Rng2p is required for both the normal process of contractile ring formation from precursor nodes and an alternative mechanism by which rings form from strands of actin filaments, as well as for ring constriction. Systematic analysis of domain deletion mutants establishes how the four domains of Rng2p contribute to cytokinesis.
Abstract
Eukaryotic cells require IQGAP family multidomain adapter proteins for cytokinesis, but many questions remain about how IQGAPs contribute to the process. Here we show that fission yeast IQGAP Rng2p is required for both the normal process of contractile ring formation from precursor nodes and an alternative mechanism by which rings form from strands of actin filaments. Our work adds to previous studies suggesting a role for Rng2p in node and ring formation. We demonstrate that Rng2p is also required for normal ring constriction and septum formation. Systematic analysis of domain-deletion mutants established how the four domains of Rng2p contribute to cytokinesis. Contrary to a previous report, the actin-binding calponin homology domain of Rng2p is not required for viability, ring formation, or ring constriction. The IQ motifs are not required for ring formation but are important for ring constriction and septum formation. The GTPase-activating protein (GAP)–related domain is required for node-based ring formation. The Rng2p C-terminal domain is the only domain essential for viability. Our studies identified several distinct functions of Rng2 at multiple stages of cytokinesis.
INTRODUCTION
IQGAP proteins have been implicated in cytokinesis in animals and fungi, but even in the best-characterized cases, remarkably little is known about mechanisms. RNA interference depletion of IQGAP1 caused defects in germline cytokinesis in early Caenorhabditis elegans embryos (Skop et al., 2004), and IQGAP1 localized to cytokinetic rings in mouse embryo cells (Bielak-Zmijewska et al., 2008), but much more is known about the roles of mammalian IQGAP1 in exocytosis and mitogen-activated protein kinase signaling than in cytokinesis (Shannon, 2012). Fortunately, genetic and molecular biology experiments on fungi revealed much more about the participation of IQGAPs in cytokinesis. For example, fission yeast have a single, essential IQGAP, the product of the rng2+ gene, and the temperature-sensitive rng2‑D5 mutation arrests the cell cycle with aberrant contractile rings or clusters of contractile ring proteins (Chang et al., 1996). Rng2p accumulates in precursors of the contractile ring called nodes and subsequently concentrates in the contractile ring along with actin filaments and myosin II throughout its formation, maturation, and constriction. Fission yeast cells are ideal to address questions about the roles of IQGAP-family proteins in cytokinesis because the process is similar to that in higher eukaryotes and has been studied in detail (Pollard and Wu, 2010).
Fission yeast form contractile rings from precursors called nodes, which assemble around the center of the cell during mitosis. Interactions of actin filaments and myosin II condense the nodes into a compact contractile ring (Vavylonis et al., 2008). If mutations compromise nodes, a contractile ring may form from strands of actomyosin (Hachet and Simanis, 2008; Huang et al., 2008; Saha and Pollard, 2012). Additional proteins join the ring during a maturation phase (Wu et al., 2003), followed by constriction and disassembly of the contractile ring, a process tightly coordinated with formation of new cell wall material, or septum.
Rng2p is a large IQGAP protein with four domains (Figure 1A), consistent with a role as a scaffold. Characterizing the activities of each domain of such a large protein is essential to understanding its biological function. The N-terminal calponin homology domain (CHD) binds actin filaments (Takaine et al., 2009). This domain is followed by a stretch of 6–11 predicted IQ motifs, short alpha helical repeats that bind EF hand–family proteins, Cdc4p in the case of Rng2p (D'Souza et al., 2001). IQGAP-family proteins contain GTPase-activating protein (GAP)–related domains (GRDs), which are typically inactive owing to the absence of the catalytic arginine that would activate GTP hydrolysis by target proteins. Instead, mammalian IQGAP1 stabilizes Cdc42-GTP (Hart et al., 1996; Kuroda et al., 1996). Budding yeast IQGAP Iqg1p/Cyk1p binds Cdc42p weakly (Epp and Chant, 1997; Osman and Cerione, 1998) and also binds the GTPase Tem1p (Shannon and Li, 1999). No evidence exists as to whether Rng2p binds any GTPases such as Cdc42p. The C-terminal domain of Rng2p is the least well characterized but has sequence homology to the C-terminal domains of RasGAPs (PROSITE).
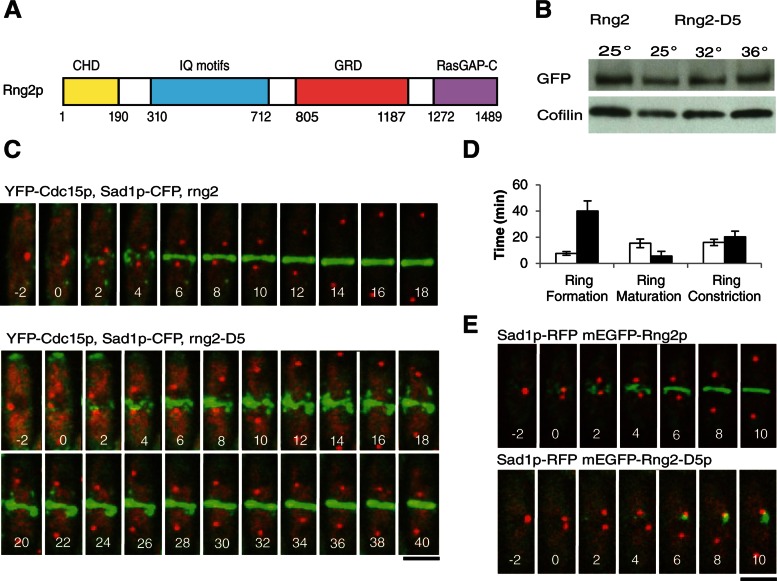
FIGURE 1:
Rng2-D5 cells assemble contractile rings more slowly at a semipermissive temperature than do wild-type cells. (A) Schematic of Rng2p domains. (B) Western blots with antibodies to GFP or cofilin of lysates of cells expressing mEGFP-Rng2p or mEGFP-Rng2-D5 at the permissive (25°C), semipermissive (32°C), or restrictive (36°C) temperature. (C) Time series of fluorescence micrographs at 2-min intervals at 32°C of wild-type (top row) and rng2-D5 (bottom row) cells expressing Sad1-CFP (red) to mark spindle pole bodies and YFP-Cdc15p (green) to mark nodes and contractile rings. Time in minutes is shown in white, with spindle pole body separation defined as time zero. (D) Average times (±1 SD) required for wild-type (open bars) and rng2-D5 (filled bars) cells to assemble (n = 27 and 9, respectively), mature (n = 25 and 21, respectively), and constrict (n = 21 and 19, respectively) contractile rings using YFP-Cdc15p as the marker for contractile rings. (E) Time series of fluorescence micrographs at 2-min intervals at 32°C of cells expressing Sad1-RFP (red) and (top) either mEGFP-Rng2p or (bottom) mEGFP-Rng2-D5 (green). Scale bars, 5 μm.
The role of these conserved domains has been studied best in budding yeast. Budding yeast IQGAP Iqg1p/Cyk1p is required for cytokinesis (Epp and Chant, 1997) and targets myosin II Myo1p to the contractile ring during later stages of cytokinesis. The CHD of Iqg1p is not required for this role but is necessary for constriction of the resulting Myo1p ring (Fang et al., 2010) and the recruitment of actin to the contractile ring (Shannon and Li, 1999). The CHD, the IQ motifs, and the C-terminal half of the protein (containing the GRD and the RasGAP-C domain) are all essential for Iqg1p function and viability in budding yeast. The IQ motifs are required for the localization of Iqg1p to the contractile ring, and the C-terminal half of the protein (containing the GRD and the RasGAP-C domain) of Iqg1p is required for contractile ring constriction (Shannon and Li, 1999).
Previous studies used a variety of approaches to investigate the roles of Rng2p in node and ring formation. One study overexpressed Rng2p constructs from plasmids to study the localization of Rng2p lacking each putative domain in fixed cells (Takaine et al., 2009). A second study (Laporte et al., 2011) used experiments on rng2-D5 cells at a restrictive temperature to identify proteins that recruit Rng2p to nodes (anillin Mid1p) and those it in turn recruits (myosin II, Myo2). A third study used rng2-M1 with three point mutations in the GRD and C-terminal domain to show that interaction of Rng2p with anillin Mid1p was important for ring formation from nodes (Padmanabhan et al., 2011). This study also used a combination of Rng2p domain–deletion constructs either in the native locus or overexpressed from plasmids to determine which domains are required to form nodes. These studies did not investigate the role of Rng2p late in cytokinesis, as all known mutations affect ring formation.
We studied the contributions of Rng2p to entire process of cytokinesis by time-lapse microscopy of live cells and by carefully constructing mutant strains with only one copy of Rng2p, expressed from the genome under the control of the native promoter. In contrast to previous work (Takaine et al., 2009), we found that the CHD of Rng2p is not required for viability, ring formation, or ring constriction. Cells depending on Rng2p constructs lacking all 11 potential IQ repeats are viable and form contractile rings with both type II myosins. More important, we found the IQ motifs of Rng2p have a previously unidentified role in contractile ring constriction and septum formation. Cells dependent on Rng2p lacking the IQ motifs accumulate the primary cell wall synthase, Bgs1p, at the division site but fail to accumulate normal amounts of Sid2p kinase and its regulatory subunit Mob1p, two components of the septation initiation network (SIN), which coordinates cytokinesis with the formation of new cell wall material in the cleavage furrow (Johnson et al., 2012). Viability depends on the Rng2p RasGAP-C domain but not the GRD. However, formation of nodes and their normal condensation into a ring requires both the GRD and the RasGAP-C domain.
RESULTS
Strains used to study the roles of Rng2p in contractile ring function
We used live cells expressing fluorescent fusion proteins from their native genomic loci to study the contributions of IQGAP Rng2p to cytokinesis. Monomeric enhanced green fluorescent protein (mEGFP)–Rng2p is a fully functional fusion of mEGFP with Rng2p (Wu et al., 2003). YFP-Cdc15p is a fully functional fusion of yellow fluorescent protein (YFP) with the F-BAR protein Cdc15p (Arasada and Pollard, 2011). mEGFP-Myo2p is a fully functional fusion of mEGFP with the heavy chain of myosin II, the main motor protein in nodes and contractile rings (Wu et al., 2003). Sad1p–cyan fluorescent protein (CFP) marked spindle pole bodies (SPBs), which we used to put each cell in the same cell cycle time frame with time zero when the SPB divided as the cells entered prophase (Wu et al., 2003).
We avoided a number of synthetic interactions between Rng2p mutations and fluorescently tagged proteins. The temperature-sensitive rng2-D5 mutation interacted strongly with tagged Rlc1p (rlc1-3GFP) at 25°C and with rlc1-mCherry at 32°C only. Cells dependent on mEGFP-Rng2pΔIQ or mEGFP-Rng2pΔGRD showed synthetic interactions with mCherry-Cdc15p, although rng2-D5 cells complemented with either deletion construct did not show synthetic interactions even at 32°C. Cells dependent on mEGFP-Rng2pΔIQ, but not untagged Rng2pΔIQ, were synthetically lethal with mCherry-Cdc15p.
The rng2-D5 mutation slowed contractile ring assembly at a semipermissive temperature
Fission yeast cells with the rng2-D5 mutation arrested at 36°C without forming a contractile ring but assembled contractile rings at the semipermissive temperature of 32°C. The rng-D5 mutation is a Rng2p G1032E substitution in the GRD (Takaine et al., 2009). A homology model of the Rng2p based on a structure of the IQGAP1 GRD (Kurella et al., 2009) has G1032 in the core of the domain, so substitution of a charged, space-filling residue such as glutamic acid would likely alter folding of the domain or even of the whole protein. The Rng2p-D5 polypeptide was stable in cells for several hours at 32 or 36°C (Figure 1B), so the phenotypes were not due to the degradation of the protein.
Contractile ring formation in rng2-D5 cells.
At the semipermissive temperature of 32°C, wild-type cells formed contractile rings reliably by 12 min after SPB separation (cell cycle time +12 min), but by time +50 min only 43% of rng2-D5 cells formed rings, and the process was highly variable (Figure 1, C and D, and Supplemental Figures 1A and 2, A and B). At the permissive temperature of 25°C rng2-D5 cells also formed contractile rings more slowly than wild-type cells (Supplemental Table S1 and Supplemental Figure S1B). The time course of all stages of cytokinesis depends on temperature, being about twice as fast at 32°C than at 25°C (Wu et al., 2003).
Node formation in rng2-D5 cells.
Wild-type fission yeast cells assemble contractile ring precursors called nodes during interphase (Martin and Berthelot-Grosjean, 2009; Moseley et al., 2009). As a cell enters mitosis, nodes concentrate around the equator and mature by accumulating anillin Mid1p, myosin II Myo2 (consisting of the Myo2p heavy chain, essential light-chain Cdc4p, and regulatory light-chain Rlc1p), IQGAP Rng2p, F-BAR protein Cdc15p, formin Cdc12p, and other proteins. Mid1p is considered to be a scaffold protein. Rng2p is proposed to link Mid1p to Myo2, whereas Cdc15p is believed to link Mid1p to Cdc12p (Laporte et al., 2011). Soon after SPB division, formin Cdc12p arrives in nodes and initiates actin polymerization.
At 32°C wild-type cells began to concentrate mEGFP-Rng2p (Figure 1E) and its downstream ligand mEGFP-Myo2p (Supplemental Figure S1C) in nodes at about time −10 min (Wu et al., 2003), but neither protein concentrated in nodes at any time in rng2-D5 cells (Figure 1E and Supplemental Figure S1C). Laporte et al. (2011) observed similar defects in rng2-D5 cells at the fully restrictive temperature of 36°C. This failure of Myo2 and Rng2p to concentrate in nodes was not due to the absence of nodes, since rng2-D5 cells at 32°C have nodes containing Blt1p (Supplemental Figure S1D), and Mid1p localizes to nodes in rng2-D5 cells even at the fully restrictive temperature of 36°C (Laporte et al., 2011). Over time at 32°C a small amount of mEGFP-Rng2p-D5 accumulated in strands or clumps around the center of some cells (Figure 1E), along with mEGFP-Myo2p (Supplemental Figure S1C). Even though Cdc15p is presumed to be on a pathway parallel to that depending on Rng2p (Laporte et al., 2011), the localization of YFP-Cdc15p was defective in rng2-D5 cells at 32°C (Figure 1C). Although some YFP-Cdc15p appeared to localize to nodes, most of the protein formed strands or clumps before SPB separation and during ring formation, much like mEGFP-Myo2p in rng2-D5 cells at 32°C (Supplemental Figure S1C). This defect may be due in part to the failure of the positive feedback mechanism by which the presence of Rng2p in nodes promotes recruitment of additional Mid1p (Laporte et al., 2011). In addition wild-type Rng2p is required for normal expression levels of Mid1p (Padmanabhan et al., 2011), and so reduced Mid1p in nodes of rng2-D5 cells may compromise Cdc15p localization.
Pathway of contractile ring formation in rng2-D5 cells.
Rather than forming contractile rings by condensation of equatorial nodes starting at time +1 min like wild-type cells, rng2-D5 cells formed rings from strands or uneven clumps containing Blt1p-mEGFP (Supplemental Figure S1D), YFP-Cdc15p (Figure 1C), mEGFP-Myo2p (Supplemental Figure S1C), and, eventually, mEGFP-Rng2-D5p (Figure 1E). These strands marked by YFP-Cdc15p always formed around the equators of rng2-D5 cells. Cells lacking Mid1p form strands containing Myo2 and Rng2p (Saha and Pollard, 2012) that merge to form a contractile ring (Hachet and Simanis, 2008; Huang et al., 2008; Saha and Pollard, 2012).
Contractile ring constriction in rng2-D5 cells.
Completed contractile rings in wild-type cells normally mature for ∼15 min at 32°C before constricting (Figure 1D), but the contractile rings that formed in less than half of rng2-D5 cells at 32°C began constricting soon after they formed (Figure 1D). Thus events that normally occur during the maturation of a contractile ring proceed during prolonged ring assembly in rng2-D5 cells. Rings constricted more slowly in rng2-D5 cells than in wild-type cells at both 32°C (p = 0.0008; Figure 1D) and 25°C, although the difference was not statistically significant at 25°C (p = 0.49; Supplemental Figure S1B).
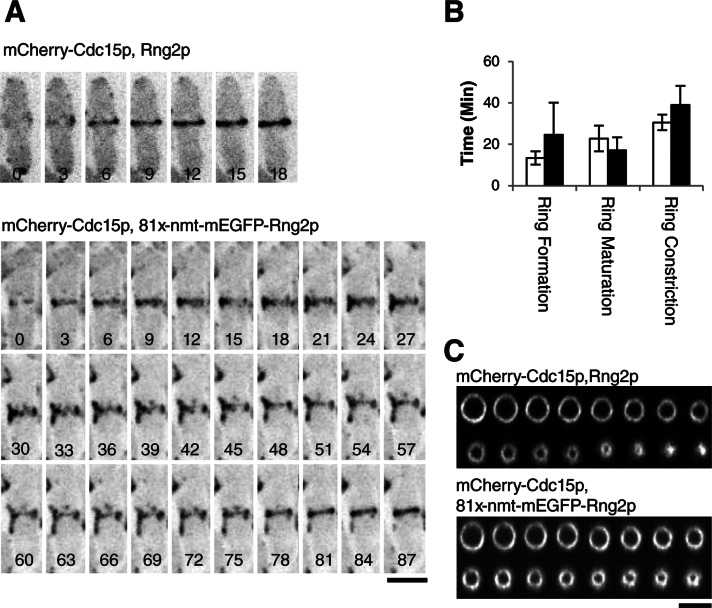
Contractile ring assembly and constriction in cells depleted of Rng2p
Because we do not know how the rng2-D5 mutation compromises Rng2p function, we tested the effect of depleting Rng2p. We replaced rng2+ with mEGFP-rng2 controlled by the thiamine-repressible 81xnmt1 promoter in the native locus. After 24 h in thiamine these cells contained 20 ± 14% as much mEGFP-Rng2p protein as cells with mEGFP-Rng2p in the native locus under the control of the rng2+ promoter (Supplemental Figure S3). About half of 81xnmt1-rng2 cells depleted of Rng2p formed rings at close to wild-type rates through the condensation of nodes, but one-third failed to form rings in 50 min. The remaining repressed cells formed rings from strands slowly over a wide range of times (Figure 2, A and B), similar to rng2-D5 cells at the semipermissive temperature. The wide range of defects in contractile ring formation may reflect cell-to-cell variations in mEGFP-Rng2p concentrations.
FIGURE 2:
Cells depleted of Rng2p assemble and constrict rings more slowly than do wild-type cells. (A) Time series of negative-contrast fluorescence micrographs at 3-min intervals at 25°C of (top) wild-type and (bottom) 81xnmt1-mEGFPrng2 cells expressing mCherry-Cdc15p. Repressed 81xnmt1-rng2 cells were grown in 10 μg/ml thiamine for 24 h to repress expression of Rng2p and imaged with 10 μg/ml thiamine in the medium. The initial time when mCherry-Cdc15p appeared near the middle of the cell is the first image in each series. (B) Average times (±1 SD) required for (open bars) rng2 and (filled bars) 81x-nmt-rng2 cells to assemble (n = 13 and 27, respectively), mature (n = 10 and 19, respectively), and constrict (n = 11 and 25, respectively) contractile rings. (C) Time series of three-dimensional reconstructions of fluorescence micrographs at 3-min intervals at 25°C of wild-type and (top) and 81xnmt1-mEGFPrng2 (bottom) cells expressing mCherry-Cdc15p. The images are Z-sections in the plane of the contractile ring. In both series, the second image corresponds to the onset of constriction. Scale bars, 5 μm.
Contractile ring constriction was unreliable in the repressed 81xnmt1-rng2 cells that formed rings; constriction failed to initiate in 9.4% of these cells and was slow or incomplete in others. Among the 76% of repressed 81xnmt1-rng2 cells that completed constriction in 50 min or less, the average time required to constrict a ring was longer and much more variable (39.0 ± 9.3 min) than for wild-type cells (30.5 ± 3.8 min), a statistically significant difference (p = 0.006; Figure 2B). We did not include in the mean constriction rate cells that persisted as an incompletely constricted ring for >9 min and failed to complete constriction (Figure 2C), so constriction was compromised much more severely than indicated by the mean rate.
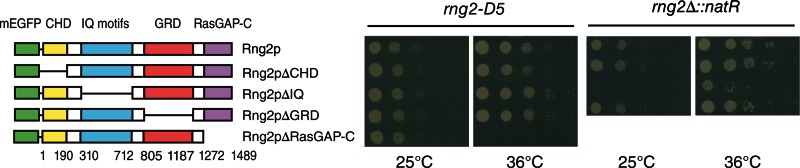
Complementation of temperature-sensitive and deletion mutations of rng2+
Control experiments verified that expression of mEGFP-Rng2p from the leu1 locus under the control of the native rng2 promoter completely rescued the viability of both rng2-D5 cells at 36°C and rng2∆ deletion mutant at 25°C (Figure 3). Expression of full-length mEGFP-Rng2p also fully corrected the cytokinesis defects of the rng2∆ deletion strain. The mEGFP-Rng2p localized to nodes at time −5.7 min, and these nodes condensed into contractile rings at rates similar to those of wild-type cells (Figure 4, A and B, and Supplemental Movie S1). These contractile rings matured and constricted at rates close to those of wild-type cells (Figure 5, A, B, and D). In cells complemented with full-length mEGFP-Rng2p, Mid1p localized to nodes prior to SPB separation and then colocalized with mEGFP-Rng2p in rings (Figure 4E). mEGFP-Rng2p expressed from the leu1 locus also rescued the ability of rng2-D5 cells to form rings at 32°C (Supplemental Figure S4A).
FIGURE 3:
Only the Ras-GAP–related C-terminal domain of Rng2p is essential for viability. Complementation experiments with rng-D5 or rng2Δ cells expressing full-length mEGFP-Rng2p or mEGFP-Rng2p deletion constructs from the leu1 locus under the control of the rng2 promoter. Photographs of agar plates with spots containing serial 1:10 dilutions of cells incubated for 48 h at the permissive (25°C) or restrictive (36°C) temperature.
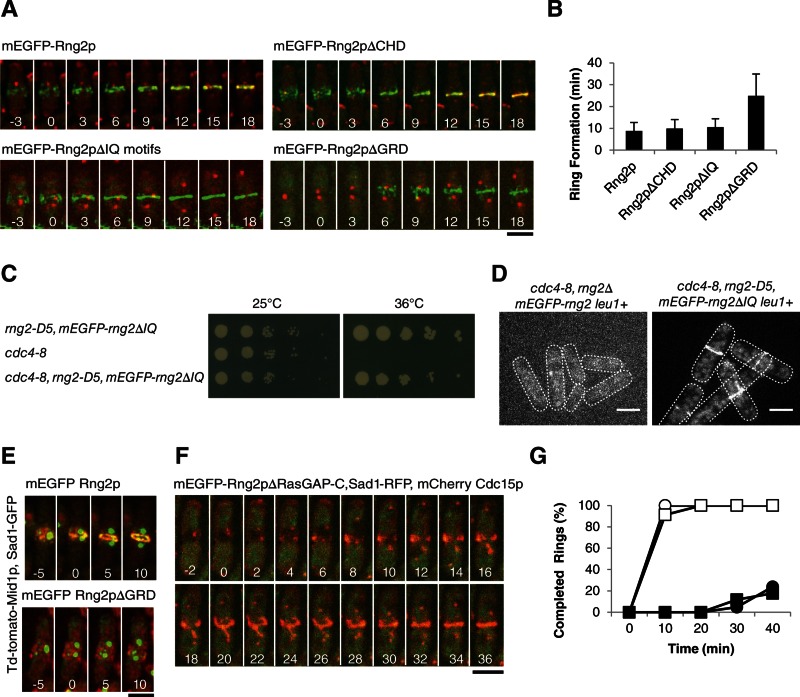
FIGURE 4:
Ability of Rng2p constructs lacking each of the first three domains to restore cytokinesis of rng2Δ cells. (A) Time series of fluorescence micrographs at 3-min intervals at 25°C of rng2Δ cells expressing Sad1-RFP (red) and complemented with full-length mEGFP-Rng2p or the indicated deletion constructs of mEGFP-Rng2p (green). mEGFP-Rng2p and mEGFP-Rng2pΔCHD cells also expressed mCherry-Cdc15p (red). Time in minutes is shown in white, with SPB separation defined as time zero throughout. (B) Average time (±1 SD) after SPB separation for each strain to form a ring using the mEGFP-Rng2p or mEGFP-Rng2p deletion construct to mark contractile rings (n = 13, 21, 8, and 27, respectively). (C) Complementation of cdc4-8 cells with mEGFP-rng2ΔIQ. (D) Fluorescence micrograph of mEGFP-Rng2p or mEGFP-Rng2pΔIQ localization in cdc4-8 cells at 32°C. (E) Time series of fluorescence micrographs at 5-min intervals at 25°C of rng2Δ cells expressing Sad1-GFP (green), Mid1p-TdTomato (red), and the indicated mEGFP-Rng2p deletion construct (green). A maximum intensity projection was formed from 13 Z-slices collected for each time point. (F) Time series of fluorescence micrographs at 2-min intervals at 32°C of rng2-D5 cells expressing Sad1-RFP (red), mCherry-Cdc15 (red), and mEGFP-Rng2pΔRasGAP-C (green). (G) Outcomes graph of percentage of cells with complete rings as a function of time after SPB separation at 32°C: (◯) rng2+ cells (n = 27); (●) rng2-D5 cells (n = 21); (□) rng2-D5, mEGFP-rng2 cells (n = 23); (■) rng2-D5, mEGFP-Rng2pΔRasGAP-C cells (n = 17). Scale bars, 5 μm.
FIGURE 5:
Rng2p constructs lacking each of the first three domains restore the ability of rng2Δ cells to constrict contractile rings. (A) Time series of fluorescence micrographs at 3-min intervals at 25°C of rng2Δ cells complemented with the indicated mEGFP-Rng2p proteins. Time in minutes is shown in white, with the time zero defined as the last time point before the initiation of constriction. (B) Average time (±1 SD) between the completion of ring assembly and onset of constriction of contractile rings in rng2Δ cells complemented with mEGFP-Rng2p (n = 14), mEGFP-Rng2pΔCHD (n = 22), or mEGFP-Rng2pΔGRD (n = 25) rings. (C) Outcomes graph of time between the completion of ring formation (time zero) to the onset of constriction of mature rings of rng2Δ cells complemented with (◯) mEGFP-Rng2p (n = 14) or (●) mEGFP-Rng2ΔIQ (n = 17). (D) Average times (±1 SD) required for complete constriction of rings in rng2Δ cells complemented with mEGFP-Rng2p (n = 9), mEGFP-Rng2pΔCHD (n = 22), or mEGFP-Rng2pΔGRD (n = 19). (E) Outcomes graph of time to finish ring constriction of rng2Δ cells complemented with (◯) mEGFP-Rng2p (n = 9) or (●) mEGFP-Rng2ΔIQ (n = 17). Scale bar, 5 μm.
Design of domain-deletion constructs
To design domain-deletion mutations for complementation experiments, we used biochemical and biophysical data to choose domain boundaries for deletions that would minimize the effect on the structure and stability of the remaining parts of the Rng2p protein (Figure 3 and Supplemental Materials and Methods). A crystal structure (PDB 1P2X) of the Rng2p CHD showed that this actin filament–binding domain extends from the N-terminus to residue 190. The region of Rng2p between the CHD and GRD is predicted to contain 6–11 α-helical IQ motifs (Wang et al., 2004). We used PROSITE and Pfam software to identify the predicted IQ motifs and deleted all 11 between residues 310 and 712. We used a crystal structure of the GAP-related domain of the mammalian IQGAP1 (PDB 3FAY) and sequence homology with other GAPs to choose residues 805 and 1187 as boundaries of the GRD. We used three criteria to choose residue 1272 and the C-terminal residue 1489 as boundaries of the RasGAP C-terminal domain with the aim of removing as much of this domain as possible without intruding on the GRD. 1) Structure: a nuclear magnetic resonance (NMR) structure of part of the RasGAP C-terminal domain from IQGAP1 (PDB 1XOH) corresponds to residues 1392–1489 of Rng2p. 2) Expression: residues 1306–1489 of Rng2p and the part of IQGAP1 corresponding to Rng2p residues 1325–1489 can be expressed in bacteria and purified (Brandt et al., 2007; Almonacid et al., 2011). 3) Domain prediction: the largest C-terminal domain of Rng2p predicted by the Pfam software was Rng2p residues 1272–1489. We also made smaller deletions in this domain. The linker region consisting of residues 191–309 was deleted in some experiments, but linker regions 713–804 and 1188–1271 were not deleted from any of our constructs.
To test for function, we expressed constructs lacking individual domains under the control of the native rng2+ promoter after insertion into the leu1 locus. The N-terminus of each construct was tagged with mEGFP, separated by a linker of four glycine residues (Figure 3). We confirmed the expression of each mEGFP-Rng2p construct by Western blotting with a polyclonal GFP antibody (Supplemental Figure S3). All of the constructs expressed at lower levels from the leu1 locus under the control of the native promoter than full-length mEGFP-Rng2p expressed from the native locus: full-length mEGFP-Rng2p (46 ± 21%); mEGFP-Rng2pΔCHD (43 ± 21%); mEGFP-Rng2pΔIQ (78 ± 11%); mEGFP-Rng2pΔGRD (31 ± 10%); and mEGFP-Rng2pΔRasGAP-C (20 ± 14%). We consider below the possible effect of these expression levels on phenotypes in the complementation experiments. We did not identify any synthetic interactions of the full-length protein expressed from the leu1 locus, although we avoided using those markers identified as being synthetically lethal with rng2-D5. In particular, mEGFP-rng2 in the leu1 locus did not interact synthetically with mCherry-cdc15, although two of the domain-deletion constructs of Rng2p described below interacted synthetically with mCherry-cdc15.
Rng2p constructs lacking each of the first three domains are generally functional
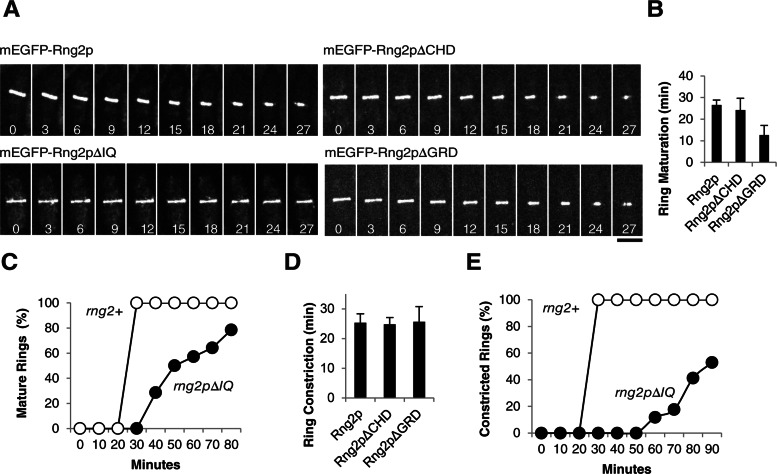
We tested the ability of four Rng2p-deletion constructs to complement the temperature-sensitive rng2-D5 mutation and a rng2∆-deletion mutation (Supplemental Table S1 and Figure 3). In spite of being expressed at low levels from the native rng2 promoter in the leu1 locus, mEGFP-Rng2pΔCHD, mEGFP-Rng2pΔIQ, and mEGFP-Rng2pΔGRD each fully rescued the viability of rng2-D5 cells at the restrictive temperature of 36°C as well as full-length mEGFP-Rng2p (Figure 3). Each deletion construct also supported the viability of the rng2∆ strain, although these null cells depending on mEGFP-Rng2pΔIQ grew more slowly than cells expressing mEGFP-Rng2p or the two other deletion constructs (Figure 3) for reasons explained below.
Role of the CHD in cytokinesis.
We found no defects in cells depending on Rng2p lacking the CHD. Cells lacking native Rng2p and depending on mEGFP-Rng2pΔCHD localized the tagged protein normally to nodes at a time of –7.1 min and formed a contractile ring normally at time of +10.5 ± 3.8 min (Figure 4, A and B), followed by a normal period of maturation and ring constriction at a rate close to that for wild-type cells (Supplemental Table S1 and Figure 5, A and B). In rng2-D5 cells at 32°C mEGFP-Rng2pΔCHD also localized to nodes and formed a ring in a time similar to that for wild-type cells (Supplemental Figure 4, B and E).
In contrast Takaine et al. (2009) reported that expression of a deletion construct missing the first 300 amino acids of Rng2p (including the CHD) from a plasmid did not support the viability of rng2-D5 cells at the restrictive temperature. We tested the hypothesis that residues 191–300 after the CHD might have a separate function from that of the CHD by constructing a strain expressing a Rng2p construct lacking residues 190–300 in the leu1 locus in rng2-D5 cells. This strain was viable at the restrictive temperature and grew at rates similar to those of wild-type cells (Supplemental Figure 5A). Because neither amino acids 1–190 nor 190–300 were essential in our assay, we tested the ability of mEGFP-Rng2pΔ1-300 to rescue the viability of rng2-D5 cells at 36°C. The viability of these cells was similar to that of wild-type cells (Supplemental Figure S5A), so we conclude that the first 300 amino acids of Rng2p are not essential for viability when Rng2p is expressed under the control of its native promoter.
We purified recombinant Rng2p CHD and measured its binding to purified muscle actin filaments. Rng2p CHD bound actin filaments with Kd ≈ 20 μM (Supplemental Figure S5B), similar to the isolated CHD of mammalian IQGAP1 (Kd = 47 μM; Mateer et al., 2004) but an order of magnitude weaker than the affinity measured for a hexahistidine (6xHis)-Rng2p-CHD construct (residues 1–189) by Takaine et al. (2009). The 6xHis tag on the CHD construct may have increased the affinity by interacting with the negatively charged actin filament (isoelectric point, 5.2).
Contributions of the IQ domain to cytokinesis.
Cells depending on Rng2p without the IQ motifs assembled contractile rings normally but had severe defects in ring constriction. mEGFP-Rng2pΔIQ localized to nodes in rng2Δ cells beginning at time −6.4 min. Because Rng2p requires Cdc4p to localize to nodes (Wu et al., 2003; Laporte et al., 2011), it was surprising that the Cdc4p-binding sites were not required for localization to nodes. This suggested that Cdc4p binding influences the structure of Rng2p, perhaps by stiffening of the IQ motifs, rather than bridging Rng2p to another protein such as Mid1p. To test this hypothesis, we asked whether Rng2pΔIQ could complement cells with the temperature-sensitive cdc4-8 mutation. If the interaction of Cdc4p with Rng2p were strictly structural, then Rng2pΔIQ would not require Cdc4p for its function, and the cdc4-8 mutation might be less detrimental in cells depending on Rng2pΔIQ. We confirmed that cdc4-8 cells failed to grow at 36°C (Nurse et al., 1976) and found that rng2-D5 mEGFP-rng2ΔIQ cdc4-8 cells grew almost as well at 36°C as rng2-D5 mEGFP-rng2ΔIQ cells (Figure 4C), although rng2-D5 mEGFP-rng2ΔIQ cdc4-8 cells showed septal defects, particularly formation of elongated cells with a single septum or multiple septa after 4 h at 36°C (Supplemental Figure 4F). Similarly, at 32°C mEGFP-Rng2pΔIQ localized to rings in 18% of cdc4-8 cells (n = 221) but mEGFP-Rng2p concentrated in only 3% of rings of cdc4-8 cells (n = 170; Figure 4D). Abnormal structures such as clumps of both mEGFP-Rng2p and mEGFP-Rng2pΔIQ were observed as well, likely reflecting attempts by these cells to form contractile rings. Cdc4p is also required for the localization and function of Myo2p (Lord and Pollard, 2004; Laporte et al., 2011), and so we specifically chose the cdc4-8 mutation for this experiment, as Cdc4p-8 supports the ability of Myo2p to glide actin filaments in vitro at normal rates (Lord and Pollard, 2004). The ability of Rng2pΔIQ to rescue the cdc4-8 phenotype is consistent with our hypothesis that Cdc4p has a structural role but not with the hypothesis that Cdc4p bridges Rng2p to another protein, such as Mid1p, to recruit Rng2p to nodes.
The nodes containing mEGFP-Rng2pΔIQ condensed into rings by time +10.5 ± 3.9 min, similar to wild-type cells (Figure 4, A and B, and Supplemental Movie S2), but the onset of constriction was delayed in many cells. Only 79% of rings depending on mEGFP-Rng2pΔIQ began to constrict within 80 min after they formed, whereas 100% of rng2Δ cells with full-length mEGFP-Rng2p began constricting within 30 min of ring formation (Figure 5C). Contractile rings in cells depending on mEGFP-Rng2pΔIQ constricted much more slowly than rings in wild-type cells. Only 53% of mEGFP-Rng2pΔIQ contractile rings constricted completely in 90 min, with an average rate of 0.04 μm/min, whereas rings in all wild-type cells constricted in 40 min or less, with an average rate of 0.12 μm/min (Figure 5, A and E). We obtained similar results with mEGFP-Rng2pΔIQ in rng2-D5 cells at 32°C, where the deletion construct increased the rate of ring formation but with high variability (Supplemental Figure S4, C and E). Ring constriction defects visualized with mEGFP-Myo2p were similar in rng2Δ cells complemented with untagged Rng2pΔIQ, so this phenotype was not due to the mEGFP tag on the fusion protein (Figure 6A).
FIGURE 6:
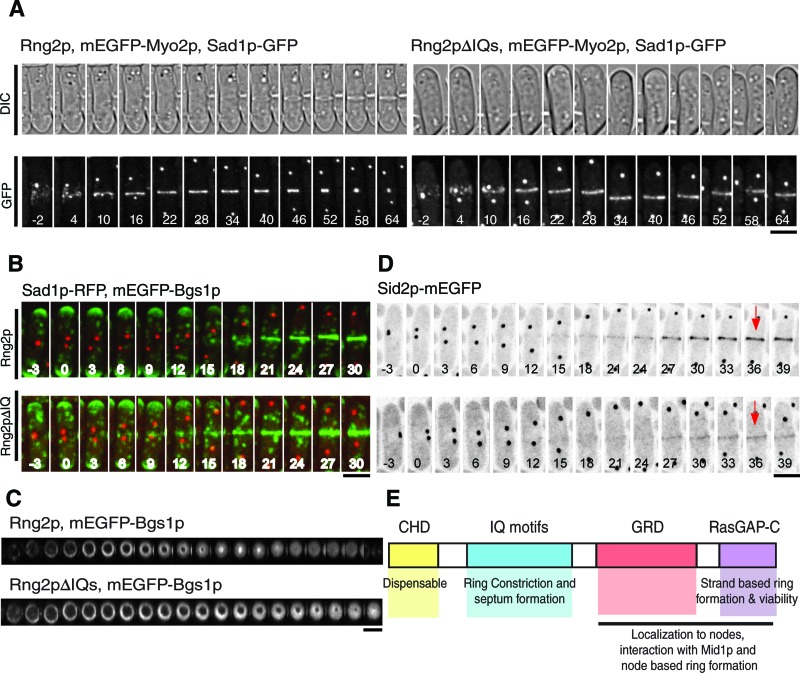
Localization of mEGFP-Myo2p, mEGFP-Bgs1p, and Sid2p-mEGFP in the contractile rings of cells complemented with Rng2pΔIQ and full-length Rng2p. (A) Time series of fluorescence and DIC micrographs at 6-min intervals at 25°C of rng2Δ cells expressing Sad1-GFP and mEGFP-Myo2p (both shown in white) complemented with (left) full-length Rng2p or (right) Rng2pΔIQ. Time in minutes is shown in white, with SPB separation defined as time zero. (B) Time series of fluorescence micrographs at 3-min intervals at 25°C of rng2Δ cells complemented with Rng2p (top) or Rng2pΔIQ (bottom) expressing mEGFP-Bgs1p (green), and Rlc1-RFP (red). Maximum intensity projections were formed from 20 Z-slices collected at each time point. Time in minutes is shown in white, with SPB separation defined as time zero. (C) Time series of Z-sections in the plane of the contractile ring reconstructed from Z-stacks of fluorescence micrographs recorded at 3-min intervals at 25°C of rng2Δ cells expressing mEGFP-Bgs1p and complemented with (top) Rng2p or (bottom) Rng2pΔIQ. In each series, the second image corresponds to the onset of constriction. (D) Time series of negative-contrast fluorescence micrographs at 3-min intervals at 25°C of rng2Δ cells expressing Sid2p-mEGFP complemented with (top) Rng2p or (bottom) Rng2pΔIQ. The fluorescence intensities of the rings were measured at the time points indicated with red arrows. (E) Summary of the contributions of the four domains of Rng2p to cytokinesis. Scale bars, 5 μm.
We asked whether the deficiency in ring constriction in mEGFP-Rng2pΔIQ cells was due to failure of Myo2p to localize to nodes and rings, since Cdc4p is required for Rng2p to recruit Myo2p. We imaged rng2Δ cells dependent on untagged Rng2pΔIQ or full-length Rng2p and expressing mEGFP-Myo2p and Sad1-GFP. In both strains, mEGFP-Myo2p localized to nodes and rings (Figure 6A), although compared with cells complemented with the full-length Rng2p, the fluorescence intensity of the mEGFP-Myo2p in nodes of Rng2pΔIQ cells was 30% lower (n = 20) than at the time of SPB separation and ∼35% lower (n = 10) in rings at times +15 and +30 min.
Differential interference contrast (DIC) images showed that cells depending on Rng2pΔIQ formed new septa later (mean time, +91.5 ± 12.9 min) than cells complemented with full-length Rng2p (+48.5 ± 5.4 min; Figure 6A). Once formed, septa appeared normal with regard to both position and orientation in cells complemented with Rng2pΔIQ. The delay in septum formation was not due to a delay in the initial localization of cell wall synthase mEGFP-Bgs1, which accumulated at time +14.4 ± 2.3 min in the center of cells dependent on Rng2pΔIQ and at time +16.4 ± 2.7 min in cells dependent on full-length Rng2p (Figure 6B). Both strains accumulated similar amounts of mEGFP-Bgs1p at the center of the cell at time +36 min. However, mEGFP-Bgs1p concentrated to the leading edge of the cleavage furrow in rng2Δ cells dependent on Rng2p but had a more uniform, disk-like distribution in the slowly constricting furrows of cells dependent on Rng2pΔIQ (Figure 6C). Surprisingly, tagging Bgs1p with mEGFP in the genome partially rescued the morphological defects of Rng2pΔIQ cells, although it did not fully correct the slow rate of ring constriction.
The second type II myosin, Myp2p, is linked genetically to septation (Bezanilla et al., 1997), but YFP-Myp2p appeared in the center of cells at about the same time in cells complemented with Rng2p (mean time, +16.5 ± 6.0 min) or Rng2pΔIQ YFP-Myp2p (time, +18.9 ± 6.0; Supplemental Figure S6A). In both strains, YFP-Myp2p initially formed an evenly dispersed, faint ring, but later the fluorescence of the ring was asymmetrical (Supplemental Figure S6B).
Cells with defects in the SIN pathway form aberrant contractile rings of actin that do not constrict (Hachet and Simanis, 2008) and fail to form a septum (Nurse et al., 1976), so it was possible that the defects observed in Rng2pΔIQ cells could be attributed to defects in SIN signaling. Several SIN proteins concentrate in SPBs, but two SIN proteins, Sid2p kinase and its regulatory subunit Mob1p, also concentrate in the contractile ring after its formation. This localization was postulated to regulate ring constriction and septum formation (Salimova et al., 2000), so we examined the localization of Sid2p-mEGFP and Mob1p-mEGFP in rng2∆ cells complemented with Rng2p or Rng2pΔIQ. Cells complemented with Rng2p localized both Mob1p and Sid2p to SPBs shortly before SPB separation and to a faint ring around the center of the cell at time +24 min that strongly increased in intensity over the next 10 min (Figure 6D and Supplemental Figure S6C). In cells complemented with Rng2pΔIQ, Mob1p and Sid2p localized to SPBs normally, but less localized to rings, and the amount did not increase as dramatically over the following 10 min, so at time +36 min the Sid2p-mEGFP fluorescence in rings was only 27% as much as for cells complemented with Rng2p (Figure 6D; time point measured is indicated in this example cell, but the reported measurement reflects an average of 13 cells).
To determine whether cells depending on Rng2pΔIQ had defects in SIN signaling, we studied the localization of Cdc7p-mEGFP, since the loss of Cdc7p from one SPB is a hallmark of SIN signaling that depends on the activity of Sid2p, the most downstream SIN kinase (Feoktistova et al., 2012). In cells dependent on either Rng2p or Rng2pΔIQ, Cdc7p-mEGFP initially concentrated in both SPBs right after separation, then gradually disappeared from one SPB over 22.5 min in Rng2p cells and 17.5 min in Rng2pΔIQ cells (Supplemental Figure S6D). This behavior suggests that Sid2p is active in Rng2pΔIQ cells, and so defects in Sid2p and Mob1p localization are due to inability to interact with the contractile ring. Other contractile ring proteins involved in ring constriction may also fail to localize properly in this strain. Additional work is required to determine whether proteins other than Rng2p also contribute to SIN protein localization to the contractile ring.
Contributions of the GRD to cytokinesis.
Although rng2Δ cells complemented with mEGFP-Rng2pΔGRD grew normally, contractile ring assembly was abnormal. This strain showed a synthetic defect with tagged mCherry-Cdc15p, and so we depended on mEGFP-Rng2pΔGRD itself to follow events during cytokinesis. mEGFP-Rng2pΔGRD failed to concentrate in nodes before SPB separation, like full-length Rng2p (Figure 4A). Instead, at time +5.6 min mEGFP-Rng2pΔGRD began to accumulate in punctate structures in some cells but in strands in others (Figure 4A and Supplemental Movie S3). These diverse, abnormal structures all went on to form contractile rings in <60 min (average time, +24.9 ± 10.0 min). Once formed, these contractile rings matured more quickly than wild-type rings and constricted at normal rates (Figure 5B). When expressed in rng2-D5 cells at 32°C, mEGFP-Rng2pΔGRD initially localized to long strands that formed rings at time +20.9 ± 6.8 min, which is faster than rng2-D5 cells at 32°C (Supplemental Figure S4, D and E).
Anillin Mid1p-TdTomato localized to nodes before SPB separation in rng2Δ cells complemented with mEGFP-Rng2pΔGRD but unexpectedly did not incorporate into rings. Instead Mid1p-TdTomato remained in nodes, even as mEGFP-Rng2pΔGRD concentrated in strands near the center of the cell (Figure 4E and Supplemental Figure S7A).
We detected no binding between the C-terminal half of Rng2p (consisting of the GRD and the RasGAP-C domain) and Mid1p. We purified a construct consisting of a His tag–maltose-binding protein and Rng2p residues 805–1489 and a His-tagged construct Mid1p M1 1-3 (the first three domains, residues 1–452). Beads loaded with up to 20 μM Rng2-GRD-RasGAP-C pulled a small fraction of the total tagged Cdc15p out of lysates of unsynchronized cells (Supplemental Figure S9C) but did not deplete Mid1p-YFP from cell lysates (Supplemental Figure S9B). In the reciprocal experiment, beads with up to 15 μM Mid1p M1 1-3 did not remove any mEGFP-Rng2p from cell lysates (Supplemental Figure S9A).
Our result that cells depending on temperature-sensitive Rng2p-D5 were considerably sicker even at the permissive temperature than cells depending on Rng2pΔGRD is surprising, given that the D5 mutation lies within the GRD. Thus substitution of glutamic acid for one glycine in the Rng2p-D5 mutation must affect other domains of the protein.
The Ras-GAP–related C-terminal domain of Rng2p is essential for cell viability.
Unlike deletion constructs lacking each of the other domains, Rng2p lacking the GAP related C-terminal domain (mEGFP-Rng2pΔRasGAP-C) did not rescue the viability of rng2-D5 cells at 36°C (Figure 3). To test whether mEGFP-rng2ΔRasGAP-C can complement rng2Δ cells, we crossed mEGFP-rng2ΔRasGAP-C cells with a strain with rng2+ deleted from the endogenous locus of one chromosome (marked by resistance to Nat) and complemented with mEGFP-rng2+ in the leu1 locus. Many of the resulting spores died at 25°C, and none of 29 viable spores were both resistant to Nat and contained mEGFP-Rng2pΔRasGAP-C in the leu1 locus. This result suggests that mEGFP-Rng2pΔRasGAP-C cannot complement rng2Δ cells, and the failure of mEGFP-Rng2pΔRasGAP-C to complement rng2-D5 cells at 36°C is not due to the elevated temperature.
mEGFP-Rng2pΔRasGAP-C did not appear to contribute to contractile ring formation in the rng2-D5 strain at 32°C. Only 18% of rng2-D5 cells expressing mEGFP-Rng2pΔRasGAP-C formed rings in <40 min, similar to cells with rng2-D5 not complemented with any Rng2p construct (Figure 4G). The truncated protein did not concentrate in nodes, and little incorporated into rings (Figure 4F and Supplemental Figure S7B).
Despite our efforts to choose boundaries for the C-terminal deletion of Rng2p that would not affect the overall fold of the protein, we could not exclude the possibility that mEGFP-Rng2pΔRasGAP-C is misfolded as a result of the deletion. To address this concern, we designed additional deletions of Rng2p within the C-terminal domain (Supplemental Figure S8A). Residues 1306–end were deleted based on work showing that this portion of the recombinant protein can be isolated (Almonacid et al., 2011). Residues 1325 to the end of Rng2p were deleted based on homology to the fragment of IQGAP1 purified in Brandt et al. (2007). An NMR structure of roughly half of the C-terminal domain of IQGAP1 (PBD 1×0H) provided guidance to subdivide the C-terminal domain into two smaller fragments.
Each deletion construct was inserted into the leu1 locus in a rng2‑D5 background and assessed for viability at the restrictive temperature. None of these smaller deletions of the Rng2p C-terminal domain rescued the viability of the rng2‑D5 mutant at 36°C (Supplemental Figure S8A), indicating that the inability of mEGFP-Rng2pΔRasGAP-C to rescue viability is not due to faulty boundaries of the deletions but rather because the entire C-terminal domain is required for Rng2p function. The C-terminal domain of Rng2p alone was not sufficient to rescue the viability of rng2‑D5 cells at the restrictive temperature, however, suggesting that either some combination of the other domains or the linker regions between domains is required for viability along with the C-terminal domain (Supplemental Figure S8B).
DISCUSSION
We show that Rng2p plays two essential roles in cytokinesis, which depend on different parts of the protein. First, the C-terminal half of Rng2p is required for the formation of the contractile ring. Second, the IQ motifs of Rng2p contribute to efficient constriction of the contractile ring. Only the RasGAP‑C domain is required for viability, whereas each of the four domains of budding yeast IQGAP Cyk1/Iqg1 is required for viability (Shannon and Li, 1999). Recruitment of Iqg1p to the site of the contractile ring formation is likely to be different from recruitment of Rng2p, as no homologue of Mid1p has been identified in budding yeast. In addition, myosin II localizes before Iqg1p in budding yeast (although Iqg1 is required for maintenance of myosin II at the ring; Fang et al., 2010). Because a major role of Rng2p is to recruit Myo2p via Mid1p, a mechanism that cannot be conserved given the differences between budding yeast and fission yeast contractile ring formation, it is not surprising the domain requirements of the IQGAP proteins are different in the two organisms.
We used mutant strains each with a single copy of various rng2 alleles in the genome controlled by the native promoter. This strategy reduced expression variability and allowed us to quantify the expression of each construct. This is important because cytokinesis is normal in cells expressing Rng2p at 46% of the wild-type level but abnormal in many cells in a population expressing an average level of 20% of the wild-type level of Rng2p.
When depletion of Rng2p or mutations in rng2+ compromise the condensation of nodes, fission yeast cells survive by using the remaining functional proteins to assemble a contractile ring from strands of actomyosin. Cells respond similarly to mutations of other cytokinesis genes, including anillin mid1+ (Hachet and Simanis, 2008; Huang et al., 2008; Saha and Pollard, 2012), cofilin adf1+ (Chen and Pollard, 2011), and polo-like kinase plo1+ (Bahler et al., 1998a). In rng2-D5 cells, these strands contain Blt1p, Myo2p, and Cdc15p but lack Mid1p. Strands must retain many interactions that normally occur in nodes in spite of the absence of Mid1p and loss of function of Rng2p, two proteins with multiple interactions in contractile rings.
The CHD is not essential for Rng2p function
We show that the ability of Rng2p to bind actin filaments is not essential for viability, ring formation, or constriction. Differences in expression strategies might explain why the CHD or Rng2p(1-300) were dispensable in our experiments but not in a previous report (Takaine et al., 2009). Our experimental design resulted in protein levels lower than that for the wild-type protein, but expression of the full-length protein at this level restored function in rng2-D5 and rng2Δ cells. Takaine et al. (2009) expressed Rng2p or deletion constructs at unknown levels from pREP1 plasmids under the control of an nmt promoter. Because Rng2p overexpression is lethal (Eng et al., 1998), different protein expression levels due to plasmid copy number or variability of expression from the nmt promoter might have complicated their experiments. We believe that our result is more physiologically relevant than the previous study.
Rng2p IQ motifs are required during ring constriction
We were surprised that Rng2p lacking all 11 predicted IQ motifs (amino acids 310–712) localized to nodes, since the IQ ligand Cdc4p (D'Souza et al., 2001) is required for Rng2p localization to nodes (Laporte et al., 2011; Padmanabhan et al., 2011). Thus Cdc4p must not link Rng2p to other node proteins. Similarly, Myo2p requires Cdc4p to concentrate in nodes, but Myo2p lacking the IQ motif that binds Cdc4p localizes normally (Laporte et al., 2011). In both cases Cdc4p binding might stiffen the IQ motifs.
Septum formation was slow in mEGFP-Rng2pΔIQ cells owing to slow maturation and constriction of contractile rings. A defect in SIN signaling could compromise septum formation, which is required for cleavage furrow invagination against internal turgor pressure (Proctor et al., 2012). The SIN is required for septum formation and ring constriction (Sparks et al., 1999; Salimova et al., 2000). Despite normal SIN signaling at the SPBs, fewer SIN proteins Mob1p/Sid2p moved to contractile rings in Rng2pΔIQ cells. This did not reduce the amount of cell wall synthase mEGFP-Bgs1p recruited to the division site, so regulation of the synthase might be altered instead. Budding yeast IQGAP Iqg1p also plays a role in contractile ring constriction, although this role requires the C-terminal half of the protein (Shannon and Li, 1999) and the CHD (Fang et al., 2010) rather than the IQ motifs.
Contributions of the GRD and RasGAP-C domain
Fission yeast viability depends on the RasGAP-C domain but not the GRD; however, the two domains appear to act together through interactions with Mid1p. Mutations in either domain can prevent Rng2p from localizing to nodes and preclude formation of a contractile ring by condensation of cortical nodes (this study and Padmanabhan et al., 2011). The same phenotype occurs in Δmid1 cells, likely because Mid1p is required to localize Rng2p and other components to nodes.
Without the interaction between Mid1p and Rng2p, node function is defective, so the available contractile ring components form strands that slowly merge into rings. Formation of contractile rings from strands requires less Rng2p and fewer domains of Rng2p than formation of rings by node condensation.
Rng2p interacts physically with Mid1p, but the domains of Rng2p mediating this interaction were previously unclear. Both overexpressed (Padmanabhan et al. 2011) and native Rng2p (Laporte et al., 2011) immunoprecipitated with Mid1p, and a purified, recombinant Rng2(1306-1489), most of the RasGAP-C domain, bound directly to partially purified recombinant GST-Mid1p1-100 immobilized on glutathione–Sepharose beads (Almonacid et al. 2011). Furthermore, Rng2pΔGRD (this study and Takaine et al., 2009), Rng2pΔRasGAP-C (this study and Takaine et al., 2009), and Rng2p with point mutations in both these domains (Padmanabhan et al., 2011) failed to localize to nodes. Mid1p did not accumulate in contractile rings depending on Rng2pΔGRD, and so the GRD is required for mutually interdependent interactions of Rng2p with Mid1p and/or other proteins in nodes. These interactions with Mid1p might be weak, since we detected no direct binding of Mid1p to the C-terminal half of Rng2p. Phosphorylation of one or both proteins might influence the binding affinity, given that Mid1p phosphorylation is required for recruitment of other node proteins (Almonacid et al., 2011).
Constructs of mEGFP-Rng2p with several truncations of the RasGAP-C domain failed to rescue viability of rng2-D5 cells, so the entire domain is required for viability. Low expression might have limited the ability of mEGFP-Rng2pΔRasGAP‑C to rescue the viability of rng2-D5 cells, but the RasGAP-C domain appears to be essential for contractile ring formation from either nodes or strands.
The RasGAP-C domain must have another function beyond a nonessential role interacting with Mid1p. Previous work on Rng2p and its mammalian homologue, IQGAP1, indicate a number of possible interactions of the RasGAP-C domain with Myo2p, formin Cdc12p, lipids, or itself, each of which could be essential. Comparison of a homology model of the RasGAP-C domain (Supplemental Figure S10, A and B) with other IQGAPs identified only one conserved surface area, a cluster of basic residues on a concave surface (Supplemental Figure S10C, black arrowhead). A similar surface is proposed to mediate binding of phosphatidylinositol 3,4,5-trisphosphate to IQGAP2 (Dixon et al., 2012), and so this area might target Rng2p to membranes.
MATERIALS AND METHODS
Strain construction and yeast methods
Table 1 lists the fission yeast strains used in this study. Unless noted otherwise, genes were tagged at their endogenous locus under the control of their native promoter using a PCR-based gene-targeting method (Bahler et al., 1998b). The strain expressing Rng2p from the repressible 81xnmt1-mEGFPrng2 gene was constructed using the same PCR-based gene-targeting method, but the repressible 81xnmt1 promoter replaced the endogenous promoter.
TABLE 1:
Strain list.
| Strain | Genotype | Source |
|---|---|---|
| IRT 013 | h– rng2-D5 leu1-32, ura4-D18, ade6-M21X | This study |
| IRT 015 | leu1-32, ura4-D18, ade6-M21X, rng2-D5, rlc1-3GFP-kanMX6 ade6 leu1-32 ura4-D18 | This study |
| IRT 019 | h+ leu1-32, ura4-D18, ade6-M21X, rng2-D5, rlc1-mcherry-natMX6 | This study |
| IRT 033 | rng2-D5, leu1-32, ura4-D18, ade6-M21X, sad1-CFP kanMX6 kanMX6-YFP-cdc15 | This study |
| IRT 034 | leu1-32, ura4-D18, ade6-M21X, sad1-CFP kanMX6 kanMX6-YFP-cdc15 | This study |
| IRT 054 | h– leu1-32, ura4-D18, ade6-M21X, kanMX6-Prng2-mEGFP-rng2 | This study |
| IRT 055 | h– leu1-32, ura4-D18, ade6-M21X, kanMX6-Prng2-mEGFP-rng2-D5 | This study |
| IRT 056 | h– leu1-32, ura4-D18, ade6-M21X, kanMX6 81xnmt1-mEGFP-rng2 | This study |
| IRT 058 | h+ leu1-32, ura4-D18, ade6-M21X, kanMX6 Pcdc15-mCherry-cdc15 | This study |
| IRT 60-1 | leu1-32, ura4-D18, ade6-M21X, kanMX6-81xnmt1-mEGFP-rng2, kanMX6-Pcdc15-mCherry-cdc15 | This study |
| IRT 62-1 | leu1-32, ura4-D18, ade6-M21X, rng2-D5 sad1-GFP-kanMX6, kanMX6-GFP-myo2 rng2-D5 ade6 leu1-32 ura4-D18 | This study |
| IRT 67 | leu1-32, ura4-D18, ade6-M21X, kanMX6-Prng2-mEGFP-rng2-D5, sad1-RFP-kanMX6 | This study |
| IRT 69 | leu1-32, ura4-D18, ade6-M21X, kanMX6-Prng2-mEGFP-rng2, sad1-RFP-kanMX6 | This study |
| IRT 71 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2 leu1+ | This study |
| IRT 74 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔGRD leu1+ | This study |
| IRT 75 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔCHD leu1+ | This study |
| IRT 83 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2 leu1+, kanMX6-mCherry-cdc15, kanMX6-sad1-RFP | This study |
| IRT 84 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔGRD leu1+, kanMX6-mCherry-cdc15, kanMX6-sad1-RFP | This study |
| IRT 85 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔCHD leu1+,kanMX6-mCherry-cdc15, kanMX6-sad1-RFP | This study |
| IRT 92 | h– leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2 leu1+ | This study |
| IRT 93 | h– leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2pΔGRD leu1+ | This study |
| IRT 94 | h– leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2pΔCHD leu1+ | This study |
| IRT 95 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-rng2 leu1+ | This study |
| IRT 96 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔIQ leu1+ | This study |
| IRT 97 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔRasGAP-C leu1+ | This study |
| IRT 102 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-rng2ΔRasGAP-C leu1+ | This study |
| IRT 103 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔRasGAP-C leu1+, kanMX6-Pcdc15-mCherry-cdc15, sad1-RFP-kanMX6 | This study |
| IRT 107 | h– leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-RasGAP-C leu1+ | This study |
| IRT 108 | h– leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-Rng2 leu1+ kanMX6mCherry Cdc15, kanMX6 sad1-RFP | This study |
| IRT 110 | h– leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2pΔCHD leu1+, kanMX6-mCherry-cdc15, kanMX6-sad1-RFP | This study |
| IRT 114 | h– leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2pΔGRD leu1+, kanMX6-sad1-RFP | This study |
| IRT 119 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2pΔIQ leu1+ | This study |
| IRT 120 | leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng-mEGFP-rng2pΔ190-300 leu1+ | This study |
| IRT121 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2ΔIQ leu1+, sad1-RFP-kanMX6 | This study |
| IRT 123 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2Δ190-300 leu1+ | This study |
| IRT 124 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+ | This study |
| IRT 126 | h– leu1-32, ura4-D18, ade6-M21X, rng2D5, Prng2-mEGFP-rng2ΔC structure leu1+ | This study |
| IRT 127 | h– leu1-32, ura4-D18, ade6-M21X, rng2D5, Prng2-mEGFP-rng2Δ C coiled coil region leu1+ | This study |
| IRT 130 | h– leu1-32, ura4-D18, ade6-M21X, rng2D5, Prng2-mEGFP-rng2Δ1306-1489 leu1+ | This study |
| IRT 131 | h– leu1-32, ura4-D18, ade6-M21X, rng2D5, Prng2-mEGFP-rng2Δ1325-1489 leu1+ | This study |
| IRT 133 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+, kanMX6-mEGFP-Myo2p, sad1-mEGFP-kanMX6 | This study |
| IRT 134 | leu1-32, ura4-D18, ade6-M21X, rng2-D5, mEGFP-rng2Δ1-300 leu1+ | This study |
| IRT 136 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2 leu1+, mid1-tdtomato-kanMX6, sad1-mEGFP-kanMX6 | This study |
| IRT 137 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2ΔGRD leu1+, mid1-tdtomato-kanMX6, sad1-mEGFP-kanMX6 | This study |
| IRT 152 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+ | This study |
| IRT 153 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, mob1-GFP-kanMX6 | This study |
| IRT 154 | leu1-32, ura4-D18, ade6-M21X,rng2-D5, Prng2-rng2ΔGRD leu1+ | This study |
| IRT 155 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, kanMX6-mEGFP-myo2, sad1-mEGFP-kanMX6 | This study |
| IRT 156 | h+ leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔCHD leu1+ | This study |
| IRT 157 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔGRD leu1+ | This study |
| IRT 160 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+, kanMX6-YFP-myp2, sad1-CFP-kanMX6 | This study |
| IRT 162 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, kanMX6-YFP-myp2, sad1-CFP-kanMX6 | This study |
| IRT 165 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, kanMX6-Pbgs1-mEGFP-bgs1, sad1-RFP-hphMX6 | This study |
| IRT 167 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+, kanMX6-Pbgs1-mEGFP-bgs1, sad1-RFP-hphMX6 | This study |
| IRT 169 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, mob1-GFP-kanMX6 | This study |
| IRT 170 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+, mob1-GFP-kanMX6 | This study |
| IRT 171 | leu1-32, ura4-D18, ade6-M21X, rng2-D5, bltI-mEGFP-kanMX6, sad1-RFP-kanMX6 | This study |
| IRT172 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, sid2-mEGFP-kanMX6 | This study |
| IRT173 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+, sid2-mEGFP-kanMX6 | This study |
| IRT 174 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2 leu1+, cdc7-mEGFP-kanMX6 | This study |
| IRT 175 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-rng2ΔIQ leu1+, cdc7-mEGFP-kanMX6 | This study |
| IRT 187 | ura4-D18, ade6-M21X cdc4-8 | This study |
| IRT 191 | leu1-32, ura4-D18, ade6-M21X, rng2-D5, Prng2-mEGFP-rng2ΔIQ leu1+, cdc4-8 | This study |
| IRT 193 | leu1-32, ura4-D18, ade6-M21X, rng2Δ::natMX6, Prng2-mEGFP-rng2 leu1+, cdc4-8 | This study |
| JG 3 | leu1-32, ura4-D18, ade6-M21X, bltI-mEGFP-kanMX6, sad1-RFP-kanMX6 | Lab stock |
| JW 800 | h– leu1-32, ura4-D18, ade6-M210, kanMX6-mEGFP-myo2, sad1-mEGFP-KanMX6 | Lab stock |
| JW 1039 | h– leu1-32, ura4-D18, ade6-M210, kanMX6-Pcdc15-mYFP-cdc15 | Lab stock |
| JW 1089 | h–, leu1-32 ura4-D18, ade6-M210, mid1-mYFP-kanMX6 | Lab stock |
All cells expressing Rng2p or Rng2p deletion constructs were made by integration into the leu1 locus by the lithium acetate method (Bahler et al., 1998b). The endogenous rng2 locus was deleted from cells complemented with Rng2p or viable Rng2p deletions by replacing the gene with a cassette for Nat resistance by the lithium acetate integration method. The resulting deletion was confirmed by PCR across the deleted region. Some deletion strains were created by crossing a strain complemented in the leu1 locus with an existing rng2Δ strain and performing a tetrad dissection.
Microscopy and data analysis
Cells were grown for 36 h in liquid YE5S at 25°C before imaging. Cells were prepared and imaged as previously described (Wu et al., 2003), except that images were acquired with an IX71 inverted microscope (Olympus, Tokyo, Japan) with a Yokogawa CSU-X1 spinning-disk confocal scanning unit and an iXon electron-multiplying charge-coupled device camera (Andor Technology, South Windsor, CT). Cells imaged at room temperature were washed twice with EMM5S and once with EMM5S with N-propyl-gallate, placed on gelatin pads, and sealed with valap. Cells were imaged at 32°C on agar pads rather than gelatin pads. To deplete Rng2p, 81xnmt1-rng2 cells were grown in 10 μg/ml thiamine for 24 h and imaged in media containing thiamine.
Unless noted otherwise, images were obtained in Z-stacks of 18 or 20 slices at 0.36-μm intervals and visualized as maximum intensity projections using ImageJ (National Institutes of Health, Bethesda, MD). Room-temperature movies were generally acquired at 3-min intervals, and movies at 32°C were acquired at 2-min intervals. Fluorescence intensity was quantified after images were corrected for camera noise and uneven illumination, and the slices were summed (Wu and Pollard, 2005). A rectangular area containing a ring was used to measure its intensity after correcting for background fluorescence of the cytoplasm and photobleaching.
Viability assays
Cells were grown in liquid culture at 25°C for 36 h. Equal numbers of cells were serially diluted 10-fold four times, and 1.5-μl samples were spotted on YE5S agar plate and grown for 36–72 h.
Immunoblotting
GFP fusion proteins were detected on immunoblots (Wu and Pollard, 2005) with a 1:4000 dilution of anti-GFP antibody (ab290; Abcam, Cambridge, MA), followed by 1:15,000 dilution of a horseradish peroxidase–conjugated anti-rabbit secondary antibody (4050-05; Southern Biotech, Birmingham, AL). Band intensities were normalized by comparison to a nonspecific band or a blot with anti-cofilin (Pollard lab stock). Cells used to test the stability of Rng2-D5p at elevated temperatures were grown at either 32°C for 2 h, the conditions used for microscopy experiments, or at 36°C for 4 h, to visualize a more extreme condition, before preparation for immunoblotting.
Homology model
We used SWISS-Model to create two homology models of Rng2p. The homology model of the GRD (residues 805–1187) was built using the structure of IQGAP1 GRD (3FAY) as the starting structure. The homology model of residues 1392–1489 of Rng2p was based on the structure of part of the RasGAP-C domain of IQGAP1 (PBD 1×0H).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Research Grant GM026132. We thank Rajesh Arasada, Caroline Laplante, and John Goss for assistance with microscopy and image analysis; Christopher Jurgenson for help with homology models; and Naomi Courtemanche for advice on cloning and protein purification.
Abbreviations used:
- 6xHis
hexahistidine
- CFP
cyan fluorescent protein
- CHD
calponin homology domain
- DIC
differential interference contrast
- GAP
GTP-activating protein
- GRD
GAP-related domain
- GST
glutathione S-transferase
- mEGFP
monomeric enhanced green fluorescent protein
- NMR
nuclear magnetic resonance
- SIN
septation initiation network
- SPB
spindle pole body
- YFP
yellow fluorescent protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-10-0775) on April 24, 2013.
I.R.T. and T.D.P. planned experiments, analyzed results, and wrote the manuscript. I.R.T. performed the experiments.
The authors declare that they have no conflict of interest.
REFERENCES
- Almonacid M, Celton-Morizur S, Jakubowski JL, Dingli F, Loew D, Mayeux A, Chen JS, Gould KL, Clifford DM, Paoletti A. Temporal control of contractile ring assembly by Plo1 regulation of myosin II recruitment by Mid1/anillin. Curr Biol. 2011;21:473–479. doi: 10.1016/j.cub.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Distinct roles for F-BAR proteins Cdc15p and Bzz1p in actin polymerization at sites of endocytosis in fission yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Steever AB, Wheatley S, Wang Y, Pringle JR, Gould KL, McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J Cell Biol. 1998a;143:1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998b;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Bezanilla M, Forsburg SL, Pollard TD. Identification of a second myosin-II in Schizosaccharomyces pombe: Myp2p is conditionally required for cytokinesis. Mol Biol Cell. 1997;8:2693–2705. doi: 10.1091/mbc.8.12.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielak-Zmijewska A, Kolano A, Szczepanska K, Maleszewski M, Borsuk E. Cdc42 protein acts upstream of IQGAP1 and regulates cytokinesis in mouse oocytes and embryos. Dev Biol. 2008;322:21–32. doi: 10.1016/j.ydbio.2008.06.039. [DOI] [PubMed] [Google Scholar]
- Brandt DT, Marion S, Griffiths G, Watanabe T, Kaibuchi K, Grosse R. Dia1 and IQGAP1 interact in cell migration and phagocytic cup formation. J Cell Biol. 2007;178:193–200. doi: 10.1083/jcb.200612071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chen Q, Pollard TD. Actin filament severing by cofilin is more important for assembly than constriction of the cytokinetic contractile ring. J Cell Biol. 2011;195:485–498. doi: 10.1083/jcb.201103067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Souza VM, Naqvi NI, Wang H, Balasubramanian MK. Interactions of Cdc4p, a myosin light chain, with IQ-domain containing proteins in Schizosaccharomyces pombe. Cell Struct Funct. 2001;26:555–565. doi: 10.1247/csf.26.555. [DOI] [PubMed] [Google Scholar]
- Dixon MJ, et al. IQGAP proteins reveal an atypical phosphoinositide (aPI). binding domain with a pseudo C2 domain fold. J Biol Chem. 2012;287:22483–22496. doi: 10.1074/jbc.M112.352773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng K, Naqvi NI, Wong KC, Balasubramanian MK. Rng2p, a protein required for cytokinesis in fission yeast, is a component of the actomyosin ring and the spindle pole body. Curr Biol. 1998;8:611–621. doi: 10.1016/s0960-9822(98)70248-9. [DOI] [PubMed] [Google Scholar]
- Epp JA, Chant J. An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr Biol. 1997;7:921–929. doi: 10.1016/s0960-9822(06)00411-8. [DOI] [PubMed] [Google Scholar]
- Fang X, Luo J, Nishihama R, Wloka C, Dravis C, Travaglia M, Iwase M, Vallen EA, Bi E. Biphasic targeting and cleavage furrow ingression directed by the tail of a myosin II. J Cell Biol. 2010;191:1333–1350. doi: 10.1083/jcb.201005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova A, Morrell-Falvey J, Chen JS, Singh NS, Balasubramanian MK, Gould KL. The fission yeast septation initiation network (SIN) kinase, Sid2, is required for SIN asymmetry and regulates the SIN scaffold, Cdc11. Mol Biol Cell. 2012;23:1636–1645. doi: 10.1091/mbc.E11-09-0792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet O, Simanis V. Mid1p/anillin and the septation initiation network orchestrate contractile ring assembly for cytokinesis. Genes Dev. 2008;22:3205–3216. doi: 10.1101/gad.1697208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a rasGAP-related domain, is a potential effector for cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yan H, Balasubramanian MK. Assembly of normal actomyosin rings in the absence of Mid1p and cortical nodes in fission yeast. J Cell Biol. 2008;183:979–988. doi: 10.1083/jcb.200806151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AE, McCollum D, Gould KL. Polar opposites: fine-tuning cytokinesis through SIN asymmetry. Cytoskeleton (Hoboken) 2012;69:686–699. doi: 10.1002/cm.21044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurella VB, Richard JM, Parke CL, Lecour LF, Jr, Bellamy HD, Worthylake DK. Crystal structure of the GTPase-activating protein-related domain from IQGAP1. J Biol Chem. 2009;284:14857–14865. doi: 10.1074/jbc.M808974200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–23367. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ. Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol. 2011;192:1005–1021. doi: 10.1083/jcb.201008171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459:852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
- Mateer SC, Morris LE, Cromer DA, Bensenor LB, Bloom GS. Actin filament binding by a monomeric IQGAP1 fragment with a single calponin homology domain. Cell Motil Cytoskeleton. 2004;58:231–241. doi: 10.1002/cm.20013. [DOI] [PubMed] [Google Scholar]
- Moseley JB, Mayeux A, Paoletti A, Nurse P. A spatial gradient coordinates cell size and mitotic entry in fission yeast. Nature. 2009;459:857–860. doi: 10.1038/nature08074. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Osman MA, Cerione RA. Iqg1p, a yeast homologue of the mammalian IQGAPs, mediates cdc42p effects on the actin cytoskeleton. J Cell Biol. 1998;142:443–455. doi: 10.1083/jcb.142.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan A, Bakka K, Sevugan M, Naqvi NI, D'Souza V, Tang X, Mishra M, Balasubramanian MK. IQGAP-related Rng2p organizes cortical nodes and ensures position of cell division in fission yeast. Curr Biol. 2011;21:467–472. doi: 10.1016/j.cub.2011.01.059. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Wu JQ. Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol. 2010;11:149–155. doi: 10.1038/nrm2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proctor SA, Minc N, Boudaoud A, Chang F. Contributions of turgor pressure, the contractile ring, and septum assembly to forces in cytokinesis in fission yeast. Curr Biol. 2012;22:1601–1608. doi: 10.1016/j.cub.2012.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha S, Pollard TD. Anillin-related protein Mid1p coordinates the assembly of the cytokinetic contractile ring in fission yeast. Mol Biol Cell. 2012;23:3982–3992. doi: 10.1091/mbc.E12-07-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Shannon KB. IQGAP family members in yeast, Dictyostelium, and mammalian cells. Int J Cell Biol. 2012;2012:894817. doi: 10.1155/2012/894817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon KB, Li R. The multiple roles of Cyk1p in the assembly and function of the actomyosin ring in budding yeast. Mol Biol Cell. 1999;10:283–296. doi: 10.1091/mbc.10.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaine M, Numata O, Nakano K. Fission yeast IQGAP arranges actin filaments into the cytokinetic contractile ring. EMBO J. 2009;28:3117–3131. doi: 10.1038/emboj.2009.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O'Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319:97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- Wang CH, Balasubramanian MK, Dokland T. Structure, crystal packing and molecular dynamics of the calponin-homology domain of Schizosaccharomyces pombe Rng2. Acta Crystallogr D Biol Crystallogr. 2004;60:1396–1403. doi: 10.1107/S0907444904012983. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD. Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell. 2003;5:723–734. doi: 10.1016/s1534-5807(03)00324-1. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.