Two microtubule cross-linkers of the major MAP65/PRC1/Ase1 family are found to modify the mechanical properties of dynamic microtubules (e.g., decrease the flexural rigidity of microtubules). This finding points to a role for these proteins in the formation of specific microtubule arrays in eukaryotic cells.
Abstract
Microtubules (MTs) are dynamic cytoskeletal elements involved in numerous cellular processes. Although they are highly rigid polymers with a persistence length of 1–8 mm, they may exhibit a curved shape at a scale of few micrometers within cells, depending on their biological functions. However, how MT flexural rigidity in cells is regulated remains poorly understood. Here we ask whether MT-associated proteins (MAPs) could locally control the mechanical properties of MTs. We show that two major cross-linkers of the conserved MAP65/PRC1/Ase1 family drastically decrease MT rigidity. Their MT-binding domain mediates this effect. Remarkably, the softening effect of MAP65 observed on single MTs is maintained when MTs are cross-linked. By reconstituting physical collisions between growing MTs/MT bundles, we further show that the decrease in MT stiffness induced by MAP65 proteins is responsible for the sharp bending deformations observed in cells when they coalign at a steep angle to create bundles. Taken together, these data provide new insights into how MAP65, by modifying MT mechanical properties, may regulate the formation of complex MT arrays.
INTRODUCTION
Microtubules (MTs) are major components of the cytoskeleton involved in fundamental cellular processes. They are rigid, polar, and dynamical elements that polymerize and depolymerize at both ends. The stochastic switching between growing and shortening phases is termed dynamic instability (Mitchison and Kirschner, 1984). In living cells, they form arrays that either radiate from structured MT-organizing centers such as the centrosome or are self-organized into linear bundles that assume different configurations, depending on the cell type (Bartolini and Gundersen, 2006). Within these arrays, MTs can be straight or highly curved (deformed MTs), presumably depending on the functions in which they are engaged (Rusan and Wadsworth, 2005; Brangwynne et al., 2006; Wasteneys and Ambrose, 2009). An example of MT deformations in cells includes the formation of MT bundles. Indeed, to create bundles, physical collisions between MTs at angles up to 40° might result in bending deformations so that they coalign (Shaw et al., 2003; Dixit and Cyr, 2004). Furthermore, beside individual MTs, small groups of MTs such as bundles also assume deformations. In plant cells that display box-like geometry, with sharp edges marking the boundaries between adjacent faces, MT bundles wind around the cell cortex and are thus deformed to adapt their organization to these physical constraints (Wasteneys and Ambrose, 2009; Ambrose et al., 2011). In fission yeast, MT bundle deformations have also been observed in correlation with nucleus positioning (Tran et al., 2001). These observations suggest that in living cells, the mechanical behavior of MTs/MT bundles is variable and must be locally regulated, depending of the functions in which they are engaged. Because MTs are highly rigid structures, with a persistence length (Lp) of 1–8 mm for naked single MTs (Hawkins et al., 2010), one essential question to understand is how they can be deformed at a scale of a few micrometers. In vivo, MT Lp was estimated to be ∼30 μm in animal culture cells (Gardel et al., 2008). In these cells, MT bending was suggested to be due to forces exerted by actin filaments and myosin motors or MT molecular motors (Brangwynne et al., 2006; Gardel et al., 2008; Bicek et al., 2009), all of which acting alone or together with thermal fluctuations (Bicek et al., 2009). In vitro studies determined that MT Lp can be modulated by various factors, including MT assembly/disassembly rates (Janson and Dogterom, 2004), the nucleotide bound to tubulin (Mickey and Howard, 1995), the length of MTs (Pampaloni et al., 2006), and/or the protofilament number (Gittes et al., 1993). Microtubule-associated proteins (MAPs) have also been suggested to alter MT flexibility. Of identified MAPs, only two have been reported to modify MT flexural rigidity in vitro, namely the neuronal MAPs tau and MAP2, which stiffen them (Mickey and Howard, 1995; Felgner et al., 1997). In contrast, increased flexibility of MTs due to MAPs has not been described. In this study, to explore whether MT deformations observed in living cells could be due to MAPs, we focus on MT cross-linkers since, as discussed, MT deformations might be a key process in generating MT bundles in eukaryotic cells (Dixit and Cyr, 2004; Bartolini and Gundersen, 2006; Bratman and Chang, 2008).
Beside molecular motors, the major MAPs that cross-link MTs in cells are members of the conserved MAP65 family (Mollinari et al., 2002; Schuyler et al., 2003; Smertenko et al., 2004; Stoppin-Mellet et al., 2013). Gene analysis reveals nine isoforms in the plant model Arabidopsis thaliana (MAP65-1 to -9; Hussey et al., 2002) and one each in vertebrates (PRC1; Mollinari et al., 2002) and yeast (Ase1; Schuyler et al., 2003). Ase1/MAP65-1 are associated with the cortical MTs during interphase (Schuyler et al., 2003; Lucas et al., 2011), and Ase1/PRC1/MAP65-1 play key roles in organizing antiparallel MTs at the midzone during mitosis (Mollinari et al., 2002; Smertenko et al., 2004; Janson et al., 2007; Gaillard et al., 2008). To explore whether MAP65 modify MT mechanical properties, we developed an original assay based on an MT micropatterning method (Portran et al., 2013) and hydrodynamic flux (Venier et al., 1994). We demonstrate that MAP65-1/Ase1 drastically increase the flexibility of both single and cross-linked MTs. This effect is independent of MT dynamical changes and is directly mediated by the MAP65 MT-binding domain. To determine whether the softening effect of MAP65 on MTs was determinant in creating bundles, we reconstituted physical collisions between growing MTs/MT bundles and found that the decrease in MT flexural rigidity mediated by MAP65/Ase1 is required to enable the bending and coalignment of MTs at steep angles. Taken together, these data provide new insights into how these key proteins, by modifying MT mechanical properties, control MT–MT encounters and result in the formation of complex MT bundle arrays. More generally, they suggest that MT softening by MAPs might represent an original mechanism that could regulate the plasticity of MT networks in cells.
RESULTS
MAP65-1/Ase1 increase the flexibility of individual MTs
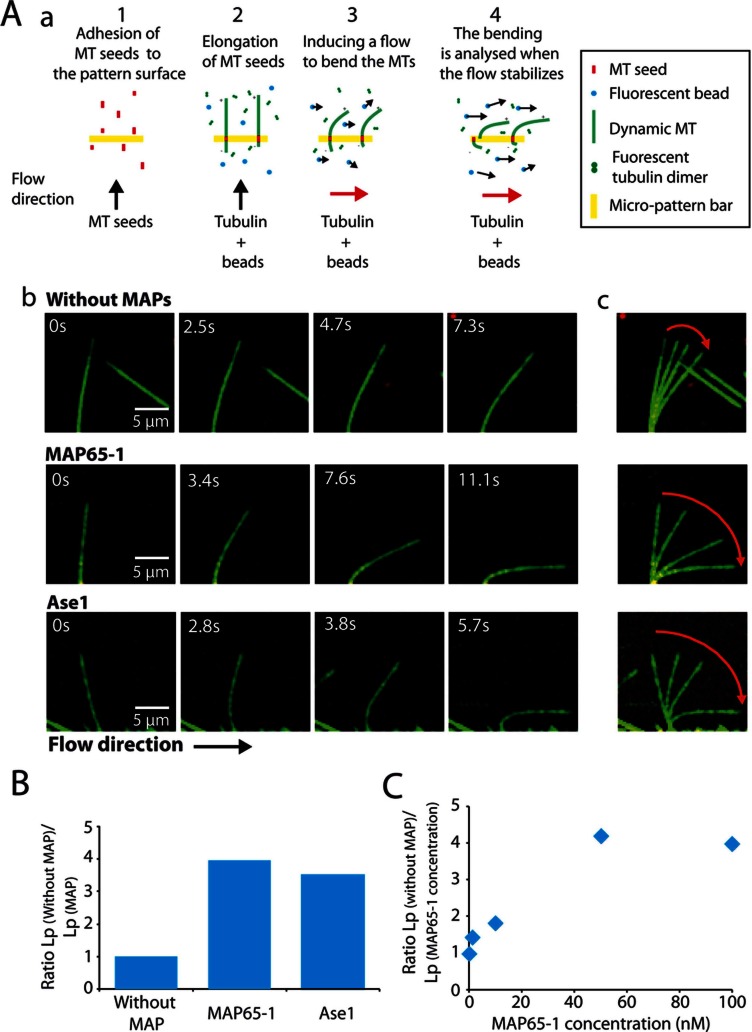
To evaluate the effect of MAP65-1 and Ase1 on MT bending stiffness, we measured Lp for MTs growing in the presence of these two MAP65s. We set up an assay in which fluorescent MTs elongate from seeds immobilized on a bar-shaped micropatterned surface (Portran et al., 2013) and are bent by hydrodynamic flow applied perpendicular to the growing MTs (Figure 1A and Supplemental Movies S1 and S2). To measure MT rigidity from flow-induced MT deformations, we adapted the equations for elastic filaments (Berro et al., 2007) and determined the MT Lp using the best-fit solution to the MT configuration (Supplemental Methods). MTs growing in the absence of MAPs have Lp = 1.58 ± 0.62 mm (Table 1 and Figure 1B), which is consistent with previous reported values (Hawkins et al., 2010). In the presence of 100 nM MAP65-1 or Ase1 (with apparent Kd of, respectively, 21 ± 2 and 41 ± 7 nM; Supplemental Figure S1, A and B), MT Lp decreases by a factor of 4 and 3.2, respectively (Table 1, Figure 1B, and Supplemental Movie S2). This effect is concentration dependent and is already observed at 1 nM MAP65-1 (Figure 1C).
FIGURE 1:
MAP65-1/Ase1 decrease the flexural rigidity of individual MTs. (A) Measurement of MT persistence length. (a) Experimental setup. MT seeds are introduced in a flowthrough chamber composed of a micropattern slide (bar shape) saturated with NeutrAvidin and a glass support. They are aligned on functionalized bar patterns by the flow and attached on the micropattern surface via biotin–NeutrAvidin link (step 1). MT seeds are further elongated by the addition of Alexa-labeled tubulin in the presence or absence of MAP65 and in the presence of fluorescent beads (step 2). When MTs reach a length of 10 μm on average, the elongation mix is perfused into the flow chamber perpendicular to the elongating MTs in order to bend them (step 3). When the flow speed reaches its maximum and when it is stabilized, MT bending is measured (step 4). (b) Time series of bending MTs that elongate in the absence or presence of 100 nM MAP65-1/Ase1. MTs are in green; MT seeds and beads are in red. (c) Superposition of the images in (b), showing the amplitude of the MT bending (red arrows). (B) Histograms of the ratio between the Lp of single MTs grown in the absence of MAPs and the Lp of MTs grown in the presence of 100 nM MAP65-1 or Ase1. MAP65-1 and Ase1 significantly decrease MT Lp. (C) Plot of the MT Lp in the presence of different concentrations of MAP65-1 (1–100 nM).
TABLE 1:
MT persistence length in the presence of MAP65.
| Condition | Number of MTs | MT length (μm) | Lp (mm) | Lp without MAP/Lp MAP65 |
|---|---|---|---|---|
| Without MAP65 | 9 | 10.39 ± 0.65 | 1.58 ± 0.62 | 1.00 |
| Ase1 | 5 | 10.50 ± 0.80 | 0.45 ± 0.18 | 3.22 |
| MAP65-1 | 13 | 11.62 ± 1.17 | 0.39 ± 0.16 | 4.05 |
| MAP65-1(MBD) | 4 | 8.30 ± 0.63 | 0.81 ± 0.34 | 1.79 |
| MAP65-4 | 4 | 13.81 ± 2.17 | 1.39 ± 0.47 | 1.04 |
| Chimera 1-4 | 6 | 13.00 ± 2.42 | 1.29 ± 0.65 | 1.12 |
Effective number, average length, and calculated Lp for single, growing MTs in the absence or presence of 100 nM MAP65.
Parameter values are reported with SD.
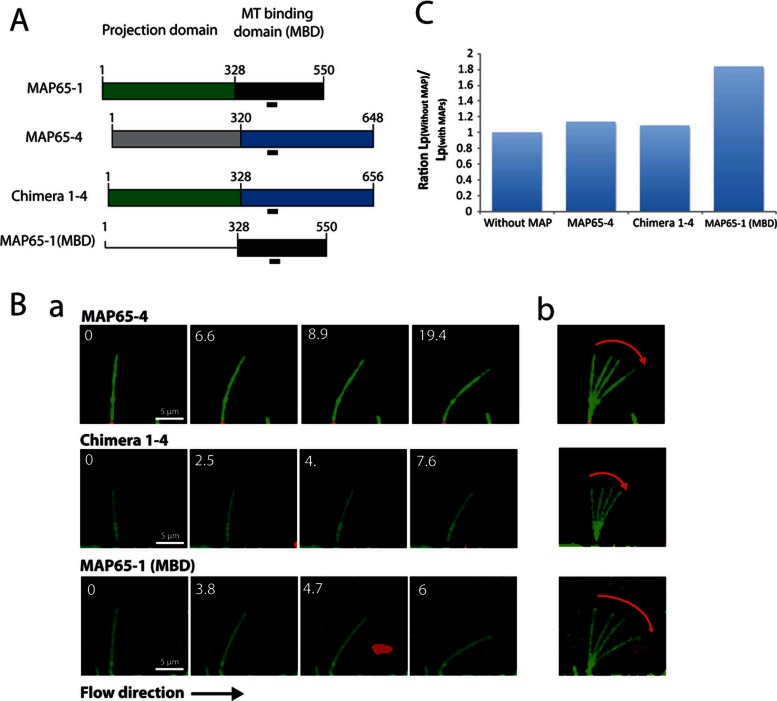
To investigate whether the increase in MT flexibility is unique to MAP65-1 and Ase1, we next determined the effect of another member of the plant MAP65 family on MT Lp. Indeed the plant MAP65, in contrast to the case for mammals and yeast, contains several members (Hussey et al., 2002). Among them, we chose MAP65-4, which associates with kinetochore fibers during mitosis and organizes MTs in bundles in vitro but differently than MAP65-1: MAP65-4 is oriented perpendicular to the MT lattice (vs. oblique for MAP65-1) and coaligns MTs with 15-nm MT interspacing (vs. 30 nm for MAP65-1; Gaillard et al., 2008; Fache et al., 2010). In the presence of 100 nM MAP65-4 (apparent Kd of 19 ± 2 nM; Supplemental Figure S1C), MT Lp = 1.39 ± 0.47 mm, which is not significantly different from that for MTs in the absence of MAPs (Table 1 and Figure 2, B and C).
FIGURE 2:
The MT-binding domain of MAP65-1 increases MT flexibility. (A) MAP65-1, MAP65-1(MBD), MAP65-4, and Chimera 1-4 constructs. MAP65-1 and MAP65-4 are divided into two domains: the projection and the MT-binding domain. The most conserved motif is underlined. Chimera 1-4 was obtained by replacing the projection domain of MAP65-4 with the projection domain of MAP65-1. (B) (a) Time series of bending MTs that elongate in the presence of 100 nM MAP65-4/Chimera 1-4/MAP65-1(MBD). MTs are in green; MT seeds and beads are in red. (b) Superposition of the images in (a) showing the amplitude of the bending (red arrows). (C) Histogram of the ratio between the Lp of single MTs grown in the absence of MAPs and the Lp of MTs grown in the presence of 100 nM MAP65-4, Chimera 1-4, and the MT-binding domain of MAP65-1.
MAP65 MT-binding domain is required to modulate MT stiffness
We next examined the relative contributions of the functional domains of MAP65-1 to increased MT flexibility. Indeed, MAP65-1, like other MAP65-family proteins, exhibits a C-terminal MT-binding domain and an N-terminal coiled-coil region (called the projection domain) required for its homodimerization (Figure 2A). The MT-binding domain is responsible for the preferential antiparallel orientation of the bundled MTs, whereas the projection domain length determines MT interspacing (Gaillard et al., 2008; Tulin et al., 2012). In contrast to Ase1 and PRC1, MAP65-1 binds to MTs as a monomer and further dimerizes when it encounters another MAP65-1 bound to an adjacent MT (Gaillard et al., 2008; Tulin et al., 2012). First, we generated a fragment carrying the MT-binding domain of MAP65-1 (named MAP65-1(MBD); Figure 2A). At a concentration of 100 nM, MAP65-1(MBD) decreases the Lp of growing MTs (0.83 ± 0.34 mm vs. 1.58 ± 0.62 nm without MAP65) but with a lower efficiency than full-length MAP65-1 (Table 1 and Figure 2, B and C). This lower efficiency could be related to its lower MT-binding affinity, which is 100-fold weaker than that of full-length MAP65-1 (apparent Kd = 2.1 ± 4 μM; Supplemental Figure S1D). The lower Kd value of the MT-binding domain compared with that of the full-length MAP65-1 might result from 1) incorrect folding due to the absence of the projection domain and/or 2) its binding to MTs as a monomer, in contrast to full-length MAP65-1, which binds bundled MTs as a dimer.
We next generated chimeras with the MT-binding domain of MAP65-1 and the dimerization domain of MAP65-4 and vice versa (referred to as Chimeras 4-1 and Chimera 1-4, respectively; Figure 2A). Chimera 4-1 hardly bundled MTs and thus could not be tested in this study, whereas Chimera 1-4 had an affinity for MTs similar to that of MAP65-1 and MAP65-4 (Supplemental Figure S1E). In the presence of 100 nM Chimera 1-4, MT Lp = 1.29 ± 2.42 mm, a value not significantly different from the one in the absence of MAPs (Table 1 and Figure 2, B and C). Thus Chimera 1-4, like MAP65-4, had no effect on MT rigidity. These data show that the MT-binding domain of MAP65-1 mediates its MT-softening effect.
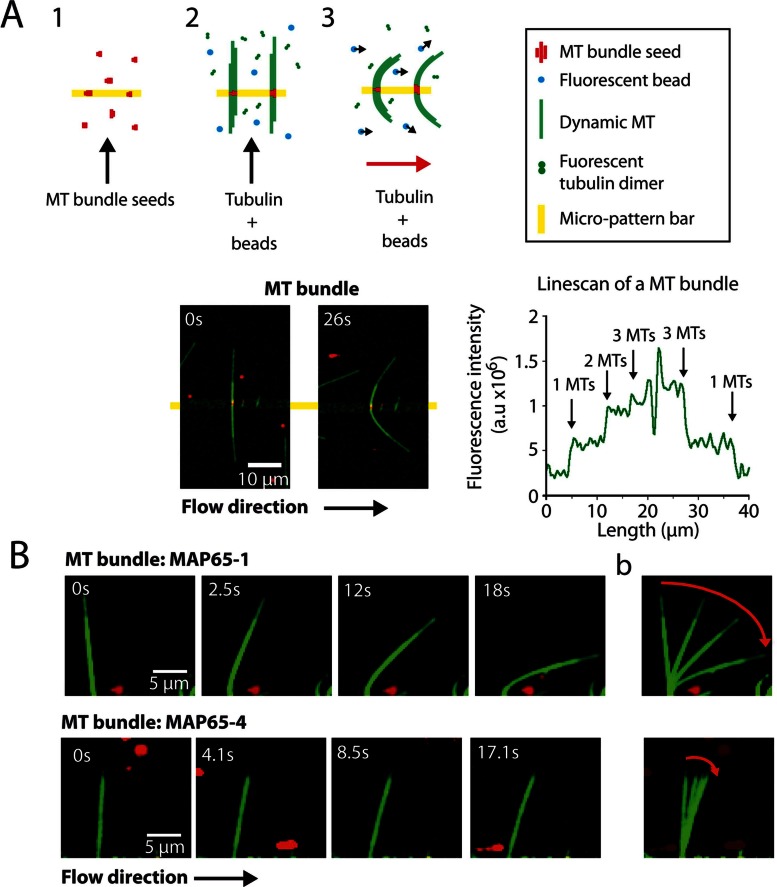
MAP65-1/Ase1 increase MT-bundle flexibility
To determine whether MAP65-1 and Ase1 also modulate the global mechanical properties of cross-linked MTs, we compared the Lp of growing MT bundles generated by MAP65-1, Ase1, MAP65-4, and Chimera 1-4 (Figure 3 and Supplemental Movies S3 and S4). We used the same experimental procedure as before but immobilized MT seed bundles, instead of individual MT seeds, on the micropatterned surface (Figure 3A). To measure the bundle's Lp, we considered only bundles having a maximum of three MTs, because with more, we could not ascertain that the number of cross-linked MTs was the same all along the bent, growing bundle. First, we focused on MAP65-4 and Chimera 1-4, which do not affect the mechanical properties of single MTs (Table 1). Bundles composed of two MTs had Lp = 3.49 ± 0.5 (MAP65-4) and 2.57 ± 0.6 mm (Chimera1-4), respectively (Table 2). The Lp of bundles with three MTs further increases, to 4.16 (MAP65-4) and 4.07 mm (Chimera1-4; Table 2). In contrast, the rigidity of bundles assembled in the presence of MAP65-1 or Ase1 remains constant, as the number of MTs increases from two to three (Lp = 1.3 mm [two or three MTs] for MAP65-1; Lp = 1.05 [two MTs] and 1.16 mm [three MTs] for Ase1; Table 2).
FIGURE 3:
MT bundle flexibility is differently regulated, depending on MAP65 cross-linker. (A) Experimental setup (top). MT seed bundles are immobilized on functionalized bar-shape patterns via biotin–NeutrAvidin link (step 1), and further elongated by perfusing tubulin in presence of MAP65 and fluorescent beads as described in Figure 1 (step 2). The hydrodynamic flow is applied when growing bundles have an average length between 10 and 20 μm (middle pattern; step 3). Time series of a bending bundle that elongates in the presence of tubulin, 100 nM MAP65-4, and fluorescent beads (bottom, left). MTs are in green; MT seeds and beads are in red. Line scan of the MT bundle (bottom, right). MT number in the bundle is determined by the level of fluorescence (right). (B) (a) Time series of bending bundles that elongate in the presence of tubulin and 100 nM MAP65-1. (b) Superposition of images showing the amplitude of the bending (red arrows).
TABLE 2:
MT bundle persistence length in the presence of MAP65.
| Condition | Number of bundles | Bundle length (μm) | Lp (mm) of bundles of two MTs | Number of bundles | Bundle length (μm) | Lp (mm) of bundles of three MTs |
|---|---|---|---|---|---|---|
| MAP65-4 | 6 | 16 ± 3.97 | 3.49 ± 0.98 | 3 | 14.35 ± 1.06 | 4.16 ± 1.24 |
| Chimera 1-4 | 9 | 15.75 ± 2.93 | 2.58 ± 1.07 | 7 | 15.59 ± 3.68 | 4.07 ± 1.15 |
| MAP65-1 | 2 | 16.16 ± 3.95 | 1.31 ± 0.57 | 7 | 14.91 ± 1.3 | 1.30 ± 0.80 |
| Ase1 | 13 | 14.94 ± 4.31 | 1.05 ± 0.44 | 6 | 16.59 ± 3.8 | 1.16 ± 0.66 |
Effective number, average length, and calculated Lp for growing MT bundles in the presence of 100 nM MAP65. Bundles are composed of two or three MTs. Parameter values are reported with SD.
The Lp of bundles depends on three factors: 1) the orientation of the bundle with respect to the flow, 2) the MT arrangement in the bundle, and 3) the length of the link between MTs, as illustrated in Supplemental Figure S2, a–f. Because we could hardly extract the two first parameters from our experimental data, we computed the flexural rigidity (Lp times kBT, where kB is the Boltzmann constant and T is the absolute temperature) of these different possible bundle configurations (Supplemental Figure S2 and Supplemental Methods). First, we assumed that MTs are linearly arranged inside bundles with a relative orientation toward the flow as illustrated by configurations (a) and (c) in Supplemental Figure S2. In these bundles, the predicted stiffness scales linearly with the number of MTs (Supplemental Figure S2), in agreement with the Lp measured for bundles induced by MAP65-4 and Chimera 1-4 (Table 2). However, for bundles induced by MAP65-1 and Ase1, we measured a value of Lp that is constant and independent of the number of MTs in the bundle (Table 2). These data show that the softening effect of these two MAP65 on single MTs is maintained when they are bundled. Second, we turned to the other possible configurations for MTs in the bundles, for which one predicts larger stiffness (Supplemental Figure S2, b and d–f). Because we did not find a large variability in bundle Lp, we conclude that these configurations are less likely to happen, indicating that configurations (a) and (c) are favored in our experimental setup.
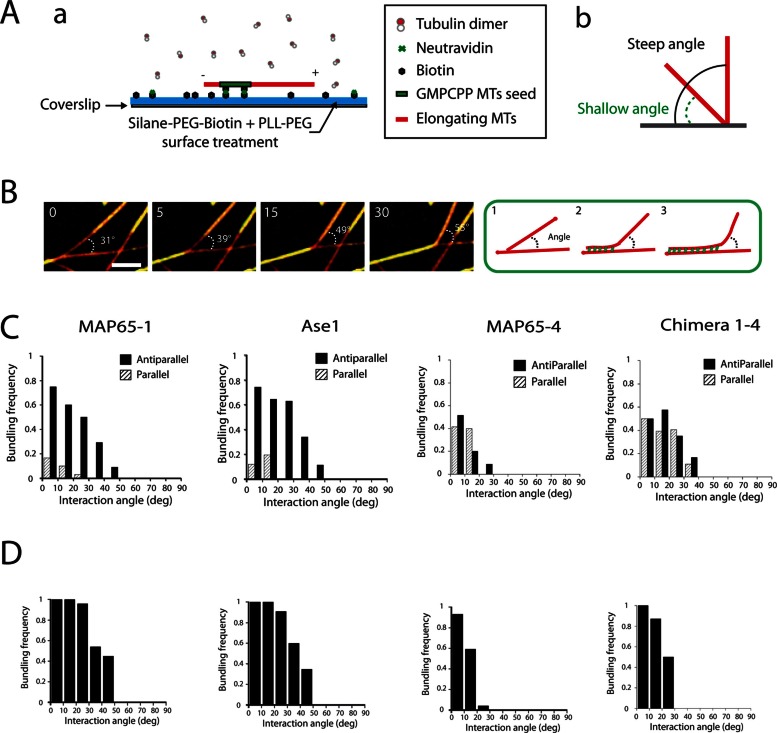
Encountering events
To investigate whether the MT-softening effect of MAP65-1 and Ase1 was determinant in creating bundles, we reconstituted physical collisions between single, growing MTs using a biomimetic system and total internal reflection fluorescence (TIRF) microscopy (Figure 4A and Supplemental Movie S5). We were able to mimic the MT collision events observed in living cells (Shaw et al., 2003; Dixit and Cyr, 2004), that is, crossover, touch-and-run, and bundling (Supplemental Figure S3 and Supplemental Movies S6–S8). The frequency of each of these outcomes in the presence of MAP65 was determined as a function of MT collision angles and MT polarity (Figure 4C and Supplemental Figure S3), with MT polarity being based on measurement of MT dynamics. In the presence of MAP65-1 and Ase1, the MT bundling was restricted to antiparallel MTs that encounter one another at angles up to 45°, in agreement with previous observations (Janson et al., 2007; Tulin et al., 2012) and result in a sharp bend of the incident MTs at the site of the collision (Figure 4, B and C, and Supplemental Figure S3). In contrast, in the presence of MAP65-4, MTs were not able to bundle MTs at collision angle >22° (Figure 4C). Below this value, bundling occurred whatever the polarity of MTs, in agreement with previous data demonstrating the nonselectivity of MAP65-4 for MT polarity (Fache et al., 2010). Finally, in the presence of Chimera 1-4, which does not modify MT Lp, we observed MT bundling at angles up to 39° whatever the polarity of MTs (Figure 4C), whereas the bundling efficiency at angles >25° was low compare to that observed for MAP65-1 (Supplemental Figure S3D). Overall these results show that the ability of MAP65-1 to bundle MTs at steep angles requires an increase in MT flexibility directly controlled by its MT-binding domain.
FIGURE 4:
MT-encountering events between single or bundled MTs. (A) (a) Experimental setup. MT seeds are introduced in a flow chamber coated with silane-PEG-biotin on a glass coverslip and silane-PEG on the glass. MT seeds are attached by biotin–NeutrAvidin link to the glass coverslip. They are further elongated by the addition of Alexa 561–labeled tubulin in the absence or the presence of MAP65. (b) Angles as defined in the text. (B) Time lapses of single elongating MTs in the presence of GFP-MAP65-1 (50 nM) and Alexa 568–labeled tubulin (22 μM) that coalign after collision in order to generate a bundle. MTs are in red; MAP65-1 are in green. Arrowheads indicate the growing ends of colliding MTs that encounter resident MTs. (C, D) Histograms representing the frequencies of bundling events between two individual MTs (C) or two bundles (D) as a function of their angle of interaction and of MT polarity in the presence of 50 nM MAP65-1, Ase1, MAP65-4, and Chimera 1-4 (with range of 10°, from 0° to 90°). Encountering frequencies are the ratio of coaligned MTs over the total number of encountering events. Red and blue bars correspond, respectively, to interactions between antiparallel and parallel MTs.
In line with the softening effect of MAP65-1 and Ase1 on MT bundles (Figure 3), we reconstituted physical collisions between growing MT bundles in the presence of these proteins. In this assay, MTs elongate from MT seed bundles as described in Figure 3. The outcomes between growing MT bundles resulted in the same events as observed for individual MTs (Figure 4D and Supplemental Figure S4). In particular, MT bundles induced by MAP65-1 and Ase1 coaligned very efficiently at angles of up to 45° in order to generate a thicker bundle, whereas those induced by MAP65-4 and Chimera 1-4 were not able to coalign at angles >25° (Figure 4D and Supplemental Figure S4). Of interest, in the presence of MAP65-1, the steep angle collisions between bundles (>45°) were sometimes associated with a local deformation of the resident bundles over a length of 5 μM (Supplemental Figure S5). However, the occurrence of these events was too scarce to be reliably quantified. Conversely, such deformations were never observed in the presence of MAP65-4 or Chimera 1-4.
DISCUSSION
In this study, we demonstrate that two MT cross-linkers involved in interphase and mitotic MT organization—the yeast Ase1 and the plant MAP65-1—have a strong and specific MT-softening effect mediated by their MT-binding domain.
Only two MAPs have been found to alter the flexibility of MTs: the neuronal MAPs tau and MAP2, which increase the MT rigidity probably by strengthening lateral and/or longitudinal interactions between tubulin dimers along the MT wall (Mickey and Howard, 1995; Felgner et al., 1997; Kar et al., 2003; Santarella et al., 2004). How MAP65-1 and Ase1 soften MTs remains to be explored. Previous studies described a direct relationship between MT dynamics and mechanical properties (Hawkins et al., 2010). In particular, high polymerization rates and/or frequent transitions from shrinkage to growth have been proposed to increase MT flexibility (Janson and Dogterom, 2004; VanBuren et al., 2005). Here we exclude such an explanation because MAP65-1 does not affect growth or shrinkage rates of individual MTs in vitro (Stoppin-Mellet et al., 2013), and this has also been suggested for MAP65-1 and Ase1 in cells (Loïodice et al., 2005; Lucas et al., 2011). Furthermore, the MT-binding domain by itself decreases MT rigidity. Thus we suggest that the increase of MT Lp instead involves structural modifications in the MT lattice induced by the direct interaction of the MAP65-1/Ase1 MT-binding domain during MT assembly. In this context, the question to be solved now is how this domain, which comprises a high conserved sequence with a spectrin fold and an unstructured part (Li et al., 2007; Kapitein et al., 2008; Subramanian et al., 2010), potentially modifies MT lattice organization is such as way as to allow decreased MT rigidity. Recent cryo–electron microscopic reconstructions with PRC1 revealed that only the spectrin fold interacts linearly along a single tubulin dimer (Subramanian et al., 2010). With this in mind, it is tempting to speculate that this domain, when bound to MTs, induces important structural changes within and/or between dimers that increase the freedom of interdimer motion during the bending of MTs. In contrast, the neuronal MAP tau binds MTs through multiple sites along but also across protofilaments (Santarella et al., 2004), suggesting that this MAP could increase MT rigidity by bridging protofilaments both laterally and longitudinally, thus limiting tubulin interdimer motions. These observations point out the importance of further study of the physical mechanisms underlying changes in MT lattice in the presence of MAPs.
Besides modifying the mechanical properties of single MTs, MAP65-1 and Ase1 also increase the flexibility of cross-linked MTs and consequently might not prevent MT sliding inside the bundle during their deformation. This suggests that either MAP65 diffuses between cross-linked MTs or MAP65 adopts different configurations. Indeed, recent studies show that Ase1, although decreasing MT sliding velocities generated by motors, does not inhibit it (Braun et al., 2011). Furthermore, Kapitein et al. (2008) showed that Ase1 diffuses between cross-linked MTs. In this study, we predicted that the bundle Lp should scale with the length of the MAP65 bonds (Supplemental Figure S2 and Supplemental Table S3), which is, respectively, 6 nm (Ase1; Schuyler et al., 2003), 15 nm (MAP65-4; Fache et al., 2010), and 30 nm (MAP65-1; Gaillard et al., 2008). However, our experimental data indicate a different picture. In particular, MAP65-1 and Ase1, which generate very different MT-interspace lengths, induce bundles having comparable Lp values.
Our results reveal intrinsic biophysical properties of MAP65 that are likely important for a wide range of cellular processes. In particular, MT encounters at steep contact angles have been suggested to be sufficient to explain the proper self-organization of MTs into ordered cortical bundles in acentrosomal plant cells (Allard et al., 2010; Eren et al., 2010). Because MAP65-1 and its homologue, MAP65-2, induce MT bundling in vivo (Lucas et al., 2011; Dhonukshe et al., 2012), we propose that these two MAP65, by softening and bundling MTs, are the key proteins involved in these processes. Although these mechanisms have been extensively observed in plant cells, we suggest that they may also apply to a wide range of differentiated eukaryotic cells that create self-ordered bundle arrays. Furthermore, the MT bundle softness due to MAP65-1 might facilitate their growth between adjacent cell faces to create the typical cortical arrays that bound different faces of the plant cells and be involved in regulating cell expansion. In line with our observations, recent in vivo studies from Dhonukshe et al. (2012) suggested that the transfacial MT bundles could be guided through MAP65-mediated CLASP localization. Beside MT cortical deformations in interphase, the regulation of MT mechanical properties by MAP65-1/Ase1 might also have implications for organizing the mitotic midzone. Indeed, to build the core of the midzone, which gives the spindle structural integrity, MAP65-1/Ase1/PRC1 cross-link antiparallel overlapping MTs that grow from the two opposite poles (Mollinari et al., 2002; Gaillard et al., 2008; Bieling et al., 2010; Subramanian et al., 2010; Portran et al., 2013). An increase in MT flexibility must allow them to coalign after encountering at steep angles (up to 40°), favoring the bundling of numerous MTs and subsequent midzone formation (Portran et al., 2013). More generally, MT buckling and bending are commonly observed in the cytoplasm of various eukaryotic cells (Brangwynne et al., 2006). However, the MT curvature that is observed at a scale of a few micrometers in cells is far from compatible with the MT Lp measured in vitro (1–8 mm; Bicek et al., 2009). In this study, by showing that MAPs significantly soften MTs in vitro, we suggest that this type of protein could be at the origin of MT deformations observed in the cytoplasm of various living cells and thus are able to regulate the local mechanical properties of MTs. Beside MAPs, other molecular mechanisms such as posttranslational modifications of tubulin could affect the MT lattice and thus alter MT flexural rigidity.
To conclude, MAP-induced MT flexibility might have numerous cell biological consequences, since it represents an alternative and simpler way for MT arrays to organize compared with assembly/disassembly processes. Hence an increase in MT flexibility should allow them to explore the cellular space laterally for binding partners, bend, and continue to grow in a new direction when encountering obstacles and/or coalign with other MTs. Thus MT softening by MAPs is a new mechanism to regulate the plasticity of MT networks in cells.
MATERIALS AND METHODS
Protein purification
Bovine brain tubulin was purified according to Vantard et al. (1994) in BRB80 buffer (80 mM 1,4-piperazinediethanesulfonic acid, pH 6.8, 1 mM ethylene glycol tetraacetic acid, and 1 mM MgCl2). Fluorescent tubulin (Alexa 488–labeled tubulin and Alexa 568–labeled tubulin) and biotinylated tubulin were prepared according to Hyman et al. (1991). Recombinant MAP65-1/4, Chimera 1-4, and MAP65-1(MBD) were purified according to Gaillard et al. (2008), and Ase1 (kindly provided by Marcel Janson, Laboratory of Cell Biology, Wageningen, The Netherlands) according to Janson et al. (2007). Chimera 1-4 is a fusion of the projection domain of MAP65-1 (amino acids [aa] 1–340) and the MT-binding domain of MAP65-4 (aa 320–648).
Determination of the apparent Kd of MAP65
Taxol-stabilized MTs were prepared by incubating purified tubulin with equimolar concentrations of taxotere in BRB80 buffer supplemented with 1 mM GTP for 30 min at 37°C. Various concentrations of green fluorescent protein (GFP)–MAP65 (in MAP buffer: 50 mM NaPi, pH 7.9, 0.1 M NaCl, 0.5 mM dithiothreitol [DTT]) were mixed with 2 μM MTs in binding buffer (1× BRB80/0.5× MAP buffer). After 30 min at room temperature, samples were centrifuged during 20 min at 100,000 × g at 25°C. Supernatant was kept, and the pellet was resuspended in binding buffer supplemented with 0.5 M NaCl to detach MAP65 from MTs. The amount of GFP-MAP65 in the pellet and the supernatant was measured by spectrofluorimetry (excitation, 490 nm; emission, 512 nm). All tubes were silanized to limit nonspecific adsorption of MAP65 (Sambrook et al., 1989). Ase1 Kd was determined according to Portran et al. (2013).
In vitro microtubule-encountering assays
Perfusion chambers.
Glass and coverslips were cleaned by successive chemical treatments as described (Portran et al., 2013). After exposure to air plasma (2 min at 60 W), coverslips were functionalized overnight in a solution of Silane-PEG-Biotin (3.5 kDa, 1 mg/ml in ethanol, 96.5%, plus 30 mM of HCl; SuSoS, Dübendorf, Switzerland) at room temperature. Glasses were passivated overnight in a solution of Silane-PEG (30 kDa, 1 mg/ml in ethanol, 96.5%, plus 30 mM HCl; Creative PEGworks, Winston Salem, NC) at room temperature. Glasses and coverslips were successively washed in ethanol and ultrapure water, dried with filtered air, and stored at 4°C away from dust. A flow cell chamber with an approximate volume of 20 μl was constructed with double-sided tape (70 μm height) between the glass and the coverslip. The perfusion chamber was coated with NeutrAvidin (25 μg/ml in 1% bovine serum albumin [BSA] in BRB80; Pierce, Rockford, IL), washed with 300 μl of 1% BSA in BRB80, and further passivated for 1 min with PLL-g-PEG (2 kDa, 0.1 mg/ml in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.4; Jenkem, Allen, TX).
MT seed preparation.
MT seeds were obtained by polymerizing 20 μM tubulin (80% biotinylated tubulin and 20% Alexa 568–labeled tubulin) in the presence of 0.5 mM GMPCPP in BRB80 at 37°C for 1 h. Next, 2 μM taxotere was added, and MT seeds were further incubated at room temperature for 30 min. MT seeds were then centrifuged for 5 min at 300,000 × g and resuspended in an equal volume of BRB80 supplemented with 0.5 mM GMPCPP and 2 μM taxotere. They were stored in liquid nitrogen and quickly warmed at 37°C before use. Seed bundles were obtained by incubating 0.3 μM MT seeds with 0.1 μM MAP65-1, MAP65-4, Ase1, or Chimera 1-4 for 10 min at room temperature.
In vitro encountering assays.
MT seeds were incubated for 5 min in the perfusion chamber. They were then elongated with an elongation mix containing 22 μM tubulin (30% Alexa 568–labeled tubulin and 70% unlabeled tubulin), 1 mM GTP, an oxygen scavenger cocktail (120 μg/ml glucose, 8 μg/ml catalase, and 40 μg/ml glucose oxidase), 20 μM DTT (Sigma-Aldrich, St. Louis, MO), 1% BSA, and 0.025% methyl cellulose (1500 CP; Sigma-Aldrich) in mix buffer (1× BRB80/0.5× MAP buffer) in the presence or the absence of MAP65. MT dynamics was visualized using an objective-based azimuthal ilas2 TIRF microscope (Nikon Eclipse Ti [Melville, NY], modified by Roper Scientific [Tucson, AZ]) and Evolve 512 camera (Photometrics, Tucson, AZ). The microscope stage was kept at 32°C using a warm stage controller (MC60; Linkam Tadworth, United Kingdom). Excitation was achieved using 491- and 561-nm lasers (Roper Scientific, France). Time-lapse recording (one frame every 5 s) was performed for 30 min using MetaMorph software (version.7.7.5; Molecular Devices, Sunnyvale, CA). Movies were processed to improve signal/noise ratio (equalize light, low pass, and flatten background filters of MetaMorph software). MT bundle elongation and dynamics were analyzed using kymographs generated by MetaMorph and analyzed with ImageJ (National Institutes of Health, Bethesda, MD). The statistical significance was determined using Student's t test. The contact angle was measured between two MTs or two MT bundles preceding the behavior of encountering. The frequency for each behavior was calculated for nine contact angle ranges of 10° from 0 to 90°. MT polarity was determined by measuring MT growth rates, since MT minus ends grow at lower rates than MT plus ends.
Microtubule flexibility measurements
Micropatterning technique and flow chamber for flexibility.
The micropatterning technique was adapted from Portran et al. (2013). Lines 3 μm thick were micropatterned on silane-PEG-passivated coverslips by exposure for 20 s under deep ultraviolet (185 nm; UVO Cleaner Model 144AX-220; Jelight, Irvine, CA) through a photomask (Toppan, Round Rock, TX). Micropatterns were functionalized by NeutrAvidin (25 μg/ml) to allow attachment of biotinylated stabilized MT seeds as described. A flow cell (30 μl volume) with two perpendicular channels was constructed with double-sided tape (70 μm height) between the glass and the patterned coverslip. The MT biotinylated seeds (2 nM) were attached perpendicular to the micropatterned line by continuous flow in the first channel (see scheme in Figure 1A). The nonattached MT seeds were washed away by perfusing buffer (1× BRB80/0.5× MAP buffer, 1% BSA). They were further elongated as described. When the polymerized MTs were ∼10 μm long, the perpendicular flow was applied using a peristaltic pump sucking 60 μl of the elongation mix at the entry of the second channel with a speed flow between 30 and 60 μm/min (measured by the bead speed at the TIRFm focal plane). MT growth and bending were visualized at 32°C using TIRF microscopy as described. Time-lapse microscopy (one frame every 315 ms) was performed for 90 s using MetaMorph software (version.7.7.5).
Image acquisition and velocity measurement.
To extract MT position during bending experiments, we used the plug-in JFilament of ImageJ. To measure the fluid velocity we used the plug-in MTrackJ of ImageJ. We collected the position of the beads and proceeded to the next image by connecting points associated with the same bead. We only took into account beads that were as bright as the seeds, assuming that these beads were in the same z-position as MTs. Therefore the bead velocity was the same as the velocity of the fluid experienced by MTs. We calculated the average velocity of the flow for each frame. If beads could not be seen for a few frames (∼1–10), we interpolated the velocity between frames.
Measurement of single MTs/MT bundle rigidity.
See Supplementary Methods and Supplemental Figure S6 for a detailed description.
Supplementary Material
Acknowledgments
We thank Marcel Janson for the gift of Ase1 cDNA and Manuel Théry and Laurent Blanchoin for critical reading of the manuscript. We thank François Nédélec for constructive remarks. We acknowledge financial support of the Institute of Complex Systems (IXXI; Lyon, France) to M.Z and the French education minister for financial support to D.P.
Abbreviations used:
- Lp
persistence length
- MAP
microtubule-associated proteins
- MT
microtubule
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0141) on April 24, 2013.
REFERENCES
- Allard JF, Ambrose JC, Wasteneys GO, Cytrynbaum EN. A mechanochemical model explains interactions between cortical microtubules in plants. Biophys J. 2010;99:1082–1090. doi: 10.1016/j.bpj.2010.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose C, Allard JF, Cytrynbaum EN, Wasteneys GO. A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat Commun. 2011;16:430–442. doi: 10.1038/ncomms1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- Berro J, Michelot A, Blanchoin L, Kovar DR, Martiel JL. Attachment conditions control actin filament buckling and the production of forces. Biophys J. 2007;92:2546–2558. doi: 10.1529/biophysj.106.094672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicek AD, Tüzel E, Kroll DM, Odde D. Anterograde microtubule transport drives microtubule bending in LLC-PK1 epithelial cells. Mol Biol Cell. 2009;20:2943–2953. doi: 10.1091/mbc.E08-09-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieling P, Telley IA, Surrey TA. Minimal midzone protein module controls formation and length of antiparallel microtubule overlaps. Cell. 2010;142:420–432. doi: 10.1016/j.cell.2010.06.033. [DOI] [PubMed] [Google Scholar]
- Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol. 2006;173:733–741. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratman SV, Chang F. Mechanisms for maintaining microtubule bundles. Trends Cell Biol. 2008;18:580–586. doi: 10.1016/j.tcb.2008.09.004. [DOI] [PubMed] [Google Scholar]
- Braun M, Lansky Z, Fink G, Ruhnow F, Diez S, Janson ME. Adaptive braking by Ase1 prevents overlapping microtubules from sliding completely apart. Nat Cell Biol. 2011;13:1259–1264. doi: 10.1038/ncb2323. [DOI] [PubMed] [Google Scholar]
- Dhonukshe P, et al. A PLETHORA-auxin transciption module controls cell division plane rotation through MAP65 and CLASP. Cell. 2012;149:2071–2093. doi: 10.1016/j.cell.2012.02.051. [DOI] [PubMed] [Google Scholar]
- Dixit R, Cyr R. Encounters between dynamic cortical microtubules promote ordering of the cortical array through angle-dependent modifications of microtubule behavior. Plant Cell. 2004;16:3274–3284. doi: 10.1105/tpc.104.026930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren EC, Dixit R, Gautam N. A three-dimensional computer simulation model reveals the mechanisms for self-organization of plant cortical microtubules into oblique arrays. Mol Biol Cell. 2010;21:2674–2684. doi: 10.1091/mbc.E10-02-0136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fache V, Gaillard J, Van Damme D, Geelen D, Neumann E, Stoppin-Mellet V, Vantard M. Arabidopsis kinetochore fiber-associated MAP65-4 cross-links microtubules and promotes microtubule bundle elongation. Plant Cell. 2010;22:3804–3815. doi: 10.1105/tpc.110.080606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felgner H, Frank R, Biernat J, Mandelkow EM, Mandelkow E, Ludin B, Matus A, Schliwa M. Domains of neuronal microtubule-associated proteins and flexural rigidity of microtubules. J Cell Biol. 1997;138:1067–1075. doi: 10.1083/jcb.138.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard J, Neumann E, Van Damme D, Stoppin-Mellet V, Ebel C, Barbier E, Geelen D, Vantard M. Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol Biol Cell. 2008;19:4534–4544. doi: 10.1091/mbc.E08-04-0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardel ML, Kasza KE, Brangwynne CVP, Liu J, Weitz DA. Mechanical response of cytoskeletal networks. Methods Cell Biol. 2008;89:487–519. doi: 10.1016/S0091-679X(08)00619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins T, Mirigian M, Selcuk Yasar M, Ross JL. Mechanics of microtubules. J Biomech. 2010;43:23–30. doi: 10.1016/j.jbiomech.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Hussey PJ, Hawkins TJ, Igarashi H, Kaloriti D, Smertenko A. The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol Biol. 2002;50:915–924. doi: 10.1023/a:1021236307508. [DOI] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Janson ME, Dogterom MA. Bending mode analysis for growing microtubules: evidence for a velocity-dependent rigidity. Biophys J. 2004;87:2723–2736. doi: 10.1529/biophysj.103.038877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loïodice I, Fu C, Brunner D, Nédélec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Janson ME, van den Wildenberg SM, Hoogenraad CC, Schmidt CF, Peterman EJ. Microtubule-driven multimerization recruits ase1p onto overlapping microtubules. Curr Biol. 2008;18:1713–1717. doi: 10.1016/j.cub.2008.09.046. [DOI] [PubMed] [Google Scholar]
- Kar S, Fan J, Smith MJ, Goedert M, Amos LA. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003;22:70–77. doi: 10.1093/emboj/cdg001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Mao T, Zhang Z, Yuan M. The AtMAP65-1 cross-bridge between microtubules is formed by one dimer. Plant Cell Physiol. 2007;48:866–874. doi: 10.1093/pcp/pcm059. [DOI] [PubMed] [Google Scholar]
- Loïodice I, Staub J, Setty TG, Nguyen NP, Paoletti A, Tran PT. Ase1p organizes antiparallel microtubule arrays during interphase and mitosis in fission yeast. Mol Biol Cell. 2005;16:1756–1768. doi: 10.1091/mbc.E04-10-0899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JR, Courtney S, Hassfurder M, Dhingra S, Bryant A, Shaw SL. Microtubule-associated proteins MAP65-1 and MAP65-2 positively regulate axial cell growth in etiolated Arabidopsis hypocotyls. Plant Cell. 2011;23:1889–1903. doi: 10.1105/tpc.111.084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickey B, Howard J. Rigidity of microtubules is increased by stabilizing agents. J Cell Biol. 1995;130:909–917. doi: 10.1083/jcb.130.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL. PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol. 2002;157:1175–1186. doi: 10.1083/jcb.200111052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampaloni F, et al. Thermal fluctuations of grafted microtubules provide evidence of a length-dependent persistence length. Proc Natl Acad Sci USA. 2006;103:10248–10253. doi: 10.1073/pnas.0603931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portran D, Gaillard J, Vantard M, Thery M. Quantification of MAP activity on geometrically controlled MT network. Cytoskeleton. 2013;70:2–23. doi: 10.1002/cm.21081. [DOI] [PubMed] [Google Scholar]
- Rusan NM, Wadsworth P. Centrosome fragments and microtubules are transported asymmetrically away from division plane in anaphase. J Cell Biol. 2005;168:21–28. doi: 10.1083/jcb.200409153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santarella RA, Skiniotis G, Goldie KN, Tittmann P, Gross H, Mandelkow EM, Mandelkow E, Hoenger A. Surface-decoration of microtubules by human tau. J Mol Biol. 2004;339:539–553. doi: 10.1016/j.jmb.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D. The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol. 2003;160:517–528. doi: 10.1083/jcb.200210021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S, Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300:1715. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Chang HY, Wagner V, Kaloriti D, Fenyk S, Sonobe S, Lloyd C, Hauser MT, Hussey PJ. The Arabidopsis microtubule-associated protein AtMAP65-1: molecular analysis of its microtubule bundling activity. Plant Cell. 2004;16:2035–2047. doi: 10.1105/tpc.104.023937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V, Fache V, Portran D, Martiel JL, Vantard M. MAP65 coordinate microtubule growth during bundle formation. Plos One. 2013;8:e56808. doi: 10.1371/journal.pone.0056808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian R, Wilson-Kubalek EM, Arthu CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran PT, Marsh L, Doye V, Inoué S, Chang FA. Mechanism for nuclear positioning in fission yeast based on microtubule pushing. J Cell Biol. 2001;16:397–411. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A, McClerklin S, Huang Y, Dixit R. Single-molecule analysis of the microtubule cross-linking protein MAP65-1 reveals a molecular mechanism for contact-angle-dependent microtubule bundling. Biophys J. 2012;102:802–809. doi: 10.1016/j.bpj.2012.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren V, Cassimeris L, Odde DJ. Mechanochemical model of microtubule structure and self-assembly kinetics. Biophys J. 2005;89:2911–2926. doi: 10.1529/biophysj.105.060913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vantard M, Peter C, Fellous A, Schellenbaum P, Lambert AM. Characterization of a 100-kDa heat-stable microtubule-associated protein from higher plants. Eur J Biochem. 1994;220:847–853. doi: 10.1111/j.1432-1033.1994.tb18687.x. [DOI] [PubMed] [Google Scholar]
- Venier P, Maggs MF, Carlier MF, Pantaloni D. Analysis of microtubule rigidity using hydrodynamic flow and thermal fluctuations. J Biol Mech. 1994;269:13353–13360. [PubMed] [Google Scholar]
- Wasteneys GO, Ambrose JC. Spatial organization of plant cortical microtubules: close encounters of the 2D kind. Trends Cell Biol. 2009;19:62–71. doi: 10.1016/j.tcb.2008.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.