Overexpression of C-terminally tagged PGAP3 causes predominant production of lysoGPI-APs in MDCK cells. In these cells, produced lysoGPI-APs are not incorporated into detergent-resistant membranes but are still delivered apically. The apical transport of both fully remodeled and lysoGPI-APs is sensitive to cholesterol depletion and ablation of N-glycosylation.
Abstract
Most glycosylphosphatidylinositol-anchored proteins (GPI-APs) are located at the apical surface of epithelial cells. The apical delivery of GPI-APs is believed to result from their association with lipid rafts. We find that overexpression of C-terminally tagged PGAP3 caused predominant production of lysoGPI-APs, an intermediate precursor in the GPI lipid remodeling process in Madin–Darby canine kidney cells. In these cells, produced lysoGPI-APs are not incorporated into detergent-resistant membranes (DRMs) but still are delivered apically, suggesting that GPI-AP association with DRMs is not necessary for apical targeting. In contrast, apical transport of both fully remodeled and lyso forms of GPI-APs is dependent on N-glycosylation, confirming a general role of N-glycans in apical protein transport. We also find that depletion of cholesterol causes apical-to-basolateral retargeting not only of fully remodeled GPI-APs, but also of lysoGPI-APs, as well as endogenous soluble and transmembrane proteins that would normally be targeted to the apical membrane. These findings confirm the essential role for cholesterol in the apical protein targeting and further demonstrate that the mechanism of cholesterol-dependent apical sorting is not related to DRM association of GPI-APs.
INTRODUCTION
In eukaryotes glycosylphosphatidylinositol (GPI) is a conserved glycolipid that anchors numerous proteins to the plasma membrane (Kinoshita et al., 2008). A preassembled GPI anchor is added to the C-terminus of the precursor protein in the endoplasmic reticulum (ER) to form a GPI-anchored protein (GPI-AP). The lipid moiety of the GPI anchor is then remodeled in the Golgi apparatus first by PGAP3, which removes the unsaturated acyl chain at the sn-2 position of the phosphatidylinositol moiety to produce a lysoGPI, followed by the addition of a saturated acyl chain–mediated PGAP2 (Figure 1A; Tashima et al., 2006; Maeda et al., 2007).
FIGURE 1:
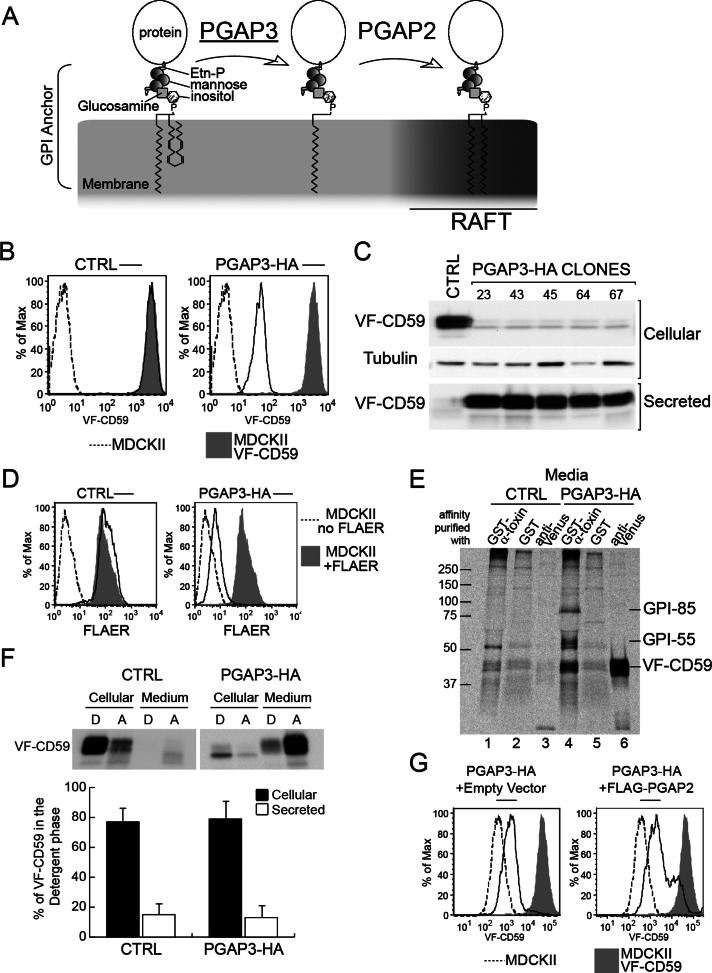
Overexpression of PGAP3-HA results in loss of GPI-APs from the surface of MDCK II cells. (A) GPI lipid remodeling in mammalian cells. PGAP3 and PGAP2 mediate removal of an unsaturated fatty acid and reacylation with a saturated fatty acid, respectively. (B) Flow cytometric analysis of surface VF-CD59 in cells expressing VF-CD59 and transfected either with empty vector (CTRL) or with a vector expressing PGAP3-HA. Shaded area, MDCK II cells expressing VF-CD59; dotted line, MDCK II cells. The x and y axes show the amount of surface VF-CD59 and the percentage of the cell population normalized to the highest peak, respectively. (C) Western blot analysis of cellular and secreted VF-CD59 in control (CTRL) and PGAP3-HA cells. VF-CD59 was detected by using an anti-Venus antibody. The loading control was probed with an anti–α-tubulin antibody. Secreted VF-CD59 was precipitated with anti-FLAG M2 affinity gel. (D) Flow cytometric analysis of the surface expression of endogenous GPI-APs in parental MDCK II cells transfected with empty vector (CTRL) or PGAP3-HA and then stained with FLAER. Shaded area, parental MDCK II cells stained with FLAER. Dotted line, MDCK II cells without FLAER staining. The x and y axes represent the surface expression of endogenous GPI-APs stained by FLAER, and the percentage of the cell population normalized to the highest peak, respectively. (E) Affinity purification of released endogenous GPI-APs and VF-CD59 detected by binding to α-toxin from C. septicum. Control cells (lanes 1–3) and PGAP3-HA–expressing MDCK II cells (lanes 4–6) stably expressing VF-CD59 and grown on plastic dishes were pulse labeled and then chased for 2 h. The media were collected and affinity purified with either GST–α-toxin (lanes 1 and 4) or GST alone (lanes 2 and 5) bound to glutathione–Sepharose or with anti-Venus antibody and rProtein A Sepharose (lanes 3 and 6). The precipitates were separated by SDS–PAGE and visualized by PhosphorImager analysis. Two endogenous GPI-APs migrating around 55 and 85k Da are marked GPI-55 and GPI-85, respectively. (F) Secreted VF-CD59 does not partition into the detergent phase. Phase partitioning of cellular and secreted VF-CD59 from control and PGAP3-HA cells into the detergent phase (D) and aqueous phase (A) of a solution of Triton X-114. After partition, VF-CD59 in each fraction was precipitated with anti-FLAG M2 affinity gel and detected by Western blotting with an anti-Venus antibody. (G) The partial restoration of surface VF-CD59 in PGAP3-HA cells transiently transfected with empty vector (left) or vector encoding FLAG–PGAP2 (right) measured by flow cytometry. Similar data were obtained by staining with mouse monoclonal antibodies against CD59 and GFP, which recognizes the GFP variant Venus.
In contrast to mammalian cells, this lipid remodeling occurs in the ER of yeast. Yeast strains deficient for GPI lipid remodeling are defective in concentrating GPI-APs at ER exit sites, resulting in delayed ER-to-Golgi transport (Bosson et al., 2006; Fujita et al., 2006; Castillon et al., 2009). Furthermore, it is likely that this lipid remodeling is a part of ER quality control of GPI-APs in yeast (Castillon et al., 2011).
Lipid rafts are proposed to be specific membrane domains enriched in sphingolipids and cholesterol. They are suspected to function in various cellular processes, including signal transduction and membrane traffic (Simons and Ikonen, 1997; Simons and Sampaio, 2011). However, the size of these microdomains or nanodomains makes it difficult to confirm their existence in living cells, and their proposed functions are often tested by modifying cellular lipid contents, which likely affects not only lipid raft composition, but also variety of cellular components and pathways (Kwik et al., 2003; Zidovetzki and Levitan, 2007; Guan et al., 2009). Therefore the nature and precise biophysical properties of these rafts are the subject of long-standing debate (Munro, 2003; Hanzal-Bayer and Hancock, 2007). Lipid rafts are frequently defined by their resistance to extraction by detergents in vitro. Whether detergent-resistant membranes (DRMs) truly reflect lipid rafts in vivo has been questioned, but this technique is commonly used to reveal the potentiality of proteins to associate with specific lipid environment (Brown and Rose, 1992; Schuck et al., 2003; Morris et al., 2011). GPI-APs, through hydrophobic interactions with the saturated fatty acyl chains of the GPI anchor, are found in DRM (Schroeder et al., 1994) and therefore are generally considered as model raft proteins. GPI-APs expressed in lipid remodeling–deficient cells are no longer partitioned in the DRM fraction (Bosson et al., 2006; Fujita et al., 2006; Maeda et al., 2007; Murakami et al., 2012; Seong et al., 2013). In polarized epithelial cells, the association of GPI-APs with lipid rafts is one of the determinants for transport to the apical plasma membrane (Simons and Ikonen, 1997; Simons and Sampaio, 2011). This notion is supported by the fact that either cholesterol depletion or inhibition of sphingolipid biosynthesis affects apical sorting of GPI-APs in an epithelial cell line, Madin–Darby canine kidney (MDCK) II (Mays et al., 1995; Paladino et al., 2004). The validity of such approaches is questionable since they likely cause various indirect consequences to the cells (Kwik et al., 2003; Zidovetzki and Levitan, 2007; Guan et al., 2009). Moreover additional observations do not support the requirement of lipid raft association for apical targeting of GPI-APs: GPI anchor does not seem to be the essential apical determinant for some GPI-APs, since the luminal domain of several endogenous GPI-APs are secreted apically without GPI anchor (Brown et al., 1989; Lisanti et al., 1989; Powell et al., 1991; Pang et al., 2004). Furthermore, it has been observed that certain GPI-APs localize to either both apical and basolateral membrane domains or exclusively to the basolateral domain (Benting et al., 1999; McGwire et al., 1999; Paladino et al., 2004; Pang et al., 2004). Alternatively, N-glycosylation has been shown to be involved in apical targeting for some but not all GPI-APs in MDCK cells (Benting et al., 1999; Pang et al., 2004; Catino et al., 2008).
In this study, we establish MDCK cells defective in the GPI lipid-remodeling step in order to examine the contribution of lipid remodeling and DRM association in the apical transport of GPI-APs without affecting cellular lipid composition. We also address the effect of cholesterol depletion and the contribution of N-glycan in the apical transport of GPI-APs in these MDCK cells.
Results
GPI-APs are secreted into the medium by cells overexpressing PGAP3-HA
To test the requirement of GPI-AP lipid remodeling in apical sorting, we first depleted PGAP3 using small interfering RNAs (siRNAs). Despite a reduction of mRNA to levels <20% of the control, GPI-APs were still partitioned into DRMs (data not shown). Based on the previously described phenotype of different PGAP3 mutant cells (Fujita et al., 2006; Maeda et al., 2007; Murakami et al., 2012), this suggests that the remaining PGAP3 activity is sufficient to support complete GPI-AP remodeling. In parallel, we also established MDCK II cell lines stably expressing dog PGAP3 tagged at the C-terminus with enhanced green fluorescent protein (EGFP; PGAP3-EGFP). Surprisingly, these cells showed decreased GPI-AP expression (data not shown), as did cells expressing human PGAP3 tagged at the C-terminus with hemagglutinin (HA; PGAP3-HA). The cell surface amounts of the model GPI-AP—CD59 tagged with Venus and FLAG (VF-CD59)— were reduced to <2% of the parental cells when measured by flow cytometry in cells expressing PGAP3-HA (Figure 1B). Of interest, this reduction in surface protein levels was not due to decreased protein expression, as we found that increased amounts of VF-CD59 were secreted into the medium in all clones expressing PGAP3-HA (Figure 1C).
Using two bacterial toxins that recognize the glycan part of the GPI anchor, we found that endogenous GPI-APs were also released efficiently from PGAP3-HA cells. Cell surface–expressed endogenous GPI-APs were detected by fluorescently labeled aerolysin (FLAER), a bacterial toxin that binds GPI-APs specifically (Diep et al., 1998; Brodsky et al., 2000). Flow cytometry showed that FLAER labeling of the cell surface in PGAP3-HA–expressing cells was <5% of the level observed in the parental MDCK II cell line (Figure 1D). We also collected endogenous GPI-APs from the culture medium by affinity purification with recombinant glutathione S-transferase (GST)–tagged α-toxin, another bacterial toxin that binds to the GPI anchor (Hong et al., 2002). In addition to VF-CD59, two endogenously expressed GPI-APs that migrate around 55 kDa (GPI-55) and 85 kDa (GPI-85) were precipitated from the culture medium of PGAP3-HA–expressing cells but not detected in the secreted fraction of the control cells (Figure 1E). Both proteins can be found among the list of endogenous GPI-APs in MDCK cells previously identified with different methods (Lisanti et al., 1988; McGwire et al., 1999).
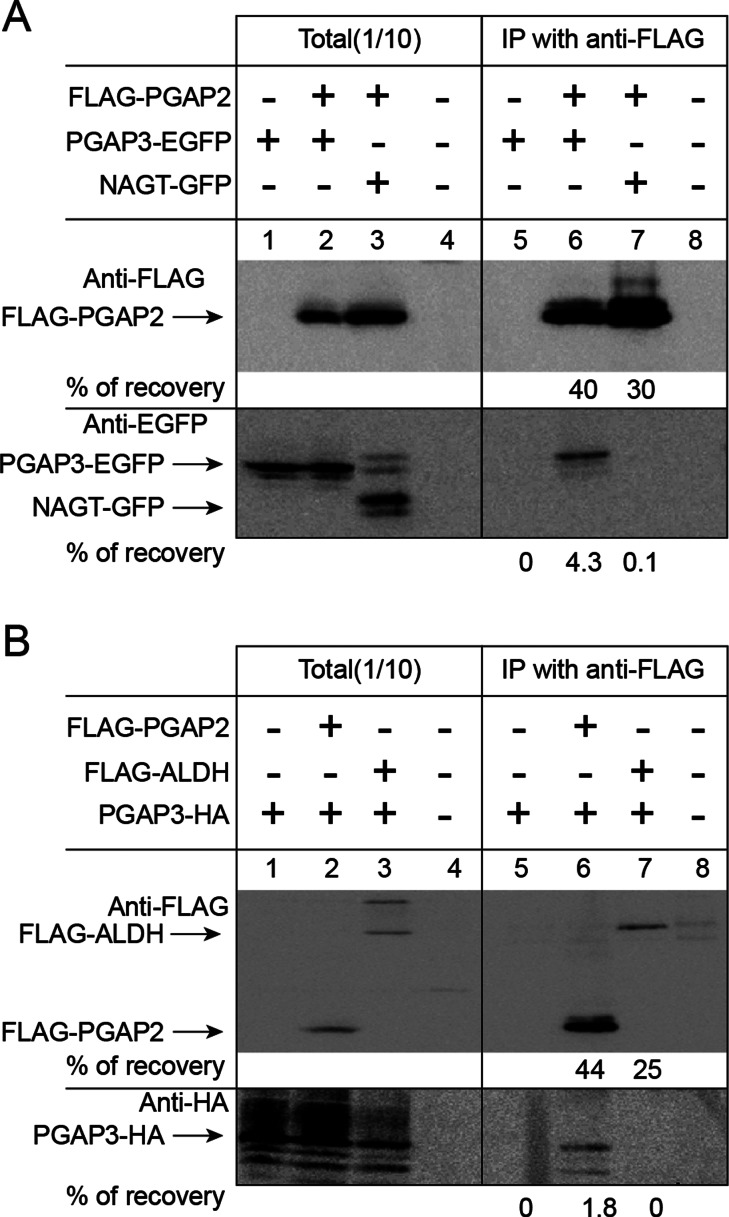
The released GPI-APs did not partition into the detergent phase of TX-114, suggesting that they had lost the acyl chains of the GPI anchor (Figure 1F). This phenotype of PGAP3-HA cells—a severe reduction of cell surface GPI-APs and release of GPI-APs lacking fatty acyl chain—is reminiscent of the PGAP2 mutant that is defective in the reacylation step, resulting in the generation of lysoGPI-APs that are secreted into the medium via the action of phospholipase D (Tashima et al., 2006). Of interest, the expression of nontagged PGAP3 did not cause this phenotype (data not shown), suggesting that addition of the C-terminal tag to PGAP3 specifically affects the reacylation step mediated by PGAP2. This hypothesis was supported by the fact that transient overexpression of PGAP2 partially restored the levels of GPI-APs at the cell surface in PGAP3-HA cells (Figure 1G). One possible explanation is that PGAP3 and PGAP2 form a protein complex to mediate their two reactions in coupled manner, and tagging at the C-terminal of PGAP3 disrupts this synergy. More directly we found that both C-terminally tagged PGAP3 constructs, PGAP3-EGFP and PGAP3-HA, can be specifically coprecipitated by FLAG-tagged PGAP2 (Figure 2, A and B). Taken together, our data suggest that PGAP2 activity is directly or indirectly affected by overexpression of PGAP3-HA, leading to the preferential production of the lysoGPI-APs, the lipid-remodeling intermediate (see details in Discussion).
FIGURE 2:
Physical interaction of PGAP3 and PGAP2. (A) Western blot analysis of the association of FLAG-PGAP2 with PGAP3-EGFP. CHO cells were transfected with FLAG-PGAP2 together with PGAP3-EGFP (lanes 2 and 6) and NAGT-GFP (lanes 3 and 7). NAGT-GFP is a nonrelevant Golgi transmembrane protein used as a control (Shima et al., 1997). (B) Western blot analysis of the association of FLAG-PGAP2 with PGAP3-HA. CHO cells were transfected with PGAP3-HA together with FLAG-PGAP2 (lanes 2 and 6) and FLAG-ALDH (lanes 3 and 7). FLAG-ALDH is a nonrelevant ER transmembrane protein used as a control (Masaki et al., 1996). (A, B) After solubilization of cells with lysis buffer containing 1% NP-40, FLAG-tagged proteins were affinity precipitated with anti-FLAG beads. Coprecipitated proteins were separated by SDS–PAGE and Western blotted against anti-FLAG, anti-EGFP, and anti-HA antibodies. Identities of bands and percentage of recovery are indicated.
In PGAP3-HA cells, produced lysoGPI-APs are not incorporated into DRMs
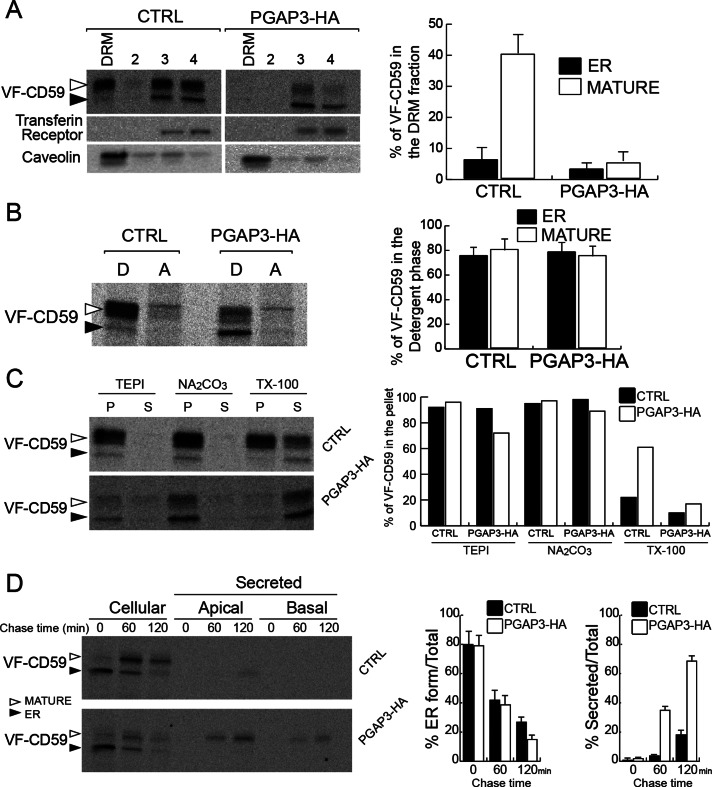
Numerous studies have shown that lipid remodeling in the Golgi apparatus confers on GPI-APs the ability to concentrate in DRMs (Bosson et al., 2006; Fujita et al., 2006; Maeda et al., 2007; Murakami et al., 2012). To examine whether lysoGPI-APs produced in PGAP3-HA cells are incorporated into DRMs, we pulse labeled cells with radiolabeled amino acids and analyzed the cells after a 1-h chase. The highly glycosylated mature form of VF-CD59 produced in the Golgi apparatus was found in the DRM fraction of control cells, as expected. In contrast, in PGAP3-HA cells, VF-CD59 cofractionated with the transferrin receptor, a marker of the glycerophospholipidic regions of the membrane (Figure 3A). This was not due to the loss of the DRM fraction, since a canonical raft marker, caveolin, still floated at the top of the Nycodenz gradient. Of importance, cellular lysoGPI-APs still partitioned into the detergent phase of TX-114, a classic criterion for membrane association (Figure 3B), and remained associated with membranes even after treatment with carbonate at high pH, a treatment that releases peripheral proteins from membranes (Figure 3C). These data demonstrate that intracellular lysoGPI-APs remain membrane anchored and exclude the possibility that lysoGPI-APs are not incorporated into DRMs due to weak association with membranes. Pulse-chase experiments showed that the kinetics of production and transport from the ER to the Golgi apparatus of GPI-APs in the control and PGAP3-HA cells are similar (Figure 3D). The appearance of mature GPI-APs in the medium was concomitant with their disappearance from PGAP3-HA cells, suggesting that lysoGPI-APs were secreted extracellularly soon after arrival at the cell surface (Figure 3D).
FIGURE 3:
Overexpression of PGAP3-HA results in loss of GPI-APs from DRM. (A) DRM assay for VF-CD59 in control and PGAP3-HA cells and quantification of the proportion of VF-CD59 in the DRM fraction. Radiolabeled VF-CD59 was immunoprecipitated from the density gradient fractions (DRM [top], 2, 3, and 4 [bottom]). Caveolin and the transferrin receptor were revealed by Western blotting. (B) Control and PGAP3-HA cells were grown on plastic dishes, pulse labeled, and chased for 1 h. Phase partitioning of newly synthesized VF-CD59 in control and PGAP3-HA cells into the detergent phase (D) and aqueous phase (A) of a Triton X-114 solution. (C) Membrane association of newly synthesized VF-CD59 prepared as in B. After homogenization of the cells and incubation of the extracts in buffer alone (TEPI), 0.1 M Na2Co3, pH 11, and 1% (wt/vol) Triton X-100, ultracentrifugation was performed to separate the membrane fraction (P) and the soluble fraction (S). (D) Cells grown on Transwell filters for 6 d were pulse labeled and chased for various times, as indicated. The cells and the media from the upper (apical) and lower (basal) chambers were analyzed. ER-to-Golgi transport was assessed by the disappearance of the ER form (left histogram). Secretion was expressed as percentage of the sum of apically and basolaterally secreted VF-CD59 compared with the total VF-CD59 signal (right histogram). (A, B, D) The data shown are means of three independent experiments. Error bars, 1 SD from the mean. (A–D) Newly synthesized VF-CD59 was detected by immunoprecipitation and PhosphorImaging after SDS–PAGE. The black arrowheads indicate the ER form and the white arrowheads the mature form of VF-CD59.
Fully remodeled GPI-APs and lysoGPI-APs are both targeted to the apical domain
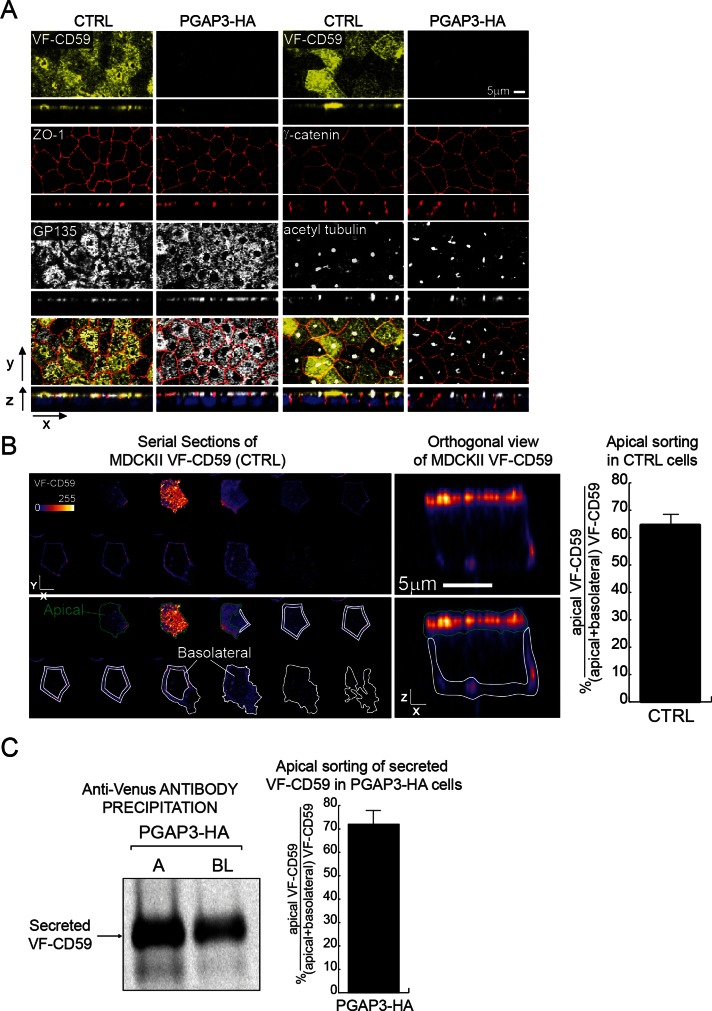
The PGAP3–HA cells grown on filters formed a polarized epithelial monolayer, and the expression and localization of the tight junction protein ZO-1, the apical transmembrane protein GP135, the lateral protein γ-catenin, and the primary cilium marker acetyl-tubulin were indistinguishable from those of control cells (Figure 4A). These results demonstrate that GPI-AP surface expression is not required for polarized monolayer formation in MDCK II cells.
FIGURE 4:
Apical targeting of GPI-APs in control and PGAP3-HA–expressing MDCK II cells grown on filters for 6 d. (A) Confocal microscopy of control (CTRL) and PGAP3-HA cells grown on filters. Apical (top) and transversal views (bottom) in each row show intrinsic fluorescence signal (VF-CD59) or staining for markers (ZO-1, GP135, γ-catenin, and acetyl tubulin). Bottom row, merged images. DAPI staining is shown in blue. (B) Serial optical sections and orthogonal views of VF-CD59 in individual control cells. The signal intensity was measured by immunofluorescence staining of VF-CD59 after surface labeling with an anti-Venus antibody. Only every third section in the stack is displayed. Top, fluorescence staining of Venus on the cell surface. Bottom, the regions bound in green and white show the areas used for quantification of the apical and basolateral surfaces, respectively. The means and 1 SD for 20 cells from two independent experiments are shown. The micrographs are pseudocolored by using the Look Up Table Fire in ImageJ. (C) Polarized release of VF-CD59 into the medium in PGAP3-HA cells. VF-CD59 released into the media in the apical (A) and basolateral (BL) chambers during a 2-h chase after pulse labeling was analyzed. The histogram shows mean apical sorting calculated from two independent experiments, and the visible part of the bar indicates the range of values.
The PGAP3-HA cells thus provided us with a model system to study the apical targeting of non–DRM-associated GPI-APs. First, we followed the trafficking of VF-CD59 expressed in control MDCK II cells as an authentic DRM-associated protein. These cells were mixed at a 1:10 ratio with MDCK II cells that did not express VF-CD59 in cocultures on filters. The mosaic culture allowed us to visualize by fluorescence microscopy isolated VF-CD59–expressing cells and quantify the signal from VF-CD59 in basolateral membranes without contribution from neighboring cells (Figure 4B). Surface VF-CD59 was specifically labeled with an anti-Venus antibody, and z-stacks of the entire cell volume were acquired by confocal microscopy with the pinhole aperture adjusted to the z-stack spacing. This analysis showed that 64.7 ± 4.0% of the total fluorescence signal was distributed on the apical membrane. Next we quantified the polarized targeting of lysoGPI-APs in PGAP3-HA cells. Because lysoGPI-APs are released into the medium in these cells, we analyzed polarized transport by quantifying the amounts of lysoGPI-APs secreted into the apical (A) and basolateral (B) media. After pulse labeling followed by a 2-h chase, 72.0 ± 6.1% of secreted VF-CD59 was released from the apical surface in PGAP3-HA cells (Figure 4C). Surprisingly, this figure was similar to that obtained in control cells for the percentage of apically localized VF-CD59 (64.7 ± 4.0%), indicating that the DRM association of GPI-APs has little or no influence on their apical targeting. To the best of our knowledge, this is the first demonstration that the intrinsic ability of GPI-APs to associate with DRM is not necessary for apical transport in polarized epithelial cells.
N-glycosylation is required for the apical targeting of fully remodeled and lysoCD59
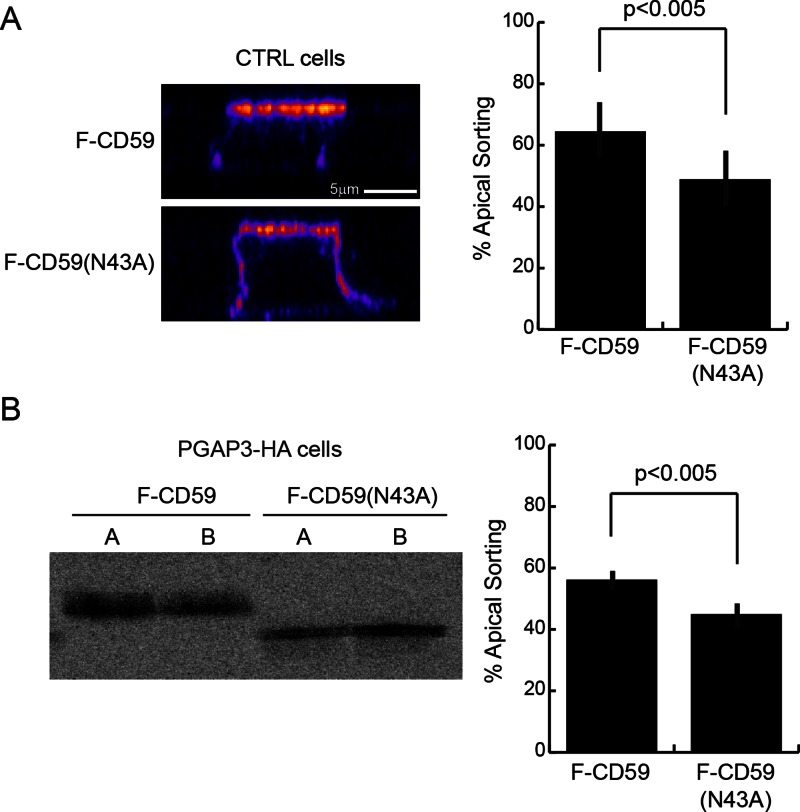
N-glycan has been shown to be involved in apical sorting of the naturally N-glycosylated GPI-anchored membrane dipeptidase and rat growth hormone engineered to contain two N-glycosylation sites and a GPI anchor (by the addition of anchoring signal of decay-accelerating factor) in MDCK cells (Benting et al., 1999; Pang et al., 2004). Because CD59 is a naturally N-glycosylated GPI-AP, we next examined the role of this N-glycan in CD59 sorting. We first compared polarized sorting of FLAG-tagged CD59 protein (F-CD59) and an N-glycan–free (N43A) variant (F-CD59(N43A)) in control cells (Figure 5A). We found that the apical localization of F-CD59(N43A) (48 ± 9.1% apical) was slightly but significantly lower than that of F-CD59 (64 ± 9.2% apical) in fully polarized MDCK cells. Next, we tested the effect of the N-glycan for the polarized targeting of lysoCD59 expressed in PGAP3-HA–expressing cells. We found that 56 ± 2.3% of secreted F-CD59 was released from the apical face of PGAP3-HA cells, whereas in contrast, 45 ± 3.1% of F-CD59(N43A) was released apically (Figure 5B). These results showed that N-glycosylation of CD59 contributes to efficient apical targeting of both fully remodeled and lyso forms of F-CD59.
FIGURE 5:
N-glycosylation is required for proper sorting of fully remodeled GPI-APs and lysoGPI-APs. (A) Orthogonal views of control cells expressing F-CD59 or F-CD59(N43A) grown on filters for 6 d. The signal intensity was measured by immunofluorescence staining of F-CD59 after fixation in 100% methanol at −20°C for 20 min and labeling with an anti-CD59 antibody. The means of apical sorting (percentage of apical signal/total signal) + SD for 20 cells from two independent experiments are shown. The micrographs are pseudocolored as in Figure 4B. (B) Polarized release of F-CD59 or F-CD59(N43A) into the apical (A) and basolateral (BL) chambers of PGAP3-HA cells grown on filters for 6 d during a 2-h chase after pulse labeling was analyzed using anti-FLAG M2 beads. The means of apical sorting (percentage of apical signal/total signal) + SD from five independent experiments are shown.
Cholesterol depletion impairs apical targeting of both fully remodeled and lysoGPI-APs
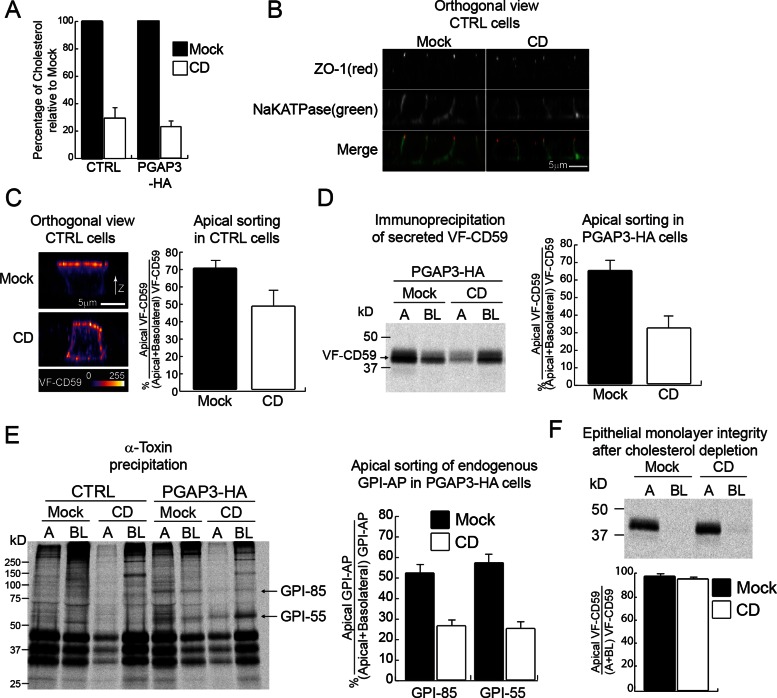
Previous studies showed that cholesterol depletion affects the DRM association of two GPI-APs, the placental alkaline phosphatase (PLAP) and GFP fused to the GPI anchor attachment signal from folate receptor in MDCK II cells, and impairs their apical sorting without affecting basolateral transport of the vesicular stomatitis virus G protein (Keller and Simons, 1998; Paladino et al., 2004). These results were interpreted as evidence that raft domains function either as a unit destined for the apical membrane or as a platform for GPI-AP oligomerization, which is required for apical targeting. This model predicts that apical targeting of lysoGPI-APs in PGAP3-HA cells should be lower than that of fully remodeled GPI-APs in control cells due to the lack of the DRM association. We found, in contrast, that apical targeting of VF-CD59 expressed in both cells was very similar, thereby questioning the DRM association as an apical sorting requirement for CD59. This prompted us to revisit the effects of cholesterol depletion on apical trafficking. We depleted total cholesterol by 71.1 ± 6.2% (control cells) and 77.1 ± 3.0% (PGAP3-HA cells) by both inhibiting cholesterol biosynthesis and removing existing cholesterol as previously described (Keller and Simons, 1998; Prydz and Simons, 2001; Paladino et al., 2004; Figure 6A). Under this condition, the localization of ZO-1 in the junctional region and the transmembrane protein Na/K ATPase at the basolateral membrane was not affected, demonstrating that junctional complexes remained intact after cholesterol depletion (Figure 6B). However, only 47.4 ± 8.9% of fully remodeled VF-CD59 distributed to the apical domain upon cholesterol depletion, as compared with 69.2 ± 3.9% in mock-treated cells (Figure 6C), confirming a predominant role for cholesterol in the apical sorting of VF-CD59 and in agreement with previous studies of other GPI-APs (Keller and Simons, 1998; Paladino et al., 2004). We further examined the effect of cholesterol depletion on the polarized release of lysoGPI-APs in PGAP3-HA cells. We found that the bulk of the protein was released basolaterally in cholesterol-depleted condition (31.3 ± 6.6% apical), whereas in mock-treated cells VF-CD59 was mostly secreted apically (64.0 ± 5.3% apical; Figure 6D). Of importance, cholesterol depletion also affected the apical sorting of two lysoGPI-APs endogenously expressed in MDCK cells. For unknown reasons, the background signal from affinity purification with α-toxin from low-density lipoprotein–depleted medium was much higher than from normal medium. Nevertheless, we could still see the two endogenous lysoGPI-APs, GPI-85 and GPI-55, released into media from PGAP3-HA–expressing cells but not from control cells as shown in Figure 1E (Figure 6E). Cholesterol depletion caused the preferential release of these two endogenous lysoGPI-APs from the basolateral surface, whereas in mock-treated cells, the proteins were released approximately equally from apical and basolateral membranes. This was not due to a defect in the monolayer integrity since in both mock and cholesterol-depleted conditions radiolabeled VF-CD59 added to the upper chamber was not recovered in the lower chamber even after 2 h of incubation (Figure 6F). Together these findings demonstrate that cholesterol depletion impairs the apical targeting of both fully remodeled GPI-APs and lysoGPI-APs. Therefore we conclude that cholesterol affects the apical transport of GPI-APs independently of their association with DRMs.
FIGURE 6:
Cholesterol depletion redirects fully remodeled and lysoGPI-APs to the basolateral membrane domain. (A) The histogram shows the mean amount of cellular cholesterol after cholesterol depletion (CD) relative to mock-depleted cells (Mock) in the parental cell line (CTRL) and in PGAP3-HA cells, calculated from three independent experiments. (B) Orthogonal views of control cells stained for markers (ZO-1, Na/K ATPase) after mock depletion (Mock) and cholesterol depletion (CD); the merged images are displayed in the bottom row. (C) Orthogonal views of control cells expressing VF-CD59 growing on filters after mock depletion (Mock) or cholesterol depletion (CD). The means of apical sorting (percentage of apical signal/total surface signal) + SD for 20 cells from two independent experiments are shown. The micrographs are pseudocolored as in Figure 4B. (D) Polarized release of VF-CD59 into the apical (A) and basolateral (BL) chambers of PGAP3-HA cells growing on filters after mock depletion (Mock) or cholesterol depletion (CD). The means of apical sorting (percentage of apical signal/total signal) + SD from three independent experiments are shown. (E) The control and PGAP3-HA cells growing on filters were depleted of cholesterol (CD) or mock depleted (Mock) and were pulse labeled and chased for 2 h. Endogenous GPI-APs released into the apical (A) and basolateral (BL) chambers were recovered by binding to recombinant α-toxin from C. septicum. The data show the mean and 1 SD from six independent experiments. (F) PGAP3-HA–expressing MDCK II cell monolayers were grown to confluency on filters and then either depleted of cholesterol (CD) or mock depleted (Mock). To determine epithelial monolayer integrity, radiolabeled VF-CD59 was added to the apical side of the monolayers. Two hours later, the apical (A) and basolateral (BL) media were recovered, VF-CD59 was immunoprecipitated, separated by SDS–PAGE, and revealed by PhosphorImager analysis (top). The data show the mean of apical sorting (percentage of apical signal/total signal) and 1 SD from three independent experiments.
Upon cholesterol depletion, apically destined membrane proteins accumulate intracellularly and are transported to the basolateral membrane
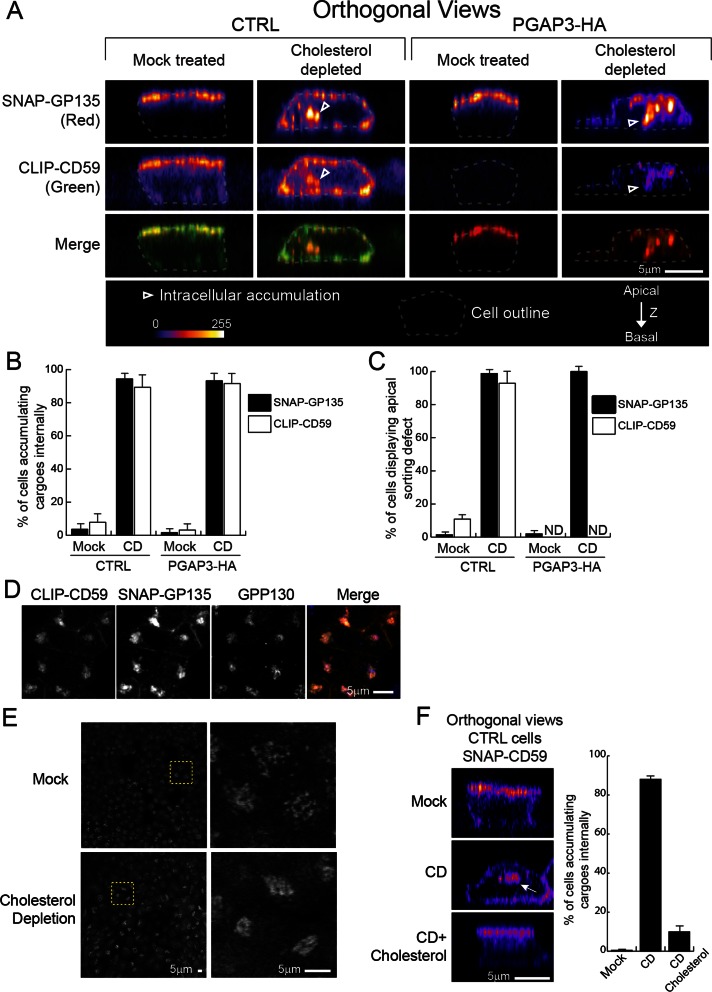
To further analyze the effect of cholesterol depletion on apical protein transport, we examined the localization of newly synthesized proteins destined for the apical membrane by using SNAP and CLIP tags. SNAP and CLIP tags are derived from the DNA repair protein O6-alkylguanine-DNA alkyltransferase and can thus be labeled covalently in living cells with O6-benzylguanine and O6-benzylcytosine derivatives, respectively (Gautier et al., 2008). We established control and PGAP3-HA cells coexpressing SNAP-tagged transmembrane protein, GP135, and CLIP-tagged CD59. The cells were seeded in mixed cultures with normal MDCK II cells (1:1 ratio) on filters. After cholesterol depletion, cells were incubated with nonfluorescent, membrane-permeable substrates to eliminate the reactivity of all existing SNAP-GP135 and CLIP-CD59. After washing, synthesis and transport of the newly synthesized proteins continued for 4 h in the presence of lovastatin. The proteins synthesized during this 4-h incubation were labeled with the fluorescent, membrane-permeable substrates and cells were fixed. In mock-treated control cells, both proteins were located at the apical cell surface, demonstrating that tagging did not affect targeting (Figure 7A). Cholesterol depletion in control cells caused redistribution of both proteins to the basolateral membrane with a concomitant increase in their intracellular localization (Figure 7, A–C). The major intracellular fraction of SNAP-GP135 and CLIP-CD59 colocalized with the cis-Golgi marker GPP130 (Figure 7D). In mock-treated PGAP3-HA cells, SNAP-GP135 localized to the apical surface, and CLIP-CD59 was undetectable at the cell surface or intracellularly (Figure 7A), presumably because it is secreted, similar to VF-CD59 (Figures 1C and 4A). By contrast, cholesterol depletion of PGAP3-HA cells resulted in accumulation of intracellular CLIP-CD59 (Figure 7, A and B), indicating that transport of lysoGPI-APs is also sensitive to cholesterol depletion, again suggesting that cholesterol depletion affects GPI-APs transport beyond its accepted role in the DRM formation. Consistent with this, cholesterol depletion was described to affect the secretory pathway (Wang et al., 2000; Stuven et al., 2003). In our experimental setup we observed that cholesterol depletion promotes compaction of the Golgi apparatus (Figure 7E). The accumulation of SNAP-CD59 in the Golgi apparatus observed in control cells after cholesterol depletion was rescued by adding back cholesterol, suggesting that cholesterol has a direct role in this process (Figure 7F). Suspecting that rather than promoting the apical targeting of some selected secretory proteins, cholesterol is more generally involved in the whole apical transport pathway, we analyzed the effect of cholesterol depletion on polarized secretion of endogenous secretory proteins in MDCK cells, using a pulse-chase protocol. We detected a number of endogenous soluble proteins, including three major proteins between 30 and 45 kDa likely corresponding to the subunits of gp80 (Kondor-Koch et al., 1985; Urban et al., 1987; Supplemental Figure S1). On cholesterol depletion, all endogenous secretory proteins detected were secreted preferentially toward basolateral membrane, as previously shown for gp80 (Keller and Simons, 1998). We conclude that cholesterol depletion causes defective secretion of all classes of proteins destined for the apical membrane, resulting in their retargeting toward the basolateral membrane independently of their association with membrane and DRMs.
FIGURE 7:
Cholesterol depletion causes accumulation into the Golgi apparatus of cargo proteins destined for the apical surface. (A) Orthogonal views of the parental cell line (CTRL) and PGAP3-HA–expressing cells, both expressing SNAP-GP135 and CLIP-CD59, growing on filters after mock depletion (Mock treated) or cholesterol depletion (Cholesterol depleted). Newly synthesized proteins were visualized by staining with membrane-permeable fluorescent substrates. The outline of each cell is indicated by a dashed line. Arrowheads point to intracellular accumulation. The micrographs are pseudocolored as in Figure 4B. The merged images are displayed in the bottom row. (B) The histogram shows the mean percentage of cells in which internal accumulation of SNAP-GP135 or CLIP-CD59 was observed. (C) The histogram shows the mean percentage of cells in which the proteins were observed in basolateral membranes. (B, C) Error bars, 1 SD. From 50 to 80 cells were analyzed for each condition. (D) Colocalization of CLIP-CD59 (green) and SNAP-GP135 (red) with GPP130 (blue). Control cells were depleted for cholesterol, and CLIP-CD59 and SNAP-GP135 were labeled as in A. (E) Golgi morphology revealed by immunofluorescence of GPP130 in MDCKII cells after mock treatment or cholesterol depletion. Left, the entire image; right, the zoom-in of the yellow selection. (F) Control MDCK II cells (CTRL) expressing SNAP-CD59 were grown on filters and mock depleted (Mock), depleted for cholesterol (CD), or depleted and then replenished with cholesterol by treatment with a complex of with 2.5 mM methyl-β-cyclodextrin and cholesterol. The images are orthogonal views of the cells showing SNAP-CD59 fluorescence. The histogram shows the mean percentage of cells in which SNAP-CD59 accumulated inside the cells. Error bars, 1 SD. Between 200 and 300 cells were analyzed for each condition.
DISCUSSION
Coupling of the two GPI lipid-remodeling steps through the association of PGAP3 and PGAP2 in a protein complex
In this study, we found that the GPI lipid-remodeling reaction is inhibited by overexpression of C-terminally tagged PGAP3 and not by nontagged PGAP3. We first suspected that C-terminal tagging affects the activity of PGAP3. However, we excluded that possibility because PGAP3-HA–expressing cells mimic the PGAP2-mutant phenotype (secretion of GPI-APs) and not the PGAP2/PGAP3-mutant phenotype (Tashima et al., 2006; Maeda et al., 2007). We next hypothesized that C-terminal tagging of PGAP3 instead affects PGAP2 activity. Supporting this, we found that C-terminally tagged PGAP3 physically interacts with FLAG-tagged PGAP2. We therefore suggest that PGAP2 and PGAP3 form a functional lipid-remodeling complex to ensure almost simultaneous deacylation and reacylation of the GPI anchor, thereby minimizing the generation of lysoGPI-APs. It is important to note that although we observed that C-terminally tagged PGAP3 interacts with FLAG-PGAP2, the amount of tagged PGAP3 coprecipitated was rather low, and therefore we do not exclude that endogenous, nontagged PGAP3 interacts with PGAP2 more efficiently than C-terminally tagged PGAP3. On the basis of these considerations, we propose two nonexclusive models to explain why PGAP3-HA overexpression causes a PGAP2 mutant phenotype: 1) C-terminal tagging of PGAP3 drastically lowers the affinity against PGAP2, and therefore the lipid-remodeling step is uncoupled, resulting in preferential production of lysoGPI-APs. The amount of endogenous PGAP2 might be still sufficient to form functional complex with overexpressed nontagged PGAP3. PGAP2 overexpression in PGAP3-HA–expressing cells might then reduce the pool of uncomplexed PGAP3-HA and contributes to a partial restoration of cell surface expression of GPI-APs. 2) Within the PGAP3-HA/PGAP2 complex the presence of C-terminal tag affects the lipid reacylation activity mediated by PGAP2. Therefore the overexpression of PGAP3-HA would mainly lead to formation of the complex PGAP3-HA/PGAP2, which is incompetent for the reacylation step and produce a large amount of lysoGPI-APs. By additional overexpression of PGAP2 in PGAP3-HA cells, endogenous PGAP3 in PGAP3-HA–expressing MDCKII cells can form the functional remodeling complex PGAP3/PGAP2 (reduction of competition between endogenous PGAP3 and PGAP3-HA for interaction with PGAP2) and contributes to partial restoration of the cell surface expression of GPI-APs.
Because fatty acid remodeling is also observed among general glycerophospholipid species, as first proposed in terms of Lands’ cycle (Lands, 1958; Shindou et al., 2009), our findings suggest that a similar coupling of deacylation and acylation reactions might also occur in this general lipid-remodeling step. Furthermore, we observed a similar phenotype after overexpression of C-terminally tagged PGAP3 in CHO cells, suggesting that this method to perturb enzyme activity could be used to address the roles of lipid remodeling and the function of cell surface GPI-APs in any cell type by simple expression of one construct.
Role of DRM association in apical targeting of GPI-APs in polarized epithelial cells
LysoCD59 in PGAP3-HA–expressing MDCK cells is not incorporated into DRMs, in contrast to fully remodeled CD59 in control cells. The fact that the apical targeting efficiency of lysoCD59 is not affected despite its lack of DRM association in PGAP3-HA cell contradicts the previous finding that disruption of DRM association of GPI-APs impairs apical targeting of GPI-APs (Keller and Simons, 1998; Paladino et al., 2004). The critical difference between the previous and the present study is that we affect the intrinsic ability of GPI-APs to associate with DRMs without manipulating the cellular cholesterol content, which most likely affects general apical transport, as shown by others (Prydz and Simons, 2001) and in this study. We used detergent insolubility to examine potential lipid raft association in the cells, similar to most previous studies elucidating roles of lipid rafts. However, one can still hypothesize that lysoGPI-APs might loosely bind to lipid rafts in vivo, and this association would not be recovered after treatment with the detergent in vitro. The effect of cholesterol depletion on apical sorting of lysoGPI-APs would not disagree with this hypothesis. Owing to the lack of assays unambiguously determining the association of proteins with lipid rafts in vivo, our interpretation of our data relies on the simplest hypothesis that lysoGPI-APs are not enriched into a specific membrane environment. Therefore we speculate that apical sorting of lysoGPI-APs and possibly fully remodeled GPI-APs does not depend on lipid raft association. Recently, a PGAP3-knockout mouse was generated, and altered immune response in this mutant mouse was reported (Murakami et al., 2012). In these cells, GPI-APs still contain one unsaturated fatty acid in sn-2 position in their lipid moieties and are likely not in the lipid raft in the cells. The localization of endogenous GPI-APs in different polarized tissues in this mouse will further verify the contribution of lipid raft association for apical sorting.
The N-glycosylation requirement for apical sorting of fully remodeled and lysoCD59
We found that loss of N-glycosylation causes an apical sorting defect of CD59 in both control and PGAP3-HA cells, suggesting that the N-glycan is involved in apical sorting of CD59 independently of DRM association. Even though it is significant, the degree of missorting is not as prominent, as seen for other N-glycosylated GPI-APs, such as GPI-anchored membrane dipeptidase (Pang et al., 2004). Because CD59 is also O-glycosylated (Rudd et al., 1997), the O-glycan might also contribute to the apical transport of CD59, as seen for the neutrophin receptor p75NTR, an apical transmembrane protein (Yeaman et al., 1997). Galectins constitute a family of soluble lectins that are released from cells via a nonclassic secretion mechanism, bind to cell-surface proteins, and can be internalized into an endosomal compartment (Lindstedt et al., 1993; Delacour et al., 2009). Among them, galectin-3 and -4 are proposed to be involved in the apical localization of detergent-soluble transmembrane protein such as p75NTR (Delacour et al., 2006) and detergent-insoluble membrane proteins, including two GPI-APs, carcinoembryonic antigen and CD59, respectively, in polarized epithelial cells (Delacour et al., 2005; Stechly et al., 2009). Further analysis to determine the requirement of these galectins for CD59 in control and PGAP3-HA–expressing cells will clarify the roles of sugar modification of GPI-APs in relation to their DRM association.
Cholesterol-dependent apical sorting does not reflect a requirement for GPI-APs to associate with DRMs
We observed that cholesterol depletion reduced the apical targeting of fully remodeled CD59 as expected, as well as of the lyso form of several GPI-APs, including two endogenously expressed GPI-APs. Cholesterol depletion also caused intracellular accumulation of both fully remodeled and lyso forms of CD59 (Figure 7A), which is in stark contrast to the lack of accumulation of CD59 intracellularly after specifically interfering with the remodeling of the GPI anchor (Figures 4A and 7A). These results emphasize that cholesterol depletion likely affects apical transport of GPI-APs beyond their exclusion from DRMs. Previously it was shown that cholesterol depletion reduces apical transport of DRM-associated proteins, including HA, two GPI-APs, PLAP, and GFP-GPI (GFP fused to the GPI anchor attachment signal from the folate receptor), several apical secreted proteins, including glycosylated gp 80, and sulfated macromolecules (Keller and Simons, 1998; Prydz and Simons, 2001; Paladino et al., 2004). We further found that secretion of all endogenous secretory proteins detected by our assay occurred mainly toward the basolateral domain after cholesterol depletion. Cholesterol depletion is known to cause defects in several specific membrane traffic steps, including intra-Golgi transport (Stuven et al., 2003) and secretory vesicle formation at the trans-Golgi network (Wang et al., 2000). Under these conditions, however, in polarized cells, basolateral secretion continues, even carrying apically destined soluble and membrane proteins. Further study is needed to identify which machineries are responsible for this mixed traffic and might help to reveal the mechanism of the cholesterol dependence of apical transport. However, cholesterol depletion likely has broad effects, as the treatment will change general membrane biophysical properties. In such a context, the observed disturbance of the apical trafficking pathway might be of indirect consequence.
MATERIALS AND METHODS
Cells and reagents
MDCK II cells were purchased from the European Collection of Cell Cultures (Salisbury, United Kingdom; catalogue no. 00062107). These cells were routinely cultured in DMEM Glutamax-I (Sigma-Aldrich, St. Louis, MO) supplemented with 10% fetal calf serum (FCS), 1× MEM nonessential amino acids (Sigma-Aldrich), and antibiotics at 37°C in an atmosphere of 5% CO2. Clones of MDCK II cells stably expressing VF-CD59, F-CD59, F-CD59(N43A), SNAP-CD59, and CLIP–CD59 were obtained by transfecting the cells with the expression vectors pME-puro-Venus-FLAG-CD59 (Rivier et al., 2010), pME-puro-FLAG-CD59, pME-puro-FLAG-CD59(N43A), pME-puro-SNAP-FLAG-CD59 (this study), and pME-puro-CLIP-FLAG-CD59 (this study) in Lipofectamine 2000 (Invitrogen, Carlsbad, CA) and selecting with 2 μg/ml puromycin in complete DMEM. Clones of MDCK II cells stably expressing SNAP-GP135 were obtained by transfecting with pCDNA3.1(+)-myc-SNAP-GP135 (this study) and selecting with 750 μg/ml G418 in complete DMEM. Clones of MDCK II cells overexpressing PGAP3-HA were obtained by transfecting with pME-hygro-hPGAP3–3HA (this study) and selecting with 400 μg/ml hygromycin in complete DMEM. The cells were cultured either on plastic dishes to 80–90% confluency or on filters in polycarbonate Transwell permeable supports (105 cells per 0.33 cm2 filter or 1.5 × 106 cells per 4.67 cm2 filter; Corning, Tewksbury, MA) for the number of days indicated. PGAP2 and PGAP3 double-mutant CHO cells (Maeda et al., 2007) were provided by T. Kinoshita (Research Institute for Microbial Diseases, Osaka University, Osaka, Japan). CHO cells and their transformants were routinely cultured in Ham's F-12 medium (Sigma-Aldrich) supplemented with 10% FCS and antibiotics at 37°C in an atmosphere of 5% CO2. The antibodies used were as follows: rat monoclonal antibody against HA (clone 3F10) from Roche Applied Bioscience (Indianapolis, IN), rabbit antibody against ZO-1 (40-2200) and mouse antibody against the transferrin receptor (13-6800) from Invitrogen, rabbit antibody against γ-catenin (sc-7900) from Santa Cruz Biotechnology (Santa Cruz, CA), mouse monoclonal antibodies against acetylated tubulin (clone 6-11B-1), tubulin (clone B-5-1-2), and FLAG (M2) from Sigma-Aldrich, rabbit antibody against caveolin (610059) from BD Biosciences (San Diego, CA), mouse antibody against Na/K ATPase (05-382) from Millipore (Billerica, MA), and rabbit antibody against GPP130 from Covance (Berkeley, CA). Mouse monoclonal antibody against GP135 was provided by G. K. Ojakian (SUNY Downstate Medical Center, New York, NY). Mouse monoclonal antibody against CD59 (clone 5H8) was provided by T. Kinoshita. Rabbit anti-Venus antibody was produced by immunizing rabbits with recombinant GST-Venus protein. FLAER, the Alexa 488–labeled variant of aerolysin, was purchased from Pinewood Scientific Services (Victoria, Canada). All SNAP and CLIP substrates were purchased from New England BioLabs (Ipswich, MA).
Plasmids
The pME-puro-HisFLAG-ratPGAP2, pGEX-α-toxin (R398G), pME-hPGAP3-3HA, pME-puro-FLAG-CD59, and pME-puro-FLAG-CD59(N43A) plasmids were obtained from T. Kinoshita. The pCNG2-encoding N-terminal domain (amino acids 1–103) of N-acetylglucosaminyltransferase I (NAGT I), including cytoplasmic tail, membrane-spanning domain, and stalk domain fused with GFP (Shima et al., 1997), was obtained from J. Gruenberg. To construct the p-dogPGAP3-EGFPN1, we first obtained sequence information on full-length dogPGAP3 cDNA by 3′ rapid amplification of cDNA ends, using cDNA prepared from MDCK II cells (European Nucleotide Archive, accession number: HF937279). Next, cDNA encoding dogPGAP3 was amplified by PCR with cDNA prepared from MDCK II cells by upper PCR primer containing the XhoI site and lower PCR primer containing BamHI, and it was then integrated into pEGFP-N1 (Clontech, Mountain View, CA). To construct pME-hygro-hPGAP3-3HA, a BamHI fragment containing cDNA encoding hPGAP3 fused to a triple-HA tag at its C-terminus, derived from pME-hPGAP3-3HA (Maeda et al., 2007), was ligated with a BamHI fragment of pME-hygro (+) provided by Y. Maeda (Osaka University, Osaka, Japan). To construct pME-puro-SNAP- or CLIP-FLAG-CD59, the cDNA encoding the SNAP or CLIP tag was amplified by PCR with pSNAP-tag(m)-E30R or pCLIP-tag(m)-E30R (provided by K. Johnsson, Ecole Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) by using upper and lower PCR primers both containing SbfI sites and was then integrated into pME-puro-FLAG-CD59 at the SbfI site in the plasmid. To construct pCDNA3.1(+)-myc-SNAP-GP135, the cDNA encoding the SNAP tag was amplified by PCR with pSNAP-tag(m)-E30R by using upper and lower PCR primers both containing XhoI sites and then integrated into the vector part of pCDNA3.1(+)-myc-Gp135 after XhoI digestion of pCDNA3.1(+)-myc-EGFP-Gp135 provided by Tzuu-Shuh Jou (National Taiwan University Hospital and National Taiwan University College of Medicine, Taipei, Taiwan). The dogPGAP3, SNAP, and CLIP tag cDNAs made by PCR amplification were verified by sequencing.
Flow cytometry
Cells stained with mouse anti-FLAG antibody (Sigma-Aldrich) and allophycocyanin-conjugated donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, West Grove, PA) were analyzed by flow cytometry on a FACSCalibur (BD Biosciences) or Gallios (Beckman Coulter, Brea, CA) machine. The data were analyzed with FlowJo software (Tree Star, Ashland, OR). FLAER staining was performed according to the manufacturer's protocols (Brodsky et al., 2000).
Analysis of the protein complexes
CHO cells derived from one 14-cm dish were electroporated with 10 μg each of plasmid at 260 V and 960 μF in a Gene Pulse (Bio-Rad, Hercules, CA). Cells were grown for 24 h to allow protein expression and then harvested using a scraper. One-tenth of cells were kept as total fraction, and the rest were solubilized with 1 ml of NP-40 lysis buffer (1% NP-40, 50 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA) and protease inhibitors (PIs; 10 μg/ml leupeptin, 5 μg/ml pepstatin A, and 2 μg/ml aprotinin) in a cold room for 2 h. The total cell lysates were centrifuged at 20,000 × g for 20 min at 4°C to remove cell aggregates and/or insoluble material, and the supernatant fractions were incubated with 20 μl (bed volume) of anti-FLAG M2 beads (Sigma-Aldrich). The precipitates were washed three times with NP-40 lysis buffer, mixed with 2× sample buffer containing 10% 2-mercaptoethanol, and then separated by SDS–PAGE and analyzed by Western blotting.
Metabolic labeling and immunoprecipitation
Before metabolic labeling, cells grown on filters in polycarbonate Transwell permeable supports or on plastic dishes (at 60–80% confluency) were preincubated with DMEM lacking l-methionine and l-cysteine (Sigma-Aldrich) supplemented with 10% dialyzed FCS, 1 mM sodium pyruvate, and 2 mM glutamine for 30 min and then pulsed with 100 μCi/ml [35S]methionine and [35S]cysteine (Easy Tag Expres35s; PerkinElmer, Waltham, MA) for 30 min. After pulse labeling, cells were cultured in fresh complete medium containing 1.7 mM each of l-methionine and l-cysteine for the indicated times. At the end of the chase period, metabolism was immediately stopped by addition of 10 mM NaF and 10 mM NaN3. The media from the upper and lower chambers of the Transwell supports were collected individually, and the cells on the filters were solubilized with RIPA buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) supplemented with protease inhibitors. The cells on plastic dishes were harvested by scraping and used for various biochemical experiments. Before immunoprecipitation, all fractions were centrifuged at 20,000 × g for 15 min at 4°C to remove cell aggregates and/or insoluble material, and the supernatant fractions were incubated with anti-Venus antibody and rProtein A Sepharose Fast Flow (GE Healthcare, Piscataway, NJ). The precipitates were washed three times with RIPA buffer, mixed with 2× sample buffer containing 10% 2-mercaptoethanol, and then separated by SDS–PAGE and analyzed with a PhosphorImager (Packard Instruments, Meriden, CT).
Isolation of GPI-APs from the medium by affinity purification with recombinant Clostridium septicum α-toxin
GST-tagged mutated Clostridium septicum α-toxin (Hong et al., 2002) expressed in BL21(DE3) Escherichia coli cells was purified on Glutathione Sepharose 4 Fast Flow affinity medium (GE Healthcare) according to the manufacturer's protocol. The media from the upper and lower chambers of pulse-labeled cells chased for 2 h were divided into two parts. One was incubated with GST–α-toxin bound to Glutathione Sepharose 4 Fast Flow, and the other was incubated with anti-Venus antibody and rProtein A Sepharose Fast Flow (GE Healthcare). The precipitates were washed three times with PBS(+) (phosphate-buffered saline [PBS] supplemented with 0.5 mM MgCl2 and 1 mM CaCl2), separated by SDS–PAGE, and analyzed with a PhosphorImager.
Detergent-resistant membrane assay
This assay was performed essentially as described (Schuck et al., 2003). Radiolabeled cells were resuspended in 500 μl of TNE (150 mM NaCl, 2 mM EDTA, 50 mM Tris-HCl, pH 7.4) supplemented with protease inhibitors. The suspension was homogenized by passage through a 22-gauge needle 20 times. A 320-μg amount of protein was resuspended in 180 μl of TNE + PIs, and then 20 μl of 10% (wt/vol) TX-100 was added, mixed gently, and incubated for 30 min on ice. After this incubation, we added 400 μl of 60% OptiPrep density gradient medium (Sigma-Aldrich) and transferred the whole mixture to 2-ml Ultra-Clear centrifuge tubes (Beckman Coulter), overlaid it with 1.2 ml of 30% OptiPrep in TNE plus PIs and 0.2 ml of TNE plus PIs, and centrifuged it at 55,000 rpm for 2 h at 4°C in a TLS55 rotor (Beckman Coulter). Four fractions of 0.5 ml were taken, starting from the top of the tubes. A 20-μl amount of each fraction was used for Western blotting, and 200 μl of each fraction was diluted into 2 ml of RIPA buffer supplemented with PIs for immunoprecipitation with anti-Venus antibody and rProtein A Sepharose Fast Flow.
GPI anchor attachment assay
This assay was performed essentially as described (Watanabe et al., 2002). The radiolabeled cells (500 μg) were suspended in TE (100 mM Tris-HCl, pH 7.5, 10 mM EDTA) supplemented with protease inhibitors (TEPI) and homogenized by passing through a 22-gauge needle 20 times. The cell extract was solubilized with Triton X-114 at a final concentration of 1% in TEPI. The insoluble materials were removed by centrifugation at 20,000 × g for 15 min at 4°C. After incubation at 32°C, followed by centrifugation at 3000 × g for 3 min at room temperature, the upper, aqueous and lower, detergent phases were collected separately. After two rounds of partitioning into detergent and aqueous phases, the detergent phase was diluted with 10 volumes of RIPA buffer with PIs, and 10× RIPA buffer plus PIs was added to the aqueous phase. Both phases were then processed for immunoprecipitation.
Membrane association assay
This assay was performed essentially as described (Watanabe et al., 2002). Radiolabeled cells (500 μg) were suspended in TEPI and homogenized by 20 strokes with a 22-gauge needle before treatment. Each cell extract was divided into three parts. The parts were mixed with an equal volume of TEPI, 0.2 M Na2Co3, pH 12, in TEPI (to a final pH of 11), or 2% (wt/vol) Triton X-100 in TEPI and incubated on ice for 30 min. Soluble and pellet fractions were prepared by centrifugation at 65,000 rpm for 30 min at 4°C in a TLA100.3 rotor (Beckman Coulter). The soluble fractions from 0.1 M Na2Co3 treatment were precipitated with 10% trichloroacetic acid, and other fractions were solubilized with RIPA buffer supplemented with protease inhibitors. All were processed for immunoprecipitation.
Immunofluorescence microscopy
MDCK II cells grown on Transwell filters for the indicated times were fixed with either 100% methanol at –20°C for 20 min (Figure 5A) or 4% paraformaldehyde for 15 min. In the case of fixation with paraformaldehyde, after quenching with 10 mM glycine in PBS(+), the filters were removed from their supports, and the cells were permeabilized with 0.1% Triton X-100 in PBS(+) for intracellular labeling (as in Figure 4A) or not, in cases in which surface immunolabeling was performed (as in Figures 4B and 6C). Cells on filters were blocked with 3% BSA in PBS(+) and then incubated for at least 2 h in the first antibody diluted in 1% BSA in PBS(+). After washing twice with PBS(+), the cells were incubated with donkey Cy3- or Cy5-conjugated anti-rabbit or mouse IgG (1:200; Jackson ImmunoResearch) in 3% BSA in PBS(+) for 1 h. After washing three times with in PBS(+), filters were mounted in Prolong Gold antifade reagent with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). The labeled cells were observed, and images were acquired with a 63×/1.4 numerical aperture oil objective lens and a Zeiss 510 Meta confocal microscope (Zeiss, Jena, Germany) at a stack spacing of 500 nm. Images of Venus-CD59– or F-CD59–expressing MDCK II cells in Figures 4B, 5A, and 6C were acquired with a 63×/1.4 numerical aperture oil objective lens with a Leica TCS STED CW confocal microscope at a stack spacing of 400 nm. Quantification of the images was performed by using ImageJ software (National Institutes of Health, Bethesda, MD).
Cholesterol depletion
Cholesterol depletion was carried out basically as described previously, with slight modification (Keller and Simons, 1998; Prydz and Simons, 2001; Paladino et al., 2004). MDCK II cells stably expressing VF–CD59 were seeded on filters in Transwell supports in DMEM supplemented with 5% FCS, 1× MEM nonessential amino acids, and antibiotics. After 24 h, the cells on filters were washed twice with DMEM alone, and the medium was changed to DMEM supplemented with 5% lipoprotein-deficient FCS, 1× MEM nonessential amino acids, and antibiotics with (for cholesterol depletion) or without (for mock treatment) 4 μM lovastatin and 0.25 mM mevalonate. After 48 h, the cells were washed twice with warm DMEM alone and then cultured in DMEM supplemented with 1× MEM nonessential amino acids (Sigma-Aldrich) and antibiotics with (for cholesterol depletion) or without (for mock treatment) 10 mM methyl-β-cyclodextrin, 4 μM lovastatin, and 0.25 mM mevalonate for 60 min at 37°C in an atmosphere of 5% CO2. We observed significant relocalization of VF-CD59 after cholesterol depletion when we used lipoprotein-deficient FCS but not normal FCS. Therefore we use lipoprotein-deficient FCS for both the cholesterol depletion and mock treatments. For measurement of intracellular cholesterol content, cells growing on filters were washed three times with PBS(+) scraped in HB buffer (3 mM imidazole, pH 7.3, 8.5% sucrose plus PIs), and the amount of total cholesterol (free and esterified) was measured by using the Amplex Red Cholesterol Assay Kit (Molecular Probes, Eugene, OR) according to the manufacturer's protocols. Data were normalized to the amount of protein to estimate the efficiency of cholesterol depletion.
Analyses of newly synthesized SNAP- and CLIP-tagged protein in mock-treated or cholesterol-depleted conditions
MDCK II cells stably coexpressing CLIP-CD59 and SNAP-GP135 with or without PGAP3-HA were seeded with normal MDCK II cells (1:1 ratio) on filters in Transwell supports and cultured as described. After mock or methyl-β-cyclodextrin treatment, all existing SNAP- or CLIP-tagged proteins were blocked by incubation in 1.4 μM SNAP–Cell Block and CLIP–Cell Block for 30 min at 37°C. After washout of the Cell Block reagent with warm DMEM without FCS, cells were incubated in DMEM supplemented with 5% lipoprotein-deficient FCS, 1× MEM nonessential amino acids, and antibiotics with (for cholesterol depletion) or without (for mock treatment) 4 μM lovastatin and 0.25 mM mevalonate for 4 h at 37°C. For the cholesterol replenishment experiment, methyl-β-cyclodextrin:cholesterol complexes were prepared at molar ratio of 5:1, as previously described (Christian et al., 1997), and cells were cultured in DMEM supplemented with 5% lipoprotein-deficient FCS, 1× MEM nonessential amino acids, antibiotics, and 2.5 mM methyl-β-cyclodextrin:cholesterol complex in the presence of 4 μM lovastatin and 0.25 mM mevalonate. The proteins synthesized during this period were fluorescently labeled by incubation with 0.3 μM SNAP–Cell TMR and 0.5 μM CLIP–Cell 505 for 1 h at 37°C in the appropriate medium. After washing three times with the appropriate medium at 37°C, cells were fixed in 100% methanol at −20°C for 20 min and washed three times with PBS(+). The filters were removed from the Transwell supports and mounted in Prolong Gold antifade reagent with DAPI. Images were acquired with a 63×/1.4 numerical aperture oil objective lens with a Leica TCS STED CW confocal microscope at a stack spacing of 400 nm. Quantification of the images was performed by using ImageJ software.
Supplementary Material
Acknowledgments
We thank J. Gruenberg, C. Scott, H. Riezman, Y. Maeda, and M. Fujita for comments; P. Couleru-Burriat for technical assistance; T. Kinoshita, G. K. Ojakian, Tzuu-Shuh Jou, and K. Johnsson for reagents; G. Lukinavicius and F. Vacca for advice; D. Wohlwend for assistance with the flow cytometry; and C. Bauer and J. Bosset for assistance with bioimaging. This study was supported by the Swiss National Science Foundation through a professorship to R.W.
Abbreviations used:
- DRM
detergent-resistant membrane
- GPI-AP
glycosylphosphatidylinositol-anchored protein
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E13-03-0160) on April 24, 2013.
The authors declare that they have no conflict of interest.
REFERENCES
- Benting JH, Rietveld AG, Simons K. N-glycans mediate the apical sorting of a GPI-anchored, raft-associated protein in Madin-Darby canine kidney cells. J Cell Biol. 1999;146:313–320. doi: 10.1083/jcb.146.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosson R, Jaquenoud M, Conzelmann A. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol Biol Cell. 2006;17:2636–2645. doi: 10.1091/mbc.E06-02-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky RA, Mukhina GL, Li S, Nelson KL, Chiurazzi PL, Buckley JT, Borowitz MJ. Improved detection and characterization of paroxysmal nocturnal hemoglobinuria using fluorescent aerolysin. Am J Clin Pathol. 2000;114:459–466. doi: 10.1093/ajcp/114.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–544. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- Brown DA, Crise B, Rose JK. Mechanism of membrane anchoring affects polarized expression of two proteins in MDCK cells. Science. 1989;245:1499–1501. doi: 10.1126/science.2571189. [DOI] [PubMed] [Google Scholar]
- Castillon GA, Aguilera-Romero A, Manzano-Lopez J, Epstein S, Kajiwara K, Funato K, Watanabe R, Riezman H, Muñiz M. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol Biol Cell. 2011;22:2924–2936. doi: 10.1091/mbc.E11-04-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon GA, Watanabe R, Taylor M, Schwabe TME, Riezman H. Concentration of GPI-anchored proteins upon ER exit in yeast. Traffic. 2009;10:186–200. doi: 10.1111/j.1600-0854.2008.00857.x. [DOI] [PubMed] [Google Scholar]
- Catino MA, Paladino S, Tivodar S, Pocard T, Zurzolo C. N- and O-glycans are not directly involved in the oligomerization and apical sorting of GPI proteins. Traffic. 2008;9:2141–2150. doi: 10.1111/j.1600-0854.2008.00826.x. [DOI] [PubMed] [Google Scholar]
- Christian AE, Haynes MP, Phillips MC, Rothblat GH. Use of cyclodextrins for manipulating cellular cholesterol content. J Lipid Res. 1997;38:2264–2272. [PubMed] [Google Scholar]
- Delacour D, Cramm-Behrens CI, Drobecq H, Le Bivic A, Naim HY, Jacob R. Requirement for galectin-3 in apical protein sorting. Curr Biol. 2006;16:408–414. doi: 10.1016/j.cub.2005.12.046. [DOI] [PubMed] [Google Scholar]
- Delacour D, et al. Galectin-4 and sulfatides in apical membrane trafficking in enterocyte-like cells. J Cell Biol. 2005;169:491–501. doi: 10.1083/jcb.200407073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delacour D, Koch A, Jacob R. The role of galectins in protein trafficking. Traffic. 2009;10:1405–1413. doi: 10.1111/j.1600-0854.2009.00960.x. [DOI] [PubMed] [Google Scholar]
- Diep DB, Nelson KL, Raja SM, Pleshak EN, Buckley JT. Glycosylphosphatidylinositol anchors of membrane glycoproteins are binding determinants for the channel-forming toxin aerolysin. J Biol Chem. 1998;273:2355–2360. doi: 10.1074/jbc.273.4.2355. [DOI] [PubMed] [Google Scholar]
- Fujita M, Umemura M, Yoko-o T, Jigami Y. PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol Biol Cell. 2006;17:5253–5264. doi: 10.1091/mbc.E06-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier A, Juillerat A, Heinis C, Correa IR, Jr, Kindermann M, Beaufils F, Johnsson K. An engineered protein tag for multiprotein labeling in living cells. Chem Biol. 2008;15:128–136. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Guan XL, et al. Functional interactions between sphingolipids and sterols in biological membranes regulating cell physiology. Mol Biol Cell. 2009;20:2083–2095. doi: 10.1091/mbc.E08-11-1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer MF, Hancock JF. Lipid rafts and membrane traffic. FEBS Lett. 2007;581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- Hong Y, Ohishi K, Inoue N, Kang JY, Shime H, Horiguchi Y, van der Goot FG, Sugimoto N, Kinoshita T. Requirement of N-glycan on GPI-anchored proteins for efficient binding of aerolysin but not Clostridium septicum alpha-toxin. EMBO J. 2002;21:5047–5056. doi: 10.1093/emboj/cdf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P, Simons K. Cholesterol is required for surface transport of influenza virus hemagglutinin. J Cell Biol. 1998;140:1357–1367. doi: 10.1083/jcb.140.6.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. 2008;144:287–294. doi: 10.1093/jb/mvn090. [DOI] [PubMed] [Google Scholar]
- Kondor-Koch C, Bravo R, Fuller SD, Cutler D, Garoff H. Exocytotic pathways exist to both the apical and the basolateral cell surface of the polarized epithelial cell MDCK. Cell. 1985;43:297–306. doi: 10.1016/0092-8674(85)90035-2. [DOI] [PubMed] [Google Scholar]
- Kwik J, Boyle S, Fooksman D, Margolis L, Sheetz MP, Edidin M. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc Natl Acad Sci USA. 2003;100:13964–13969. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lands WE. Metabolism of glycerolipides; a comparison of lecithin and triglyceride synthesis. J Biol Chem. 1958;231:883–888. [PubMed] [Google Scholar]
- Lindstedt R, Apodaca G, Barondes SH, Mostov KE, Leffler H. Apical secretion of a cytosolic protein by Madin-Darby canine kidney cells. Evidence for polarized release of an endogenous lectin by a nonclassical secretory pathway. J Biol Chem. 1993;268:11750–11757. [PubMed] [Google Scholar]
- Lisanti MP, Caras IW, Davitz MA, Rodriguez-Boulan E. A glycophospholipid membrane anchor acts as an apical targeting signal in polarized epithelial cells. J Cell Biol. 1989;109:2145–2156. doi: 10.1083/jcb.109.5.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisanti MP, Sargiacomo M, Graeve L, Saltiel AR, Rodriguez-Boulan E. Polarized apical distribution of glycosyl-phosphatidylinositol-anchored proteins in a renal epithelial cell line. Proc Natl Acad Sci USA. 1988;85:9557–9561. doi: 10.1073/pnas.85.24.9557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y, Tashima Y, Houjou T, Fujita M, Yoko-o T, Jigami Y, Taguchi R, Kinoshita T. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol Biol Cell. 2007;18:1497–1506. doi: 10.1091/mbc.E06-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masaki R, Yamamoto A, Tashiro Y. Membrane topology and retention of microsomal aldehyde dehydrogenase in the endoplasmic reticulum. J Biol Chem. 1996;271:16939–16944. doi: 10.1074/jbc.271.28.16939. [DOI] [PubMed] [Google Scholar]
- Mays RW, Siemers KA, Fritz BA, Lowe AW, van Meer G, Nelson WJ. Hierarchy of mechanisms involved in generating Na/K-ATPase polarity in MDCK epithelial cells. J Cell Biol. 1995;130:1105–1115. doi: 10.1083/jcb.130.5.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGwire GB, Becker RP, Skidgel RA. Carboxypeptidase M, a glycosylphosphatidylinositol-anchored protein, is localized on both the apical and basolateral domains of polarized Madin-Darby canine kidney cells. J Biol Chem. 1999;274:31632–31640. doi: 10.1074/jbc.274.44.31632. [DOI] [PubMed] [Google Scholar]
- Morris RJ, Jen A, Warley A. Isolation of nano-meso scale detergent resistant membrane that has properties expected of lipid “rafts.”. J Neurochem. 2011;116:671–677. doi: 10.1111/j.1471-4159.2010.07076.x. [DOI] [PubMed] [Google Scholar]
- Munro S. Lipid rafts: elusive or illusive. Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- Murakami H, Wang Y, Hasuwa H, Maeda Y, Kinoshita T, Murakami Y. Enhanced response of T lymphocytes from Pgap3 knockout mouse: insight into roles of fatty acid remodeling of GPI anchored proteins. Biochem Biophys Res Commun. 2012;417:1235–1241. doi: 10.1016/j.bbrc.2011.12.116. [DOI] [PubMed] [Google Scholar]
- Paladino S, Sarnataro D, Pillich R, Tivodar S, Nitsch L, Zurzolo C. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J Cell Biol. 2004;167:699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Urquhart P, Hooper NM. N-glycans, not the GPI anchor, mediate the apical targeting of a naturally glycosylated, GPI-anchored protein in polarised epithelial cells. J Cell Sci. 2004;117:5079–5086. doi: 10.1242/jcs.01386. [DOI] [PubMed] [Google Scholar]
- Powell SK, Lisanti MP, Rodriguez-Boulan EJ. Thy-1 expresses two signals for apical localization in epithelial cells. Am J Physiol. 1991;260:C715–C720. doi: 10.1152/ajpcell.1991.260.4.C715. [DOI] [PubMed] [Google Scholar]
- Prydz K, Simons K. Cholesterol depletion reduces apical transport capacity in epithelial Madin-Darby canine kidney cells. Biochem J. 2001;357:11–15. doi: 10.1042/0264-6021:3570011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier A-S, Castillon GA, Michon L, Fukasawa M, Romanova-Michaelides M, Jaensch N, Hanada K, Watanabe R. Exit of GPI-anchored proteins from the ER differs in yeast and mammalian cells. Traffic. 2010;11:1017–1033. doi: 10.1111/j.1600-0854.2010.01081.x. [DOI] [PubMed] [Google Scholar]
- Rudd PM, Morgan BP, Wormald MR, Harvey DJ, van den Berg CW, Davis SJ, Ferguson MA, Dwek RA. The glycosylation of the complement regulatory protein, human erythrocyte CD59. J Biol Chem. 1997;272:7229–7244. doi: 10.1074/jbc.272.11.7229. [DOI] [PubMed] [Google Scholar]
- Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. Resistance of cell membranes to different detergents. Proc Natl Acad Sci USA. 2003;100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong J, Wang Y, Kinoshita T, Maeda Y. Implication of lipid moiety in oligomerization and immunoreactivities of GPI-anchored proteins. J Lipid Res. 2013;54:1077–1091. doi: 10.1194/jlr.M034421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima DT, Haldar K, Pepperkok R, Watson R, Warren G. Partitioning of the Golgi apparatus during mitosis in living HeLa cells. J Cell Biol. 1997;137:1211–1228. doi: 10.1083/jcb.137.6.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindou H, Hishikawa D, Harayama T, Yuki K, Shimizu T. Recent progress on acyl CoA: lysophospholipid acyltransferase research. J Lipid Res. 2009;50(suppl):S46–S51. doi: 10.1194/jlr.R800035-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- Simons K, Sampaio JL. Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol. 2011;3:a004697. doi: 10.1101/cshperspect.a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stechly L, et al. Galectin-4-regulated delivery of glycoproteins to the brush border membrane of enterocyte-like cells. Traffic. 2009;10:438–450. doi: 10.1111/j.1600-0854.2009.00882.x. [DOI] [PubMed] [Google Scholar]
- Stuven E, Porat A, Shimron F, Fass E, Kaloyanova D, Brugger B, Wieland FT, Elazar Z, Helms JB. Intra-Golgi protein transport depends on a cholesterol balance in the lipid membrane. J Biol Chem. 2003;278:53112–53122. doi: 10.1074/jbc.M300402200. [DOI] [PubMed] [Google Scholar]
- Tashima Y, Taguchi R, Murata C, Ashida H, Kinoshita T, Maeda Y. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol Biol Cell. 2006;17:1410–1420. doi: 10.1091/mbc.E05-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban J, Parczyk K, Leutz A, Kayne M, Kondor-Koch C. Constitutive apical secretion of an 80-kD sulfated glycoprotein complex in the polarized epithelial Madin-Darby canine kidney cell line. J Cell Biol. 1987;105:2735–2743. doi: 10.1083/jcb.105.6.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Thiele C, Huttner WB. Cholesterol is required for the formation of regulated and constitutive secretory vesicles from the trans-Golgi network. Traffic. 2000;1:952–962. doi: 10.1034/j.1600-0854.2000.011205.x. [DOI] [PubMed] [Google Scholar]
- Watanabe R, Funato K, Venkataraman K, Futerman AH, Riezman H. Sphingolipids are required for the stable membrane association of glycosylphosphatidylinositol-anchored proteins in yeast. J Biol Chem. 2002;277:49538–49544. doi: 10.1074/jbc.M206209200. [DOI] [PubMed] [Google Scholar]
- Yeaman C, Le Gall AH, Baldwin AN, Monlauzeur L, Le Bivic A, Rodriguez-Boulan E. The O-glycosylated stalk domain is required for apical sorting of neurotrophin receptors in polarized MDCK cells. J Cell Biol. 1997;139:929–940. doi: 10.1083/jcb.139.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zidovetzki R, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim Biophys Acta. 2007;1768:1311–1324. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.