have three or more SMN2 copies, and some individuals with four or five SMN2 genes have been found to be phenotypically normal (Lefebvre et al., 1995; McAndrew et al., 1997; Prior et al., 2004).
SMN is degraded through the ubiquitin proteasome system (UPS; Chang et al., 2004; Burnett et al., 2009; Kwon et al., 2011). In this system, proteins destined for degradation are tagged by linkage to ubiquitin through the action of three enzymes: E1 (ubiquitin-activating), E2 (ubiquitin-conjugating), and E3 (ubiquitin ligase) (Petroski, 2008). E3 ligases confer the substrate specificity in this process and thus have been considered candidates for targeted inhibition of protein degradation.
We have shown that SMN protein levels are increased in cell culture and animal models with inhibition of the proteasome (Burnett et al., 2009; Kwon et al., 2011). Because protein degradation by the UPS is an essential process in cellular homeostasis and general inhibition may have undesired effects, we sought to identify and characterize the E3 ligase(s) responsible for specifically targeting SMN for degradation. We chose to evaluate mind bomb 1 (Mib1) as a candidate, based on its role in inhibiting the outgrowth of neurites in cultured neurons (Choe et al., 2007). Loss of Drosophila melanogaster Mib1 increases the number of synaptic boutons at neuromuscular junctions (NMJs), producing synaptic overgrowth, while reduction of SMN reduces the number of NMJ boutons in D. melanogaster and results in aberrantly truncated motor neurons in Danio rerio (McWhorter et al., 2003; Choe et al., 2007; Chang et al., 2008). Mib1 is a multidomain enzyme with three carboxyl-terminal really interesting new gene (RING) finger domains (Jin et al., 2002; Itoh et al., 2003; Yoo et al., 2006; Zhang et al., 2007). Its best-characterized role is in activating the Delta-Notch signaling pathway, which controls differentiation by regulating cell–cell communication (Itoh et al., 2003; Chen and Casey Corliss, 2004; Barsi et al., 2005; Koo et al., 2005a; Lai et al., 2005). Other functions of Mib1 have also emerged. Mib1 has been identified in postsynaptic densities, in which it has been shown to function as a binding partner of the CDK5 complex in regulating morphogenesis in postmitotic neurons (Choe et al., 2007) and in regulating apoptosis by promoting the proteasomal degradation of the anti-apoptotic factor death-associated protein kinase (DAPK; Jin et al., 2002). Recently overexpression of Mib1 was shown to promote the ubiquitination of receptor-like tyrosine kinase (RYK), reducing its steady-state levels at the plasma membrane (Berndt et al., 2011).
In this study, we show that SMN interacts with Mib1 and that overexpression of Mib1 in cultured cells increases SMN ubiquitination and degradation. Importantly, Mib1 knockdown results in increased SMN levels in cultured cells and ameliorates the neuromuscular pharyngeal pumping defect in Caenorhabditis elegans deficient in SMN, indicating a physiological role for Mib1 in modulating SMN.
RESULTS
Mib1 increases SMN ubiquitination and protein turnover
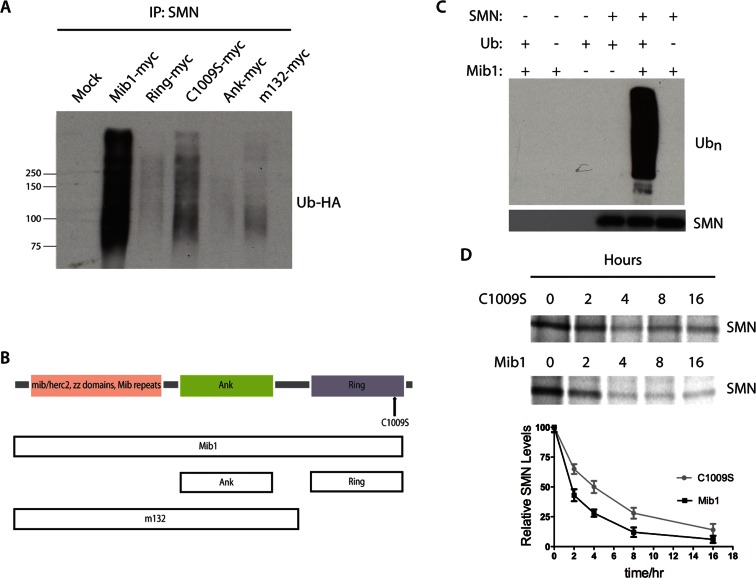
E3 ligases promote protein degradation by catalyzing the transfer of ubiquitin molecules from the E2 enzyme onto substrate proteins. To determine whether Mib1 ubiquitinates SMN, we cotransfected the motor neuron–derived hybrid cell line, NSC34, with hemagglutinin (HA)-tagged ubiquitin and full-length or selected domains of myc-tagged Mib1. The cells were then lysed in buffer containing ubiquitin aldehyde to inhibit deubiquitination and immunoprecipitated with an antibody to SMN. To ensure that the ubiquitin-positive bands on the Western blot were ubiquitinated SMN and not ubiquitinated proteins associated with SMN, we disassociated SMN from its binding partners before immunoprecipitation by denaturing them with 1% SDS, followed by renaturation in 4.5% Triton X-100. These conditions were sufficient to dissociate SMN from known binding partners (Supplemental Figure S1). Immunoblots of immunoprecipitated SMN were probed with anti-HA antibody to detect ubiquitinated SMN. The ubiquitination of endogenous SMN, as indicated by a high-molecular-weight, ubiquitin-positive smear, is increased in cells expressing full-length, but not truncated or active-site mutant forms of Mib1 (Figure 1, A and B). In contrast, overexpressing the E3 ligase parkin did not increase SMN ubiquitination, ruling out the possibility that overexpressing any E3 ligase would indiscriminately increase SMN ubiquitination (Figure S2). We then performed an in vitro ubiquitination assay to determine whether Mib1 directly ubiquitinates SMN. Purified recombinant Mib1 and SMN proteins were incubated in reaction buffer containing ubiquitin, ubiquitin-activating enzyme (E1), and the ubiquitin-conjugating enzyme (E2) UBCH5b. Mib1 ubiquitinates SMN in this cell-free system, as seen by Western blots probed with an antibody to polyubiquitinated proteins, consistent with the results in cultured cells (Figure 1C). Given that Mib1 ubiquitinates SMN, we next sought to quantify the effect of Mib1 on SMN protein turnover. We performed pulse–chase analysis using HEK-293T cells transfected with wild-type Mib1-myc or an active-site mutant, Mib1-C1009S-myc, to determine whether the E3 ligase activity of Mib1 alters SMN protein half-life. We found that overexpressing wild-type Mib1 decreased the half-life of newly synthesized radiolabeled SMN by half, from ∼4 to 2 h, compared with the active-site Mib1 mutant (Figure 1D). In addition, overexpressing Mib1 in the NSC34 cells reduced steady-state levels of SMN protein, and this effect was blocked by the proteasome inhibitor bortezomib (Figure 2A), indicating that Mib1 targets SMN for proteasomal degradation.
FIGURE 1:
(A) NSC-34 cells were transfected with 2 μg Mib1 and 1 μg HA-Ub cDNAs. The cells were harvested 48 h later, and endogenous SMN was immunoprecipitated. Immunoprecipitated proteins were resolved by SDS–PAGE, and the proteins were analyzed by Western blotting. The blots were probed with an HA antibody to detect ubiquitinated SMN. (B) Schematic representation of Mib1 protein domains. (C) Cell-free SMN ubiquitination assay. Recombinant SMN was incubated with E1 and E2 (UBCH5B) enzymes with or without Mib1 and ubiquitin for 1 h at 37°C. Western blots were probed with an anti-polyubiquitin antibody (FK1). (D) Pulse–chase analysis of endogenous SMN in the presence of 2 μg Mib1-myc or Mib1-C1009S-myc. The data represent mean ± SEM of three independent experiments.
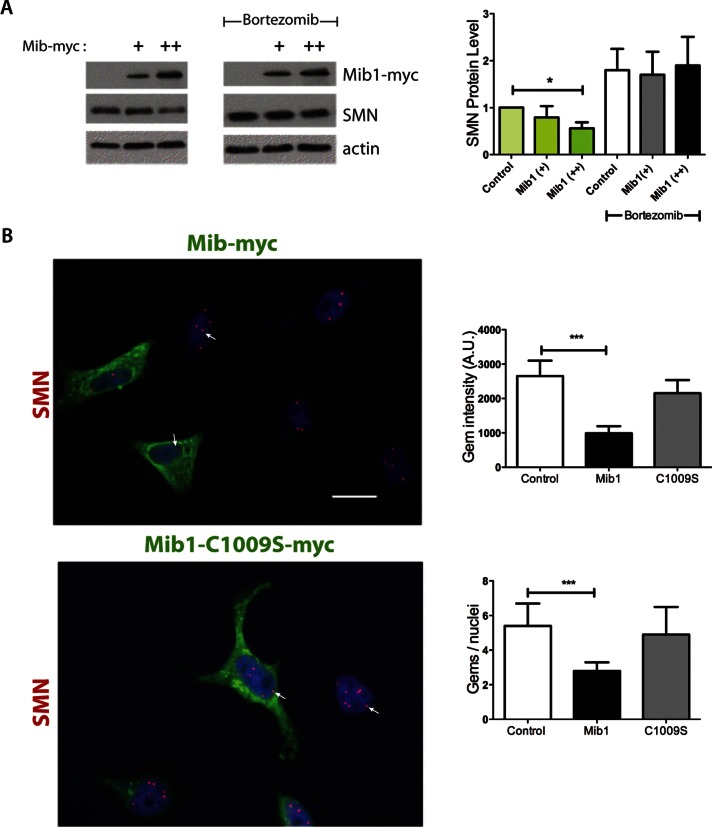
FIGURE 2:
Effects of Mib1 overexpression on SMN protein levels and gem number. (A) HEK-293T cells were transfected with either 1 or 2 μg Mib1-myc cDNA. At 24 h after transfection, cells were treated with either vehicle (water) or 1 μM bortezomib and harvested 6 h later. Cell lysates were resolved by SDS–PAGE, and densitometry of the resulting bands was performed using ImageJ software (National Institutes of Health). SMN protein levels were corrected based on actin values. The data represent mean ± SEM of three independent experiments. *, p < 0.05. (B) Overexpression of Mib1 decreases gem number and intensity in cultured cells. HeLa cells were transfected with Mib1-myc or Mib-C1009S-myc for 24 h. The cells were fixed and stained with antibodies to SMN and myc and the appropriate Alexa Fluor secondary antibodies for visualization by microscopy. The number and intensity of nuclear gems were quantitated using Nikon NIS Elements software. At least 300 cells were counted for each transfection condition. That data represent mean ± SEM of three independent experiments. ***, p < 0.001.
The SMN complex is assembled in the cytoplasm before it is imported into the nucleus, where it localizes to Cajal bodies or gems. The number of these SMN-positive nuclear bodies has been used as a measure for evaluating changes in functional SMN protein levels (Coovert et al., 1997; Jarecki et al., 2005; Wolstencroft et al., 2005). We counted the number of SMN-positive nuclear puncta per nucleus and assessed their fluorescence intensity in HeLa cells overexpressing wild-type Mib1 or mutant Mib1-C1009S. We found that the number and intensity of SMN-positive bodies were significantly reduced in cells overexpressing wild-type Mib1 compared with untransfected cells (Alexa Fluor 488–negative) and with those expressing Mib1-C1009S (p < 0.0001, Figure 2B). Together, these data demonstrate that Mib1 determines SMN protein levels by promoting its ubiquitination and subsequent degradation.
Reducing Mib1 expression increases SMN protein levels
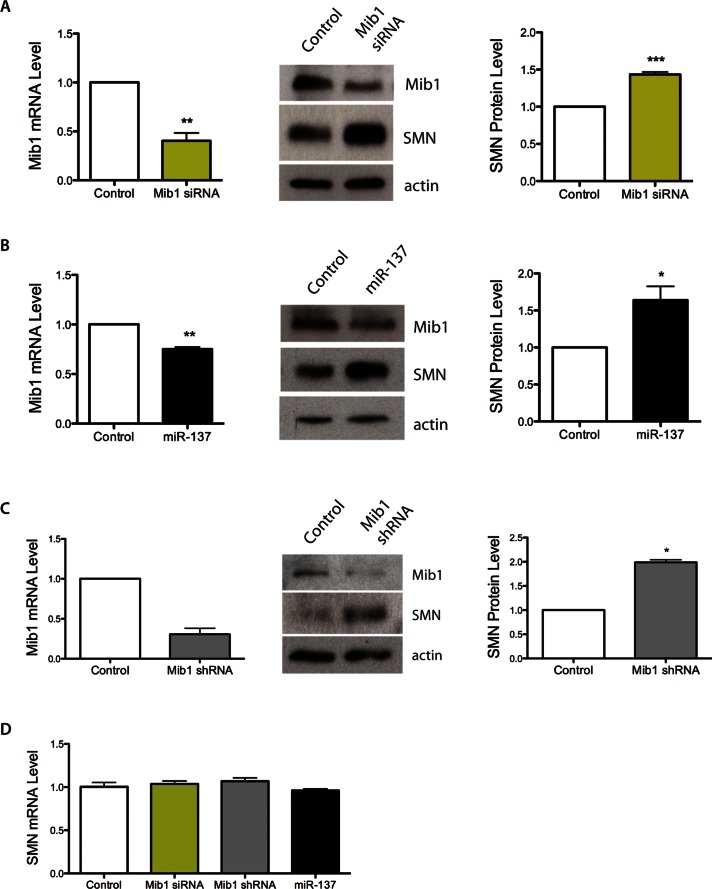
Because Mib1 overexpression decreased SMN levels we next investigated whether decreasing Mib1 increases SMN levels. We transfected HEK-293T cells with Mib1 small interfering RNA (siRNA) or a scrambled control, and we confirmed by RT-PCR that Mib1 levels were reduced by more than half 72 h after transfection with Mib1 siRNA (Figure 3A). Importantly, SMN protein levels were significantly increased when Mib1 was knocked down (p < 0.001, Figure 3A). It has previously been reported that the microRNA miR-137 targets Mib1 mRNA and regulates its protein expression (Smrt et al., 2010). We sought to determine whether overexpressing miR-137 also affects SMN protein levels. We confirmed that transfection of cells with miR-137 decreased Mib1 mRNA and protein levels (Figure 3B), and we found that SMN levels were increased in cells transfected with miR-137 compared with a scrambled microRNA control (Figure 3B). Importantly, we confirmed that short hairpin RNA (shRNA) knockdown of Mib1 increases SMN protein levels in SMA patient–derived fibroblasts (Figure 3C). SMN transcript levels were unchanged by siRNA to Mib1 or by miR-137 (Figure 3D). Thus, using three independent techniques and two cell lines we have shown that reducing Mib1 expression results in increased SMN protein levels.
FIGURE 3:
(A) HEK-293T cells were transfected with either 100 pmol of scrambled siRNA or Mib1 siRNA for 72 h. Mib1 and SMN protein levels were determined after 72 h by Western blotting. (B) HEK-293T cells were transfected with either 100 μM negative control miRNA or miR-137 for 72 h. SMN protein levels were determined by Western blotting. The data represent the mean ± SEM of three independent experiments. (C) SMA patient–derived fibroblasts (3813) were transfected with Mib1 siRNA or scrambled control. Cells were harvested 72 h post-transfection, and Mib-1 mRNA and SMN protein levels were assessed by RT-PCR and Western blot analysis, respectively. (D) HEK-293T cells were transfected with Mib1 siRNA, miR-137, or scrambled control. Cells were harvested 72 h posttransfection, and SMN mRNA level was analyzed by RT-PCR. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Interaction of Mib1 and SMN is mediated by the N-terminal domain of Mib1 and the segment of SMN encoded by exon 6
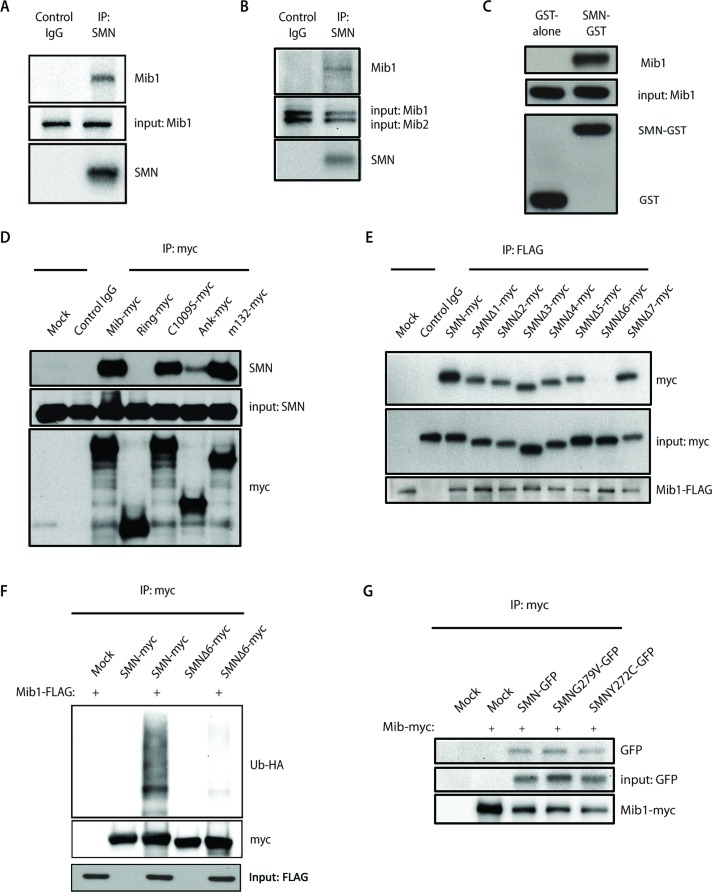
We next investigated whether Mib1 interacts with SMN. Using a monoclonal antibody to SMN, we found that endogenous Mib1 coimmunoprecipitated with endogenous SMN (Figure 4A), indicating that these proteins associate. No interaction was detectable between SMN and Mib2, a homologue of Mib1 (Koo et al., 2005b) with very similar structural domain organization (Figure 4B), demonstrating the specificity of the interaction between Mib1 and SMN. To further characterize the interaction between Mib1 and SMN, we used purified recombinant Mib1 in pull-down assays with purified glutathione S-transferase (GST)-SMN or GST alone. We found that GST-SMN but not GST alone pulled down Mib1 (Figure 4C), confirming a direct interaction between SMN and Mib1 and showing that other factors, including members of the SMN complex, are not required for this interaction.
FIGURE 4:
(A) Endogenous SMN was immunoprecipitated from HEK-293T cell lysates with an antibody to SMN, and immunoprecipitates were resolved by SDS–PAGE. Association with Mib1 was determined by Western blot analysis using an antibody to Mib1. (B) Protein lysates from HEK-293T cells were immunoprecipitated with an SMN antibody. While Mib1 coimmunoprecipitated with SMN, Mib2 was not detected by Western blot analysis. Membranes were probed with anti-SMN, anti-Mib1, and anti-Mib2 antibodies. (C) GST or GST-SMN was used to pull down purified recombinant Mib1 protein. (D) Myc-tagged Mib1 constructs containing either full-length or truncated transcripts of the Mib1 gene were transiently transfected into HEK-293T cells. Immunoprecipitation using an anti-myc antibody was performed, and interaction with endogenous SMN was detected by Western blot analysis using an anti-SMN antibody. (E) HEK-293T cells were transfected with 0.5 μg full-length SMN cDNA or various SMN constructs with a single-exon deletion, and 2 μg Mib1-FLAG. Mib1-FLAG was immunoprecipitated and resolved on a SDS–PAGE gel. Western blots were probed with antibodies to FLAG and Myc to determine binding of Mib1 to SMN. (F) HEK-293T cells were transfected with 2 μg Mib1-FLAG, 1 μg HA-Ub, and 1 μg either full-length SMN-myc or SMNΔ6-myc. The cells were harvested 48 h later, myc-tagged SMN was immunoprecipitated using an anti-myc antibody, and the proteins were separated by SDS–PAGE and detected by Western blotting. The blots were probed with an HA antibody to detect ubiquitinated SMN. (G) HEK-293T cells were transfected with 2 μg Mib1-myc and either 0.5 μg GFP-tagged wild-type SMN, SMN G279V, or SMN Y272C. The cells were harvested 48 h later, and SMN was immunoprecipitated with an anti-myc antibody and resolved on a SDS–PAGE gel. Western blots were probed with antibodies to myc and GFP to determine interaction of Mib1 and wild-type SMN or the disease-associated mutants.
We then asked which domains of Mib1 mediate the interaction with SMN. Using constructs containing various regions of the Mib1 gene, we performed coimmunoprecipitation assays to map the domains of Mib1 required for SMN binding. While the N-terminal region of Mib1 (m132-myc) coimmunoprecipitated with SMN, constructs containing the middle ankyrin repeat and C-terminal RING domain showed weak or no interaction (Figure 4D). Additionally, the C-terminal active-site mutation C1009S had no effect on SMN binding (Figure 4D). To map the regions of SMN that mediate binding with Mib1, we cotransfected a FLAG-tagged Mib1 construct with constructs expressing either full-length SMN or SMN lacking each of its seven translated exons. Full-length SMN and each deletion mutant, with the exception of SMNΔ6, coimmunoprecipitated with Mib1, indicating that the SMN exon 6 is required for interaction with Mib1 (Figure 4E). Moreover, we found that deletion of SMN exon 6 not only disrupts binding with Mib1 but markedly reduced its ubiquitination by Mib1 (Figure 4F). Because the domain encoded by exon 6 mediates SMN oligomerization, we examined whether SMN oligomerization is required for Mib1 to interact with SMN. We found that Mib1 coimmunoprecipitated with wild-type SMN as well as with the patient-derived SMN mutants SMN-G279V and SMN-Y272C, which do not efficiently self-associate or oligomerize (Figure 4G). Thus SMN oligomerization is not required for Mib1 association.
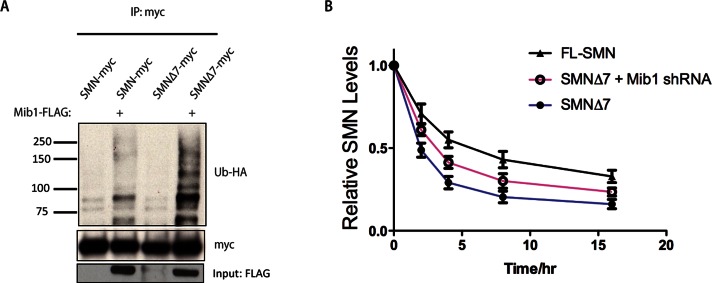
We sought to determine whether Mib1 contributes to the instability of SMNΔ7. We found that overexpressing Mib1 resulted in greater ubiquitination of SMNΔ7 than full-length SMN, and knocking down Mib1 increased SMNΔ7 stability (Figure 5, A and B). These data suggest that the instability of the SMNΔ7 is due in part to ubiquitination by Mib1.
FIGURE 5:
(A) HEK-293T cells were transfected with 2 μg full-length SMN or SMNΔ7 and 1 μg Mib1-FLAG and HA-Ub cDNAs. The cells were harvested 48 h later, SMN was immunoprecipitated and resolved by SDS–PAGE, and the proteins were analyzed by Western blotting. The blots were probed with an HA antibody to detect ubiquitinated SMN. (B) Pulse–chase analysis of full-length SMN and SMNΔ7, in the absence or presence of 2 μg Mib1-myc. The data represent the mean ± SEM of three experiments.
Loss of the C. elegans orthologue mib-1 ameliorates the neuromuscular defects caused by smn-1 loss of function
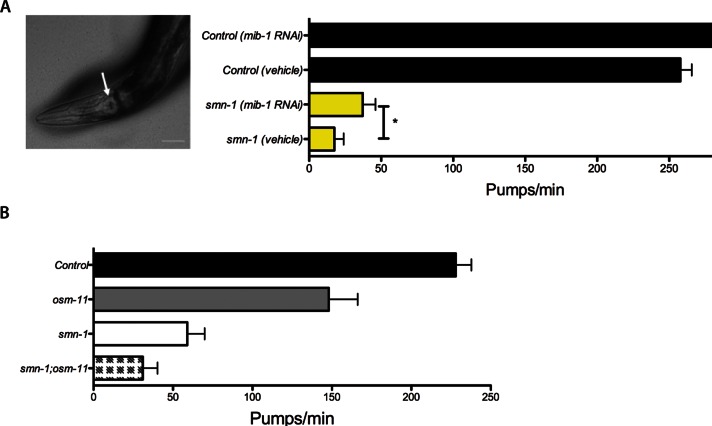
Potential cross-species conservation of the SMN-MIB1 interaction was examined in C. elegans. The C. elegans genome has a single orthologue of SMN, smn-1. Homozygous smn-1 loss of function causes developmental delay, impaired locomotion, reduced pharyngeal pumping, and larval lethality (Briese et al., 2009; Dimitriadi et al., 2010). Although complete loss of SMN function causes lethality, smn-1 animals can survive for several days due to partial maternal rescue. The C. elegans orthologue of human MIB1 is the gene mib-1 (Berndt et al., 2011). To assess genetic interactions, we decreased Mib-1 levels in smn-1–deficient animals by RNA interference (RNAi) through feeding (Timmons and Fire, 1998; Timmons et al., 2001). Knocking down the Mib-1 C. elegans orthologue increased the pharyngeal pumping rates of smn-1–deficient animals, but this did not reach statistical significance (p = 0.06, Figure S3). RNAi by feeding is generally less effective in neurons than in other tissues (Kamath et al., 2001; Timmons et al., 2001; Rual et al., 2004). To circumvent this difficulty, we increased neuronal sensitivity to RNAi feeding by ectopic neuronal expression of the SID-1 double-stranded RNA channel in smn-1 animals (Calixto et al., 2010). In this sensitized background, mib-1 RNAi significantly ameliorated the smn-1 pharyngeal pumping defects (Figure 6A; p = 0.04), suggesting that MIB-1 is a conserved cross-species modifier of SMN loss-of-function defects. Because Mib1 plays an important role in Notch function (Itoh et al., 2003; Lai et al., 2005; Le Borgne et al., 2005), we examined whether the MIB-1 impact was due to perturbations in Notch signaling. Loss of the Notch coligand function (Komatsu et al., 2008; Singh et al., 2011) in smn-1 mutants did not ameliorate the pharyngeal pumping defect of smn-1(ok355);osm-11(rt142) animals (Figure 6B). While we cannot rule out SMN-independent effects, these data suggest that MIB-1 directly impacts SMN-1 function independent of Notch signaling.
FIGURE 6:
Knockdown of the C. elegans orthologue of Mib1 ameliorates the pharyngeal pumping neuromuscular defects of smn-1 loss-of-function animals. (A) mib-1 knockdown significantly increases pharyngeal pumping in smn-1 deficient animals with SID-1 sensitization, which increases RNAi neuronal sensitivity. (B) Loss of the Notch coligand function in smn-1 mutants did not ameliorate the pharyngeal pumping defect of smn-1(ok355);osm-11(rt142) animals. Pharyngeal pumping events were measured in both control and mib-1 RNAi-fed animals. All error bars are SEM; *, p < 0.05.
DISCUSSION
Through this study, we provide evidence that the E3 ligase Mib1 ubiquitinates and catalyzes SMN protein degradation. These effects are not associated with changes in SMN mRNA expression and are not observed when a ligase-defective point mutant of Mib1 is expressed. Knocking down Mib1 increases SMN protein levels in cultured cells and partially rescues a neuromuscular defect due to SMN deficiency in C. elegans. These data indicate that Mib1 reduces SMN protein stability by targeting it for degradation by the proteasome and represents a new modifier of the SMA phenotype.
The inverse correlation between SMN2 copy number and SMA severity indicates that increasing SMN protein levels may ameliorate the disease phenotype. Mechanisms to increase SMN protein levels have been actively pursued as therapeutic options for SMA. Inhibiting the degradation of SMN is one way to increase its steady-state level. Inhibiting the proteasome increases SMN levels in cultured cells and SMA model mice (Kwon et al., 2011), and it ameliorates the disease phenotype. Identifying enzymes that specifically target SMN for degradation could thus provide new therapeutic targets. We show here that overexpression of Mib1 increases ubiquitination of SMN, as indicated by increased ubiquitin–SMN conjugates. We found that overexpressed Mib1 accelerates the rate of SMN degradation, whereas knockdown of Mib1 by either siRNAs or the microRNA mir-137 stabilizes SMN. Our data demonstrate that Mib1-mediated ubiquitination of SMN enhances its degradation and decreases steady-state SMN protein levels.
While most SMA patients have homozygous deletion of the SMN1 gene, they all retain at least one copy of SMN2. The major SMN2 gene product is the truncated SMNΔ7 protein that, based on its ability to extend the survival of SMA mice, likely retains some function but is unstable and rapidly degraded. The instability of SMNΔ7 is thought to be due to a number of factors, including its inability to self-associate (Lorson et al., 1998; Burnett et al., 2009) and the presence of a degradation signal that promotes its rapid turnover (Cho and Dreyfuss, 2010). In this study, we show that Mib1 ubiquitinates SMNΔ7 to a greater extent than full-length SMN. We propose that the increased Mib1-mediated ubiquitination of SMNΔ7 could at least in part account for the relative instability of the SMNΔ7 protein. Consistent with this, reducing Mib1 levels results in increased stability of SMNΔ7 protein.
We found that the interaction of SMN with Mib1 is likely mediated by the domain encoded by exon 6. Missense mutations in exon 6 and deletion of exon 7 disrupt SMN's ability to oligomerize (Lorson et al., 1998; Pellizzoni et al., 1999; Zhang et al., 2003; Le et al., 2005), resulting in severe SMA; this may be because SMN that fails to oligomerize is rapidly degraded (Burnett et al., 2009). A mutation in SMN exon 6 that disrupts oligomerization (Y272C) also destabilizes the SMN protein, and cells from a patient with this mutation have similar SMN levels to those with homozygous deletion of SMN1 (Lefebvre et al., 1997). One possibility is that Mib1 interacts only with SMN that is oligomerized, as exon 6 mediates SMN oligomerization. However, this is not likely because Mib1 interacts with SMN that has the patient-derived mutations G279V and Y272C, which do not efficiently oligomerize (Figure 4G). Thus it is more likely that the inability of SMN to oligomerize increases the likelihood of binding to Mib1. Because the exon 6 domain mediates SMN self-association (Lorson et al., 1998), this domain may not be available for interaction with Mib1 when SMN oligomerizes or is in a complex. This could explain the increased stability of complexed SMN, which is inaccessible to Mib1 and thus unlikely to be ubiquitinated and degraded.
While this article was in preparation, Usp9x (also known as fat facets, FAM), a deubiquitinating enzyme (DUB), was found to stabilize SMN by reducing its ubiquitination (Han et al., 2012). Interestingly, Usp9x/FAM also interacts with Mib1 (Choe et al., 2007), which points to the possibility that Usp9x and Mib1 act in concert to regulate SMN protein levels.
Two genetic modifiers of SMA have been reported in humans: SMN2 and plastin3. A genome-wide RNAi screen in C. elegans and Drosophila deficient in SMN identified other genes that modify the disease phenotype in these model systems (Dimitriadi et al., 2010). SMN deficiency in C. elegans is characterized by marked reduction in the rate of pharyngeal pumping. We show here that Mib1 knockdown by RNAi partially rescues the SMN-deficient phenotype in C. elegans, indicating that Mib1 may be a modifier of SMA. Our biochemical data suggest that Mib1 promotes SMN degradation by the proteasome. However, we cannot rule out the possibility that SMN-independent pathways involving other functions of Mib1 are responsible for the partial rescue of the neuromuscular phenotype. Nonetheless, the identification of a new modifier of the SMA phenotype is of particular importance, given the currently limited number of targets with a favorable therapeutic index.
The steady-state level of a protein is governed by the balance between protein synthesis and degradation. Strategies to increase SMN protein synthesis by up-regulating the gene, stabilizing the mRNA, and altering splicing to promote inclusion of exon 7 have been pursued in SMA therapeutic development. That the SMN2 gene produces some full-length SMN protein suggests that strategies to slow its degradation may also be therapeutic options. Because SMN is ubiquitinated and degraded by the proteasome, retarding its turnover either through blocking the proteasome or inhibiting SMN ubiquitination may be useful in treating SMA. We have shown that SMA mice treated with the proteasome inhibitor bortezomib show reduced pathology and improved motor function (Kwon et al., 2011). However, the toxicity of available proteasome inhibitors may preclude chronic use in SMA patients. While small molecule inhibitors of the proteasome have been developed for research and clinical use, E3 ubiquitin ligases are emerging as targets for disease treatment, because they confer the substrate specificity of the UPS. Drugs that inhibit E3 ligases involved in cell cycle regulation and cell proliferation are currently under study to treat various types of cancer. One such E3 ligase, murine double minute 2 (MDM2), regulates the tumor suppressor p53 and represses its role in cell cycle arrest and apoptosis (Fakharzadeh et al., 1993; Haupt et al., 1997). Inhibitors of the human homologue of MDM2 have been shown to reduce tumor growth and induce tumor cell apoptosis in vitro, providing an incentive for targeting MDM2 function to treat cancer in vivo (Issaeva et al., 2004; Koblish et al., 2006). High-throughput small molecule compound screens have been used to identify inhibitors of E3 ligases (Davydov et al., 2004; Huang et al., 2005). These screens may be adapted to identify molecules that disrupt the interaction between a ligase and its protein substrate, which would confer greater selectivity and reduce off-target effects. Nutlin, which inhibits the interaction between MDM2 and p53, was identified in a small molecule screen (Vassilev et al., 2004) and is currently being evaluated in early-phase clinical trials. While new, less toxic, general proteasome inhibitors are being developed, targeting an E3 ligase that selectively tags SMN for degradation may be safer and more effective. Small molecule screens to identify inhibitors of Mib1 may thus lead to novel therapeutics for SMA.
MATERIALS AND METHODS
Plasmids and miRNA
Plasmids encoding human full-length and truncated Mib1 were constructed as previously described (Itoh et al., 2003). Mib1 shRNA and nonsilencing control were purchased from SABiosciences (KM26177G) and pre-miR miRNA precursors (control and miR-137) were purchased from Ambion (PM10513). Myc-Parkin was obtained from the Chitnis lab.
Cell culture, transfection, and Western blotting
Human embryonic kidney HEK-293T and NSC34 cells were maintained at 37°C with 5% CO2 in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS and penicillin/streptomycin/glutamine (Invitrogen). Cells were plated into a six-well culture plate and transiently transfected with 0.5–4 μg of plasmid DNA per well using FuGENE HD transfection reagent (Roche Applied Science, Indianapolis, IN) according to the manufacturer's protocol. The total amount of DNA used for transfection was kept constant in each experiment by adding an appropriate amount of empty vector plasmid. Cells were harvested 24–48 h posttransfection.
For siRNA and miRNA transfections, HEK-293T cells were plated subconfluently in a six-well culture dish and transfected with 100 pmol of siRNA/miRNA and 10 μl of Lipofectamine 2000 (Invitrogen). After 48 h of transfection, cells were again transfected with siRNA/miRNA for 24 h and were harvested after a total 72 h of transfection.
Western blotting, immunoprecipitation, and pull-down assays
Transfected cells were lysed in either NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, pH 8.0) or RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 150 mM NaCl, 0.1% SDS, pH 8.0) and quantified using the BCA Protein Assay Kit (Pierce, Rockford, IL) for Western blotting. Equal amounts of protein lysate (15 μg) were run and separated on a 10% SDS–PAGE (Invitrogen) and transferred to a polyvinylidene fluoride membrane (Invitrogen). They were then probed with the appropriate antibodies in 5% milk and washed with Tris-buffered saline and Tween 20. Antibodies: anti-SMN (BD Transduction), anti-Mib1 (Epitomics, Burlingame, CA), and anti-Mib2 (Sigma-Aldrich, St. Louis, MO) were used at a 1:1000 dilution; anti-FLAG M2 (Sigma-Aldrich), anti-Myc 9E10 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-HA (Sigma-Aldrich), and anti-actin (Sigma-Aldrich) were used at a 1:5000 dilution. Anti-Mib1 serum was a gift from Junmin Peng (St. Jude Children's Research Hospital). The signal was visualized with a secondary antibody (anti-mouse or anti-rabbit immunoglobulin-horseradish peroxidase, both at 1:3000–10,000 dilution) using a chemiluminescence detection system (Perkin Elmer-Cetus, Waltham, MA).
For immunoprecipitation experiments, cell lysates were clarified by centrifugation and precleared with pan-mouse immunoglobulin G (IgG) Dynabeads (Invitrogen) for 1 h at 4°C. The lysates were then incubated with 1 μg of antibody for 2 h to overnight and then incubated with pan-mouse IgG Dynabeads for 1 h on a rotator at 4°C. The beads were washed with NP-40 buffer four to five times and boiled in SDS gel-loading buffer. Eluted proteins were run on an SDS–PAGE for Western blotting.
For the pull-down assay, 5 μg purified recombinant Mib1 protein purchased from Origene (Rockville, MD; TP321377) was incubated with GST or SMN-GST beads at 4°C on a rotator overnight. Samples were spun down, and the beads were washed five times with NP-40 buffer. Following the last wash, the beads were boiled in SDS gel-loading buffer for 5 min, and eluted proteins were processed for Western blotting.
RNA extraction and quantification
Cells were homogenized in 500 μl TRIzol reagent (Invitrogen) and spun for 10 min at 4°C. One hundred microliters of chloroform was added to the supernatant, and the mixture was vortexed and spun down again. After the aqueous upper phase was transferred to another tube, 250 μl of isopropanol and 50 μl 3M sodium acetate were added. The RNA was then pelleted, washed with 70% ethanol, and resuspended in 100 μl DNase- and RNase-free water. Total RNA was quantified by absorption at 260 nm, and 450 ng of each RNA sample was converted to cDNA using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems, Bedford, MA). Aliquots of 20 μl quantitative PCRs were run in triplicate using the ABI Prism 7900 sequence detection system (Applied Biosystems). The levels of hypoxanthine phosphoribosyltransferase (HPRT; control), SMN, and Mib1 transcripts were quantified by the threshold cycle with primers purchased from Applied Biosystems.
Immunohistochemical analysis of gem numbers
The gem number was assayed 24 h after transfection with myc-Mib1 or myc-Mib1-C1009S. The cells were fixed, permeabilized, and immunostained with anti-SMN and anti-myc antibodies. Blinded counts of gems were performed in more than 300 nuclei for each condition.
In vitro ubiquitination
Recombinant SMN protein (Enzo Life Sciences, Farmingdale, NY) was incubated with ubiquitin, an E1 ubiquitin-activating enzyme (Boston Biochem, Cambridge, MA), E2 ubiquitin-conjugating enzyme, UBCH5B, and recombinant Mib1 (OriGene Technologies, Rockville, MD) in reaction buffer (50 mM Tris-HCl, pH 7.5, 2 mM ATP, 4 mM MgCl2, 2 mM dithiothreitol [DTT]) for 1 h at 37°C. Reactions were quenched with 30 μl of 5× Laemmli sample buffer (200 mM Tris, pH 6.8, 32% [vol/vol] glycerol, 6.4% [wt/vol] SDS, 0.32% [wt/vol] bromophenol blue, and 200 mM DTT), boiled for 5 min, and separated via SDS–PAGE. SMN ubiquitination was analyzed by Western blotting, using antibody against polyubiquitin (FK1; Enzo Life Sciences).
Pulse–chase protein labeling
Pulse–chase analysis was performed as previously described (Burnett et al., 2009). In brief, HEK-293T cells were incubated in cysteine-methionine–free medium (Sigma-Aldrich) for 2 h or 12 h; this was followed by incubation in cysteine-methionine–free medium containing 100 μCi of [35S]-labeled cysteine-methionine (GE Healthcare, Piscataway, NJ) for 1 h. After labeling, the cells were washed once with culture medium containing a 10-fold excess of unlabeled methionine and cysteine (5 mM each) and then incubated further in the same medium. The cells were collected at the indicated time points and processed for immunoprecipitation with anti-SMN antibody (BD Transduction Laboratories). Immunoprecipitations were carried out for 6 h at 4°C with antibodies bound to protein G-Sepharose (Sigma-Aldrich). After three washes with lysis buffer (100 mM NaCl, 10 mM Tris-HCl, pH 7.4, 2.5 mM MgCl2, 0.1% NP-40, and protease [Roche]) and phosphatase inhibitor cocktails (Sigma-Aldrich), bound proteins were eluted by boiling in SDS–PAGE sample buffer. Immunocomplexes were separated on 10% SDS–PAGE. The gels were dried and exposed to a phosphorimager screen, and the signal was visualized with a Storm phosphorimager system (Molecular Dynamics, Piscataway, NJ). Densitometric analysis of the protein bands was carried out using ImageQuant PhosphorImager software (Molecular Dynamics).
C. elegans strains
TU3311 uIs60 [unc-119p::yfp, unc-119p::sid-1] (Calixto et al., 2010), HA2258 smn-1(ok355)/hT2[bli-4(e937) let-?(q782) qIs48] (I;III); uIs60 (Briese et al., 2009), and HA2201 smn-1(ok355)/hT2[bli-4(e937) let-?(q782) qIs48] (I;III); osm-11(rt142)X were maintained at 20°C according to standard protocols (Brenner, 1974).
Construction of RNAi feeding clone for Y47D3A.22/mib-1
A 796–base pair PCR product corresponding to the C. elegans mib-1 cDNA (Berndt et al., 2011) was cloned into the L4440 RNAi feeding vector; this was followed by transformation into the HT115(DE3) bacterial strain (Timmons and Fire, 1998; Timmons et al., 2001). Primers for amplification were ACH72 5’-tgatgctagcgcagcttctcctgttgcgtg-3’ and ACH73 5’-tgatctcgaggcgggagccgatttgaactg-3’.
C. elegans pharyngeal pumping assay
The pharyngeal pumping assay was performed as previously described (Dimitriadi et al., 2010). Briefly, eggs hatched on L4440 control vector (L4440 [RNAi]) or (Y47D3A.22/mib-1[RNAi]) bacterial feeding strains (Kamath and Ahringer, 2003) were reared after 2 d at 25°C and 1 d at 20°C. Pumping rates were determined using an AxioCam ICc1 camera on a Zeiss Stemi SV11 microscope (Carl Zeiss, Jena, Germany). A pumping event was scored as a pharyngeal grinder movement in any axis, and average pumping rates (± SEM) were derived in a blinded manner from at least three independent trials (n ≥ 30 animals in total). Heterozygous and homozygous smn-1 loss-of-function animals were distinguished by green fluorescent protein (GFP) fluorescence. Both wild-type (N2) and smn-1 loss-of-function heterozygous animals showed pumping rates that were not significantly different. Hence, heterozygous smn-1 loss-of-function animals were used as controls in this study.
Statistics
The biochemical data were analyzed with the GraphPad Prism software package (GraphPad Software, San Diego, CA) and compared statistically by either t test or one-way analysis of variance, with the Newman-Keuls multiple-comparison post hoc correction. To compare differences among the three groups, we performed a nonparametric equality of medians test, because the data were not normally distributed. If this was statistically significant, then a pairwise comparison between the two treatment groups was done using a Mann-Whitney test.
Supplementary Material
Acknowledgments
We thank Liu Yang (University of Arkansas) for the myc-tagged FL-SMN and deletion mutant cDNAs. This work was supported by intramural research funds from the National Institute of Neurological Disorders and Stroke and from National Institutes of Health–National Institute of Neurological Disorders and Stroke (NS066888) to A.C.H.
Abbreviations used:
- Mib1
mind bomb 1
- NMJ
neuromuscular junction
- RING
really interesting new gene
- SMA
spinal muscular atrophy
- SMN
survival of motor neuron
- UPS
ubiquitin proteasome system
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-01-0042) on April 24, 2013.
The authors declare that they have no conflict of interest.
REFERENCES
- Barsi JC, Rajendra R, Wu JI, Artzt K. Mind bomb1 is a ubiquitin ligase essential for mouse embryonic development and Notch signaling. Mech Dev. 2005;122:1106–1117. doi: 10.1016/j.mod.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Berndt JD, Aoyagi A, Yang P, Anastas JN, Tang L, Moon RT. Mindbomb 1, an E3 ubiquitin ligase, forms a complex with RYK to activate Wnt/β-catenin signaling. J Cell Biol. 2011;194:737–750. doi: 10.1083/jcb.201107021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese M, Esmaeili B, Fraboulet S, Burt EC, Christodoulou S, Towers PR, Davies KE, Sattelle DB. Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum Mol Genet. 2009;18:97–104. doi: 10.1093/hmg/ddn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett BG, Munoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH. Regulation of SMN protein stability. Mol Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods. 2010;7:554–559. doi: 10.1038/nmeth.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, et al. Modeling spinal muscular atrophy in Drosophila. PLoS One. 2008;3:e3209. doi: 10.1371/journal.pone.0003209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HC, Hung WC, Chuang YJ, Jong YJ. Degradation of survival motor neuron (SMN) protein is mediated via the ubiquitin/proteasome pathway. Neurochem Int. 2004;45:1107–1112. doi: 10.1016/j.neuint.2004.04.005. [DOI] [PubMed] [Google Scholar]
- Chen W, Casey Corliss D. Three modules of zebrafish Mind bomb work cooperatively to promote Delta ubiquitination and endocytosis. Dev Biol. 2004;267:361–373. doi: 10.1016/j.ydbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Cho S, Dreyfuss G. A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 2010;24:438–442. doi: 10.1101/gad.1884910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe EA, Liao L, Zhou JY, Cheng D, Duong DM, Jin P, Tsai LH, Peng J. Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci. 2007;27:9503–9512. doi: 10.1523/JNEUROSCI.1408-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AH. The survival motor neuron protein in spinal muscular atrophy. Hum Mol Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- Davydov IV, Woods D, Safiran YJ, Oberoi P, Fearnhead HO, Fang S, Jensen JP, Weissman AM, Kenten JH, Vousden KH. Assay for ubiquitin ligase activity: high-throughput screen for inhibitors of HDM2. J Biomol Screen. 2004;9:695–703. doi: 10.1177/1087057104267956. [DOI] [PubMed] [Google Scholar]
- Dimitriadi M, et al. Conserved genes act as modifiers of invertebrate SMN loss of function defects. PLoS Genet. 2010;6:e1001172. doi: 10.1371/journal.pgen.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakharzadeh SS, Rosenblum-Vos L, Murphy M, Hoffman EK, George DL. Structure and organization of amplified DNA on double minutes containing the mdm2 oncogene. Genomics. 1993;15:283–290. doi: 10.1006/geno.1993.1058. [DOI] [PubMed] [Google Scholar]
- Han KJ, Foster DG, Zhang NY, Kanisha K, Dzieciatkowska M, Sclafani RA, Hansen KC, Peng J, Liu CW. Ubiquitin-specific protease 9x deubiquitinates and stabilizes the spinal muscular atrophy protein—survival motor neuron. J Biol Chem. 2012;287:43741–43752. doi: 10.1074/jbc.M112.372318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- Huang J, Sheung J, Dong G, Coquilla C, Daniel-Issakani S, Payan DG. High-throughput screening for inhibitors of the e3 ubiquitin ligase APC. Methods Enzymol. 2005;399:740–754. doi: 10.1016/S0076-6879(05)99049-6. [DOI] [PubMed] [Google Scholar]
- Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321–1328. doi: 10.1038/nm1146. [DOI] [PubMed] [Google Scholar]
- Itoh M, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jarecki J, Chen X, Bernardino A, Coovert DD, Whitney M, Burghes A, Stack J, Pollok BA. Diverse small-molecule modulators of SMN expression found by high-throughput compound screening: early leads towards a therapeutic for spinal muscular atrophy. Hum Mol Genet. 2005;14:2003–2018. doi: 10.1093/hmg/ddi205. [DOI] [PubMed] [Google Scholar]
- Jin Y, Blue EK, Dixon S, Shao Z, Gallagher PJ. A death-associated protein kinase (DAPK)-interacting protein, DIP-1, is an E3 ubiquitin ligase that promotes tumor necrosis factor-induced apoptosis and regulates the cellular levels of DAPK. J Biol Chem. 2002;277:46980–46986. doi: 10.1074/jbc.M208585200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koblish HK, et al. Benzodiazepinedione inhibitors of the Hdm2:p53 complex suppress human tumor cell proliferation in vitro and sensitize tumors to doxorubicin in vivo. Mol Cancer Ther. 2006;5:160–169. doi: 10.1158/1535-7163.MCT-05-0199. [DOI] [PubMed] [Google Scholar]
- Komatsu H, et al. OSM-11 facilitates LIN-12 Notch signaling during Caenorhabditis elegans vulval development. PLoS Biol. 2008;6:e196. doi: 10.1371/journal.pbio.0060196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo BK, et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development. 2005a;132:3459–3470. doi: 10.1242/dev.01922. [DOI] [PubMed] [Google Scholar]
- Koo BK, Yoon KJ, Yoo KW, Lim HS, Song R, So JH, Kim CH, Kong YY. Mind bomb-2 is an E3 ligase for Notch ligand. J Biol Chem. 2005b;280:22335–22342. doi: 10.1074/jbc.M501631200. [DOI] [PubMed] [Google Scholar]
- Kwon DY, Motley WW, Fischbeck KH, Burnett BG. Increasing expression and decreasing degradation of SMN ameliorate the spinal muscular atrophy phenotype in mice. Hum Mol Genet. 2011;20:3667–3677. doi: 10.1093/hmg/ddr288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai EC, Roegiers F, Qin X, Jan YN, Rubin GM. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development. 2005;132:2319–2332. doi: 10.1242/dev.01825. [DOI] [PubMed] [Google Scholar]
- Le TT, Pham LT, Butchbach ME, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AH. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum Mol Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Remaud S, Hamel S, Schweisguth F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 2005;3:e96. doi: 10.1371/journal.pbio.0030096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Androphy EJ. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum Mol Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Strasswimmer J, Yao JM, Baleja JD, Hahnen E, Wirth B, Le T, Burghes AH, Androphy EJ. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AH. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am J Hum Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McWhorter ML, Monani UR, Burghes AH, Beattie CE. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- Pellizzoni L, Charroux B, Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc Natl Acad Sci USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD. The ubiquitin system, disease, and drug discovery. BMC Biochem. 2008;9:S7. doi: 10.1186/1471-2091-9-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior TW, Swoboda KJ, Scott HD, Hejmanowski AQ. Homozygous SMN1 deletions in unaffected family members and modification of the phenotype by SMN2. Am J Med Genet A. 2004;130:307–310. doi: 10.1002/ajmg.a.30251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rual JF, et al. Toward improving Caenorhabditis elegans phenome mapping with an ORFeome-based RNAi library. Genome Res. 2004;14:2162–2168. doi: 10.1101/gr.2505604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K, et al. C. elegans Notch signaling regulates adult chemosensory response and larval molting quiescence. Curr Biol. 2011;21:825–834. doi: 10.1016/j.cub.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrt RD, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Wolstencroft EC, Mattis V, Bajer AA, Young PJ, Lorson CL. A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels. Hum Mol Genet. 2005;14:1199–1210. doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- Yoo KW, Kim EH, Jung SH, Rhee M, Koo BK, Yoon KJ, Kong YY, Kim CH. Snx5, as a Mind bomb-binding protein, is expressed in hematopoietic and endothelial precursor cells in zebrafish. FEBS Lett. 2006;580:4409–4416. doi: 10.1016/j.febslet.2006.07.009. [DOI] [PubMed] [Google Scholar]
- Zhang C, Li Q, Jiang YJ. Zebrafish Mib and Mib2 are mutual E3 ubiquitin ligases with common and specific delta substrates. J Mol Biol. 2007;366:1115–1128. doi: 10.1016/j.jmb.2006.11.096. [DOI] [PubMed] [Google Scholar]
- Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.