Although a clear picture of the in vitro assembly process is established for vimentin, the role of associated partner proteins and their effect on intermediate filament assembly has not been fully examined. This study finds delayed dynamics of desminopathy-linked mutant desmin in myocytes and hindered assembly when associated to nebulin.
Abstract
Desmin intermediate filaments (DIFs) form an intricate meshwork that organizes myofibers within striated muscle cells. The mechanisms that regulate the association of desmin to sarcomeres and their role in desminopathy are incompletely understood. Here we compare the effect nebulin binding has on the assembly kinetics of desmin and three desminopathy-causing mutant desmin variants carrying mutations in the head, rod, or tail domains of desmin (S46F, E245D, and T453I). These mutants were chosen because the mutated residues are located within the nebulin-binding regions of desmin. We discovered that, although nebulin M160–164 bound to both desmin tetrameric complexes and mature filaments, all three mutants exhibited significantly delayed filament assembly kinetics when bound to nebulin. Correspondingly, all three mutants displayed enhanced binding affinities and capacities for nebulin relative to wild-type desmin. Electron micrographs showed that nebulin associates with elongated normal and mutant DIFs assembled in vitro. Moreover, we measured significantly delayed dynamics for the mutant desmin E245D relative to wild-type desmin in fluorescence recovery after photobleaching in live-cell imaging experiments. We propose a mechanism by which mutant desmin slows desmin remodeling in myocytes by retaining nebulin near the Z-discs. On the basis of these data, we suggest that for some filament-forming desmin mutants, the molecular etiology of desminopathy results from subtle deficiencies in their association with nebulin, a major actin-binding filament protein of striated muscle.
INTRODUCTION
The cytoskeleton is a dynamic three-dimensional (3D) network that coordinates many critical cell behaviors. To thrive, cells rely on their cytoskeleton to respond to their environment. The cytoskeleton is composed of three types of filamentous proteins: actin, tubulins, and intermediate filament proteins. These proteins generate filament systems whose concerted operations allow cells to absorb mechanical stress, modulate the position and motility of cytoplasmic organelles, transmit force between their neighbors, and control cell movement and division.
The intermediate filaments (IFs) of the cytoskeleton and nucleoskeleton are a family of proteins characteristically expressed in all metazoan tissues (Karabinos et al., 2001; Zhong et al., 2010; Mencarelli et al., 2011). IFs maintain cell shape and mechanical integrity, regulate organelle anchorage and movement, and participate in cell migration and adhesion (Helfand et al., 2003; Schoumacher et al., 2010; Nekrasova et al., 2011). The tissue-specific expression of IF proteins is accompanied by distinct posttranslational modifications (Omary et al., 2006). For example, phosphorylation of vimentin is a key event during epithelial tumor cell migration, and increased phosphorylation of desmin occurs during fasting and muscle atrophy (Mendez et al., 2010; Cohen et al., 2012). Moreover, p21-activated kinase–mediated phosphorylation of desmin interferes with its normal assembly process (Ohtakara et al., 2000).
IFs display a tripartite molecular blueprint composed of non–α-helical “head” and “tail” domains with a central α-helical rod domain. The rod domain in type III IFs is subdivided into a presumptive α-helical pre–coiled-coil domain, coil 1A, coil 1B, and coil 2, interrupted by two linkers, L1 and L12 (Chernyatina et al., 2012). Desmin intermediate filaments (DIFs) integrate myofibrils in three dimensions by forming a honeycomb-like network that connects each Z-disc and results in the exquisite, near-crystalline arrangement of sarcomeres within mature skeletal myofibers (Granger and Lazarides, 1979). A highly interwoven desmin cytoskeleton remains after high-salt extraction of actin and myosin filaments in skeletal muscle (Granger and Lazarides, 1978; Wang and Ramirez-Mitchell, 1983).
More than 60 distinct mutations of the desmin gene in humans cause desminopathy, a type of hereditary and sporadic myofibrillar myopathy (Clemen et al., 2013). Although desminopathy shows clinical heterogeneity and variable age of onset, recurrent symptoms of this disease include progressive distal myopathy and muscle weakness, arrhythmia, conduction defects, heart failure, cardiomyopathy, dysphagia, and respiratory insufficiency (van Spaendonck-Zwarts et al., 2010). Of interest, desminopathy is also linked to vocal cord palsy and pharyngeal distal myopathy (Albertyn et al., 2010). Recognized histological phenotypes such as cytosolic protein aggregates, myofibril dissolution, rubbed-out fibers, and rimmed vacuoles are characteristically found in biopsies from human muscle tissues (Shinde et al., 2008; Claeys et al., 2009; McLendon and Robbins, 2011).
Although a single underlying mechanism for desminopathy is unlikely, studies propose that a number of desmin mutations cause disease by interfering with the assembly of filaments from tetramers (Bar et al., 2005b, 2006). In some cases it is clear that mutant desmin fails to make filaments either because protein assembly arrests before unit-length filaments (ULFs) are formed or because mutant proteins fail to make filaments of appropriate length or width (Dalakas et al., 2003). Other studies show that mutant desmin is capable of making seemingly normal filaments in vitro. In these cases, however, inappropriate binding of desmin to its ligands in muscle cells significantly contributes to desminopathy (Conover et al., 2009).
The interior of myocytes is filled with precisely organized sarcomeres, the cell's smallest force-generating unit, which is composed of interdigitated myosin and actin filaments. Biochemical data show that nebulin is a major sarcomeric actin-binding protein and binds to DIFs near the Z-discs and participates in a complex that mechanically integrates myofibers (Bang et al., 2002; Conover et al., 2009; Tonino et al., 2010). Nebulin is abundant in skeletal myocytes and is expressed in low amounts in cardiomyocytes (Wang and Williamson, 1980; Kruger et al., 1991; Fock and Hinssen, 2002; Kazmierski et al., 2003; Bang et al., 2006). In addition, nebulin binds to tropomodulin and CapZ, stabilizing thin filaments and Z-discs (McElhinny et al., 2001; Pappas et al., 2008, 2010). Apart from a solely structural role, studies suggest that nebulin regulates muscle contraction and calcium handling (Bang et al., 2009; Chandra et al., 2009; Gokhin et al., 2009). New evidence suggests that nebulin cooperates with the neural Wiskott–Aldrich syndrome protein, a major activator of the Arp2/3 complex, to induce branched actin filaments in skeletal muscle (Takano et al., 2010).
The focus of this study is to define further the mechanisms used by DIFs to associate with sarcomeres and hence increase our understanding of how alterations in mutant desmin and actin thin filaments contribute to desminopathy. We investigate the effect of nebulin binding during distinct stages of desmin filament assembly and assess how and when nebulin integrates with desmin filaments during assembly. Our hypothesis is that a number of filament-forming mutants of desmin inappropriately bind to nebulin and contribute thereby to the underlying molecular etiology of desminopathy. To test our hypothesis, we combine electron microscopy and cosedimentation assays with live-cell imaging using fluorescence recovery after photobleaching (FRAP). We determine binding alterations of nebulin to mutant desmin filaments in early and late stages of filament assembly. In addition, we report that mutant desmin E245D is significantly less mobile than controls in live myocytes. Taken together, our results support a model in which assembly-competent mutant desmin contributes to desminopathy through untimely and abnormal binding to nebulin.
RESULTS
Nebulin module M164 closely codistributes with desmin around the Z-discs of the skeletal muscle
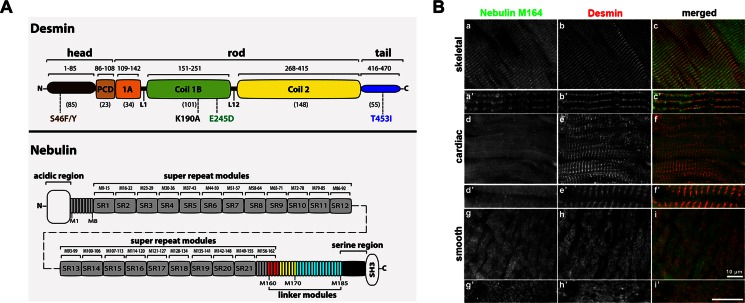
Desmin is an ∼53-kDa IF protein abundant in all types of muscle cells. It has a central ∼45-nm α-helical rod flanked by non–α-helical head and tail regions (Herrmann et al., 1996). One of its binding partners, nebulin, a large (∼700–900 kDa), multimodular muscle protein, extends from the Z-discs of the sarcomeres near the pointed ends of the actin thin filaments (Labeit and Kolmerer, 1995; Wang et al., 1996; Bang et al., 2002; Littlefield and Fowler, 2008). Modules M160–164 of nebulin bind with high affinity to the coil 1B region of desmin. These modules are not likely to be the only ones involved in binding to desmin, however, as other regions in the C-terminal region of nebulin also appear to contribute (Bang et al., 2002; Conover et al., 2009). Both desmin and nebulin form complex filament systems in muscle cells that synergistically collaborate to integrate the myofibril contractile machinery to the IF cytoskeletal network (Figure 1A).
FIGURE 1:
Nebulin module 164 borders one side of the Z-disc in cardiomyocytes. (A) The layouts of desmin and nebulin. Desmin is composed of an α-helical coiled-coil rod central region bordered by non–α-helical head and tail regions. It is assumed that the rod of desmin is subdivided into a pre-coil domain (pcd) and coil 1A, linker L1, coil 1B, linker L12, and coil 2 domains, as reported for vimentin (Nicolet et al., 2010). Shown is the localization of desminopathy desmin mutations, S46F/Y (head), E245D (coil 1B), and T453I (tail), and the random mutation K190A (coil 1B) used in this study. Nebulin contains 184 actin-binding single modules (M), short segments formed by 35–amino acid tandem repeat containing the SDxxYK motif. The modules are organized into superrepeats formed by seven modules containing the WLKGIGW motif. The N-terminal end of nebulin is rich in glutamic acid (acidic region), with a SH3 domain near its C-terminus. The nebulin modules that are important for interaction with desmin are shown in yellow (Bang et al., 2002), and the ones that have the highest affinity for desmin are shown in red (Conover et al., 2009). (B) Canine muscle tissues were costained with nebulin M164 (green) and desmin (red). Although desmin was detected in all three types of muscle analyzed (b, e, h), these images show that nebulin's intensity is the highest in skeletal muscle (a) and is expressed in low amounts in cardiac or smooth muscles (d, g). Bar, 10 μm.
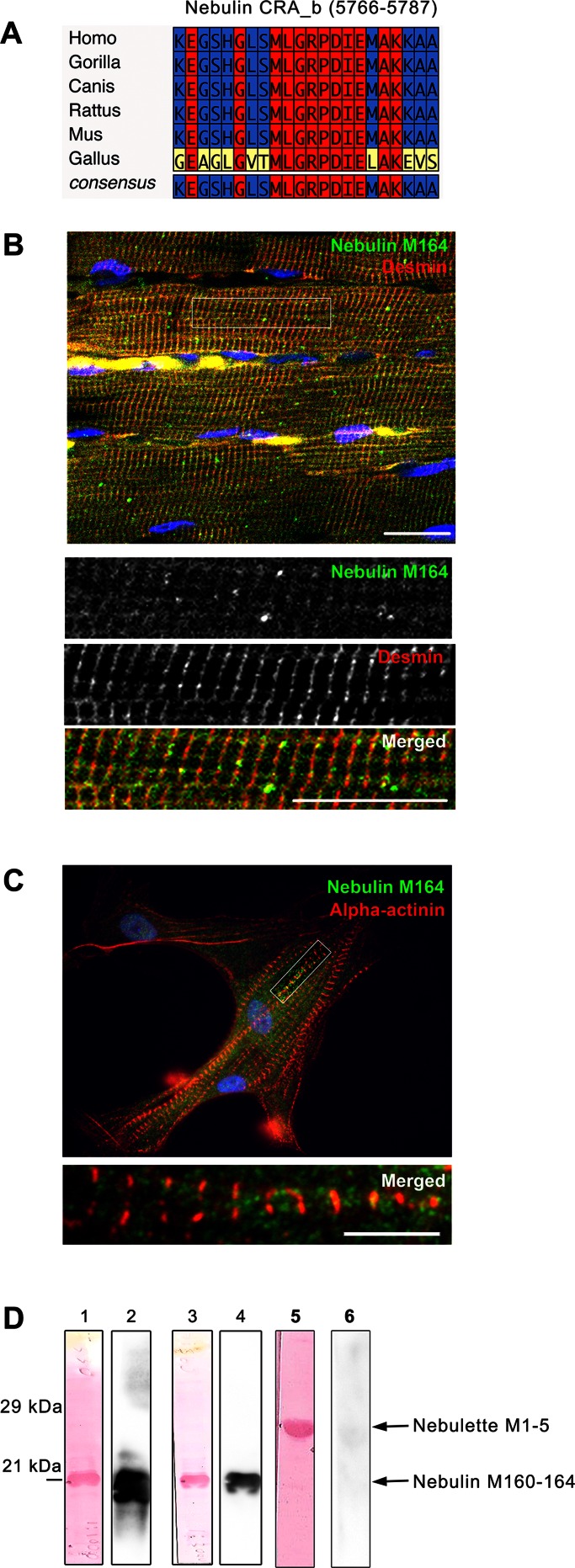
To determine the localization of nebulin module M164 in striated muscle, a high-affinity binding site for desmin, we designed peptide antibodies against human nebulin with little cross-reactivity to nebulette based on primary sequence analyses. Nebulette is a modular cardiac protein that belongs to the nebulin family of actin-binding proteins with high degree of homology to nebulin (Moncman and Wang, 2002). Online multiple alignment ClustalW of the peptide region used to generate the antibodies showed that the 22-mer peptide in human nebulin (5766–5787) does not have homology to nebulette and is identical among humans, chimps, dogs, and rodents but only displays 54% identical residues when aligned to chick nebulin (Figure 2A). Immunoblot analyses showed negligible cross-reactivity to nebulette recombinant protein (Figure 2D).
FIGURE 2:
Distribution patterns of desmin and nebulin in primary myocytes and muscle tissues. (A) Primary sequence analysis of the 22mer peptide within nebulin's module 164 (5766–5787) used to generate the custom peptide antibody. Blastp and ClustalW (version 1.83) alignments of this peptide show a highly conserved region in nebulin for the different species analyzed (human, apes, canines, and rodents). In contrast, this peptide only had 54% identical amino acids when aligned to chick nebulin. (B) Rat psoas skeletal muscle showed striking desmin (red) striations that appear to encircle each Z-disc, whereas nebulin M164 (green) is closely located on or along these desmin striations (see inset). Bar, 10 μm. (C) Primary cardiomyocytes costained for nebulin M164 (green) and Z-disc marker α-actinin (red) show intense nuclear staining. Moreover, the α-actinin staining shows clear striations at the Z-discs of the sarcomeres of muscle cells, whereas nebulin M164 was enriched at the periphery of the Z-discs. The inset shows filaments of nebulin projecting outward in both directions from the Z-discs. Bar, 10 μm. (D) Recombinant nebulin M160–164 is recognized by peptide antibody against nebulin module 164 and does not cross-react with nebulette M1–5. Immunoblots and corresponding total protein stain (Ponceau S, lanes 1, 3, and 5) of N-terminal His-tagged nebulin M160–164 recombinant protein (lanes 1–4) or nebulette M1–5 (lanes 5 and 6) were probed using standard Western blot conditions with the following antibodies: anti–nebulin M164 antibodies (lanes 2 and 6) or commercial anti-His antibodies (lane 4). Blots detect a ∼21-kDa band corresponding to the expected size of the recombinant nebulin M160–164 fragmentin strips containing nebulin, but little or no reactivity was found in the strip containing nebulette M1–5.
Examination of canine biopsy tissues showed that nebulin M164 exhibited a striated pattern surrounding desmin, more prominently in the skeletal muscles analyzed, and that nebulin M164 was occasionally organized as “doublets” around desmin (Figure 1B, a–c, arrows in insets a′ and c′). Of note, we also observed a moderate expression of nebulin M164 in cardiac and smooth muscles that was loosely associated with desmin filaments (Figure 1B, d–i). Unexpectedly, we found a moderate coexpression of nebulin M164 and desmin in brain and liver, whereas no significant expression was detected in lung or spleen tissues (Supplemental Figure S1, a–f).
In an evaluation of the distribution of the nebulin M164 epitope in rat psoas muscle, confocal image analyses showed clear nebulin M164 striations intimately interwoven with desmin striations (Figure 2B). In contrast, mature primary cardiomyocytes displayed bright nuclear and punctate cytoplasmic staining for nebulin M164 without consistently showing a striated pattern surrounding the Z-discs (Figure 2C). Nevertheless, occasionally we observed short filaments of nebulin in the vicinity of α-actinin extending out from the Z-disc (Figure 2C, inset). Taken together, our data show a close codistribution of nebulin and desmin in some tissues but not in others. Furthermore, our results are consistent with the original description for desmin localization at the periphery of the muscle Z-disc (Lazarides, 1978) and refine our understanding of nebulin distribution in the vicinity of the Z-discs (Millevoi et al., 1998; Pappas et al., 2008).
Nebulin associates with desmin in a complex, nonlinear manner
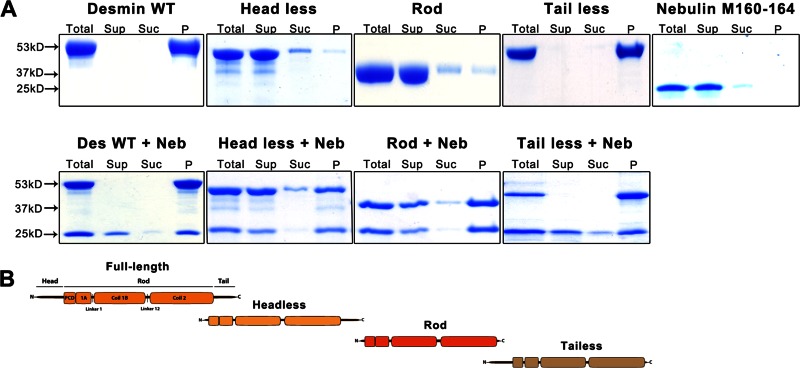
To study the kinetics of desmin–nebulin association, we used high-speed cosedimentation assays (for details, see Supplemental Figure S2) to determine whether nebulin preferentially bound to a particular desmin domain. Using this approach, we tested full-length desmin's ability to bind to nebulin modules M160–164 and compared it to desmin-truncated proteins: headless, rod, and tailless. Western blot analysis confirmed the specificity of the recombinant nebulin M160–164 (Figure 2D). Our data demonstrate that all desmin fragments tested were capable of sedimenting nebulin to similar extents (Figure 3). Band densitometry analysis of the pellet fractions showed that equimolar mixtures of nebulin and the respective desmin fragment yielded the following sedimentation values: wild-type (WT) desmin ∼59%, headless ∼57%, rod ∼70%, and tailless ∼29%. As expected, desmin pelleted efficiently to ∼97% during assembly conditions, whereas insignificant sedimentation ∼1% was detected for the nebulin alone (Figure 2A, top). Because nebulin M160–164 is a basic fragment with an isoelectric point of 9.44 and desmin has an acidic isoelectric point of 4.96, we examined the possibility that the observed binding between these proteins occurred solely because of electrostatic interaction. To rule out this possibility, we tested by cosedimentation assay whether the soluble acidic protein bovine serum albumin (BSA) associated with nebulin (Supplemental Figure S3). No significant interaction occurred by electrostatic molecular interactions between these proteins: both remained in the supernatant, and neither protein was detected in the pellet fractions.
FIGURE 3:
Desmin contains multiple binding sites for nebulin. High-speed cosedimentation was used to determine whether a specific desmin domain preferentially bound to a nebulin fragment. Four recombinant proteins comprising one (rod, headless, tailless) or all desmin domains were purified and coassembled with a nebulin recombinant fragment encompassing modules 160–164 for 1 h at 37°C under buffer conditions that facilitated desmin filament assembly. (A) Coomassie-stained SDS–PAGE gels show each sample fractionated into total (T), supernatant (Sup), sucrose (Suc), and pellet (P). Whereas headless, rod, and nebulin are largely soluble, both full-length and tailless desmin sedimented in the pellet (top). When nebulin M160–164 was added to full-length desmin or any of the desmin truncation proteins tested, all of them bound, as demonstrated by nebulin cosedimenting with each protein. (B) Schematic representation of desmin fragments used in this assay.
Our data suggest that desmin binds nebulin M160–164 via multiple binding sites. This is consistent with a previous peptide blot desmin array, which identified multiple binding sites within desmin for nebulin M160–170 (Conover and Gregorio, 2011). In addition, our results suggest either that the rod domain contains higher-affinity binding sites for nebulin or this domain is more exposed in the molecule than in headless or tailless desmin. Nevertheless, our data clearly show that nonhelical head and tail desmin domains still contain binding sites for nebulin. Although some differences in the interaction between nebulin and a particular desmin domain were detected using this approach, the possibility of transient interactions during IF assembly in cells cannot be excluded. Based on the recent crystal structure deduced for vimentin (Chernyatina et al., 2012), desmin is also believed to form extended, parallel, left-handed coiled-coil dimers. This conformation is predicted to allow desmin to accommodate multiple nebulin binding contacts, allowing direct linkage to the contractile sarcomere units.
Nebulin alters the sedimentation behavior of desmin prefilament oligomers
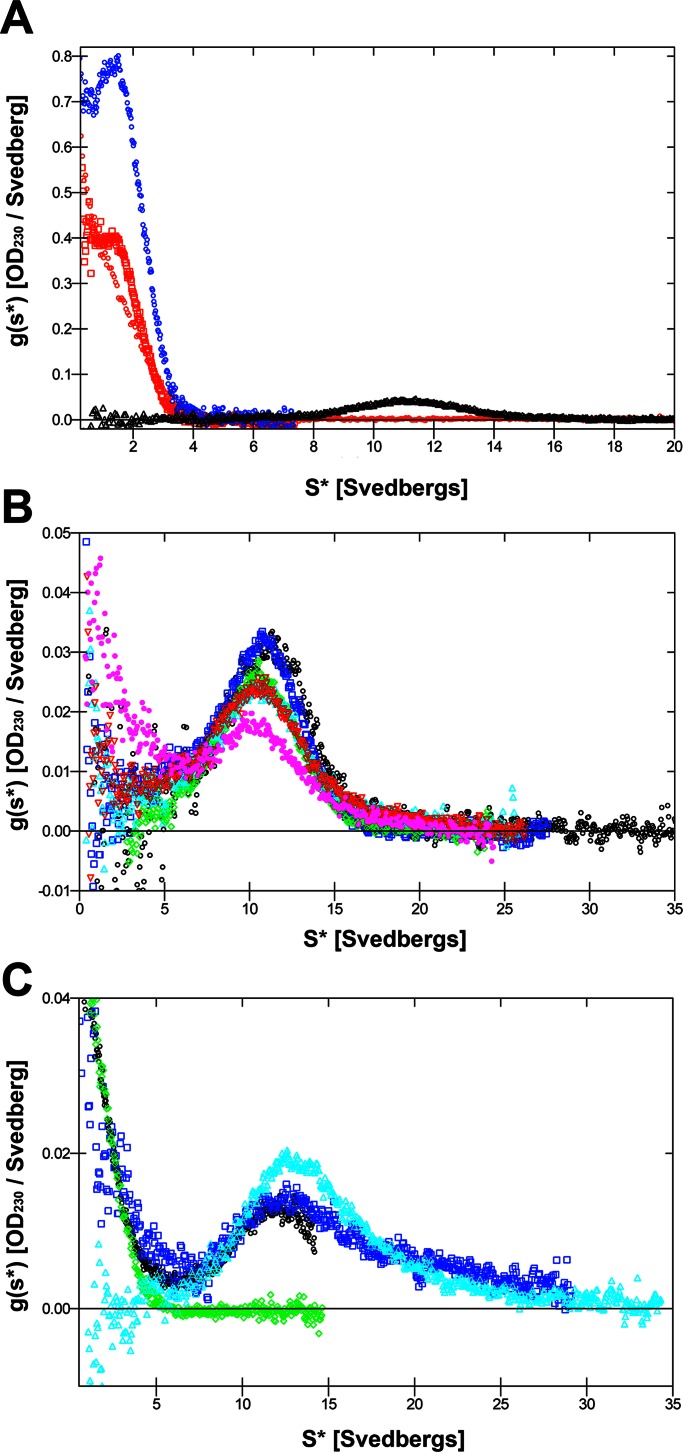
To further characterize the desmin–nebulin molecular complexes in solution, we used the well-established method of analytical ultracentrifugation (Harding and Rowe, 2010). To discern how many WT desmin oligomers interact with nebulin M160–164, we used sedimentation velocity centrifugation, comparing the profiles for mixtures of desmin and nebulin at different molar ratios under low-salt conditions (Figure 4). Our results showed that nebulin M160–164 sedimented at an average sedimentation coefficient of ∼1.45 S, whereas WT desmin sedimented with a sharp peak of ∼11 S at 20°C. Of interest, when nebulin M160–164 was added in 10-fold excess relative to desmin at a sedimentation velocity of 40,000 rpm, strong interaction between the two recombinant proteins was detected (Figure 4A). Approximately half of the total nebulin in solution bound to WT desmin, as indicated from ∼50% decrease in the amount of nebulin recovered in the mixed sample as compared with nebulin alone (Figure 4A). Moreover, the desmin–nebulin complex sediments at a higher sedimentation coefficient, which extends beyond 20 S.
FIGURE 4:
Nebulin M160–164 binds to desmin oligomers. The association between nebulin M160–164 and desmin oligomers was evaluated by sedimentation velocity analysis at 20°C in a 0.2 mM sodium phosphate (pH 7.5) buffer system. (A) At 40,000 rpm, a strong interaction between desmin and nebulin M160–164 was detected when a 10-fold excess of nebulin in relation to desmin (100 μM nebulin plus 9.3 μM desmin) was used. This is shown by ∼50% decrease for nebulin in the area under the curve for the mixed sample (red plot) as compared with nebulin alone (blue plot). Under these conditions desmin sediments at ∼11 S, as previously reported (black). (B) At 20,000 rpm, different molar ratios of nebulin in relation to desmin alone (black) were used as follows: 0.9 (blue), 2.3 (green), 4.7 (cyan), 9.3 (red), and 0 (pink) μM. A twofold excess of nebulin in relation to desmin significantly reduced the area under the desmin curve at ∼11 S (pink), as compared with a 1:1 ratio (red), suggesting a decrease in the concentration of free desmin oligomers. (C) At 30,000 rpm, for a molar ratio of desmin to nebulin of 1:2 (7.5 μM desmin to 16 μM nebulin), the desmin–nebulin complex exhibited a broad sedimentation velocity between ∼20 and 30 S, as evidenced from the dark blue curve imposed above the sedimentation profile for desmin alone (aqua). For comparison, we also evaluated nebulin alone (green).
To evaluate the effect of nebulin binding to desmin oligomers sedimenting at ∼11 S, we tested the following range of molar ratios with a fixed concentration of desmin while increasing the amounts of nebulin in solution: 1:0.1, 1:0.25, 1:0.5, 1:1, and 1:2. At a low sedimentation velocity of 20,000 rpm, we observed that the interaction between desmin and nebulin was detected when nebulin was present in twofold excess, as indicated by the corresponding drop in the amount of desmin under the peak at ∼11 S, whereas we found a modest decrease at a 1:0.25 desmin-to-nebulin ratio and none at a 1:0.1 ratio (Figure 4B). Next, at an intermediate sedimentation velocity of 30,000 rpm using a 1:2 M ratio of desmin to nebulin M160–164, under low-salt conditions, we readily detected a desmin–nebulin complex sedimenting with a spread between 20 and 30 S, as shown by the area under the dark blue curve being clearly elevated as compared with desmin alone (Figure 4C). Taken together, our data from analytical ultracentrifugation demonstrate that there is a strong interaction between protofibrils of desmin and soluble nebulin M160–164 and that the desmin–nebulin complex sediments faster than either desmin or nebulin alone.
Desmin and desminopathy-associated mutants strongly interact with nebulin during different assembly stages
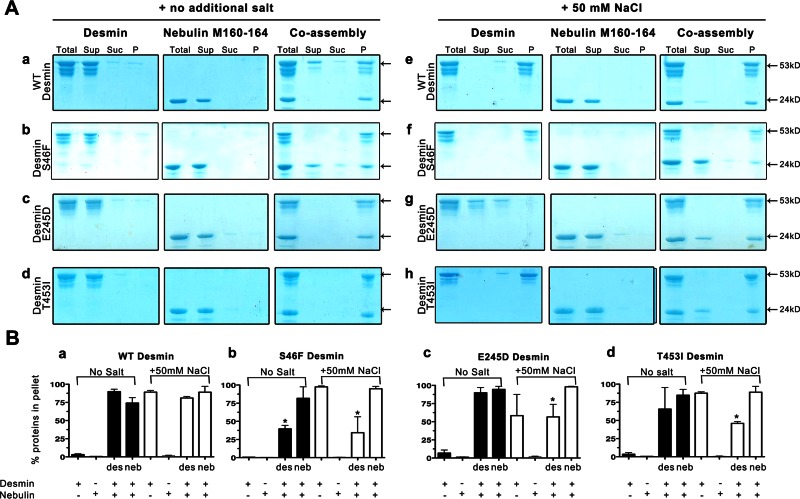
We used high-speed cosedimentation assays to determine the extent and assembly stage at which desmin associates to nebulin. Three samples (supernatant, sucrose, pellet) were collected after ultracentrifugation for SDS–PAGE analysis (Supplemental Figure S2). We used well-documented buffer conditions to initiate filament assembly (25 mM Tris-HCl, 50 mM NaCl, pH 7.5, 37°C) and quantified the relative band densities for each protein in each experiment using ImageJ. Under low-salt conditions, it is well known that WT desmin does not form filaments but instead is mostly found as a mixture of dimers and tetramers (Huiatt et al., 1980; Renner et al., 1981). Under these conditions, a significant amount of WT desmin partitioned into the supernatant and sucrose fractions (Supplemental Figure S4a), whereas negligible amounts were recovered in the pellet fractions (Figure 5, A, a, and B, a; compare first black bars). When in a salt-containing buffer, desmin assembles into filaments and is mostly insoluble, which causes it to pellet (Figure 5, A, e, and B, a; first white bar). In contrast, with or without salt, nebulin M160–164 is mostly soluble, because we did not recover protein in the pellet fraction (Figure 5, A, a, and B, a; second black and white bars).
FIGURE 5:
Nebulin binding delays the assembly of mutant desmin into filaments. Cosedimentation assays evaluated the efficiency of WT and mutant desmin interaction with nebulin under conditions that promote desmin dimer and tetramer formation or filament assembly. (A) Right, a cosedimentation assay in proteins dialyzed in 5 mM Tris (pH 7.4) at a molar ratio of 1:1 without salt; left, a parallel assay under salt conditions that promote desmin filament assembly (+100 mM NaCl). In both cases, the proteins were incubated for 1 h at 37°C before ultracentrifugation. (B) The amount of desmin or nebulin recovered in pellets was quantified by band densitometry using ImageJ software. Plotted are the averaged and standard deviations obtained from three independent experiments. Bar graphs show the percentage of protein recovered in the pellets in the absence (black bars) or presence (white bars) of salt. Coassembled fractions are plotted in the third bar (intensity measured for desmin) and fourth bar (intensity measured for nebulin) for each condition.
To determine whether soluble desmin can bind to nebulin, we coassembled both proteins for 1 h without salt. Under these conditions, desmin was soluble, presumably present mostly as dimers and tetramers rather than as ULFs or elongated filaments (Herrmann et al., 1996). Unexpectedly, our results showed that desmin tetramers and prefilament oligomers are capable of binding to nebulin to some extent (Figures 4 and 5, A, a, and B, a; fourth black bar). When mutant desmin S46F or desmin T453I was coassembled with nebulin, nebulin pelleted with both of these mutants to a similar extent without salt (Figure 5B; compare third black bars in b and d to a). Under high-salt conditions, unlike WT desmin, only ∼25% of these mutant desmins were recovered from the pellets (Figure 5B; compare third white bars in b and d to a).
Without additional salt in the buffer, mutant desmin E245D tetramers associated more nebulin relative to WT desmin, whereas in assembly conditions that promote filament formation less mutant desmin was recovered in the pellet fraction (Figure 5B; compare fourth black bars, c to a). Our results showed that under salt conditions that promote desmin filament assembly, up to ∼95% of total nebulin M160–164 was recovered in the pellet when coassembled with WT or mutant desmin E245D (Figure 5B; compare fourth white bars in a and c). Only ∼25% of assembled mutant desmin S46F was associated with ∼95% of nebulin, however, suggesting severely perturbed, hindered, or delayed DIF binding partner associations (Figure 5B, b, third and fourth white bars). Furthermore, our analysis revealed that ∼50% of assembled mutant desmin T453I is associated with ∼95% of nebulin (Figure 5B, d).
Our data indicate that nebulin M160–164 are capable of binding to WT and mutant desmin oligomers and filaments in different degrees. We conclude that assembled mutant desmin prevents proper stoichiometry of nebulin binding because we saw a markedly decreased amount of mutant desmin recovered in the pellets in the presence of nebulin (Figure 5B, compare white third bars in b–d to a). This finding might be attributable to differences in nanostructure or surface variations of mutant desmin that affect nebulin-binding specificity. Taken together, our results show that binding interactions between WT desmin and nebulin are sensitive to desmin's assembly state because mutations in desmin significantly perturb the extent of interaction. Therefore our data strongly suggest that desmin's assembly state is important for nebulin recruitment.
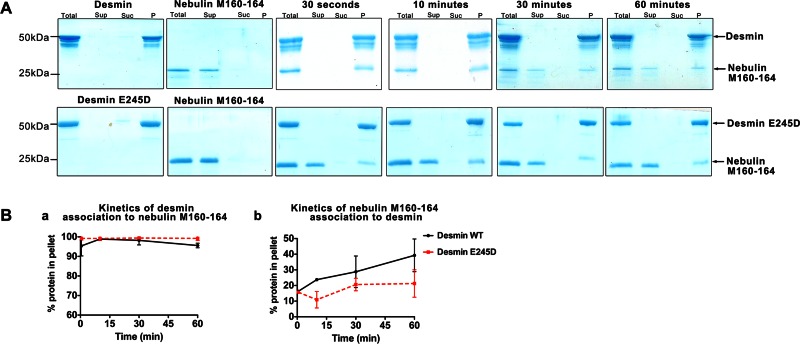
Delayed interaction kinetics of mutant desmin E245D to nebulin
To determine the kinetics at which the nebulin interacts with WT desmin and evaluate whether there are assembly delays or other alterations when nebulin M160–164 binds to mutant desmin E245D, we used a high-speed cosedimentation assay under salt conditions that favor desmin filament elongation (Figures 6 and Supplemental Figure S2). Equimolar samples of desmin and nebulin were assembled for 30 s or 10, 30, or 60 min at 37°C. After 30 s of assembly, it was evident that WT and mutant desmin E245D bound to nebulin to different degrees (Figure 6B). Our analysis revealed that ∼11% of mutant desmin bound to nebulin, in contrast to ∼25% of WT desmin. At later time points, the amount of nebulin that bound WT desmin reached ∼35% of total protein, in contrast to mutant desmin E245D, which stabilized at ∼20%. Our results show a delayed binding kinetic profile for desminopathy-linked mutant desmin E245D to nebulin. This finding constitutes a previously unrecognized underlying factor in desminopathy caused by filament-forming desmin mutations.
FIGURE 6:
Delayed kinetic profile of mutant desmin E245D associated with nebulin M160–164. Recombinant nebulin M160–164 binds strongly to WT and mutant desmin E245D filaments assembled in vitro. (A) Kinetic cosedimentation assays were performed using assembly conditions that promoted desmin filament formation. (B) Gel band densities in the pellet were measured for desmin (a) and nebulin bands (b) using ImageJ and plotted as a function of time. The pelleting behavior of nebulin when coincubated with mutant desmin E245D was significantly delayed as compared with WT desmin (compare red to black plots in Figure 4B, b). Values plotted show the average of three independent experiments.
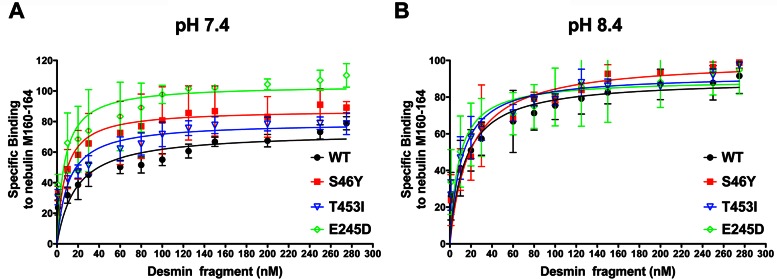
Enhanced binding affinity and capacities of mutant desmin for nebulin M160–164
To determine whether there are differences in binding of the mutant desminopathy-linked mutations (S46Y, E245D, and T453I) to the nebulin M160–164 fragment and further elucidate the biochemical basis for the desmin–nebulin association in this disorder, we compared the binding affinities and capacities of these proteins at pH 7.4 and 8.4 by enzyme-linked immunosorbent assays (ELISAs). These desmin mutations were selected for this study because they were found within known desmin nebulin-binding regions (Conover et al., 2009; Conover and Gregorio, 2011). Mutant desmin S46Y, located in the head region of desmin, was previously reported to form filaments with wider diameters than WT in vitro (Sharma et al., 2009), whereas the desmin E245D mutation located in the coil 1B of desmin formed seemingly normal filaments as visualized by transmission electron microscopy (Bar et al., 2005a). In contrast, recent data show that T453I mutation, located in the tail of desmin, forms a mixture of filaments and aggregates in C2C12 skeletal muscle cell lines (Chourbagi et al., 2011).
Our data show that all of the desmin mutants tested in this assay bound to nebulin M160–164 recombinant fragment with a stronger binding affinity (Kd) and capacity (Bmax) at pH 7.4 than for full-length WT desmin (Figure 7A and Table 1). In contrast, our results show no significant binding differences when the protein pairs are tested at pH 8.4 (Figure 7B and Table 1). This analysis reveals subtle but reproducible binding alterations of mutant desmin binding to nebulin at pH 7.4. When compared with the binding to WT desmin, we obtain an average binding affinity (Kd ≈ 18 nM), whereas the head mutant desmin S46Y yields approximately twofold higher binding affinity (Kd ≈ 8 nM), the coil 1B mutant desmin E245D mutant has approximately threefold stronger binding affinity (Kd ≈ 2 nM), and the tail mutant desmin T453I exhibits approximately twofold greater binding affinity (Kd ≈ 11 nM). With respect to the binding capacity of the interaction between desmin and nebulin, at pH 7.4, we found ∼21, ∼42, and ∼8% increased Bmax for mutant desmin S46F, E245D, and T453I, respectively, over WT desmin. Unexpectedly, we found no significant changes in this assay when we probed nebulin binding to these mutant desmins when these proteins were dialyzed out of urea at pH 8.4. In conclusion, our data are consistent with a model in which all three mutant desmins contributed to desminopathy by altering the stoichiometry of binding between thin filaments and the IF network in a pH-dependent manner. Furthermore, we speculate that the tighter association caused by the higher binding affinity displayed by each of the mutant desmins assayed (S46F, E245D, and T453I) to the actin-thin filament binding protein nebulin progressively perturbs or at least delays the dynamics of myosin–actin cross-bridge interactions in muscle cells. Further studies will discern whether this is the case.
FIGURE 7:
Mutant desmin binds to nebulin M160–164 with higher affinity and capacity in a pH-dependent manner. The binding of a nebulin M160–164 to WT desmin or mutant desmin (S46Y, E245D, and T453I) was compared in ELISAs at pH 7.4 and 8.4. Graphs show the normalized averaged binding to nebulin for WT and each mutant under the same experimental conditions. Curves show the nonlinear fitted averages of seven or eight experiments for WT desmin and two or three experiments for mutant desmin using the Michaelis–Menten equation. Values show the means ± SD. (A) ELISAs show increased binding capacities and affinities for all mutants tested as compared with WT when desmin proteins were dialyzed to 5 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, pH 7.4. (B) No difference in binding capacities and affinities for nebulin were found for mutants as compared with WT when the indicated desmin proteins were dialyzed to 5 mM Tris-HCl, 1 mM EDTA, 1 mM EGTA, pH 8.4. Table 1 lists the averages of all the measured Kd and Bmax values obtained for each experiment.
TABLE 2:
Primers used in this study.
| Primer sequence (5′ to 3′) | Gene and species | Enzyme |
|---|---|---|
| (Forward) AAAGTGCATGAAGACGAGAT CCGTGAGTTG | Homo sapiens pDS5-desmin E245D | None |
| (Reverse) CTCACGGATCTCGTCTTCATG CACTTT | H. sapiens pDS5-desmin E245D | None |
| (Forward) cgcgaa AGGCCT CCTCAGATCCTGCTGGCGAAG | H. sapiens nebulin M160–164 cloned into pET15b (Novagen) | StuI |
| (Reverse) gatgat CTCGAG TCATTA TGGATTGTATTTTGGTGT | H. sapiens nebulin M160–164 cloned into pET15b (Novagen) | XhoI |
| (Forward) gga AGATCTGATGTCCTACAGACGAGAGCTGGAG | Mus musculus tropomodulin 1 cloned into pEGFP-C1 | BglII |
| (Reverse) ccg GAATTC CTAGACACCGCTGCGGCACTT | M. musculus tropomodulin 1. cloned into pEGFP-C1 | EcoRI |
| (Forward) cgcgaa AGGCCT CCTCAGATCCTGCTGGCGAAG | H. sapiens nebulin M160–164 cloned into pET15b (Novagen) | StuI |
| (Reverse) gatgat CTCGAG TCATTA TGGATTGTATTTTGGTGT | H. sapiens nebulin M160–164 cloned into pET15b (Novagen) | XhoI |
Restriction enzyme site used is underlined.
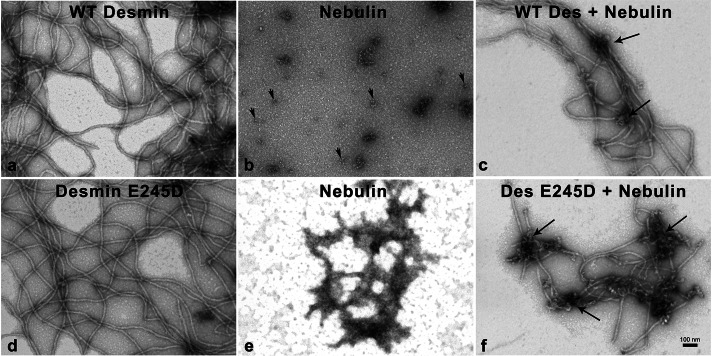
Nebulin M160–164 threads desmin filaments
To analyze the interaction of nebulin and desmin IFs at an ultrastructural level qualitatively, we evaluated negative-stained samples of assembled proteins in vitro in reconstitution assays by transmission electron microscopy. Nebulin recombinant fragments were added at 1:1 M ratio to preformed desmin IFs 30 min after the initiation of assembly. At this time, both WT and mutant desmin E245D formed elongated 10- to 12-nm-wide filaments (Figure 8, a and d). Structurally, nebulin appeared to form mixtures of monomers and, less frequently, higher-order structures consisting of small, “peanut”-shaped structures (Figure 8, b and e, respectively). Higher magnification of these nebulin structures showed donut-like monomers (data not shown). When DIFs and nebulin were combined, we observed that nebulin readily incorporated within the desmin filaments (Figure 8, c and f). Intriguingly, nebulin appeared to align the DIFs, as we observed near-parallel individual filaments connected by knob-like proteins frequently present on the grids. Furthermore, we observed increased amounts of nebulin bound to mutant desmin E245D filaments (Figure 8f). These results are consistent with a previous report that showed that this particular mutant desmin bound nebulin with an enhanced binding affinity (Conover et al., 2009). In conclusion, these data support a direct protein–protein interaction between nebulin and desmin filaments. Subtle differences in the binding properties between nebulin and mutant desmins probably have profound consequences for correctly winding desmin filaments around the Z-discs in myocytes.
FIGURE 8:
Nebulin associates with WT and mutant desmin filaments. Electron micrographs of negatively stained assembly products of WT desmin and mutant desmin E245D (a and d) show their interaction with nebulin M160–164 (c and f). Desmin proteins were renatured into 5 mM Tris-HCl (pH 8.4) and assembled by addition of 20 mM Tris-HCl plus 50 mM NaCl (pH 7.0) buffer for 30 min at 37°C. Assembly products were allowed to attach to the grid for 10 s. The recombinant nebulin fragment was incubated with assembled mature IFs at molar ratio of 1:1 of WT desmin for 30 min (c) or desmin E245D (f) 10 min. The nebulin fragment produced small structures (arrowheads, b) and sparingly made microaggregates (e). The nebulin fragment incorporated into the desmin filaments, threading the filaments (arrows, c). A tight association of nebulin to mutant desmin E245D filaments was observed, causing knotted filaments (arrows, f). Bar, 100 nm.
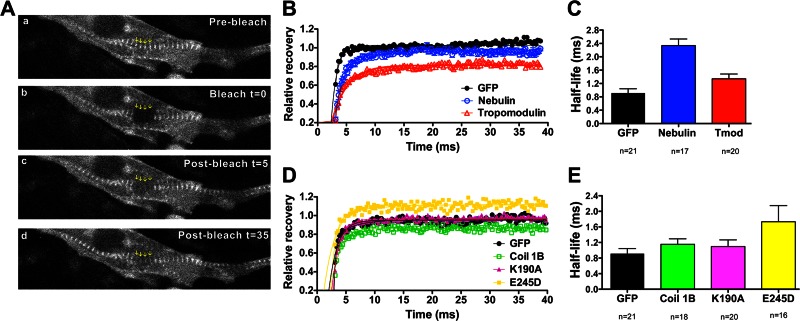
Dynamics of WT and mutant desmin in living cells
In live epithelial cells, vimentin readily incorporates into the endogenous IF network and is highly dynamic, as short fibrils “squiggles” are constantly lengthening or shortening in video recordings (Yoon et al., 1998). In fibroblasts, vimentin nonfilamentous particles regulate the formation of lamellipodia during migration in a phosphorylation-dependent manner (Goldman et al., 2008). Little is known about desmin IFs assembly dynamics in live myocytes. Our previous research suggested that the coil 1B of desmin efficiently incorporates within sarcomere Z-discs and that the coil 1B mutants (K190A and E245D) are significantly displaced from Z-discs, accumulating as intracellular cytoplasmic aggregates (Conover et al., 2009; Conover and Gregorio, 2011). We chose to use the K190A desmin mutation because it is not known to be pathogenic in humans, and yet it maps within the coil 1B region of desmin, close to the desminopathy-linked mutation E245D, and exhibits significantly reduced binding capacity to nebulin. The rationale used to select the coil 1B region of desmin rather than the full-length molecule in this analysis is that this domain targets about twice as efficiently to the Z-discs in cardiomyocytes, closely resembling the assembly pattern for endogenous desmin in these cells. Furthermore, the desmin coil 1B domain contains a high-affinity nebulin-binding site that likely is also missing binding sites for other desmin IFAP.
Although it was reported that both desmin green fluorescent protein (GFP)–coil 1B (K190A and E245D) proteins appeared to form nonfilamentous particles in cardiomyocytes to some extent, no information is available regarding their relative mobility in cardiomyocytes. To begin to investigate the mobility and molecular dynamics of desmin in living cells and study how the muscle cellular milieu modifies the relative mobility of desmin and its mutants, we expressed WT and mutant desmin GFP-tagged coil 1B in live cardiomyocytes and studied their dynamics by FRAP using time-lapse confocal fluorescence microscopy. The recoveries of nebulin and tropomodulin, muscle thin filament proteins, were also compared with desmin dynamics in these cells. Strikingly, our data show a significant increase in desmin E245D dynamics (Figure 9, D and E). The increase in the number of exchangeable mutant desmin E245D could be attributable to this mutant binding to the dark, immobile protein fraction more tightly or caused, indirectly, by its altered interactions to muscle binding partners.
FIGURE 9:
Dynamics of desmin and thin filament proteins in live cardiomyocytes. FRAP compares the dynamics of GFP-nebulin M163-170 to GFP-tropomodulin (Tmod), and desmin GFP-coil 1B to its mutants K190A and E245D in live cardiomyoctes. (A) The live image of a cardiomyocyte shows the recovery of GFP-Tmod fluorescence after photobleaching. The arrows point to the photobleached region of interest on a myofibril, at the pointed ends of the thin filaments, near the M-lines of the sarcomeres. (B, D) Fluorescence intensities were normalized to bleach level of zero, and the recoveries of fluorescence were plotted as a function of time after bleaching. The best-fit values of the fluorescence recoveries were a one-phase association equation from which half-lives (t1/2) were calculated. (C, E) Bar graphs comparing the averaged half-lives (t1/2) obtained for each protein. The calculated mean recovery half-life is as follows: for GFP, 0.9 ms; for the nebulin fragment, 2.3 ms; for Tmod, 1.3 ms; for coil 1B, 1.2 ms; for coil 1B K190A, 1.1 ms; and for coil 1B E245D, 1.7 ms. The data show that nebulin, tropomodulin, and mutant desmin GFP-coil 1B E245D are significantly slower relative to cells expressing GFP alone, whereas insignificant differences were measured for GFP-coil 1B or its K190A mutant. The number of cells analyzed per sample is indicated (n) under each column. Values show the means ± SD. Asterisks indicate significant differences relative to cells expressing GFP alone (*p < 0.05; **p < 0.01; Student's unpaired t test).
Comparison of the dynamics of thin filament proteins showed reduced exchange for the nebulin fragment as compared with tropomodulin and significantly slower mobility with respect to GFP control (Figure 9B; compare red and blue to black plot). No significant changes were found between the calculated half-lives of desmin GFP-coil 1B and mutant desmin K190A relative to GFP controls (Figure 9, D and E; compare green and purple to black plot). Remarkably, the mutant desmin GFP-coil 1B E245D reached a higher plateau than all the other proteins examined (Figure 9D; compare the yellow plot to the others). The increase in the plateau might indicate a slower turnover for this mutant in cardiomyocytes compared with GFP-coil 1B or mutant GFP-coil 1B K190A or, alternatively, that this mutation causes a binding protein partner to significantly retard its mobility and incorporation into the endogenous desmin network. Therefore it is likely that nebulin or members of the nebulin family of actin proteins retard this mutant desmin by promoting its binding to its immobile, dark fractions. Furthermore, these results support our previous in vitro finding that mutant desmin coil 1B E245D binds to nebulin with a approximately threefold higher binding capacity than nonmutated coil 1B (Conover et al., 2009).
DISCUSSION
The assembly of IF proteins in different tissues requires the recruitment of a complex set of cell type–specific associated proteins (Herrmann and Aebi, 2000). In muscle cells, one of these proteins is nebulin, a protein we propose directly interconnects the desmin IF network to the actin thin filaments. A detailed structural understanding of the association of desmin with nebulin is therefore essential for understanding the molecular processes derailed in desminopathy. To achieve this goal, this study determined how desmin assembly kinetics responded to nebulin binding and the dynamics of mutant desmin assembly in cardiomyocytes.
At what stage of desmin assembly does nebulin bind?
On the basis of what is known about IF assembly, we hypothesized that the docking efficiency of nebulin depends on the stage of desmin assembly. A widely accepted in vitro model for IF assembly predicts that this process proceeds as follows: tetramers, whose structure were elucidated recently in full detail, are the precursors of unit-length filaments (ULFs), as on average eight of them laterally associate as a result of the increase of ionic strength. For filament elongation, ULFs undergo longitudinal annealing and radial compaction, resulting in 10- to 12-nm filaments (Herrmann and Aebi, 1999, 2004; Herrmann et al., 1999, 2007). We characterized three desminopathy-associated mutants (S46F, E245D, and T453I), mapping within the known nebulin-binding peptides, reported in the literature to be linked to human desminopathy. Our data indicate that WT desmin and these mutants strongly interact with nebulin early and late during desmin assembly in vitro (Figure 5). The finding that nebulin binds desmin ULFs and short IFs is consistent with a report that showed that coil 1 of vimentin interacts with assembly-competent vimentin tetramers (Kirmse et al., 2007). In contrast to WT desmin, we report in this study that significantly less mutant desmin S46F and T453I was sedimented when bound to nebulin (Figure 5), suggesting that these mutations may trap nebulin, either rendering the molecule immobile or restricting its dynamics along the thin filaments in cells. Sedimentation profiles showed that mutant desmin E245D bound nebulin with delayed kinetics as compared with WT desmin (Figure 6B). Remarkably, we found evidence of delayed interaction kinetics of mutant desmin E245D to nebulin, supporting the idea that binding alterations with muscle proteins underlie a subset of desminopathy cases.
Nebulin M160–164 tangles desmin filaments
Our qualitative transmission electron microscopy studies suggested that the association of nebulin M160–164 appears to tangle desmin filaments and that its interaction with mutant desmin E245D binds distinctly increasing amounts of protein. Furthermore, our results are reminiscent of plakophilin interaction with IF proteins and mature IFs, as various types of IFs exhibited massive formation of thick bundles upon addition of plakophilin or recombinantly generated fragments of it (Hofmann et al., 2000). Besides nebulin, there are several binding partners that mediate the association of desmin IFs to muscle Z-discs. A number of mutations in desmin impeded their assembly in the form of heteropolymers with synemin, an IF protein expressed in neurons and myocytes (Chourbagi et al., 2011). Synemin depletion in cardiomyocytes decreased desmin targeting to the Z-discs, indicating that this protein also likely participates in the organization of desmin within these structures (Lund et al., 2012). Much has been learned regarding the 3D structure of a four-layer Z-disc by studying fish fin muscles, and future studies should address the fine 3D structural aspects of how the nebulin–desmin complex integrates within the Z-disc central muscle signaling and stretch sensor structures (Luther, 2000, 2009).
Delayed assembly of mutant desmin E245D with Z-discs in living cardiomyocytes
To assess the differences in recovery of fluorescence between WT and mutant GFP-tagged desmin and begin to define the network of protein interactions modulated by desmin in live cardiomyocytes, we used FRAP to rapidly bleach a small area in the myofibrils present in the cytosol of these cells (Figure 9). Of importance, we observed that most of the proteins tested by this approach exhibited near complete recovery (∼90–97%), suggesting that very few of the proteins within the bleached area stably bound to the immobile fractions. Surprisingly, we found a significant increase in the mean recovery half-life of mutant GFP-desmin E245D and GFP-nebulin M163-170. The slower mobility suggests a reduction in the number of molecules that are able to exchange. Such behavior is consistent with nebulin acting as a tethering factor for desmin. These changes were specific to this mutant, because the mean mobile fractions of GFP-coil 1B and its K190A mutant were nearly identical to that observed in cells expressing GFP alone (Figure 9, D and E). We propose a model by which desmin coil 1B provides a high local concentration of nonfilamentous particles that are required for the rapid intercalated exchange of desmin subunits in cardiomyocytes. Consistent with our model is the report that cells coexpressing vimentin-tagged fluorescent proteins (GFP and mCherry) are able to make alternating green and red filaments using an intercalated exchange mechanism in fibroblasts (Colakoglu and Brown, 2009).
Earlier FRAP studies suggested that different factors likely regulate vimentin and keratin IFs dynamic properties in the same epithelial cell (Yoon et al., 1998, 2001). Thus, although it is likely that nebulin, an integral thin filament–binding partner, stabilizes Z-disc–associated desmin, it is also possible that other mechanisms regulate desmin dynamics in a beating myocardium. In animal models, vimentin and keratin nonfilamentous particles and squiggles move with microtubules and actin-based motors and serve as signaling scaffolds (Yoon et al., 1998, 2001; Helfand et al., 2004; Windoffer et al., 2006). Recently it was reported that vimentin filaments disassemble into particles upon p21 kinase phosphorylation at Ser-38, directly modulating the formation of lamellipodia of migrating fibroblasts (Helfand et al., 2011). Similar to vimentin, it is possible that desmin is posttranslationally modified by phosphorylation, which might in turn modulate desmin assembly and disassembly within living cells. These exciting possibilities remain to be investigated in future studies.
Role of the desmin–nebulin system in desminopathy
A clinical study of patients affected by myofibrillar myopathies found that most likely only a subset of stress-responsive Z-disc proteins, rather than an unspecified generalized sarcomere myopathy, is responsible for the muscle dysfunction observed (Claeys et al., 2009). We predict that this set of proteins includes desmin, nebulin, and other Z-disc–associated desmin-binding proteins such as plectin 1d, syncoilin, and synemin (Bellin et al., 2001; Newey et al., 2001; Konieczny et al., 2008). Mutations in desmin cause desminopathy, a muscle disease that has devastating consequences for the normal function of striated and smooth muscle and can lead to heart failure and wheelchair dependence (Goldfarb et al., 2008).
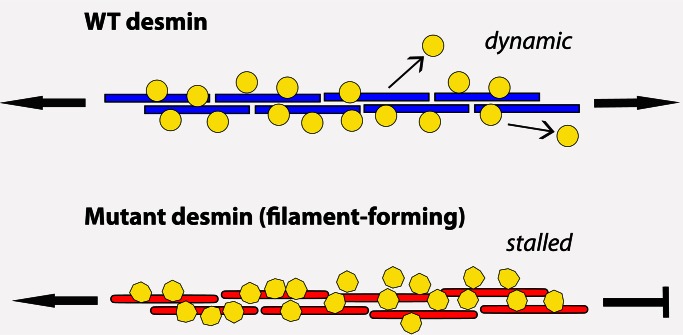
In this study, on the basis of our biochemical and live-cell imaging findings, we propose a working model by which mutant desmin, compared with WT desmin, binds more tightly with markedly increased stoichiometry to nebulin in cells (Figure 10). Because nebulin strongly binds to these mutants at the Z-disc, this interaction, in turn, slows the dynamics of desmin turnover and restricts nebulin mobility at the thin-filament barbed ends. Taken together, these effects lead to the progressive loss of actin filament stability because nebulin molecules are unable to slide freely on the thin filaments, severely limiting their normal interactions with tropomyosin and troponin complexes. Ultimately, we speculate that the desmin mutations interfere with optimal actin–myosin cross-bridge kinetics, leading to progressive heart failure. Studies are required to elucidate the exact temporal and spatial contributions of all Z-disc desmin IF partner complexes and the role of the small heat-shock protein and chaperone αB-crystallin to fully understand the underlying molecular effects of inherited filament-forming desmin mutations on desminopathy.
FIGURE 10:
A model for the association of nebulin binding to mutant desmin. In this model, mutant desmins competent for filament assembly bind strongly to nebulin with altered stoichiometry, slowing the dynamics of desmin turnover, retaining and retarding nebulin mobility at the ends of the actin thin filament near the Z-bands of the sarcomeres (DIFs are depicted as cylinders). We predict that nebulin changes its conformation upon binding to mutant desmin (note that nebulin is represented as yellow circles when bound to WT desmin and as hexagons when bound to mutant desmin). We propose that the altered association indirectly and ultimately leads to a progressive loss of actin filament stability, interfering with actin–myosin cross-bridge kinetics. For clarity, the WT desmin (blue) and mutant desmin rods (red) and the nebulin M160–164 modules (rather than the multimodular full-length molecule) are drawn.
MATERIALS AND METHODS
Cloning and mutagenesis
The sequences of the primers used in this study are listed in Table 2. To generate an N-terminal histidine (His)-tagged recombinant nebulin, encompassing modules 160–164, the insert was amplified by PCR using Phusion High-Fidelity Polymerase (Finnzymes Thermo Scientific, Vantaa, Finland) as an StuI–XhoI insert from cDNA (4368814; Applied Biosystems, Foster City, CA), produced from human skeletal RNA (540030; Stratagene, Santa Clara, CA), and cloned into pET15b (Novagen, Gibbstown, NJ). Mutant desmin S46F/Y and T453I were cloned and purified as described previously (Sharma et al., 2009; Bar et al., 2010). The E245D mutation was introduced into human pDS5-desmin (Bar et al., 2005a) using a PCR-based QuikChange site-directed mutagenesis kit (Stratagene). The PCR product was digested with DpnI and transformed into XL1-blue Escherichia coli. All constructs were verified by sequencing.
TABLE 1:
Binding parameters obtained in Figure 5.
| Protein | Binding affinity Kd (nM) | Binding capacity Bmax |
|---|---|---|
| WT desmin | 18 ± 7 (7.4), n = 8 14 ± 4 (8.4), n = 7 | 73 ± 5 (7.4) 90 ± 4 (8.4) |
| Desmin S46Y | 8 ± 2 (7.4), n = 3 19 ± 4 (8.4), n = 2 | 88 ± 4 (7.4) 100 ± 5 (8.4) |
| Desmin E245D | 7 ± 2 (7.4), n = 3 8 ± 3 (8.4), n = 2 | 104 ± 5 (7.4) 90 ± 5 (8.4) |
| Desmin T453I | 11 ± 4 (7.4), n = 3 11 ± 3 (8.4), n = 2 | 79 ± 4 (7.4) 92 ± 4 (8.4) |
Boldface indicates significant changes of mutant desmin compared with WT desmin. Parentheses show the pH at which desmin was dialyzed. n, number of experiments averaged per sample.
ELISAs
Triplicate wells in ELISA-coated 96-well plates (BD Biosciences, Franklin Lakes, NJ) were coated with 200 nM His-nebulin M160–164 in 0.1 M carbonate buffer, pH 9.6, overnight at 4°C. Wells were washed and blocked for 1 h with binding buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4, 80 mM KCl, 2 mM MgCl2, 0.05% Tween-20, 0.2% BSA). WT or mutant desmin was added to each in solution dissolved in binding buffer at concentrations ranging from 1 to 300 nM for 1 h at room temperature. After five washes with binding buffer, the plate was incubated with mouse monoclonal anti-desmin (D1033; Sigma-Aldrich, St. Louis, MO) for 1 h. After washes, anti-mouse horseradish peroxidase–conjugated antibody (Bio-Rad, Hercules, CA) was added for 1 h at room temperature. Dissolved substrate (2,2-azino-bis 3-ethylbenzothiazoline-6-sulfonic acid diammonium salt; A9941; Sigma-Aldrich) in 0.05 M phosphate–citrate buffer (pH 5, P4809; Sigma-Aldrich) was incubated for 30 min at 37°C. The binding interaction was determined by a colorimetric reaction at A405 on a BMG plate reader (BMG Labtech, Ortenberg, Germany). The Bmax and Kd values were determined using Prism, version 5.0 (GraphPad Software, La Jolla, CA), using a one-site binding hyperbola nonlinear regression fit.
Desmin purification from inclusion bodies and ion exchange chromatography
For the expression and purification of desmin protein, desmin cDNA was subcloned into pDS5 vector to make full-length, headless, rod, and tailless desmin (Sharma et al., 2009). Each clone was expressed in BL-21 pLYs DE3 or JM109 chemically competent E. coli (Stratagene). Purification using an inclusion body protocol and ion exchange chromatography was carried out as described (Herrmann et al., 2004). After resuspension of cells in a Dounce homogenizer (Sartorius, Bohemia, NY), 200 μl of 10% NP-40, 0.2% Triton X-100, and 10 mM MgCl2 were added. Subsequently, proteins were purified using two-step ion exchange purification: DEAE-Sepharose followed by CM Sepharose Fast Flow (GE Healthcare, Piscataway, NJ). Each column was equilibrated in 10 mM Tris (pH 7.5) and 1 mM dithiothreitol (DTT) with 8 M urea. A peristaltic pump (Bio-Rad) was set to a flow rate of 1.17 ml/min to elute proteins with a 0–0.3 M KCl salt gradient. Each fraction was analyzed by SDS–PAGE, and pooled samples were applied to a CM Sepharose column. Because of the presence of salt in the loading sample and the relatively high pH used, pH 7.5, most proteins present in the DEAE-column eluate did not bind to the CM Sepharose Fast Flow matrix. For the binding of headless desmin to CM Sepharose Fast Flow, the protein eluted from the DEAE column was dialyzed extensively against 30 mM sodium formate, 1 mM EDTA, and 1 mM ethylene glycol tetraacetic acid (EGTA), pH 4.0 (Herrmann et al., 1996).
Protein renaturation and cosedimentation assays
WT and mutant desmin were dialyzed in four sequential steps. Briefly, protein stocks were diluted to 0.5 g/l and placed into 50,000 MWCO dialysis tubing (Spectrum Laboratories, Rancho Dominguez, CA). Dialysis tubing was placed into 8 M urea in dialysis buffer (5 mM Tris, 1 mM EDTA, 0.1 mM EGTA, 1 mM DTT), and the pH was adjusted to 8.4 or 7.4 as indicated. Proteins were serially dialyzed out from 8 to 4 to 2 to 1 M urea into dialysis buffer for 20 min each step at room temperature on a plate stirrer. Dialysis tubing was placed into a plastic beaker with new dialysis buffer for overnight dialysis at 4°C. For densitometry measurements, images were saved as 8-bit TIFF files, and their background was subtracted with a rolling ball of radius 50 pixels using ImageJ software (National Institutes of Health, Bethesda, MD). A rectangular tool was used to draw equal band areas each band for analysis. Gaussian density plots for each band were quantified. The relative percentage for each fraction was normalized relative to the sum of all the gray values measured in the supernatant, sucrose, and pellet fractions. Values plotted show the means ± SD from data obtained in three independent experiments.
Isolation of cardiomyocyte primary cultures
Embryonated chicken eggs were purchased from Texas A&M Poultry Department, and beating hearts from 6-d-old embryos were dissected and collected in Hank's saline solution. Embryonic hearts were digested with trypsin, followed by multiple washes before plating at a density of 5 × 105 cells/ml in 35-mm cell culture dishes on #1.5 coverslips (0.17 mm) in MEM with Earle's salts (SH30244.01; Hyclone, Logan, UT) and supplemented with 5% fetal bovine serum, 2 mM l-glutamine, 1% penicillin, and streptomycin mix. The next day, to minimize fibroblast proliferation, cardiomyocytes were fed with media lacking l-glutamine. After 24 h of incubation at 37°C and 5% CO2, cells were transfected with EndoFree DNA diluted to 0.5 μg/μl using the lipid-based Effectene transfection reagent (Qiagen, Valencia, CA).
Immunofluorescence and immunohistochemistry analyses
After 4 d in culture, myocytes were washed in phosphate-buffered saline (PBS), relaxed (150 mM KCl, 5 mM MgCl2, 10 mM 3-(N-morpholino)propanesulfonic acid, 1 mM EGTA, 4 mM ATP, pH 7.4) for 15 min, fixed in 3% paraformaldehyde for 30 min at room temperature, and rinsed three times with 1× PBS. Before staining, the cells were permeabilized in 0.2% Triton X-100 in PBS for 5 min and blocked with 2% BSA and 1% normal goat serum in 1× PBS for 30 min at room temperature. Canine and biopsy tissues were obtained from the pathology laboratory at Texas A&M College of Veterinary Medicine and Biomedical Sciences. Rat psoas tissue was a kind gift of G. Stoica (Texas A&M University). Paraffin-embedded sections (∼5 μm) were deparaffinized and heated in 10 mM citrate buffer (pH 6.0) for 10 min, blocked with 5% normal serum in Tris-saline buffered saline (100 mM Tris, 150 mM NaCl, pH 7.4), and incubated for 1 h with the indicated primary antibodies. After washing, sections were incubated with secondary antibodies and imaged as described. Tissue samples were mounted in Vectashield Mounting Media (Hard-set H1400; Vector Laboratories, Burlingame, CA).
Both chick cardiomyocytes and canine tissues were stained with an affinity-purified custom peptide rabbit antibody against human nebulin peptide (Ac-KEGSHGLSMLGRPDIEMAKKAC-amide). This antibody was immunodepleted with a nebulette peptide (C-Ahx-YSAELDRPDIKMAT-amide) to increase cardiac nebulin specificity. Incubation was followed by secondary rabbit immunoglobulin G–conjugated 0.2 μg/ml Alexa 594 (Life Technologies, Carlsbad, CA) and 0.05 μg/ml anti–α-actinin (A7811; Sigma-Aldrich), or anti–human desmin clone D33 (M0760; Dako, Carpinteria, CA), sarcomeric actin (Clone Alpha-Sr-1; Dako) antibodies raised in mice, followed by secondary 0.1 μg/ml goat anti–mouse Alexa 488 (Life Technologies). Coverslips were stained with nuclear stain 4′,6-diamidino-2-phenylindole (1 ng/ml) in distilled H2O at room temperature for 10 min as indicated. Coverslips were mounted in Aqua Poly/Mount (Polysciences, Warrington, PA 18606) for wide-field or confocal analyses. Cells were imaged using an Olympus IX-70 (Olympus, Tokyo, Japan) for wide-field microscopy or a Zeiss LSM 510 (Carl Zeiss, Jena, Germany) for confocal microscopy. Images were pseudocolored red and green for presentation with Photoshop CS6 (Adobe, San Jose, CA).
Analytical ultracentrifugation
Analyses were performed between 20,000 and 40,000 rpm at 20°C in a 0.2 mM sodium phosphate (pH 7.5) buffer system. Desmin was dialyzed out of 8 M urea by serial dialysis into sodium phosphate buffer. Nebulin M160–164 was added at the indicated molar ratios, based on concentrations obtained by Bradford assays. The sedimentation coefficient (s value), reported as svedberg (S) units, is a function of the individual or complex protein mass. The change of sample concentration as a function of radius and time was measured using an Optima XL-A analytical ultracentrifuge (Beckman Coulter, Brea, CA), using DCDT+ software to calculate a differential apparent sedimentation coefficient g(s*) in sedimentation velocity assays. The g(s*) distribution (y-axis) was determined by extrapolating the distribution back to time zero, accounting for the radial dilution effects (Mucke et al., 2004). Thus the total area under the g(s*) curve represents the loading concentration of any one species in a mixture.
FRAP
Cardiomyocytes were transfected with varying GFP constructs using Effectene transfection reagent. After 16 h of incubation, dishes (155380; Lab-Tek, Nalge Nunc International, Rochester, NY) were washed and subsequently fed every 48 h. After 5 d, dishes were imaged and bleached to determine recovery using an Olympus IX81 FV1000 laser scanning confocal microscope equipped with a 60×/0.7 air (dry) objective. The fluorescence of each GFP construct was detected using a 488-nm argon laser with an area of ∼1.5 × 1.5 μm. The intensity of the laser ranged from 3 to 7%, which was activated after up to 20 scanned frames to photobleach an area above and away from the nucleus of the cell. Image series of up to 250 frames at 260-ms intervals were collected with a photomultiplier detector. Image size was 164 × 172 pixels (unidirectional scanning, 0.165 μm/pixel), and images were processed using the Olympus software (FV-10 viewer) by selecting three regions of interest: a bleached region, an unbleached region, and a region outside the cell to account for background. The mean intensities of each of these three regions were calculated, and the average background was subtracted from the average photobleached area. To normalize the data, the intensity was subtracted from the intensity right after bleaching (T0), divided by the intensity at plateau (TR), and then subtracted from the average photobleached area. The recovery half-time from the mobile fractions was calculated using F(t) = a(1 − e−t/τ) + c and t1/2 = ln(2) τ. Both equations were plugged into the solver tool (Excel; Microsoft, Redmond, WA) using the least-squares method by allowing a, c, and τ to vary in the first equation.
Electron microscopy
Protein samples were mounted for transmission electron microscopy imaging. Samples were diluted to 0.1 g/l in 5 mM Tris-HCl (pH 7.4), buffered, and mounted on carbon film on 200 square mesh copper grids (CF200-Cu-50; Electron Microscopy Sciences, Hatfield, PA) and fixed for 10 s with in 5 mM Tris, 20 mM NaCl, and 0.1% glutaraldehyde. The fixed protein sample was removed from the grid using blotting paper. Grids were rinsed with 0.1 μm of filtered distilled H2O for 5 s, blotted dry, negatively stained with 0.2% uranyl acetate for 20 s, and blotted dry. Images were obtained using a Morgagni FEI transmission electron microscope (FEI, Hillsboro, OR).
Supplementary Material
Acknowledgments
We are grateful to C. C. Gregorio for providing cDNA constructs for this study, N. Mücke for help with analytical ultracentrifugation data analysis, C. M. Wilcox for help with drawings, and K. E. Timberlake for technical assistance. We thank H. Payne, R. Mouneimne, and S. Vitha for help in electron and confocal microscopies. We thank D. Conover and C. Wilcox for editorial help. The American Heart Association funded this research under Grant 2110057 to G.M.C.
Abbreviations used:
- DIFs
desmin intermediate filaments
- FRAP
fluorescence recovery after photobleaching
- IF
intermediate filament
- ULF
unit-length filament
- WT
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-11-0840) on April 24, 2013.
*These authors contributed equally to this study.
Present addresses: †Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX 77843;
‡Department of Otolaryngology, University Hospital Heidelberg, D-69120 Heidelberg, Germany.
REFERENCES
- Albertyn C, Maguire C, Murphy RP. PONM04 Myofibrillar myopathy caused by a mutation in the desmin gene: expanding the phenotype. J Neurol Neurosurg Psychiatry. 2010;81:e61. [Google Scholar]
- Bang ML, Caremani M, Brunello E, Littlefield R, Lieber RL, Chen J, Lombardi V, Linari M. Nebulin plays a direct role in promoting strong actin-myosin interactions. FASEB J. 2009;23:4117–4125. doi: 10.1096/fj.09-137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bang ML, Gregorio C, Labeit S. Molecular dissection of the interaction of desmin with the C-terminal region of nebulin. J Struct Biol. 2002;137:119–127. doi: 10.1006/jsbi.2002.4457. [DOI] [PubMed] [Google Scholar]
- Bang ML, Li X, Littlefield R, Bremner S, Thor A, Knowlton KU, Lieber RL, Chen J. Nebulin-deficient mice exhibit shorter thin filament lengths and reduced contractile function in skeletal muscle. J Cell Biol. 2006;173:905–916. doi: 10.1083/jcb.200603119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar H, Fischer D, Goudeau B, Kley RA, Clemen CS, Vicart P, Herrmann H, Vorgerd M, Schroder R. Pathogenic effects of a novel heterozygous R350P desmin mutation on the assembly of desmin intermediate filaments in vivo and in vitro. Hum Mol Genet. 2005a;14:1251–1260. doi: 10.1093/hmg/ddi136. [DOI] [PubMed] [Google Scholar]
- Bar H, Mucke N, Kostareva A, Sjoberg G, Aebi U, Herrmann H. Severe muscle disease-causing desmin mutations interfere with in vitro filament assembly at distinct stages. Proc Natl Acad Sci USA. 2005b;102:15099–15104. doi: 10.1073/pnas.0504568102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar H, Mucke N, Ringler P, Muller SA, Kreplak L, Katus HA, Aebi U, Herrmann H. Impact of disease mutations on the desmin filament assembly process. J Mol Biol. 2006;360:1031–1042. doi: 10.1016/j.jmb.2006.05.068. [DOI] [PubMed] [Google Scholar]
- Bar H, Schopferer M, Sharma S, Hochstein B, Mucke N, Herrmann H, Willenbacher N. Mutations in desmin's carboxy-terminal “tail” domain severely modify filament and network mechanics. J Mol Biol. 2010;397:1188–1198. doi: 10.1016/j.jmb.2010.02.024. [DOI] [PubMed] [Google Scholar]
- Bellin RM, Huiatt TW, Critchley DR, Robson RM. Synemin may function to directly link muscle cell intermediate filaments to both myofibrillar Z-lines and costameres. J Biol Chem. 2001;276:32330–32337. doi: 10.1074/jbc.M104005200. [DOI] [PubMed] [Google Scholar]
- Chandra M, Mamidi R, Ford S, Hidalgo C, Witt C, Ottenheijm C, Labeit S, Granzier H. Nebulin alters cross-bridge cycling kinetics and increases thin filament activation: a novel mechanism for increasing tension and reducing tension cost. J Biol Chem. 2009;284:30889–30896. doi: 10.1074/jbc.M109.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyatina AA, Nicolet S, Aebi U, Herrmann H, Strelkov SV. Atomic structure of the vimentin central alpha-helical domain and its implications for intermediate filament assembly. Proc Natl Acad Sci USA. 2012;109:13620–13625. doi: 10.1073/pnas.1206836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourbagi O, Bruston F, Carinci M, Xue Z, Vicart P, Paulin D, Agbulut O. Desmin mutations in the terminal consensus motif prevent synemin-desmin heteropolymer filament assembly. Exp Cell Res. 2011;317:886–897. doi: 10.1016/j.yexcr.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Claeys KG, et al. Differential involvement of sarcomeric proteins in myofibrillar myopathies: a morphological and immunohistochemical study. Acta Neuropathol. 2009;117:293–307. doi: 10.1007/s00401-008-0479-7. [DOI] [PubMed] [Google Scholar]
- Clemen CS, Herrmann H, Strelkov SV, Schroder R. Desminopathies: pathology and mechanisms. Acta Neuropathol. 2013;125:47–75. doi: 10.1007/s00401-012-1057-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Zhai B, Gygi SP, Goldberg AL. Ubiquitylation by Trim32 causes coupled loss of desmin, Z-bands, and thin filaments in muscle atrophy. J Cell Biol. 2012;198:575–589. doi: 10.1083/jcb.201110067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colakoglu G, Brown A. Intermediate filaments exchange subunits along their length and elongate by end-to-end annealing. J Cell Biol. 2009;185:769–777. doi: 10.1083/jcb.200809166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover GM, Gregorio CC. The desmin coil 1B mutation K190A impairs nebulin Z-disc assembly and destabilizes actin thin filaments. J Cell Sci. 2011;124:3464–3476. doi: 10.1242/jcs.087080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conover GM, Henderson SN, Gregorio CC. A myopathy-linked desmin mutation perturbs striated muscle actin filament architecture. Mol Biol Cell. 2009;20:834–845. doi: 10.1091/mbc.E08-07-0753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalakas MC, et al. Progressive skeletal myopathy, a phenotypic variant of desmin myopathy associated with desmin mutations. Neuromusc Disord. 2003;13:252–258. doi: 10.1016/s0960-8966(02)00271-7. [DOI] [PubMed] [Google Scholar]
- Fock U, Hinssen H. Nebulin is a thin filament protein of the cardiac muscle of the agnathans. J Muscle Res Cell Motil. 2002;23:205–213. doi: 10.1023/a:1020909902462. [DOI] [PubMed] [Google Scholar]
- Gokhin DS, Bang ML, Zhang J, Chen J, Lieber RL. Reduced thin filament length in nebulin-knockout skeletal muscle alters isometric contractile properties. Am J Physiol Cell Physiol. 2009;296:C1123–C1132. doi: 10.1152/ajpcell.00503.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb LG, Olive M, Vicart P, Goebel HH. Intermediate filament diseases: desminopathy. Adv Exp Med Biol. 2008;642:131–164. doi: 10.1007/978-0-387-84847-1_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman RD, Grin B, Mendez MG, Kuczmarski ER. Intermediate filaments: versatile building blocks of cell structure. Curr Opin Cell Biol. 2008;20:28–34. doi: 10.1016/j.ceb.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger BL, Lazarides E. The existence of an insoluble Z disc scaffold in chicken skeletal muscle. Cell. 1978;15:1253–1268. doi: 10.1016/0092-8674(78)90051-x. [DOI] [PubMed] [Google Scholar]
- Granger BL, Lazarides E. Desmin and vimentin coexist at the periphery of the myofibril Z disc. Cell. 1979;18:1053–1063. doi: 10.1016/0092-8674(79)90218-6. [DOI] [PubMed] [Google Scholar]
- Harding SE, Rowe AJ. Insight into protein-protein interactions from analytical ultracentrifugation. Biochem Soc Trans. 2010;38:901–907. doi: 10.1042/BST0380901. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Chang L, Goldman RD. The dynamic and motile properties of intermediate filaments. Annu Rev Cell Dev Biol. 2003;19:445–467. doi: 10.1146/annurev.cellbio.19.111401.092306. [DOI] [PubMed] [Google Scholar]
- Helfand BT, Chang L, Goldman RD. Intermediate filaments are dynamic and motile elements of cellular architecture. J Cell Sci. 2004;117:133–141. doi: 10.1242/jcs.00936. [DOI] [PubMed] [Google Scholar]
- Helfand BT, et al. Vimentin organization modulates the formation of lamellipodia. Mol Biol Cell. 2011;22:1274–1289. doi: 10.1091/mbc.E10-08-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filament assembly: temperature sensitivity and polymorphism. Cell Mol Life Sci. 1999;55:1416–1431. doi: 10.1007/s000180050382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments and their associates: multi-talented structural elements specifying cytoarchitecture and cytodynamics. Curr Opin Cell Biol. 2000;12:79–90. doi: 10.1016/s0955-0674(99)00060-5. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Aebi U. Intermediate filaments: molecular structure, assembly mechanism, and integration into functionally distinct intracellular Scaffolds. Annu Rev Biochem. 2004;73:749–789. doi: 10.1146/annurev.biochem.73.011303.073823. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Bar H, Kreplak L, Strelkov SV, Aebi U. Intermediate filaments: from cell architecture to nanomechanics. Nat Rev Mol Cell Biol. 2007;8:562–573. doi: 10.1038/nrm2197. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, Ku NO, Aebi U. Characterization of distinct early assembly units of different intermediate filament proteins. J Mol Biol. 1999;286:1403–1420. doi: 10.1006/jmbi.1999.2528. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Haner M, Brettel M, Muller SA, Goldie KN, Fedtke B, Lustig A, Franke WW, Aebi U. Structure and assembly properties of the intermediate filament protein vimentin: the role of its head, rod and tail domains. J Mol Biol. 1996;264:933–953. doi: 10.1006/jmbi.1996.0688. [DOI] [PubMed] [Google Scholar]
- Herrmann H, Kreplak L, Aebi U. Isolation, characterization, and in vitro assembly of intermediate filaments. Methods Cell Biol. 2004;78:3–24. doi: 10.1016/s0091-679x(04)78001-2. [DOI] [PubMed] [Google Scholar]
- Hofmann I, Mertens C, Brettel M, Nimmrich V, Schnolzer M, Herrmann H. Interaction of plakophilins with desmoplakin and intermediate filament proteins: an in vitro analysis. J Cell Sci. 2000;113:2471–2483. doi: 10.1242/jcs.113.13.2471. [DOI] [PubMed] [Google Scholar]
- Huiatt TW, Robson RM, Arakawa N, Stromer MH. Desmin from avian smooth muscle. Purification and partial characterization. J Biol Chem. 1980;255:6981–6989. [PubMed] [Google Scholar]
- Karabinos A, Schmidt H, Harborth J, Schnabel R, Weber K. Essential roles for four cytoplasmic intermediate filament proteins in Caenorhabditis elegans development. Proc Natl Acad Sci USA. 2001;98:7863–7868. doi: 10.1073/pnas.121169998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierski ST, Antin PB, Witt CC, Huebner N, McElhinny AS, Labeit S, Gregorio CC. The complete mouse nebulin gene sequence and the identification of cardiac nebulin. J Mol Biol. 2003;328:835–846. doi: 10.1016/s0022-2836(03)00348-6. [DOI] [PubMed] [Google Scholar]
- Kirmse R, Portet S, Mucke N, Aebi U, Herrmann H, Langowski J. A quantitative kinetic model for the in vitro assembly of intermediate filaments from tetrameric vimentin. J Biol Chem. 2007;282:18563–18572. doi: 10.1074/jbc.M701063200. [DOI] [PubMed] [Google Scholar]
- Konieczny P, Fuchs P, Reipert S, Kunz WS, Zeold A, Fischer I, Paulin D, Schroder R, Wiche G. Myofiber integrity depends on desmin network targeting to Z-disks and costameres via distinct plectin isoforms. J Cell Biol. 2008;181:667–681. doi: 10.1083/jcb.200711058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger M, Wright J, Wang K. Nebulin as a length regulator of thin filaments of vertebrate skeletal muscles: correlation of thin filament length, nebulin size, and epitope profile. J Cell Biol. 1991;115:97–107. doi: 10.1083/jcb.115.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labeit S, Kolmerer B. The complete primary structure of human nebulin and its correlation to muscle structure. J Mol Biol. 1995;248:308–315. doi: 10.1016/s0022-2836(95)80052-2. [DOI] [PubMed] [Google Scholar]
- Lazarides E. Comparison of the structure, distribution and possible function on desmin (100 A) filaments in various types of muscle and nonmuscle cells. Birth Defects Orig Artic Ser. 1978;14:41–63. [PubMed] [Google Scholar]
- Littlefield RS, Fowler VM. Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler. Semin Cell Dev Biol. 2008;19:511–519. doi: 10.1016/j.semcdb.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund LM, Kerr JP, Lupinetti J, Zhang Y, Russell MA, Bloch RJ, Bond M. Synemin isoforms differentially organize cell junctions and desmin filaments in neonatal cardiomyocytes. FASEB J. 2012;26:137–148. doi: 10.1096/fj.10-179408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther PK. Three-dimensional structure of a vertebrate muscle Z-band: implications for titin and alpha-actinin binding. J Struct Biol. 2000;129:1–16. doi: 10.1006/jsbi.1999.4207. [DOI] [PubMed] [Google Scholar]
- Luther PK. The vertebrate muscle Z-disc: sarcomere anchor for structure and signalling. J Muscle Res Cell Motil. 2009;30:171–185. doi: 10.1007/s10974-009-9189-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElhinny AS, Kolmerer B, Fowler VM, Labeit S, Gregorio CC. The N-terminal end of nebulin interacts with tropomodulin at the pointed ends of the thin filaments. J Biol Chem. 2001;276:583–592. doi: 10.1074/jbc.M005693200. [DOI] [PubMed] [Google Scholar]
- McLendon PM, Robbins J. Desmin-related cardiomyopathy: an unfolding story. Am J Physiol Heart Circ Physiol. 2011;301:H1220–H1228. doi: 10.1152/ajpheart.00601.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencarelli C, Ciolfi S, Caroti D, Lupetti P, Dallai R. Isomin: a novel cytoplasmic intermediate filament protein from an arthropod species. BMC Biol. 2011;9:17. doi: 10.1186/1741-7007-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez MG, Kojima S, Goldman RD. Vimentin induces changes in cell shape, motility, and adhesion during the epithelial to mesenchymal transition. FASEB J. 2010;24:1838–1851. doi: 10.1096/fj.09-151639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Trombitas K, Kolmerer B, Kostin S, Schaper J, Pelin K, Granzier H, Labeit S. Characterization of nebulette and nebulin and emerging concepts of their roles for vertebrate Z-discs. J Mol Biol. 1998;282:111–123. doi: 10.1006/jmbi.1998.1999. [DOI] [PubMed] [Google Scholar]
- Moncman CL, Wang K. Targeted disruption of nebulette protein expression alters cardiac myofibril assembly and function. Exp Cell Res. 2002;273:204–218. doi: 10.1006/excr.2001.5423. [DOI] [PubMed] [Google Scholar]
- Mucke N, Kreplak L, Kirmse R, Wedig T, Herrmann H, Aebi U, Langowski J. Assessing the flexibility of intermediate filaments by atomic force microscopy. J Mol Biol. 2004;335:1241–1250. doi: 10.1016/j.jmb.2003.11.038. [DOI] [PubMed] [Google Scholar]
- Nekrasova OE, Mendez MG, Chernoivanenko IS, Tyurin-Kuzmin PA, Kuczmarski ER, Gelfand VI, Goldman RD, Minin AA. Vimentin intermediate filaments modulate the motility of mitochondria. Mol Biol Cell. 2011;22:2282–2289. doi: 10.1091/mbc.E10-09-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newey SE, Howman EV, Ponting CP, Benson MA, Nawrotzki R, Loh NY, Davies KE, Blake DJ. Syncoilin, a novel member of the intermediate filament superfamily that interacts with alpha-dystrobrevin in skeletal muscle. J Biol Chem. 2001;276:6645–6655. doi: 10.1074/jbc.M008305200. [DOI] [PubMed] [Google Scholar]
- Nicolet S, Herrmann H, Aebi U, Strelkov SV. Atomic structure of vimentin coil 2. J Struct Biol. 2010;170:369–376. doi: 10.1016/j.jsb.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Ohtakara K, Inada H, Goto H, Taki W, Manser E, Lim L, Izawa I, Inagaki M. p21-activated kinase PAK phosphorylates desmin at sites different from those for Rho-associated kinase. Biochem Biophys Res Commun. 2000;272:712–716. doi: 10.1006/bbrc.2000.2854. [DOI] [PubMed] [Google Scholar]
- Omary MB, Ku NO, Tao GZ, Toivola DM, Liao J. “Heads and tails” of intermediate filament phosphorylation: multiple sites and functional insights. Trends Biochem Sci. 2006;31:383–394. doi: 10.1016/j.tibs.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Pappas CT, Bhattacharya N, Cooper JA, Gregorio CC. Nebulin interacts with CapZ and regulates thin filament architecture within the Z-disc. Mol Biol Cell. 2008;19:1837–1847. doi: 10.1091/mbc.E07-07-0690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas CT, Krieg PA, Gregorio CC. Nebulin regulates actin filament lengths by a stabilization mechanism. J Cell Biol. 2010;189:859–870. doi: 10.1083/jcb.201001043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner W, Franke WW, Schmid E, Geisler N, Weber K, Mandelkow E. Reconstitution of intermediate-sized filaments from denatured monomeric vimentin. J Mol Biol. 1981;149:285–306. doi: 10.1016/0022-2836(81)90303-x. [DOI] [PubMed] [Google Scholar]
- Schoumacher M, Goldman RD, Louvard D, Vignjevic DM. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J Cell Biol. 2010;189:541–556. doi: 10.1083/jcb.200909113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Mucke N, Katus HA, Herrmann H, Bar H. Disease mutations in the “head” domain of the extra-sarcomeric protein desmin distinctly alter its assembly and network-forming properties. J Mol Med. 2009;87:1207–1219. doi: 10.1007/s00109-009-0521-9. [DOI] [PubMed] [Google Scholar]
- Shinde A, Nakano S, Sugawara M, Toyoshima I, Ito H, Tanaka K, Kusaka H. Expression of caveolar components in primary desminopathy. Neuromusc Disord. 2008;18:215–219. doi: 10.1016/j.nmd.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Takano K, Watanabe-Takano H, Suetsugu S, Kurita S, Tsujita K, Kimura S, Karatsu T, Takenawa T, Endo T. Nebulin and N-WASP cooperate to cause IGF-1-induced sarcomeric actin filament formation. Science. 2010;330:1536–1540. doi: 10.1126/science.1197767. [DOI] [PubMed] [Google Scholar]
- Tonino P, Pappas CT, Hudson BD, Labeit S, Gregorio CC, Granzier H. Reduced myofibrillar connectivity and increased Z-disk width in nebulin-deficient skeletal muscle. J Cell Sci. 2010;123:384–391. doi: 10.1242/jcs.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Spaendonck-Zwarts K, van Hessem L, Jongbloed JD, de Walle HE, Capetanaki Y, van der Kooi AJ, van Langen IM, van den Berg MP, van Tintelen JP. Desmin-related myopathy: a review and meta-analysis. Clin Genet. 2010;80:354–366. doi: 10.1111/j.1399-0004.2010.01512.x. [DOI] [PubMed] [Google Scholar]
- Wang K, Knipfer M, Huang QQ, van Heerden A, Hsu LC, Gutierrez G, Quian XL, Stedman H. Human skeletal muscle nebulin sequence encodes a blueprint for thin filament architecture. Sequence motifs and affinity profiles of tandem repeats and terminal SH3. J Biol Chem. 1996;271:4304–4314. doi: 10.1074/jbc.271.8.4304. [DOI] [PubMed] [Google Scholar]
- Wang K, Ramirez-Mitchell R. A network of transverse and longitudinal intermediate filaments is associated with sarcomeres of adult vertebrate skeletal muscle. J Cell Biol. 1983;96:562–570. doi: 10.1083/jcb.96.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Williamson CL. Identification of an N2 line protein of striated muscle. Proc Natl Acad Sci USA. 1980;77:3254–3258. doi: 10.1073/pnas.77.6.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windoffer R, Kolsch A, Woll S, Leube RE. Focal adhesions are hotspots for keratin filament precursor formation. J Cell Biol. 2006;173:341–348. doi: 10.1083/jcb.200511124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KH, Yoon M, Moir RD, Khuon S, Flitney FW, Goldman RD. Insights into the dynamic properties of keratin intermediate filaments in living epithelial cells. J Cell Biol. 2001;153:503–516. doi: 10.1083/jcb.153.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M, Moir RD, Prahlad V, Goldman RD. Motile properties of vimentin intermediate filament networks in living cells. J Cell Biol. 1998;143:147–157. doi: 10.1083/jcb.143.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Wilson KL, Dahl KN. Beyond lamins other structural components of the nucleoskeleton. Methods Cell Biol. 2010;98:97–119. doi: 10.1016/S0091-679X(10)98005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.