Cse4 is posttranslationally modified in Saccharomyces cerevisiae. Ipl1 contributes to Cse4 phosphorylation in vivo and in vitro. Phosphorylation of Cse4 at centromeres is enhanced in response to nocodazole or reduced cohesion. The results suggest that phosphorylation of Cse4 ensures faithful chromosome segregation.
Abstract
The centromeric histone H3 variant (CenH3) is essential for chromosome segregation in eukaryotes. We identify posttranslational modifications of Saccharomyces cerevisiae CenH3, Cse4. Functional characterization of cse4 phosphorylation mutants shows growth and chromosome segregation defects when combined with kinetochore mutants okp1 and ame1. Using a phosphoserine-specific antibody, we show that the association of phosphorylated Cse4 with centromeres increases in response to defective microtubule attachment or reduced cohesion. We determine that evolutionarily conserved Ipl1/Aurora B contributes to phosphorylation of Cse4, as levels of phosphorylated Cse4 are reduced at centromeres in ipl1 strains in vivo, and in vitro assays show phosphorylation of Cse4 by Ipl1. Consistent with these results, we observe that a phosphomimetic cse4-4SD mutant suppresses the temperature-sensitive growth of ipl1-2 and Ipl1 substrate mutants dam1 spc34 and ndc80, which are defective for chromosome biorientation. Furthermore, cell biology approaches using a green fluorescent protein–labeled chromosome show that cse4-4SD suppresses chromosome segregation defects in dam1 spc34 strains. On the basis of these results, we propose that phosphorylation of Cse4 destabilizes defective kinetochores to promote biorientation and ensure faithful chromosome segregation. Taken together, our results provide a detailed analysis, in vivo and in vitro, of Cse4 phosphorylation and its role in promoting faithful chromosome segregation.
INTRODUCTION
The kinetochore is the chromosomal attachment site of spindle microtubules and is required for faithful chromosome segregation. Point centromeres in budding yeasts are small (∼125 base pairs), whereas regional centromeres of fission yeast and other multicellular eukaryotes are large (4–10,000 kb) and contain repetitive DNA (Burrack and Berman, 2012). The kinetochore is a large macromolecular structure of >100 proteins categorized as inner, middle, or outer kinetochore proteins, depending on their proximity to centromeric DNA or the microtubules. The middle kinetochore, containing the COMA complex (Ctf19, Okp1, Mcm21, and Ame1), serves as a linker between the inner and outer kinetochore. The outer kinetochore, containing the Ndc80 and Dam1 complexes, mediates the attachment of the kinetochores to microtubules (De Wulf et al., 2003; Lampert et al., 2010). Defects in spindle–kinetochore interactions lead to cell cycle delay dependent on the spindle assembly checkpoint (SAC), which inhibits activation of the anaphase-promoting complex (APC). The APC is inactive until all chromosomes are correctly aligned at the metaphase plate, thereby preventing premature entry into anaphase and minimizing chromosome missegregation (Clarke and Bachant, 2008).
The conserved Ipl1/Aurora B protein kinase is a regulator of the SAC that localizes to kinetochores, spindles, spindle midzone, and the midbody (Kotwaliwale et al., 2007; Cheeseman and Desai, 2008). Ipl1 ensures that inappropriate kinetochore–microtubule interactions are destabilized, leading to unattached kinetochores and activation of the SAC (Pinsky et al., 2006). Defects in biorientation are observed in Saccharomyces cerevisiae ipl1 mutants (Biggins et al., 1999; Tanaka et al., 2002). Ipl1 substrates such as Dam1 and Ndc80, together with additional targets of Ipl1, are critical for establishing chromosome biorientation (Akiyoshi et al., 2009; Kemmler et al., 2009; Tien et al., 2010).
Despite the differences in centromeres, several components of the kinetochore and posttranslational modifications (PTMs) of centromeric histone H4 are conserved across species (Kitagawa and Hieter, 2001; Choy et al., 2011; Gascoigne and Cheeseman, 2012). The conserved centromeric histone H3 variant CenH3 (Cse4 in S. cerevisiae, CENP-A in Homo sapiens) is essential for chromosome segregation (De Rop et al., 2012). CenH3 homologues have a conserved histone H3–like C-terminal region and a unique N-terminus, and both are essential for kinetochore function (Black et al., 2007). In humans, the N-terminus of CENP-A is phosphorylated by Aurora kinase, and this contributes to kinetochore function and cytokinesis (Zeitlin et al., 2001a, b; Kunitoku et al., 2003). CenH3 phosphorylation has also been reported in maize (Zhang et al., 2005). In S. cerevisiae Cse4 is ubiquitinated and methylated, and in vitro studies indicate that the C-terminal histone fold domain can be phosphorylated (Buvelot et al., 2003; Hewawasam et al., 2010; Ranjitkar et al., 2010; Samel et al., 2012). Given the important role of core histone PTMs in epigenetic regulation, CenH3 PTMs may be critical for chromosome segregation (Choy et al., 2012). We use mass spectrometry to identify PTMs of Cse4 and focus on phosphorylation. On the basis of in vivo and in vitro results, we conclude that S. cerevisiae Cse4 is a substrate of Ipl1 and phosphorylation of Cse4 serves to destabilize defective kinetochores to promote biorientation and ensure faithful chromosome segregation.
RESULTS
PTMs of Cse4 identified by mass spectrometry occur at evolutionarily conserved locations
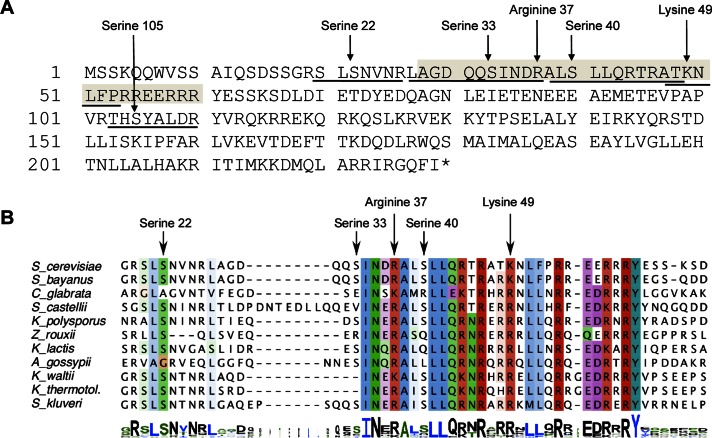
Cse4 purified from a wild-type strain expressing either His-HA-CSE4 or His-HA-cse4-15KR (Takahashi et al., 2008) was analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS). Several phosphorylation sites (Ser-22, Ser-33, Ser-40, and Ser-105), one methylation site (Arg-37), and one acetylation (Lys-49) site were identified in the N-terminus of Cse4 (Figure 1A and Table 1). Methylation of Arg-37 was recently reported (Samel et al., 2012). Five of the PTMs (Ser-22, Ser-33, Arg-37, Ser-40, and Lys-49) are contained within region that is evolutionarily conserved among different fungi with point centromeres and includes the essential N-terminal domain (Chen et al., 2000; Figure 1B). For this study, we focus on phosphorylation.
FIGURE 1:
Amino acids in Cse4 that are posttranslationally modified are evolutionarily conserved. (A) Cse4 is phosphorylated, methylated, and acetylated. The peptides identified by mass spectrometry are underlined, and the modified amino acid residues indicated (also see Table 1). The essential N-terminal domain (END) is shaded (amino acids 28–60). (B) The region containing the PTM sites within the Cse4 N-terminus is conserved. ClustalW alignment of the Cse4 END regions of point centromere yeasts. Alignment in logo format is shown at the bottom (Schneider and Stephens, 1990).
TABLE 1:
Modified peptides identified in vivo by LC-MS/MS.
| Protein | Peptide sequence | Modification |
|---|---|---|
| Cse4-15KR | 20SLS#NVNR26 | S22 phosphorylation |
| 27LAGDQQS#INDR37 | S33 phosphorylation | |
| 27LAGDQQSINDR##37 | R37 methylation | |
| 103THS#YALDR110 | S105 phosphorylation | |
| Cse4 | 27ALS#LLQR27 | S40 phosphorylation |
| 47ATK#NLFPR54 | K49 acetylation |
Centromeric association of phosphorylated Cse4 is increased in cells treated with nocodazole or depleted of cohesion
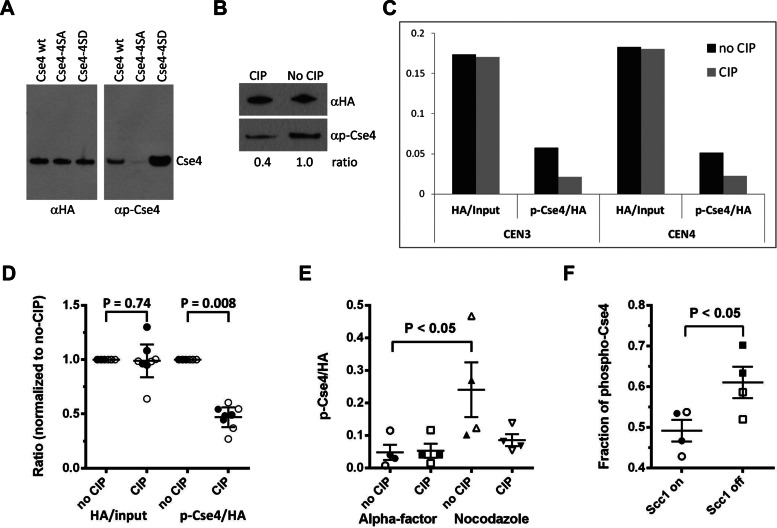
To facilitate detection of Cse4 phosphorylation, we generated a phosphoserine-specific antibody (αp-Cse4) and tested the specificity of the antibody using hemagglutinin (HA)-tagged CSE4 or cse4-4SA and cse4-4SD alleles in which Ser-22, Ser-33, Ser-40, and Ser-105 were substituted with the nonmodifiable or phosphomimetic amino acid alanine or aspartate, respectively (Supplemental Figure 1A). The antibody showed a strong reduction in reactivity to cse4-4SA compared with CSE4 and cse4-4SD (Figure 2A). This result demonstrates that αp-Cse4 exhibits differential affinities depending on the phosphorylation state of the four phosphorylated serines in Cse4. The reduced reactivity in protein samples treated with calf intestine phosphatase (CIP) validates the specificity of the αp-Cse4 for phosphorylated Cse4 (Figure 2B). The strong signal with cse4-4SD suggests that the aspartate side chain allows recognition by αp-Cse4 (Figure 2A), analogous to results from studies with a mps1 phosphomimetic mutant (Tyler et al., 2009).
FIGURE 2:
Centromeric association of phosphorylated Cse4. (A) Specificity of Cse4 phospho antibody (αp-Cse4). Western blots of Cse4 proteins purified from strains (BY4741) expressing wild-type Cse4 (pMB1601), cse4-4SA (pMB1566), or cse4-4SD (pMB1563) were probed with αHA (loading control) or αp-Cse4. (B) Phosphatase treatment of Cse4 reduces affinity of αp-Cse4 to Cse4. Cse4 protein purified from BY4741 expressing wild-type Cse4 (pMB1601) was treated with or without CIP and analyzed by Western blots probed with αHA (loading control) or αp-Cse4. (C) Phosphorylated Cse4 is associated with centromeres. ChIP of extracts treated with (CIP) or without CIP (no CIP) was performed with αHA or αp-Cse4 (YMB8378) and assayed by PCR for CEN3 and CEN4 sequences. The graph shows an example of a single experiment. Analysis of four independent experiments and statistical significance are shown in D. (D) Ratios of αHA/input DNA and αp-Cse4/αHA signals normalized to the no-CIP condition. Four independent biological replicates were performed. Error bars show the mean ± 95% confidence intervals. Statistical significance was determined by paired t test. Open symbols, CEN3; filled symbols, CEN4. (E) Centromere-associated phosphorylated Cse4 is increased in nocodazole-treated cells. ChIP samples from wild-type strain (YMB8378) treated with α-factor or nocodazole were analyzed as described in C. The αp-Cse4 signals (normalized to αHA) before and after CIP treatment were calculated. Results from two independent experiments were pooled. Lines on the scatter plots show the mean ± SE of the mean. Statistical significance was determined by one-way analysis of variance (ANOVA). Open symbols, CEN3; filled symbols, CEN4. (F) Phosphorylation of Cse4 is increased in cells depleted of Scc1. ChIP was performed on strain YMB8674 arrested in metaphase (Cdc20 OFF) with defective cohesion (Scc1 OFF). The control strain (Scc1 ON) was grown in galactose medium. Fraction of phospho-Cse4 is the fraction of the total αp-Cse4 signal removed by CIP treatment. Results from two independent experiments were pooled and plotted as in D. Significance was determined by t test. Open symbols, CEN3; filled symbols, CEN4.
To examine the association of phosphorylated Cse4 with centromeres, we used our αp-Cse4 in chromatin immunoprecipitation (ChIP) experiments (Figure 2C). Cross-linked chromatin samples treated or not treated with CIP from a strain expressing HA-tagged CSE4 from its own promoter at the endogenous chromosomal locus were analyzed using centromere-specific primers (CEN3, CEN4). Serial dilutions of the input DNA were used to determine that the PCR amplification conditions were in the linear range (Supplemental Figure 2A). No enrichment of Cse4 (αHA) or phosphorylated Cse4 (αp-Cse4) at a noncentromeric control region (actin gene) was observed (data not shown). We quantified the centromeric association of Cse4 (αHA) relative to input DNA and that of phosphorylated Cse4 (αp-Cse4) relative to total Cse4 at centromeres (αHA) in CIP-treated and untreated samples (Figure 2C). Analysis of four independent experiments and statistical significance is shown in Figure 2D. As expected, no significant difference (p > 0.74) in the level of centromeric association of Cse4 (αHA) in response to CIP treatment was observed; however, recognition of phosphorylated Cse4 by αp-Cse4 was significantly reduced after CIP treatment compared with the untreated sample (p < 0.008), indicating that phosphorylated Cse4 associates with centromeric DNA.
Given the essential role of Cse4 in kinetochore function and the centromeric association of phosphorylated Cse4, we asked whether phosphorylation is enhanced upon treatment with a microtubule-depolymerizing agent such as nocodazole or reduced tension between the sister chromatids by depletion of cohesion. ChIP experiments using cells treated with α-factor or nocodazole (Figure 2E) showed higher levels (p < 0.05) of phosphorylated Cse4 at centromeres of cells in the G2/M phase of the cell cycle (nocodazole) compared with G1 (α-factor; Figure 2E). The level of phosphorylated Cse4 at centromeres was further enhanced (p < 0.05) in cells depleted for the cohesion subunit Scc1, a condition that leads to reduced tension between the sister kinetochores (Keating et al., 2009; Figure 2F). Levels of total Cse4 (α-HA) at centromeres remained unchanged regardless of the phosphorylation status, indicating that the affinity of Cse4 for centromeres is not affected by phosphorylation. Taken together, our results show that Cse4 phosphorylation is increased at centromeres with defective kinetochore–microtubule attachment or reduced tension.
cse4 phosphomimetic mutants suppress ipl1-2, and Ipl1 contributes to phosphorylation of Cse4 in vivo and in vitro
Destabilization of inappropriate kinetochore–microtubule interactions mediated by Ipl1 phosphorylation of substrates in the kinetochore constitutes an essential step to resolve biorientation defects (Pinsky et al., 2006). Given the increased phosphorylation of Cse4 at centromeres with defective kinetochore–microtubule attachment or reduced tension and the known role of Ipl1 in ensuring biorientation, we tested a potential role of Ipl1 for Cse4 phosphorylation in vivo.
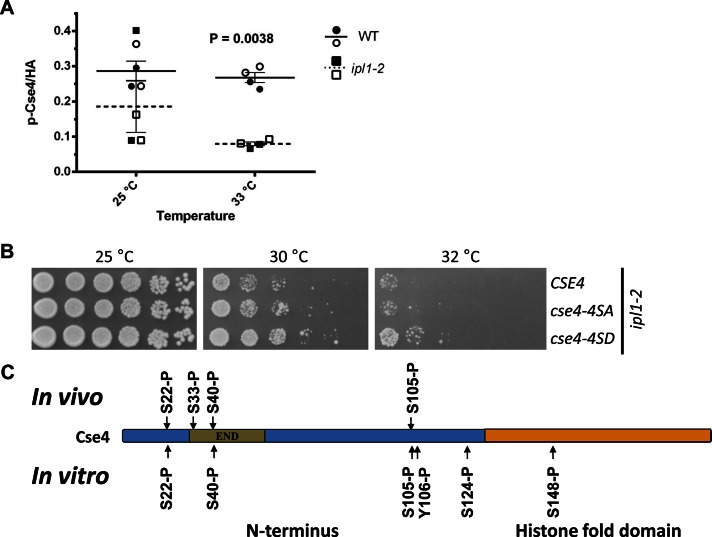
We performed ChIP experiments using ipl1-2 (Chan and Botstein, 1993) and its isogenic wild-type control strain. We observed that the phosphorylated Cse4 signal at CEN3 and CEN4 was markedly reduced (p = 0.0038) in the ipl1-2 strain compared with the wild-type strain (Figure 3A). The difference is pronounced at 33°C, the nonpermissive temperature for ipl1-2 strains. The reduced centromeric association of phosphorylated Cse4 in the ipl1-2 strain at 33°C is not due to altered expression of Cse4 in these strains (Supplemental Figure 3A).
FIGURE 3:
Ipl1 phosphorylates Cse4 in vivo and in vitro and phosphomimetic cse4-4SD suppresses temperature sensitivity of an ipl1-2 strain. (A) Ipl1 contributes to the phosphorylation of centromere-associated Cse4. ChIP experiments were performed with WT (YMB8525) and ipl1-2 (YMB8528) strains grown at 25°C and shifted to 33°C for 3 h. The p-Cse4 signal is plotted, with lines showing the mean ± SE of the mean. Results from two independent experiments were pooled. Statistical significance was analyzed by two-way ANOVA (strain, temperature), which indicated that observed variation was significant and predominately due to differences between the wild-type and ipl1-2 strains (p = 0.0038). Results of pairwise tests were corrected for multiple comparisons (Bonferroni). Open symbols, CEN3; filled symbols, CEN4. (B) Phosphomimetic cse4-4SD suppresses the temperature sensitivity of an ipl1-2 strain. Serial dilutions of ipl1-2 (KT1963) with CSE4 (YMB8528), cse4-4SA (YMB8529), or cse4-4SD (YMB8530) were plated on YPD medium and incubated at the indicated temperatures for 2–3 d. (C) Ipl1 phosphorylates Cse4 in vitro. Products of in vitro kinase assays using purified Cse4, ATP, and Ipl1-Sli15 at 30°C for 90 min were analyzed by mass spectrometry. Cse4 modification sites are shown, along with those identified by in vivo analysis (Figure 1A; also see Tables 1 and 2 and Supplemental Figure 3B).
The ipl1-2 strains exhibit a temperature-sensitive phenotype for growth. If lethality of ipl1-2 at the nonpermissive temperature is partly due to lower levels of phosphorylated Cse4, we reasoned that a phosphomimetic of Cse4, cse4-4SD, should suppress the temperature sensitivity of an ipl1-2 strain. Indeed, results from viability assays showed that the cse4-4SD phosphomimetic mutant, but not cse4-4SA, partially suppressed the growth defect of ipl1-2 at 30°C (Figure 3B).
To examine whether Ipl1 phosphorylates Cse4 directly, we performed in vitro kinase assays using Ipl1, Sli15, and Cse4 purified from Escherichia coli followed by LC-MS/MS analysis. Phosphorylation at Ser-22, Ser-40, and Ser-105 was detected, as previously by in vivo analysis (Figure 1A). In addition, the in vitro assays showed phosphorylation of Tyr-106, Ser-124, and Ser-148 (Figure 3C and Table 2). No phosphorylation was detected from control in vitro assays containing purified Cse4 but lacking Ipl1-Sli15 (data not shown). Furthermore, we performed an in vitro assay containing radiolabeled ATP. With increasing incubation time (5–90 min) we observed increased phosphorylation of Cse4 and Ipl1 (Supplemental Figure 3B). Ipl1 autophosphorylates, and this serves as internal positive control. No phosphorylation was observed after 90 min in the control sample without the kinase Ipl1-Sli15. These results support our conclusion that Ipl1 contributes to phosphorylation of Cse4 in vivo and in vitro.
TABLE 2:
Modified peptides identified in vitro by LC-MS/MS.
| Protein | Peptide sequence | Modification |
|---|---|---|
| Cse4 in vitro (Ipl1/Sli15) | 20SLS#NVNR26 | S22 phosphorylation |
| 38ALS#LLQR44 | S40 phosphorylation | |
| 103THS#YALDR110 | S105 phosphorylation | |
| 103THSY#ALDR110 | Y106 phosphorylation | |
| 122KQS#LKR127 | S124 phosphorylation | |
| 148S#TDLLISK155 | S148 phosphorylation |
cse4 phosphomimetic mutants suppress growth and chromosome segregation defects of Ipl1 substrate mutant dam1 spc34
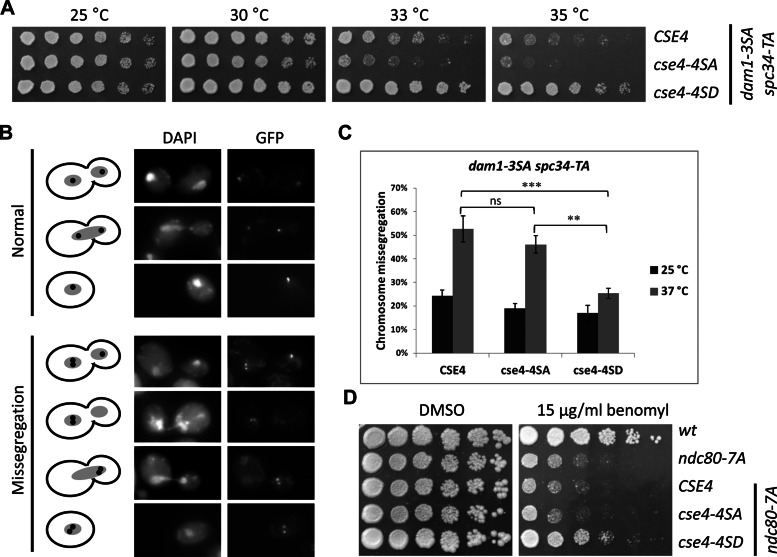
Given the results for phosphorylation of Cse4 by Ipl1, we tested whether cse4 phosphomimetic mutants could suppress mutants of Ipl1 substrates Dam1, Spc34, and Ndc80, which mediate remodeling of kinetochore–microtubule interactions (Cheeseman et al., 2002; Akiyoshi et al., 2009; Keating et al., 2009). A partially nonphosphorylatable dam1-3SA (S257A S265A S292A) strain exhibits a chromosome segregation defect when combined with spc34-TA (T199A), which encodes another subunit of the Dam1 complex (Cheeseman et al., 2002). We observed that cse4-4SD suppressed the temperature sensitivity of dam1-3SA spc34-TA strain, whereas this strain showed slightly increased temperature sensitivity when combined with cse4-4SA (Figure 4A). To further understand the molecular basis for the cse4-4SD–mediated suppression of growth we measured the segregation of a single GFP-labeled chromosome. Our results show that cse4-4SD suppresses chromosome missegregation of the dam1-3SA spc34-TA strains at 37°C by almost 50% compared with CSE4 (p < 0.001) and cse4-4SA (p < 0.01), nearly to levels observed at 25°C (p < 0.01; Figure 4, B and C). CSE4, cse4-4SA, and cse4-4SD did not show chromosome segregation defects in a wild-type strain background (Supplemental Figure 3C).
FIGURE 4:
Phosphomimetic cse4-4SD suppresses phenotypes of nonphosphorylatable dam1 spc34 and ndc80 strains. (A) Suppression of temperature sensitivity of dam1 spc34 strains by cse4-4SD. Serial dilutions of dam1-3SA scp34-TA (DDY2503) with CSE4 (YMB8390), cse4-4SA (YMB8458), or cse4-4SD (YMB8393) were assayed for growth at the indicated temperatures on YPD medium for 2–3 d. (B, C) Suppression of chromosome segregation of dam1 spc34 by cse4-4SD. Segregation of a GFP-labeled chromosome (GFP) in strains from A grown at 25 or 37°C was assayed as described in Materials and Methods. Fluorescence images in B show examples of normal or missegregation events scored and quantified in (C). DAPI staining shows the position of the nucleus in each of the cells. Statistical significance was determined by two-way ANOVA (strain, temperature), correcting for multiple comparisons (Bonferroni). ***p < 0.001, **p < 0.01; ns, not significant. Error bars represent SE of the mean. In each of three independent experiments at least 100 cells were counted for each strain. (D) Suppression of benomyl sensitivity of ndc80-7SA by cse4-4SD. Serial dilutions of ndc80-7A (SBY7123) with CSE4 (YMB8395), cse4-4SA (YMB8087), or cse4-4SD (YMB8086) were assayed for growth on YPD containing 15 μg/ml benomyl (or DMSO as control) at 30°C for 4 d.
Analogous to the results for the dam1-3SA spc34-TA, expression of cse4-4SD in an ndc80-7A strain suppressed its benomyl sensitivity; CSE4 or cse4-4SA had no effect (Figure 4D). The suppression phenotypes by cse4-4SD are not due to altered expression of the cse4 proteins, as Western blots showed that dam1-3SA spc34-TA and ndc80-7SA strains express equivalent levels of Cse4, cse4-4SA, or cse4-4SD when grown at permissive and nonpermissive temperatures (Supplemental Figure 3A). Taken together, our results show that cse4 phosphomimetic mutants suppress phenotypes of Ipl1 substrate mutants dam1 spc34 and ndc80. The suppression of growth and chromosome segregation defects of dam1 spc34 by cse4-4SD suggests that phosphorylation of Cse4 may serve to destabilize kinetochores with defective microtubule attachments and promote biorientation when other Ipl1 substrates cannot be fully phosphorylated.
cse4 phosphorylation mutants lead to growth defects and increased chromosome segregation defects in kinetochore mutants okp1 and ame1
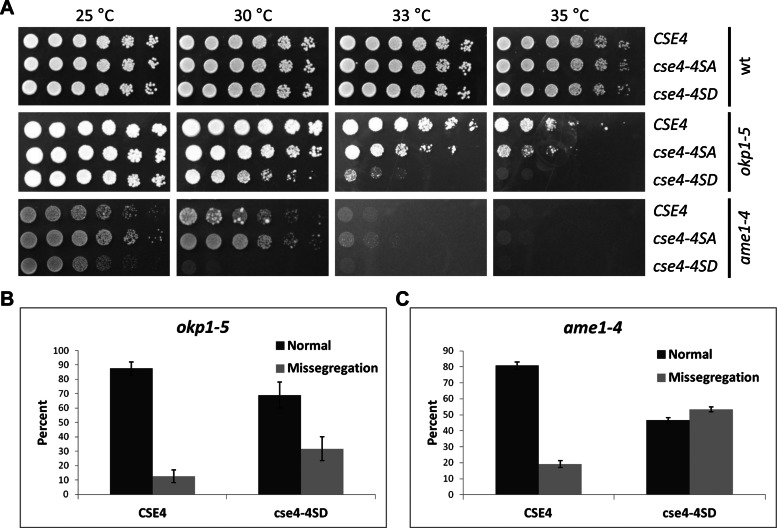
If phosphorylation of Cse4 destabilizes defective kinetochore attachments, we reasoned that cse4 phosphomimetic mutants combined with kinetochore mutants should result in defects in chromosome segregation and growth. For these studies we used mutants in the kinetochore COMA complex, which interacts with Cse4 (Ortiz et al., 1999; Bloom and Joglekar, 2010). The okp1-5 and ame1-4 mutants with cse4-4SD displayed enhanced temperature sensitivity, which was more severe for the ame1-4 strains, with little or no growth even at 30°C (Figure 5A). Wild-type strains with CSE4 or cse4-4SA or cse4-4SD did not exhibit growth defects at the different temperatures we tested (Figure 5A). The growth defect of okp1-5 cse4-4SD and ame1-4 cse4-4SD strains is not due to altered Cse4 expression, as Western blot analysis showed that levels of Cse4, cse4-4SA, and cse4-4SD are similar in okp1-5 and ame1-4 strains grown at 25 or 33°C (Supplemental Figure 3A).
FIGURE 5:
cse4 phosphorylation mutations cause growth defects and increased chromosome segregation defects in okp1 and ame1 strains. (A) Enhanced temperature sensitivity of cse4 phosphorylation mutants in okp1 and ame1 strains. Serial dilutions of wild-type strain (DDY1925), okp1-5 (YV167), or ame1-4 (YV171) with CSE4 (YMB8378, YMB8384, YMB8654), cse4-4SA (YMB8382, YMB8388, YMB8655), or cse4-4SD (YMB8379, YMB8386, YMB8656) were assayed at the indicated temperatures on YPD medium for 2–3 d. (B, C) cse4-4SD enhances the chromosome segregation defects of okp1-5 (B) and ame1-4 (C) strains. Strains expressing either CSE4 (YMB8384, YMB8672) or cse4-4SD (YMB8386, YMB8673) were arrested with α-factor and shifted to 33 and 30°C, respectively, for 3 h. Segregation of a GFP-labeled chromosome was scored in large-budded cells undergoing mitosis (n > 35), and missegregation of the GFP-labeled chromosome was quantified as described in Figure 4B and Materials and Methods. Error bars represent mean deviations for two independent experiments.
We next examined the strains for chromosome segregation defects and observed that chromosome missegregation was increased twofold in the okp1-5 cse4-4SD strain compared with okp1-5 CSE4 (Figure 5B). Consistent with previous reports (Knockleby and Vogel, 2009), the ame1-4 mutants showed higher chromosome missegregation compared with the okp1-5 strain, which was further increased by 2.5-fold with cse4-4SD (Figure 5C). On the basis of our results, we propose that destabilization of the kinetochore by constitutive phosphorylation of Cse4 is detrimental in conjunction with mutations in COMA components.
DISCUSSION
In this study we identified phosphorylation, acetylation, and methylation sites in Cse4 and showed that these sites are located within a region conserved in other point centromere yeasts. We generated a phosphoserine antibody and observed that the association of phosphorylated Cse4 with centromeres is increased in cells treated with nocodazole or with reduced cohesion between sister chromatids. Our results provide evidence that Ipl1 contributes to phosphorylation of Cse4 for faithful chromosome segregation. For example, we showed 1) phosphorylation of Cse4 using in vitro kinase assays with Ip1/Sli15 and agreement of these data with the in vivo mass spectrometry analysis, 2) significantly reduced levels of phosphorylated Cse4 at centromeres in an ipl1-2 strain at the nonpermissive temperature, 3) suppression of temperature sensitivity of ipl1-2 and Ipl1 substrate mutants dam1 spc34 and ndc80-7SA by a cse4 phosphomimetic mutant cse4-4SD, and 4) suppression of chromosome segregation defects of dam1 spc34 by cse4-4SD. Our results also show that cse4 phosphorylation mutants lead to growth defects and increased chromosome segregation defects in kinetochore mutants okp1 and ame1. On the basis of our results, we propose that phosphorylation of Cse4 destabilizes defective kinetochores to promote biorientation and faithful chromosome segregation.
Phosphorylation of CENP-A was reported almost a decade ago, and one of these studies showed that this modification plays a role in cytokinesis (Zeitlin et al., 2001a, b; Kunitoku et al., 2003). To the best of our knowledge, there have been no other reports on the function of phosphorylated CENP-A. In S. cerevisiae Cse4 was shown to be methylated and ubiquitinated, and in vitro studies indicate that the C-terminal histone fold domain can be phosphorylated (Buvelot et al., 2003; Hewawasam et al., 2010; Ranjitkar et al., 2010; Samel et al., 2012). The observed sequence similarity of the region containing most of the identified PTM sites among the different related yeasts suggests that this region has an important function and that the modifications may also be conserved (Figure 1B).
The phosphoserine antibody (αp-Cse4) that we generated provided a tool for characterization of Cse4 phosphorylation. This antibody showed a clear preference for binding to the phosphomimetic form of Cse4 (4SD) compared with the nonphosphorylatable form (4SA; Figure 2A). The recognition of cse4-4SD suggests that the aspartate chain allows the recognition by αp-Cse4. Analogous observations were made in studies using a phosphospecific antibody for Mps1 (Tyler et al., 2009). The ability of the antibody to distinguish specifically between phosphorylated and nonphosphorylated Cse4 allowed us to determine whether phosphorylated Cse4 is associated with centromeres. Using αp-Cse4, we showed that Cse4 phosphorylation is increased at centromeres with defective kinetochore–microtubule attachment or reduced cohesion between sister chromatids. Similar findings were reported for enhanced recognition of phosphoepitopes at kinetochores by a 3F3/2 antibody in response to tension defects induced by Taxol treatment (Gorbsky and Ricketts, 1993). 3F3/2 recognizes phosphoepitopes on kinetochores and spindle pole bodies in many cell types. Our study provides a first report of the association of a phosphorylated inner kinetochore protein to centromeric DNA in budding yeast.
Genetic and biochemical analysis showed that Ipl1 contributes to the phosphorylation of Cse4. The partial suppression of the temperature sensitivity of ipl1-2 by cse4-4SD (Figure 3B) is similar to the previously reported suppression of ipl1-2 by phosphomimetic dam1 at the semipermissive temperature (Cheeseman et al., 2002). That cse4-4SD only partially suppresses the ipl1-2 growth defect suggests that cse4-4SD cannot compensate for the lack of phosphorylation of all other Ipl1 substrates, such as Ndc80, Dam1, Cep3, Ipl1, Spc34, Ask1, Sli15, Mif2, and Dsn1, in the ipl1 mutant (Cheeseman et al., 2002; Westermann et al., 2003).
Our results for in vitro kinase assays using Ipl1, Sli15, and Cse4 showed phosphorylation at Ser-22, Ser-40, and Ser-105 consistent with our in vivo analysis (Figure 1A). The failure to detect phosphorylation of Ser-33 in the in vitro assay may be due to the sensitivity of the assay or the possibility that this site is a substrate for another kinase such as Mps1, Cdc5, Cdc28, or Bub1. The differences in phosphorylation sites of Cse4 identified in vitro and in vivo may also be due to the nature of the substrate (purified Cse4 vs. chromatin), the increased amount of in vitro sample, or the conditions of the in vitro assay. In addition, Ser-148 is within the Cse4 histone fold domain, which mediates histone–histone interactions in the nucleosome core, and it is possible that this site is masked in vivo. Kemmler et al. (2009) also observed additional phosphorylation sites for Dam1 in vitro compared with those observed in vivo by mass spectrometry.
Ipl1-mediated phosphorylation of Dam1 and Ndc80 leads to destabilization of inappropriate kinetochore–microtubule interactions, thereby allowing cells to reestablish proper attachments (Pinsky et al., 2006). We propose that phosphorylation of Cse4 affects the structure and conformation of the kinetochore to destabilize aberrant kinetochore and microtubule attachments for faithful chromosome segregation. A prediction of this model is the destabilization of defective kinetochore attachments and error correction by phosphomimetic cse4-4SD when Dam1 or Ndc80 cannot be phosphorylated by Ipl1. Consistent with our hypothesis, we see suppression of the temperature sensitivity of dam1 spc34 strains and benomyl sensitivity of ndc80-7SA strains by cse4-4SD. Given that chromosome segregation defects contribute to the temperature sensitivity of dam1 spc34 strains, we investigated the molecular basis for the suppression by cse4-4SD by using a cell biology approach with a GFP- labeled reporter chromosome. As expected, our results showed that cse4-4SD suppressed chromosome missegregation of the dam1 spc34 strains at 37°C by almost 50% compared with CSE4 and cse4-4SA, nearly to levels observed at 25°C. These results are quite striking, given that the assay uses only one labeled reporter chromosome, and suggest that the robust suppression of the growth phenotype by cse4-4SD may be due to error correction of more than one defective kinetochore.
Cse4 interacts with the COMA complex, which has two essential (Okp1 and Ame1) and two nonessential subunits (Mcm21 and Ctf19; De Wulf et al., 2003). Okp1 has a role in establishing tension across sister kinetochores (De Wulf et al., 2003), and okp1-5 strain exhibits synthetic lethality with cse4 mutants (Chen et al., 2000). Furthermore, Samel et al. (2012) recently reported synthetic growth defects of a methylation mutant of CSE4, namely cse4-R37A, in ame1-4 and okp1-5 strains. We observed increased temperature sensitivity and chromosome missegregation defects in cse4-4SD ame1-4 and cse4-4SD okp1-5 strains. These results suggest that constitutive phosphorylation of Cse4 may destabilize even bioriented kinetochores if the kinetochore is already weakened due to defects in the COMA complex.
In conclusion, we identified PTMs in Cse4 in a region that is evolutionarily conserved and showed that Ipl1-mediated phosphorylation of Cse4 regulates chromosome segregation. We showed that the association of phosphorylated Cse4 with centromeres is increased in cells treated with nocodazole or with reduced cohesion between sister chromatids. Our results provide evidence that Ipl1 contributes to phosphorylation of Cse4 in vivo and in vitro. On the basis of genetic interactions of cse4 with kinetochore mutants, we propose that phosphorylation of Cse4 destabilizes defective kinetochores to promote biorientation and chromosome segregation. Future studies will help us understand the role of Cse4 phosphorylation by Ipl1 or other potential kinases in destabilizing defective kinetochores to promote biorientation and maintain genome stability.
MATERIALS AND METHODS
Yeast strains and plasmids
Plasmids and yeast strains used in this study are described in Tables 3 and 4, respectively. S. cerevisiae strains are isogenic to BY4741, W303-1A, or YPH499 (S288C). For genetic interaction analysis, a PCR-based method was used to replace endogenous CSE4 with HA-tagged CSE4 or cse4 phosphorylation mutant alleles expressed from their own promoters in wild-type, okp1-5, dam1 spc34, and ndc80-7SA strains. Primers used to amplify natMX sequence were, for first PCR, mx4F3 and mxR4, and for second PCR, mx4F3 and cse4dn. Primers used to amplify CSE4 sequence were cse4up and mxR2 (for primers see Table 5). Gene replacement of CSE4 with the integrated alleles was verified by sequence analysis, and Western blots confirmed the expression of the HA-tagged protein. At least two independent strains were analyzed for each experiment. For interactions of cse4 phosphorylation mutants with ame1-4 and ipl1-2, the cse4 alleles were introduced into heterozygous ame1-4 or ipl1-2 diploids, and independently derived double mutants were obtained by tetrad dissection.
TABLE 3:
Plasmids used in this study.
| Name | Promoter, gene, and marker | Source |
|---|---|---|
| pYES2 | GAL1, 2 μ, URA3 | |
| pMB1313 | GAL-8His-HA-cse4-K15R+C (2 μm/URA3) | This study |
| pMB1345 | GAL-8His-HA-CSE4 (2 μm/URA3) | This study |
| pMB1601 | GAL-6His-3HA-CSE4 (2 μm/URA3) | This study |
| pMB1566 | GAL-6His-3HA-cse4-4SA (S22A S33A S40A S105A) (2 μm/URA3) | This study |
| pMB1563 | GAL-6His-3HA-cse4-4SD (S22D S33D S40D S105D) (2 μm/URA3) | This study |
| pRB199 | 6His-3HA-CSE4 (YCp/URA3) | This study |
| pMB1462 | 6His-3HA-cse4-4SA (S22A S33A S40A S105A)(YCp/URA3) | This study |
| pMB1463 | 6His-3HA-cse4-4SD (S22D S33D S40D S105D) (YCp/URA3) | This study |
TABLE 4:
Yeast strains used in this study.
| Strain (parent) | Relevant genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems (Huntsville, AL) |
| DDY1925 | Mata his3-∆200 ura3-52 ade2-1 HIS::pCu-lac1-GFP leu2-3112::lacO::LEU2 | Cheeseman et al. (2002) |
| DDY2503 | MATa, his3∆200, ura3-52, HIS3::pCu-LacI-GFP, leu2-3112::lacO::LEU2, dam1(S257A S265A S292A)::KanMX, spc34∆::HIS3, spc34(T199A)::URA3 | Cheeseman et al. (2002) |
| KT1113 | MATα leu2 ura3-52 his3 | Stuart et al. (1994) |
| KT1963 | MATa leu2 ura3-52 his3 ipl1-2 | Bharucha et al. (2008) |
| YMB8303 (S288C) | Matα ura3-52his3Δ200 leu2 lys2-201 ade2-201 trp1Δ63 | Brachmann et al. (1998) |
| SBY7123 (W303) | MATa ura3-1::ndc80-7A::URA3 ndc80ΔHIS3 PDS1-myc18:LEU2 | Akiyoshi et al. (2009) |
| T6984 | Mata ∆scc1::TRP leu::GAL-SCC1::LEU ade::TetR-GFP::ADE his::tetOS::HIS MET-CDC20::TRP, Ura-auxotroph | Keating et al. (2009) |
| W303-1A | MATa ade2 ura3 trp1 leu2 his3 can1 ssd1 | Thomas and Rothstein (1989) |
| YV167 | Mata okp1-5:TRP1 his3-11,15::LacI:GFP:HIS3 CEN15:LacO:URA3 SPC29:CFP:KanMX | Knockleby and Vogel (2009) |
| YV171 | Mata ame1-4:TRP1 his3-11,15::LacI:GFP:HIS3 CEN15:LacO:URA3 SPC29:CFP:KanMX | Knockleby and Vogel (2009) |
| YMB7239 (YMB7273) | MATa cse4∆::KanMX pYES2-8His-HA-CSE4 (pMB1345) | This study |
| YMB7273 (BY4741) | MATa/α cse4∆::KanMX | Open Biosystems |
| YMB8086 (SBY7123) | MATa CSE4:: 6His-3HA-cse4-4SD (S22D S33D S40D S105D)::mx4NAT | This study |
| YMB8087 (SBY7123) | MATa CSE4:: 6His-3HA-cse4-4SA (S22A S33A S40A S105A)::mx4NAT | This study |
| YMB8378 (DDY1925) | MATa CSE4:: 6His-3HA-CSE4::mx4NAT | This study |
| YMB8379 (DDY1925) | MATa CSE4:: 6His-3HA-cse4-4SD (S22D S33D S40D S105D)::mx4NAT | This study |
| YMB8382 (DDY1925) | MATa CSE4:: 6His-3HA-cse4-4SA (S22A S33A S40A S105A)::mx4NAT | This study |
| YMB8384 (YV167) | MATa CSE4:: 6His-3HA-CSE4::mx4NAT | This study |
| YMB8386 (YV167) | MATa CSE4:: 6His-3HA-cse4-4SD (S22D S33D S40D S105D)::mx4NAT | This study |
| YMB8388 (YV167) | MATa CSE4:: 6His-3HA-cse4-4SA (S22A S33A S40A S105A)::mx4NAT | This study |
| YMB8390 (DDY2503) | MATa CSE4:: 6His-3HA-CSE4::mx4NAT | This study |
| YMB8393 (DDY2503) | MATa CSE4:: 6His-3HA-cse4-4SD (S22D S33D S40D S105D)::mx4NAT | This study |
| YMB8395 (SBY7123) | MATa CSE4:: 6His-3HA-CSE4::mx4NAT | This study |
| YMB8458 (DDY2503) | MATa CSE4:: 6His-3HA-cse4-4SA (S22A S33A S40A S105A)::mx4NAT | This study |
| YMB8525 (KT1113) | MATα CSE4::6His-3HA-CSE4::mx4NAT | This study |
| YMB8528 (KT1963) | MATa CSE4:: 6His-3HA-CSE4::mx4NAT | This study |
| YMB8529 (KT1963) | MATa CSE4:: 6His-3HA-cse4-4SA (S22A S33A S40A S105A)::mx4NAT | This study |
| YMB8530 (KT1963) | MATa CSE4:: 6His-3HA-cse4-4SD (S22D S33D S40D S105D)::mx4NAT | This study |
| YMB8651 (YMB8303, YV171) | MATa/α AME1/ame1-4:TRP1 CSE4/CSE4Δ::6His-3HA-CSE4::mx4NAT | This study |
| YMB8652 (YMB8303, YV171) | MATa/α AME1/ame1-4:TRP1 CSE4/CSE4Δ::6His-3HA-cse4 4SA (S22A S33A S40A S105A)::mx4NAT | This study |
| YMB8653 (YMB8303, YV171) | MATa/α AME1/ame1-4:TRP1 CSE4/CSE4Δ::6His-3HA-cse4 4SD (S22D S33D S40D S105D)::mx4NAT | This study |
| YMB8654 (YMB8651) | MATa ame1-4:TRP1 CSE4Δ::6His-3HA-CSE4::mx4NAT his3-11,15::LacI:GFP:HIS3 SPC29:CFP:KanMX | This study |
| YMB8655 (YMB8652) | MATa ame1-4:TRP1 CSE4Δ::6His-3HA-cse4 4SA (S22A S33A S40A S105A)::mx4NAT his3-11,15::LacI:GFP:HIS3 SPC29:CFP:KanMX | This study |
| YMB8656 (YMB8653) | MATa ame1-4:TRP1 CSE4Δ::6His-3HA-cse4 4SD (S22D S33D S40D S105D)::mx4NAT his3-11,15::LacI:GFP:HIS3 SPC29:CFP:KanMX | This study |
| YMB8672 (YMB8651) | MATa ame1-4:TRP1 CSE4Δ::6His-3HA-CSE4::mx4NAT his3-11,15::LacI:GFP:HIS3 CEN15:LacO:URA3 | This study |
| YMB8673 (YMB8653) | MATa ame1-4:TRP1 CSE4Δ::6His-3HA-cse4 4SD (S22D S33D S40D S105D)::mx4NAT his3-11,15::LacI:GFP:HIS3 CEN15:LacO:URA3 | This study |
| YMB8674 (T6984) | pRB199 | This study |
TABLE 5:
Primers used in this study.
| Name | Short description | Sequence (5′–3′) |
|---|---|---|
| mx4F3 | Forward to amplify natMX sequence | AGA TCT GTT TAG CTT GCC TCG TCC |
| mxR4 | Reverse to amplify natMX sequence | GAA AAA TCG GCT CCA GCC CTG AAG ACA AAA TAT CAG AAT TCG AGC TCG TTT TCG ACA CTG |
| cse4dn | Used in second PCR with mx4F3 to add CSE4 sequence | TAA TAT GAC CTT ATA ATA ACC TTA TTT AAA ACA TTT AAA GTA CCT TAA GTC AAT ATA AAC CCC GAA AAA GGG AAA AAT CGG CTC CAG CCC |
| cse4up | Forward to amplify CSE4 sequence | AAA AAC AGT AGT AGA TAA TCT TGA GAA TTT ACT TTA TAA AAA ACA GAA GAA GGA CTG AAT |
| mxR2 | Reverse to amplify CSE4 sequence | GAC CCG GCG GGG ACG AGG CAA GCT AAA CAG ATC TGA AAT ATA GGG GAC TGC C |
| OMB244 | Forward CEN3 | GAT CAG CGC CAA ACA ATA TGG |
| OMB245 | Reverse CEN3 | AAC TTC CAC CAG TAA ACG TTT C |
| OMB764 | Forward CEN4 | GCG CAA GCT TGC AAA AGG TCA CAT G |
| OMB765 | Reverse CEN4 | CGA ATT CAT TTT GGC CGC TCC TAG GTA |
Mass spectrometry
Cse4 was purified from a wild-type strain (BY4741) overexpressing either His-HA-CSE4 (pMB1345) or His-HA-cse4-15KR (pMB1313) under conditions designed to maximize detection of PTM sites (Takahashi et al., 2008). Samples for mass spectrometry were separated on a 4–12% denaturing Bis-Tris SDS-polyacrylamide gel (Invitrogen, Carlsbad, CA) and stained with Coomassie brilliant blue. Bands corresponding to Cse4 and Cse415KR were excised, reduced, alkylated, and digested with trypsin in situ. Mass spectrometry was done as described previously with minor modifications (Waybright et al., 2008). The volume injected was 7 μl, and electrospray potential was 1.78 kV. We used the S. cerevisiae proteome database (containing 5861 protein sequences; www.expasy.org, downloaded July 2007) for analysis of the data.
Growth and chromosome segregation assays
For growth assays, a fivefold serial dilution of cells was spotted on 1% yeast extract, 2% bactopeptone, and 2% glucose (YPD), YPD with dimethyl sulfoxide (DMSO), or 15 μg/ml benomyl in DMSO. For chromosome segregation assays, dam1 spc34 strains were grown in YPD medium supplemented with a final concentration of 0.8 mM adenine to reduce background fluorescence and 250 μM CuSO4 to induce expression of the LacI-GFP fusion reporter. Cultures were grown at the permissive temperature (25°C) and then shifted to either the permissive or the nonpermissive temperature (37°C) and grown overnight. Cells were fixed for 5 min in 4% methanol-free formaldehyde and resuspended in phosphate-buffered saline (PBS) containing 10 μg/ml 4′,6-diamidino-2-phenylindole (DAPI). Only cells with resolved GFP foci or large-budded cells each with a separate nucleus (identified by DAPI fluorescence) were used for quantification purposes. To be scored as normal chromosome segregation, the GFP-labeled chromosomes colocalized within the nucleus of unbudded cells or in each of separated nuclei in large-budded cells. For scoring missegregation, we counted cells that contained more than one GFP focus within a single nucleus or large-budded cells in which one nuclear mass was devoid of GFP foci. A Zeiss Axioskop 2 (Carl Zeiss, Jena, Germany) with 100× objective numerical aperture (NA) ∼1.4 was used to image cells. Chromosome segregation assays with okp1 and ame1 strains were essentially as described previously (Knockleby and Vogel, 2009). In brief, cells were grown at 25°C overnight in SC-HIS with fivefold excess adenine. The next day, cells were diluted into SC-HIS with fivefold excess adenine and 40 mM 3-amino adipate for ∼2 h in the presence of α-factor (RP01002; GenScript, Piscataway, NJ) to arrest cells in G1. Cells were shifted to the semipermissive temperature (33°C for okp1 and 30°C for ame1) for 1 h. Cells were released into pheromone-free medium, and samples were collected ∼2 h after release, when most cells were in mitosis, fixed for 5 min in 4% methanol-free formaldehyde, and resuspended in PBS containing DAPI (10 μg/ml). Chromosome segregation was assayed in mitotic cells. For normal chromosome segregation the GFP-labeled centromeres colocalized with the nucleus in each of the large-budded cells with separated nuclei. For chromosome missegregation we quantified cells that contained more than one GFP focus within a single nucleus or cells in which one cell contains a nucleus devoid of GFP foci. A DeltaVision microscope work station (Applied Precision, Issaquah, WA) was used with 100× objective NA ∼1.4 to acquire Z-stacks. The Z-stack for each field of cells was imaged such that the entire depth of the cells was included to ensure collection of the total fluorescence from every cell. ImageJ software (National Institutes of Health, Bethesda, MD) was used to construct projections from each set of Z-stacks.
Cell cycle arrest and depletion of cohesion experiments
Cells were grown in YPD to logarithmic phase and treated with 3 μM α-factor (RP01002; GenScript) for 135 min (G1) or 20 μg/ml nocodazole (M1404; Sigma-Aldrich, St. Louis, MO) for 150 min (G2/M). To confirm cell cycle arrest, cellular morphology was monitored and DNA content determined by flow cytometric analysis, as described previously (Mishra et al., 2011; Supplemental Figure 2B). Samples were analyzed using a Becton-Dickinson FACSCalibur flow cytometer and Cell Quest software (BD Biosciences, Franklin Lakes, NJ). For experiments evaluating the centromeric association of phosphorylated Cse4 in response to reduced tension, we used a well-characterized reporter strain in which expression of Scc1 is controlled by the GAL1 promoter and that of Cdc20 by the MET3 promoter (Keating et al., 2009). ChIP experiments were carried out using cells that were arrested by α-factor (G1) in methionine-free media (Cdc20 ON) containing galactose (Scc1 ON) and then shifted to medium containing glucose (Scc1 OFF) and methionine (Cdc20 OFF). Cell biology approaches have shown lack of sister chromatid cohesion and reduced tension in this strain under the experimental conditions used (Keating et al., 2009). Controls included strains grown continuously in galactose medium containing methionine, a condition that allows expression of Scc1, while repressing Cdc20.
Protein analysis and generation of phosphospecific Cse4 antibody
We generated phosphospecific Cse4 antibodies (αp-Cse4) using peptides corresponding to the Cse4-Ser-33 site with N-terminal cysteine residues (H2N-CLAGDQQ(Sp)IND-amide). Polyclonal antibodies were raised against the conjugated phosphopeptide in rabbits by New England Peptide (Gardner, MA) and affinity purified. Secondary antibodies were horseradish peroxidase–conjugated (HRP) sheep anti-mouse immunoglobulin G (IgG; NA931V; Amersham, Piscataway, NJ) and HRP-conjugated sheep anti-rabbit IgG (NA934V; Amersham). To purify histidine-tagged Cse4 protein from S. cerevisiae, (8–16) × 108 cells were disrupted using glass beads and vortexing (75 min) in 500 μl of lysis buffer (0.1 M Tris, pH 8.0, 6 M guanidine chloride, 0.5 M NaCl). Extracts were clarified by centrifugation and incubated with nickel-charged Superflow NTA resin (Qiagen, Valencia, CA) for 1 h at room temperature and washed once with lysis buffer and three times with washing buffer (100 mM Tris-Cl, pH 8.0, 20% glycerol, 1 mM phenylmethylsulfonyl fluoride). The bound protein was eluted by boiling in 2× Laemmli sample buffer with 200 mM imidazole. Samples were resolved by SDS–PAGE on 4–12% Bis-Tris SDS-polyacrylamide gels (NP0322BOX; Novex, San Diego, CA). Blots were washed with TBST (Tris-buffered saline plus 0.1% Tween 20) three times for 10 min. Western blot analysis was done using primary antibodies α-HA (16B12; BAbCO, Richmond, CA) or αp-Cse4 (0.5 μg/ml) in TBST containing 5% (wt/vol) dried skim milk. ImageJ software was used for quantification of Western blot signals. Western blot analysis showed reactivity of the αp-Cse4 antibody to cse4-Ser-33A (data not shown).
ChIP experiments
For ChIP experiments, cultures grown to logarithmic phase or arrested with α-factor or treated with nocodazole or upon deletion of cohesion were treated with formaldehyde (final 1%) for 20 min at 30°C, followed by addition of 2.5 M glycine for 5 min at 30°C. Cells were washed twice with 1× PBS, resuspended in 0.7 ml of breaking buffer (0.1 M Tris, pH 8.0, 20% glycerol, protease inhibitor cocktail [P8215; Sigma-Aldrich]), and lysed by vortexing for 1 h at 4°C in presence of glass beads. Cell lysate was added to 1 ml FA buffer (50 mM Na–4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.6, 1 mM EDTA, 1% Triton X-100, 150 mM NaCl, 0.1% Na deoxycholate) with protease inhibitor cocktail. After centrifugation the chromatin pellet was resuspended in FA buffer and sonicated on ice at 10% output power 10 times for 15 s. The resulting soluble chromatin fraction was denoted as input and treated or not treated with 200 U/ml alkaline CIP (MA M0290L; New England BioLabs, Ipswich, MA) for 1 h at 37°C. Immunoprecipitation was carried out using Dynabeads Protein A (100.02D, Invitrogen) coated with antibodies (αHA [16B12; BAbCO], αp-Cse4 [Basrai lab], no antibody) overnight at 4°C. The immunoprecipitated DNA–protein complexes were washed for 5 min at room temperature, each wash in FA buffer (twice), FA-HS buffer (50 mM Na-HEPES, pH 7.6, 1 mM EDTA, 1% Triton X-100, 500 mM NaCl, 0.1% Na deoxycholate; twice), RIPA buffer (10 mM Tris pH 8.0, 1 mM EDTA, 250 mM LiCl, 0.5% Nonidet P40, 0.5% Na deoxycholate; twice), and TE (pH 8.0; once), and finally resuspended in elution buffer (25 mM Tris, pH 7.6, 10 mM EDTA, 0.5% SDS). For the input control, the input sample was diluted in 2× Stop buffer (20 mM Tris, pH 8.0, 100 mM NaCl, 20 mM EDTA, 1% SDS). All samples were incubated overnight at 65°C to reverse the cross-linking. After centrifugation, supernatant was treated with 0.7 mg/ml Proteinase K (AM2546; Ambion, Austin, TX) at 55°C for 4–5 h. After phenol/chloroform/isoamyl alcohol extraction, the immunoprecipitated DNA was ethanol precipitated at −80°C overnight. Twenty-microliter PCRs were performed using primer pairs for at least two different centromeric sequences (0.5 μM primers [ Table 5], 1× HotStar-Taq polymerase Master Mix [203445; Qiagen], denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 68°C for 45 s for 25–27 cycles). Ten-microliter PCR products were resolved on a 1.6% agarose gel containing ethidium bromide. Quantification of bands on agarose gels used GeneSnap software (Syngene, Frederick, MD). Linear range of quantification was verified by analyzing serial dilutions of input DNA (Supplemental Figure 2A).
In vitro assays for phosphorylation of Cse4
Cse4 was produced in E. coli and purified by Sephacryl-S200 chromatography as described (Luger et al., 1997) and was phosphorylated with purified GST-Ipl1 and GST-Sli15 (kindly donated by Trisha Davis, University of Washington, Seattle, WA). The 100-μl reaction contained 10 μg of Cse4, 0.5 μM GST-Ipl1, 0.5 μM GST-Sli15 (residues 554–698), 200 mM NaCl, 10 mM ATP, 25 mM MgCl2, and 50 mM HEPES buffer, pH 7.2. Reactions were incubated at 30°C for 90 min. Control reactions lacked GST-Ipl1 and GST-Sli15. The samples were run on 4–12% Bis-Tris SDS-polyacrylamide gels (Invitrogen) and the Cse4 bands excised and analyzed by mass spectrometry as described. For in vitro assays using radiolabeled ATP a 20-μl reaction contained 1 μg of Cse4, 0.5 μM GST-Ipl1, 0.5 μM GST-Sli15, 25 mM Tris-HCl pH 7.5, 2 mM dithiothreitol, 10 mM MgCl2, 0.5 mM EDTA, 100 μM ATP, and 1 μCi of [γ-32P]ATP. Reactions were incubated at 30°C for 5, 15, 30, and 90 min. Reactions were stopped with 5 μl of 4× NuPAGE LDS loading buffer (Life Technologies, Grand Island, NY) and boiled for 5 min at 95°C. Control reactions lacked GST-Ipl1 and GST-Sli15. The samples were run on 4–12% Bis-Tris SDS-polyacrylamide gels (Invitrogen) and stained with Coomassie blue, and radiolabeled proteins were detected using a Storm Detector (GE Healthcare, Piscataway, NJ).
Supplementary Material
Acknowledgments
We are grateful to Trisha N. Davis for purified Ipl1/Sli15, Sue Biggins for strains and advice, Domenic Esposito of the National Cancer Institute for construction of cse4 alleles, and Todd Stukenberg, Tony Hazbun, and Basrai Laboratory members for advice. We thank Katherine McKinnon of the National Cancer Institute Vaccine Branch FACS Core and Tatiana Karpova and Jim McNally of the National Cancer Institute Fluorescence Imaging Group. M.B. is supported by the Intramural Research Program of the National Cancer Institute, National Institutes of Health.
Abbreviations used:
- ANOVA
analysis of variance
- CEN3
centromere of chromosome 3
- CenH3
centromeric histone H3 variant
- ChIP
chromatin immunoprecipitation
- CIP
calf intestine phosphatase
- COMA
complex of Ctf19, Okp1, Mcm21, and Ame1
- END
essential N-terminal domain
- HA
hemagglutinin
- PTM
posttranslation modification
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0893) on May 1, 2013.
*These authors contributed equally to this work.
REFERENCES
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics. 2009;183:1591–1595. doi: 10.1534/genetics.109.109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha JP, Larson JR, Gao L, Daves LK, Tatchell K. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:1032–1045. doi: 10.1091/mbc.E07-05-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Jansen LE, Maddox PS, Foltz DR, Desai AB, Shah JV, Cleveland DW. Centromere identity maintained by nucleosomes assembled with histone H3 containing the CENP-A targeting domain. Mol Cell. 2007;25:309–322. doi: 10.1016/j.molcel.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Bloom K, Joglekar A. Towards building a chromosome segregation machine. Nature. 2010;463:446–456. doi: 10.1038/nature08912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Burrack LS, Berman J. Flexibility of centromere and kinetochore structures. Trends Genet. 2012;28:204–212. doi: 10.1016/j.tig.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S, Tatsutani SY, Vermaak D, Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- Chen Y, Baker RE, Keith KC, Harris K, Stoler S, Fitzgerald-Hayes M. The N terminus of the centromere H3-like protein Cse4p performs an essential function distinct from that of the histone fold domain. Mol Cell Biol. 2000;20:7037–7048. doi: 10.1128/mcb.20.18.7037-7048.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Acuna R, Au WC, Basrai MA. A role for histone H4K16 hypoacetylation in Saccharomyces cerevisiae kinetochore function. Genetics. 2011;189:11–21. doi: 10.1534/genetics.111.130781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy JS, Mishra PK, Au WC, Basrai MA. Insights into assembly and regulation of centromeric chromatin in Saccharomyces cerevisiae. Biochim Biophys Acta. 2012;1819:776–783. doi: 10.1016/j.bbagrm.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke DJ, Bachant J. Kinetochore structure and spindle assembly checkpoint signaling in the budding yeast, Saccharomyces cerevisiae. Front Biosci. 2008;13:6787–6819. doi: 10.2741/3189. [DOI] [PubMed] [Google Scholar]
- De Rop V, Padeganeh A, Maddox PS. CENP-A: the key player behind centromere identity, propagation, and kinetochore assembly. Chromosoma. 2012;121:527–538. doi: 10.1007/s00412-012-0386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;17:2902–2921. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Cheeseman IM. T time for point centromeres. Nat Cell Biol. 2012;14:559–561. doi: 10.1038/ncb2509. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ, Ricketts WA. Differential expression of a phosphoepitope at the kinetochores of moving chromosomes. J Cell Biol. 1993;122:1311–1321. doi: 10.1083/jcb.122.6.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewawasam G, Shivaraju M, Mattingly M, Venkatesh S, Martin-Brown S, Florens L, Workman JL, Gerton JL. Psh1 is an E3 ubiquitin ligase that targets the centromeric histone variant Cse4. Mol Cell. 2010;40:444–454. doi: 10.1016/j.molcel.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating P, Rachidi N, Tanaka TU, Stark MJ. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J Cell Sci. 2009;122:4375–4382. doi: 10.1242/jcs.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K, Hieter P. Evolutionary conservation between budding yeast and human kinetochores. Nat Rev Mol Cell Biol. 2001;2:678–687. doi: 10.1038/35089568. [DOI] [PubMed] [Google Scholar]
- Knockleby J, Vogel J. The COMA complex is required for Sli15/INCENP-mediated correction of defective kinetochore attachments. Cell Cycle. 2009;8:2570–2577. doi: 10.4161/cc.8.16.9267. [DOI] [PubMed] [Google Scholar]
- Kotwaliwale CV, Frei SB, Stern BM, Biggins S. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev Cell. 2007;13:433–445. doi: 10.1016/j.devcel.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunitoku N, Sasayama T, Marumoto T, Zhang D, Honda S, Kobayashi O, Hatakeyama K, Ushio Y, Saya H, Hirota T. CENP-A phosphorylation by Aurora-A in prophase is required for enrichment of Aurora-B at inner centromeres and for kinetochore function. Dev Cell. 2003;5:853–864. doi: 10.1016/s1534-5807(03)00364-2. [DOI] [PubMed] [Google Scholar]
- Lampert F, Hornung P, Westermann S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J Cell Biol. 2010;189:641–649. doi: 10.1083/jcb.200912021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- Mishra PK, Au WC, Choy JS, Kuich PH, Baker RE, Foltz DR, Basrai MA. Misregulation of Scm3p/HJURP causes chromosome instability in Saccharomyces cerevisiae and human cells. PLoS Genet. 2011;7:e1002303. doi: 10.1371/journal.pgen.1002303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz J, Stemmann O, Rank S, Lechner J. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 1999;13:1140–1155. doi: 10.1101/gad.13.9.1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Ranjitkar P, Press MO, Yi X, Baker R, MacCoss MJ, Biggins S. An E3 ubiquitin ligase prevents ectopic localization of the centromeric histone H3 variant via the centromere targeting domain. Mol Cell. 2010;40:455–464. doi: 10.1016/j.molcel.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samel A, Cuomo A, Bonaldi T, Ehrenhofer-Murray AE. Methylation of CenH3 arginine 37 regulates kinetochore integrity and chromosome segregation. Proc Natl Acad Sci USA. 2012;109:9029–9034. doi: 10.1073/pnas.1120968109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart JS, Frederick DL, Varner CM, Tatchell K. The mutant type 1 protein phosphatase encoded by glc7–1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol Cell Biol. 1994;14:896–905. doi: 10.1128/mcb.14.2.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Dulev S, Liu X, Hiller NJ, Zhao X, Strunnikov A. Cooperation of sumoylated chromosomal proteins in rDNA maintenance. PLoS Genet. 2008;4:e1000215. doi: 10.1371/journal.pgen.1000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tien JF, Umbreit NT, Gestaut DR, Franck AD, Cooper J, Wordeman L, Gonen T, Asbury CL, Davis TN. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J Cell Biol. 2010;189:713–723. doi: 10.1083/jcb.200910142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler RK, Chu ML, Johnson H, McKenzie EA, Gaskell SJ, Eyers PA. Phosphoregulation of human Mps1 kinase. Biochem J. 2009;417:173–181. doi: 10.1042/BJ20081310. [DOI] [PubMed] [Google Scholar]
- Waybright T, Gillette W, Esposito D, Stephens R, Lucas D, Hartley J, Veenstra T. Identification of highly expressed, soluble proteins using an improved, high-throughput pooled ORF expression technology. Biotechniques. 2008;45:307–315. doi: 10.2144/000112916. [DOI] [PubMed] [Google Scholar]
- Westermann S, Cheeseman IM, Anderson S, Yates JR 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin SG, Barber CM, Allis CD, Sullivan KF. Differential regulation of CENP-A and histone H3 phosphorylation in G2/M. J Cell Sci. 2001a;114:653–661. doi: 10.1242/jcs.114.4.653. [DOI] [PubMed] [Google Scholar]
- Zeitlin SG, Shelby RD, Sullivan KF. CENP-A is phosphorylated by Aurora B kinase and plays an unexpected role in completion of cytokinesis. J Cell Biol. 2001b;155:1147–1157. doi: 10.1083/jcb.200108125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li X, Marshall JB, Zhong CX, Dawe RK. Phosphoserines on maize centromeric histone H3 and histone H3 demarcate the centromere and pericentromere during chromosome segregation. Plant Cell. 2005;17:572–583. doi: 10.1105/tpc.104.028522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.