Transcription factor Msn2 is a primary mediator of stress response in yeast cells. We use fluorescence microscopy, genetic analysis, and computational modeling to address the mechanistic basis of Msn2 nuclear localization under different stresses. The “bursting” phase of the Msn2 response has significant functional implications.
Abstract
All cells perceive and respond to environmental stresses through elaborate stress-sensing networks. Yeast cells sense stress through diverse signaling pathways that converge on the transcription factors Msn2 and Msn4, which respond by initiating rapid, idiosyncratic cycles into and out of the nucleus. To understand the role of Msn2/4 nuclear localization dynamics, we combined time-lapse studies of Msn2-GFP localization in living cells with computational modeling of stress-sensing signaling networks. We find that several signaling pathways, including Ras/protein kinase A, AMP-activated kinase, the high-osmolarity response mitogen-activated protein kinase pathway, and protein phosphatase 1, regulate activation of Msn2 in distinct ways in response to different stresses. Moreover, we find that bursts of nuclear localization elicit a more robust transcriptional response than does sustained nuclear localization. Using stochastic modeling, we reproduce in silico the responses of Msn2 to different stresses, and demonstrate that bursts of localization arise from noise in the signaling pathways amplified by the small number of Msn2 molecules in the cell. This noise imparts diverse behaviors to genetically identical cells, allowing cell populations to “hedge their bets” in responding to an uncertain future, and to balance growth and survival in an unpredictable environment.
INTRODUCTION
All cells perceive environmental stresses and respond to them in a way that contributes to the cell's survival under adverse conditions. In yeast cells, a wide variety of stresses, including heat, oxidative agents, nutrient depletion, and high or low osmolarity, induces a common, stereotypic transcriptional response, referred to as the environmental stress response (Gasch et al., 2000). A number of sensing pathways, including those responsive to changes in temperature, osmolarity, such as the Hog1 stress-activated protein kinase pathway, and nutrient status, mediated by protein kinase A (PKA), AMP-activated kinase (SNF1), and target of rapamycin complex 1 (TORC1), provide input regarding potentially stressful environmental conditions (Zaman et al., 2008; de Nadal and Posas, 2010; de Nadal et al., 2011). These pathways converge on stress-sensitive transcription factors, two of which, encoded by functionally redundant genes MSN2 and MSN4, contribute significantly to the environmental stress response (Gasch et al., 2000). The transcriptional changes elicited by these factors promote survival not immediately to the stress provoking the response, but instead to prolonged or subsequent stresses (Giaever et al., 2002; Berry and Gasch, 2008; Berry et al., 2011; Klosinska et al., 2011).
The primary mode of regulation of Msn2 activity is subcellular relocalization. That is, in the absence of stress, Msn2 resides predominantly in the cytoplasm. After application of stress, cytoplasmic Msn2 relocates to the nucleus, where it activates a large cohort of stress-responsive genes (Görner et al., 1998; Gasch et al., 2000). This process is mediated at least in part by phosphorylation of Msn2, catalyzed by PKA and SNF1, and dephosphorylation of Msn2, catalyzed by protein phosphatases 1 and 2A (Santhanam et al., 2004; De Wever et al., 2005). Jacquet et al. (2003) observed that Msn2 nuclear localization in response to stress occurs in a concerted and oscillatory manner. That is, upon application of stress, most of the Msn2 protein in the cell enters the nucleus en masse, remains there for a short period of time, returns en masse to the cytoplasm, and continues oscillating irregularly and cell autonomously between nucleus and cytoplasm for several hours in the absence of any apparent additional external cues. This persistent but irregular nuclear–cytoplasmic shuttling was also observed by Cai et al. (2008) for the transcription factor Crz1 in response to calcium addition to cells. They proposed that this cycling facilitated a coherent response of the cohort of downstream genes, which would not be possible by a graded change in the amount of transcription factor relocated to the nucleus in proportion to the external stimulus. These investigators likened this process to frequency modulation of transcription factor activation, in contrast to the more common amplitude modulation. More recently, Hao and O'Shea (2011) reported that different stimuli elicited different dynamics of Msn2 nuclear localization, with some stimuli yielding the oscillatory behavior, or frequency modulation, and others promoting a sustained nuclear entry, or amplitude modulation, in proportion to the intensity of the stimulus. They found that different dynamics had distinct effects on the transcriptional output of Msn2-responsive genes, with frequency modulation yielding a more productive response than did amplitude modulation. Thus oscillatory or “bursting” nuclear localization of Msn2, as opposed to nuclear localization per se, appears to be critical for its efficient transcriptional activation.
The mechanism by which Msn2 or other transcription factors exhibits irregular nuclear–cytoplasmic oscillations in response to a simple stimulus step function is unresolved. Jacquet, Goldbeter, and colleagues provided a model and some supporting evidence that the oscillatory behavior of Msn2 is tied to intrinsic oscillations of PKA activity in the cell, in conjunction with stochastic fluctuations of the levels of components in the signaling pathway (Garmendia-Torres et al., 2007; Gonze et al., 2008). Although this model recapitulates much of the behavior of Msn2, it depends on a number of ad hoc assumptions about Msn2 and the PKA signaling network and, as noted in this article, fails to explain all of the properties of the Msn2 response to stress. Stochastic fluctuation or intrinsic noise in signaling pathways tied to transcription factors contributes to cell-to-cell variation in transcriptional output, sufficiently so that noise correlation provides a means of assigning the expression of individual genes as downstream of specific signaling pathways (Stewart-Ornstein et al., 2012). Thus noise permeates cellular signaling pathways.
In this article we define the signaling pathways responsible for Msn2 nuclear localization in response to different stresses. We show that Msn2 dynamics results from the activity of a signaling network comprising PKA, SNF1, protein phosphatase 1 (PP1), and Hog1, with varied contributions from the different components in response to different stresses. We examine both experimentally and computationally the mechanistic underpinnings of the irregular oscillations of Msn2, and find that they arise predominantly from stochastic noise within the system rather than oscillatory feedback in the signaling network. Moreover, our results suggest that this noise arises mainly from the signaling networks and then is amplified by the discrete number of Msn2 molecules within the cell. Given the contribution of these oscillations to the transcriptional output of Msn2 activation, our results highlight the importance of noise both in generating robust transcriptional responses to stimuli and in promoting distinct behaviors of genetically identical cells to uniform stress conditions.
RESULTS
Msn2 responds with different dynamics to different stresses
Msn2 resides predominantly in the cytoplasm in unstressed cells but rapidly concentrates in the nucleus upon stresses such as nutrient limitation, hyperosmotic shock, or heat shock. Recent work showed that the dynamics of Msn2 localization differs depending on the nature of the stress to which cells are subject (Hao and O'Shea, 2011). As a means of understanding how these different dynamical behaviors of Msn2 arise, we explored the signaling pathways that mediate nuclear localization in response to individual stresses. To do so, we examined the behavior of Msn2–green fluorescent protein (GFP), expressed from the endogenous MSN2 locus, in single wild-type or mutant cells over time after application of different stresses or their combinations. All strains carried a deletion of MSN4, unless otherwise noted. The fusion protein is indistinguishable from the wild-type protein in its ability to induce transcription in response to any of the stresses we examined (Supplemental Figure S2G). We imaged these cells by time-lapse fluorescence microscopy on a wide-field microscope after they were immobilized in a microfluidic flow cell under continuous media flow. Two media inlets to the flow cell allowed rapid switching from one medium to another. We measured intensity of nuclear GFP over time using an automated image analysis program, as described in Materials and Methods. Detailed quantification of control samples indicated that ∼5% of the total cellular Msn2-GFP resides in the nucleus under unstressed conditions. In contrast, 80–90% accumulates in the nucleus after a severe stress such as complete glucose depletion. We represent the results of these experiments in Figure 1 and elsewhere as kymographs, in which each row reports the behavior of GFP in a single cell over time, with the level of nuclear localization indicated by the intensity of color at each time point.
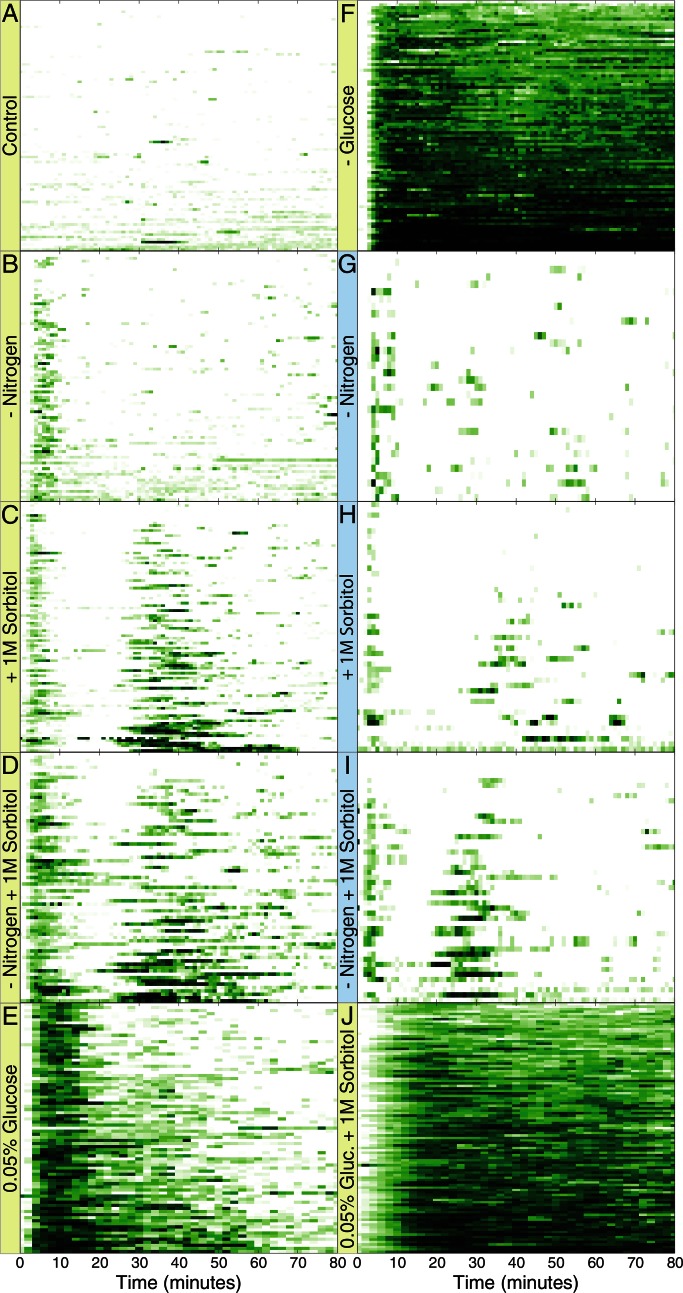
FIGURE 1:
Distinct dynamic patterns of Msn2 nuclear localization in response to different stresses. Y3630 (MSN2-GFP msn4∆ prototroph) cells were immobilized in a microfluidic flow cell perfused with SD medium, and imaged over time. At time 0, the medium was switched from SD to one of the indicated conditions: SD-nitrogen (B, G), SD-glucose (F), SD with 0.05% glucose (E), SD + 1 M sorbitol (C, H), SD-nitrogen + 1M sorbitol (D, I), SD with 0.05% glucose + 1 M sorbitol (J), and SD (A). Data are presented as kymographs representing the subcellular localization of GFP-tagged Msn2 in individual cells (y-axis) over time (x-axis) with the intensity of green color indicating the extent of nuclear occupancy by Msn2-GFP. G–I report experiments performed on a two-photon microscope, whereas a wide-field fluorescence microscope was used for A–F and J. See also Supplemental Figure S1. Frames were captured once every 90 s for E, once every minute for A–D, F, and J, and once every 20 s for G–I. For convenience of comparison, data for G–I were converted to 1-min intervals by averaging the −20-, 0-, and 20-s values for each minute.
The behavior of Msn2 in individual cells in response to a variety of stresses is depicted in Figure 1, and the behavior averaged over all cells under each condition is shown in Supplemental Figure S1. Cells bathed in SD medium (synthetic medium containing 2% glucose and 5 mM ammonium sulfate) were shifted to the same medium (Control) or to the same medium lacking glucose or ammonium sulfate, or containing 1.0 M sorbitol, or a combination of these. Cytoplasmic Msn2 relocated to the nucleus in most cells within minutes of the application of stress. At varying times thereafter, nuclear Msn2 returned to the cytoplasm and then underwent irregular bursts of concerted but short-lived periods of nuclear localization. These bursts of nuclear localization occurred only rarely in the absence of stress (Figure 1A and Supplemental Figure S1A; see also Supplemental Figure S4A). Although this overall pattern was observed for all stresses, different stresses elicited distinct variations in it. For instance, the duration and extent of the initial localization in response to a glucose downshift were dose dependent. After transfer of cells from 2 to 0.05% glucose, Msn2 localized to the nucleus in most cells and then relocated to the cytoplasm generally within 15 min of nuclear entry (Figure 1E and Supplemental Figure S1E). Transfer of cells to a medium completely lacking glucose resulted in sustained nuclear localization, however, with cells adapting only slightly after 80 min (Figure 1F and Supplemental Figure S1F). A second variation of the general pattern was observed after sorbitol addition. After the first wave of nuclear localization, Msn2 underwent a second, relatively coherent wave of nuclear localization, the timing, duration, and intensity of which were also dose dependent (Figure 1C and Supplemental Figure S1, K–N). We observed the same pattern with KCl as the osmolite (data not shown). As noted later, this second peak is an indirect consequence of translational remodeling attendant on stress activation of the high-osmolarity response (HOG) pathway.
Previous studies showed that fluorescence microscopy can produce phototoxic stress, which can itself induce nuclear localization of Msn2 (Garmendia-Torres et al., 2007; and data not shown). Accordingly, we also performed these experiments using two-photon fluorescence microscopy, which minimizes phototoxic stress. As noted in Figure 1 and Supplemental Figure S1 (compare B vs. G, C vs. H, and D vs. I), although the two-photon microscope allowed image collection every 20 s with little phototoxicity, our wide-field microscope provided images every 60 s without inducing noticeably greater levels of phototoxic stress. Because the wide-field microscope was the more sensitive of the two, we performed the majority of our experiments on this instrument.
Our observations further demonstrated that Msn2 responds to a combination of stresses in a synergistic manner. Depriving cells of nitrogen coincident with sorbitol addition resulted in a small increase in the duration of the initial response compared with sorbitol addition alone, as well as accelerating the onset of the second coherent wave of nuclear localization (Figure 1, C, D, H, and I, and Supplemental Figure S1, C, D, H, and I). Thus the concurrent application of these two stresses yielded an approximately additive response. In contrast, adding sorbitol while limiting glucose caused a delay in nuclear localization relative to either treatment alone, as well as persistent Msn2 nuclear localization not observed after either single stress (Figure 1J and Supplemental Figure S1J). In this case, the response to the dual treatment was not simply an additive combination of the responses to the individual treatments. From these observations we conclude that the nuclear entry and exit of Msn2 upon the application of stress is carefully controlled by the cell in speed, duration, frequency, and intensity.
Finally, our observations indicate that, although all cells in an experiment are genetically identical and subjected to identical treatment, the responses of individual cells can be quite distinct. Some cells exhibit little or no Msn2 nuclear localization in response to a stress, whereas others respond with robust and prolonged nuclear localization. This individual response is not due to differing positions of the cells within the microfluidic chamber, as data are collected only from the middle of the flow cell, where the media flow is uniform. Moreover, the distribution of weakly and strongly responding cells was random within a field of view. Nor is the individual response due to different cell cycle stages: budding and nonbudding cells exhibited a similar variation in their responses (data not shown). Instead, as discussed later, this difference in the degree of response to a given stress by genetically identical cells likely results from stochasticity in stress response pathways.
Correlation of nuclear localization and transcription activation
We assessed whether Msn2 localization to the nucleus is transcriptionally productive by measuring transcription of a chromosomally integrated reporter gene consisting of four STRE elements (binding sites for Msn2) inserted upstream of the LEU2 promoter driving transcription of the LacZ coding region (Supplemental Figure S2). Accumulation of LacZ mRNA after these stresses was strongly dependent on Msn2, indicating that this construct serves as a faithful reporter of Msn2 performance as a transcriptional activator (Supplemental Figure S2, A and B). In batch culture, the pattern of LacZ mRNA accumulation attendant on a particular stress was consistent with the pattern of Msn2 nuclear localization elicited by that stress. For instance, LacZ mRNA after a glucose downshift increased in proportion to the severity of the downshift (Supplemental Figure S2C). Similarly, the accumulation of LacZ mRNA after a combined sorbitol addition and glucose downshift was significantly delayed relative to that after either treatment alone (Supplemental Figure S2E). Thus our results suggest that nuclear localization of Msn2 on average results in transcriptional activation of Msn2-responsive genes.
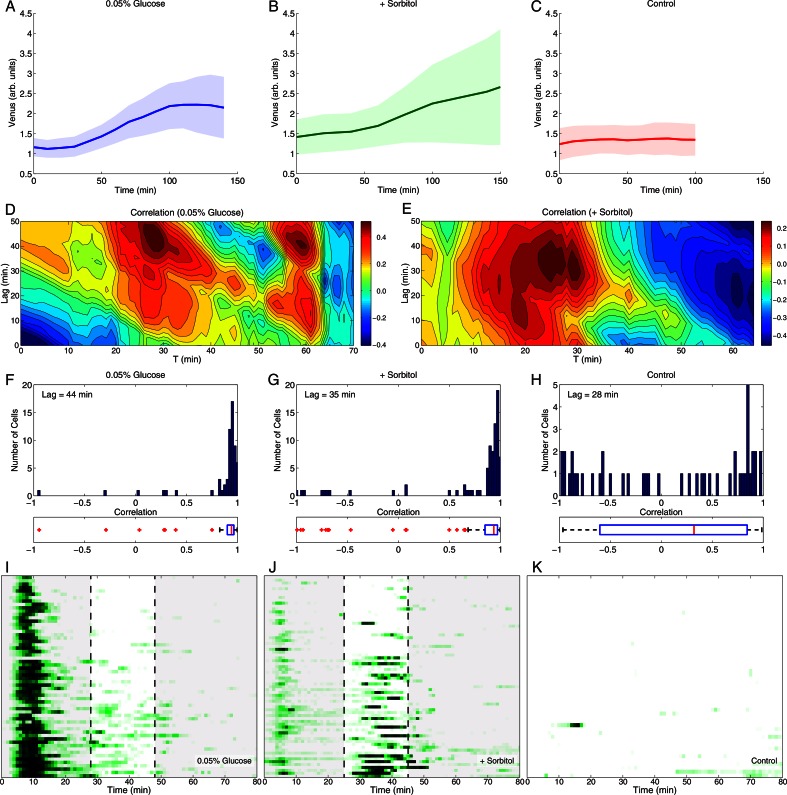
We extended these studies to examine the correlation between nuclear localization of Msn2 and subsequent transcriptional activation in individual cells by inserting a STRE(4)-PLEU2-YFP (Venus) into a strain expressing Msn2-mCherry. In bulk cultures Venus mRNA accumulated with the same kinetics and extent after stress as did LacZ mRNA expressed from the STRE(4)-PLEU2-LacZ construct (Supplemental Figure S2I). We measured Msn2-mCherry nuclear localization in individual cells and concurrently measured Venus accumulation by whole-cell fluorescence for several hours after application of various stresses. As shown in Figure 2, A and B, Venus fluorescence started to increase in cells 30–40 min after application of the stress and continued rising for approximately 2 h. No increase in fluorescence or Venus mRNA was observed in the absence of stress or in the absence of Msn2 (Figure 2C and Supplemental Figure S2H). Most cells exhibited strong correlation between integrated Msn2 nuclear localization and Venus fluorescence that accumulated after a lag (for simplicity, set to the optimal lag from Figure 2, D and E), particularly in glucose-limited cultures (Figure 2, F and G). Some cells exhibited uncorrelated behavior or even inverse correlation, however, suggesting that nuclear localization per se was not the sole determinant of productive transcriptional activation by Msn2. In Figure 2, D and E, we applied a sliding window over 20-min intervals to calculate the period of nuclear Msn2 localization that best correlated with the extent of accumulation of Venus fluorescence, as measured over 20-min intervals at various lags subsequent to Msn2 localization. As shown in Figure 2, I and J, for both sorbitol treatment and glucose deprivation that period corresponded to the secondary stage of irregular bursts rather than to the initial wave of Msn2 localization immediately after the application of stress. This determination reinforces the idea that nuclear localization is not the sole determinant of transcriptional activation and further highlights the bursting period of Msn2 behavior as the primary driver of gene activation.
FIGURE 2:
Transcriptional activation only partially correlates with Msn2 nuclear localization. Cells of strain Y3992 (MSN2-mCherry msn4∆ STRE(4)-PLEU2-YFP (Venus)) were immobilized in a microfluidic flow cell and imaged over time to measure nuclear localization of Msn2 and Venus accumulation in individual cells. (A–C) Average Venus fluorescence intensity as a function of time for a population of cells subjected at time 0 to a glucose downshift to 0.05%, addition of 1 M sorbitol, or no stress. The shaded areas demarcate the average response ± 1 SD over all cells. (D, E) Contour maps of correlation coefficients between the Msn2 nuclear localization averaged over the cell population and integrated over the 20-min interval starting at the time indicated on the x-axis and the accumulation of Venus during the 20-min interval that lags the Msn2 interval as indicated on the y-axis. The lag is computed with respect to the start of the Msn2 interval (zero lag means the two intervals coincide). The accumulation of Venus is computed as the difference between final and initial average Venus fluorescence intensity in the delayed 20-min interval. For instance, after a downshift to 0.05% glucose we observed a maximum correlation between Msn2 nuclear localization integrated over 28- to 48-min interval and Venus production in the 72- to 92-min interval (44-min delay). (F–H) Histograms of correlation coefficients for all cells. For each cell we compute the correlation between the Msn2 nuclear localization integrated over the time interval [0, t] and the level of Venus production at time t + lag. The time lag was adopted from D and E. Box plots below each histogram indicate the median correlation (red bars), the 25th and 75th percentiles (blue boxes), the range of the data points not considered outliers (dashed lines) and outliers (red crosses). Points are considered outliers if they are larger than q3 + 1.5 (q3 – q1) or smaller than q1 – 1.5 (q3 – q1), where q1 and q3 are the 25th and 75th percentiles, respectively. (I–K) Kymographs of Msn2 nuclear localization in the experiments described in A–C. Shown on the kymographs are the 20-min windows of integrated Msn2 nuclear localization that best correlate with subsequent Venus production, as described in D and E. Data were collected at 1-min intervals. See also Supplemental Figure S2.
Modulation of PKA activity is sufficient but not necessary for stress-responsive nuclear Msn2 localization
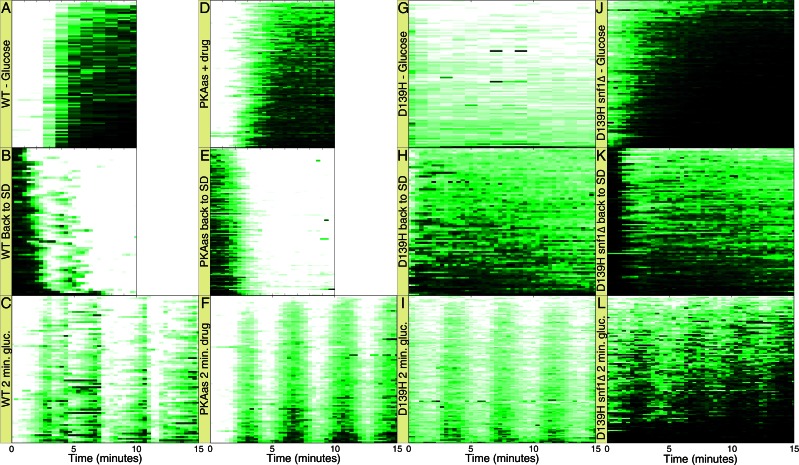
PKA affects Msn2 subcellular localization: PKA phosphorylation of four sites in the Msn2 nuclear localization sequence (NLS) and one site in the Msn2 nuclear export sequence (NES) sequesters Msn2 to the cytoplasm. Previous studies of the bursting behavior of Msn2 nuclear localization posited a feedback loop in PKA regulation that would elicit cell-autonomous oscillations of PKA activity, driving corresponding oscillations of Msn2 nuclear localization (Garmendia-Torres et al., 2007). To test the role of PKA activity on Msn2 localization, we examined how artificial modulation of PKA activity affects Msn2 nuclear residence. Switching from glucose-containing to glucose-free medium or vice versa resulted in nuclear import or nuclear export of Msn2, respectively, with a lag of 2–3 min (Figure 3, A and B). Furthermore, cycling between these two media elicited a corresponding cycle of nuclear localization of Msn2 as rapidly as every minute (Figure 3C and Supplemental Figure S3A). Consistent with a previous report (Hao and O'Shea 2011), we could recapitulate this pattern of Msn2 behavior by periodically inhibiting PKA through addition and withdrawal of the ATP analogue 1NM-PP1 to a strain, designated pka-as, in which all three TPK genes were mutated to render the encoded kinase subunits sensitive to the analogue (Zaman et al., 2009; Figure 3, D and F, and Supplemental Figure S3, C and D). Given that glucose regulates PKA activity, these observations are consistent with the hypothesis that glucose regulation of Msn2 localization proceeds through PKA.
FIGURE 3:
PKA modulation is sufficient but not necessary for regulation of Msn2 nuclear localization. (A–C) Kymographs of Y3630 (MSN2-GFP msn4∆ prototroph) cells immobilized in a microfluidic chamber switched from SD medium to 0% glucose at time 0 (A) or from SD lacking glucose to SD + 2% glucose at time 0 (B), or subjected to 2-min cycles between 0 and 2% glucose (C). (D–F) Kymographs of Y3817 (pka-as MSN2-GFP msn4∆) cells immobilized as previously and switched from SD medium to SD + 1 μM inhibitor 1NM-PP1 at time 0 (D) or from SD +1 M 1NM-PP1 to SD at time 0 (E) or subjected to 2-min cycles of SD and SD + 1 M 1NM-PP1 (F). (G–I) Kymographs of Y3845 (pka-wimpD139H MSN2-GFP msn4∆) cells treated as in A–C. (J–L) Kymographs of Y3964 (pka-wimpD139H snf1Δ MSN2-GFP msn4∆) cells treated as in A–C. Note that Msn2-GFP resides in the nucleus at time 0 in G and J due to the low levels of PKA activity in these strains. See also Supplemental Figure S3. Data were collected at 20-s intervals, except for A and G, which were collected at 1-min intervals.
Although the foregoing results document that PKA can regulate Msn2 localization, they do not establish whether PKA activity is necessary for glucose regulation of Msn2 localization. To test this, we examined nuclear localization of Msn2 in response to glucose addition in strains expressing only low-level, constitutive PKA activity. These strains, designated pka-wimp, carry deletions of BCY1 and two of the three TPK genes encoding the PKA catalytic subunit, as well as a debilitating mutation in the remaining TPK gene. Were PKA the sole mediator of glucose input to Msn2 localization, we would not expect to observe altered Msn2 localization in response to changes in glucose levels in the pka-wimp strains. Contrary to this expectation, glucose withdrawal from or addition to pka-wimp cells results in relocalization of Msn2 from the cytoplasm to the nucleus or vice versa, respectively (Figure 3, G and H). Moreover, cycling glucose addition every few minutes results in a corresponding cycling of Msn2 localization in the mutant cells (Figure 3I and Supplemental Figure S3, E and F). We obtained the same results with three different pka-wimp strains—tpk2 E235Q, tpk2 V218G, and tpk2 D139H—exhibiting residual PKA activity of ∼8, 3, and 1% of wild type, respectively (Figure 3, G–I, and Supplemental Figure S3, E, F, and I–N). These results suggest that signaling pathways other than PKA participate in Msn2 localization.
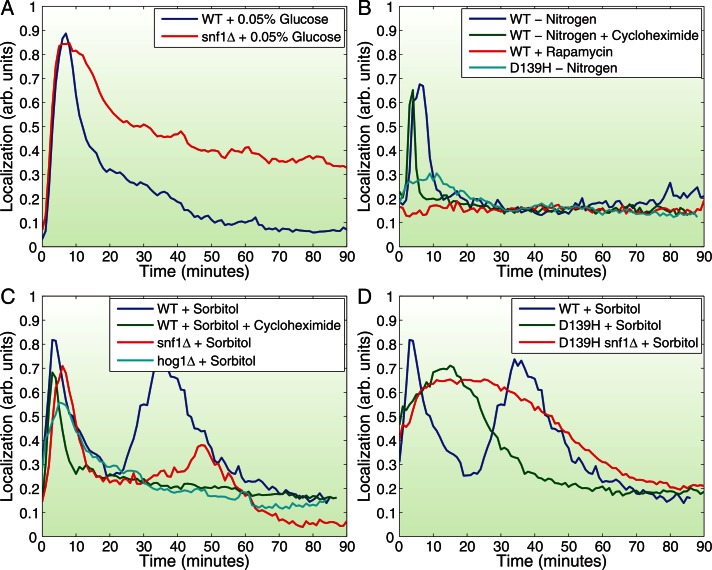
Consistent with previous work that implicated SNF1 and PP1 in this capacity (Mayordomo et al., 2002; De Wever et al., 2005), we found that deleting SNF1 in the pka-wimp strains virtually eliminated the cycling of Msn2 in and out of the nucleus during switches between control and glucose-free media. Instead, Msn2 essentially remained in the nucleus in both control and glucose-free conditions (Figure 3, J–L, and Supplemental Figure S3, G and H). We found that SNF1 also participates in Msn2 localization in response to glucose availability under normal PKA levels, although in this case the effect of SNF1 deletions manifests primarily as a delay in exodus of Msn2 from the nucleus after the initial glucose-depletion elicited nuclear entry (Figure 4A and Supplemental Figure S4), as previously reported. Thus PKA is not the sole purveyor of glucose signaling to Msn2; SNF1 provides a locus for rapid Msn2 regulation in conditions of low PKA activity and a delayed response in the presence of normal PKA activity.
FIGURE 4:
Characterization of signaling pathways controlling Msn2 localization in response to nutrient downshift and osmotic shock. (A) Average Msn2 nuclear localization in >100 Y3630 (MSN2-GFP msn4∆ [WT]) or Y3847 (snf1Δ MSN2-GFP msn4∆ [snf1∆]) cells as a function of time after transfer at time 0 from SD to SD + 0.05% glucose. (B) Average Msn2 nuclear localization in >100 Y3630 (MSN2-GFP msn4∆ [WT]) or Y3845 (pka-wimpD139H MSN2-GFP msn4∆ [D139H]) cells as a function of time after transfer at time 0 from SD to SD lacking ammonium sulfate (–Nitrogen), from SD to SD lacking ammonium sulfate plus 10 μg/ml cycloheximide (–Nitrogen +Cycloheximide), or from SD to SD + 200 nM rapamycin (+Rapamycin). (C, D) Average Msn2 nuclear localization in >100 Y3630 (MSN2-GFP msn4∆ [WT]), Y3847 (snf1Δ MSN2-GFP msn4∆ [snf1∆]), Y3989 (hog1Δ MSN2-GFP msn4∆ [hog1∆]), Y3845 (pka-wimpD139H MSN2-GFP msn4∆ [D139H]), or Y3964 (pka-wimpD139H snf1Δ MSN2-GFP msn4∆ [D139H snf1∆]) cells as a function of time after transfer from SD to SD + 1 M Sorbitol (+Sorbitol) or SD + 1 M Sorbitol + 10 μg/ml cycloheximide (+Sorbitol +Cycloheximide). See also Supplemental Figure S4.
Several signaling pathways mediate Msn2 localization to nutrient and osmotic stress
As noted, stresses such as nitrogen starvation and increased osmolarity also promote nuclear localization of Msn2. Nitrogen starvation elicits an immediate and coherent burst of nuclear localization of Msn2 in most cells, followed by a period of increased frequency of erratic bursts (Figure 1, B and G, and Supplemental Figure S1). The TORC1 pathway informs cells of nitrogen source availability, suggesting that inhibition of TORC1 should mimic nitrogen withdrawal with regard to Msn2 localization (De Virgilio and Loewith, 2006). However, previous studies yielded conflicting results of inhibiting TORC1 by addition of rapamycin (Beck and Hall, 1999; Mayordomo et al., 2002; Santhanam et al., 2004). As shown in Figure 4B, we find that rapamycin addition alone has no effect on Msn2 localization, indicating that TORC1 does not directly regulate the stress response. In contrast, attenuation of the PKA pathway in a pka-wimp strain almost completely eliminated the ability of nitrogen withdrawal to drive Msn2 into the nucleus. Thus nitrogen regulation of Msn2 nuclear entry flows through PKA rather than TORC1.
Sorbitol addition yields a complex pattern of Msn2 behavior, with an initial coherent burst of nuclear occupancy, followed by a second wave whose timing and duration are dose dependent (Figure 4C and Supplemental Figure S1N). The high-osmolarity mitogen-activated protein (MAP) kinase pathway regulates specific transcriptional responses to sorbitol addition and exhibits synthetic interactions with Msn2 (Capaldi et al., 2008). Deletion of HOG1, encoding the MAP kinase component of the HOG pathway, or treatment of cells with cycloheximide eliminates the secondary wave of Msn2 nuclear localization without significantly affecting the initial burst of localization (Figure 4C). We conclude that sorbitol activation of the HOG pathway induces the secondary response but that this response results from events downstream of the pathway requiring de novo protein synthesis. Similarly, elimination of SNF1 attenuates the secondary wave of sorbitol-induced Msn2 nuclear localization without affecting the initial response. Finally, diminished PKA signaling significantly alters the pattern of behavior of Msn2 in response to sorbitol addition, delaying and broadening the initial wave of localization and eliminating a secondary response (Figure 4D). This effect is exacerbated by the concurrent elimination of SNF1. Thus the rapid increase in nuclear localization of Msn2 in response to sorbitol and its subsequent exodus are substantially dependent on PKA and facilitated by SNF1.
A comprehensive model of Msn2 response to different stresses
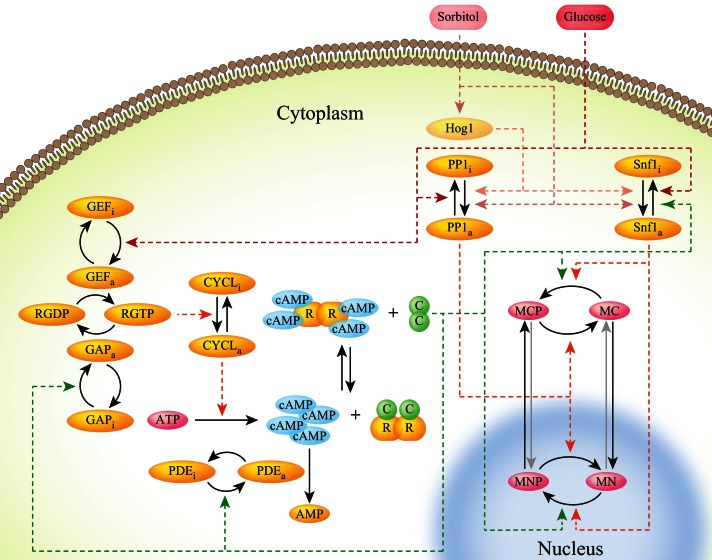
Goldbeter, Jacquet, and colleagues proposed a model of the Ras-PKA network impinging on Msn2 to attempt to account for the bursting behavior of Msn2 as a negative feedback loop within the PKA circuit (Garmendia-Torres et al., 2007). We expanded this theoretical network to include our aforementioned findings, adding Hog1, Snf1, and a phosphatase acting on Msn2, which we think likely to be PP1 (Figure 5). In this augmented network, PKA promotes cytoplasmic retention and inhibits nuclear entry of Msn2 by phosphorylating its NLS and NES sites (Görner et al., 1998, 2002). Glucose and nitrogen activate PKA by as-yet-unknown mechanisms. SNF1 inhibits Msn2 nuclear residence by phosphorylating at least the S582 site, as reported previously (De Wever et al., 2005), and is itself deactivated by glucose and activated by sorbitol, the latter pathway taking place largely through Hog1, consistent with our epistasis experiments. In addition, the model requires that SNF1 be inhibited by PKA, since the rate of SNF1-promoted exit of Msn2 from the nucleus in response to sorbitol is inversely proportional to the residual PKA activity in the various tpk-wimp strains: the lower the PKA activity, the faster is the exit (Figure 6, E and F, and data not shown). We propose in the model that glucose inhibits and sorbitol stimulates the phosphatase PP1, which itself promotes the nuclear entry of Msn2. Finally, Hog1 is the main player in the sorbitol response of Msn2 in cases when PKA is active and inhibits contribution by SNF1.
FIGURE 5:
Model of the signaling network regulating Msn2 nuclear occupancy. Our proposed Msn2 signaling network, showing the Ras/PKA branch, Snf1, PP1, and Hog1. MCP, phosphorylated cytoplasmic Msn2; MNP, phosphorylated nuclear Msn2; MC, unphosphorylated cytoplasmic Msn2; MN, unphosphorylated nuclear Msn2; C, catalytic subunits of PKA; R, regulatory subunits of PKA; RGTP and RGDP, Ras bound to GTP and GDP, respectively; CYCL, adenylyl cyclase; GAP, GTPase-activating proteins Ira1/2; GEF, GDP/GTP exchange factor Cdc25. The subscripts a and i indicate active and inactive forms of proteins, respectively. Dashed lines show catalytic reactions, and solid lines represent physical transitions.
FIGURE 6:
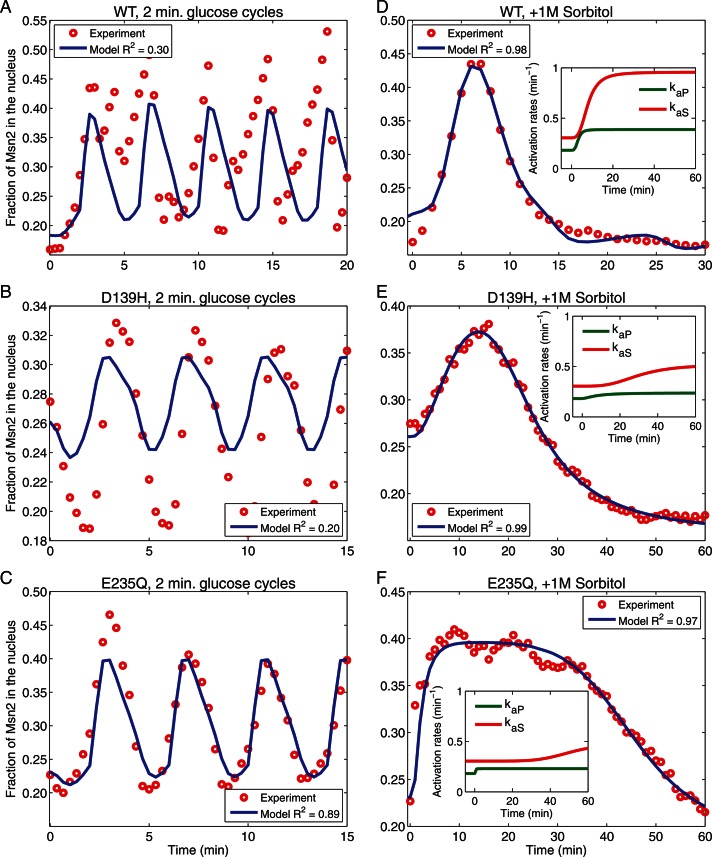
Correlation of experimental and simulated data. Simulations of Msn2 nuclear localization (blue lines) were performed by numerical integration of the reaction rate equations based on the model depicted in Figure 5, using the SimBiology toolbox from MATLAB. To simulate the glucose cycles (A–C), we used a single set of parameters either taken from Garmendia-Torres et al. (2007) or derived from optimization of the fit to data from the pka-wimp strains and the wild-type strain (see Supplemental Methods). Predictions are compared with experimental observations (red circles). Experimental data were obtained with strains Y3630 (WT; A, D), Y3845 (tpk-wimpD139H; B, E), and Y3842 (tpk-wimpE235Q; C, F) upon 2-min cycles of glucose addition and withdrawal (A–C) or after addition of 1 M sorbitol (D–F). To simulate sorbitol addition (D–F), we modify the activation rates of PP1 (kaP) and SNF1 (kaS) as shown in the insets of D–F (see Supplemental Methods). R2, coefficient of determination.
We represented this model as a system of ordinary differential equations with parameters for the PKA portion of the pathway either as previously defined or as determined from our experiments (see Supplemental Methods). We fitted the parameters that give the dynamics of SNF1 and PP1 in response to glucose oscillations using experimental data for three pka-wimp strains. We modeled the nuclear shuttling of Msn2 for wild type using these parameters and optimizing an additional set of six parameters (see Supplemental Methods). The final set of parameters provided a reasonable description of the experimental observations for multiple strains (Figure 6, A–C). To simulate the Msn2 response to sorbitol addition (Figure 6, D–F), we allow activation rates for PP1 and SNF1 to change as a consequence of the medium modification (see Supplemental Methods). Overall, we find that this expanded model reasonably describes the experiments with different strains and different conditions (Figure 6). Moreover, the model does not rely on only feedback regulation of PKA to account for the overall behavior of Msn2, since it accurately describes the dynamics of the system even if the PKA circuit is eliminated in the pka-wimp strains.
Stochastic noise in the Msn2 circuit
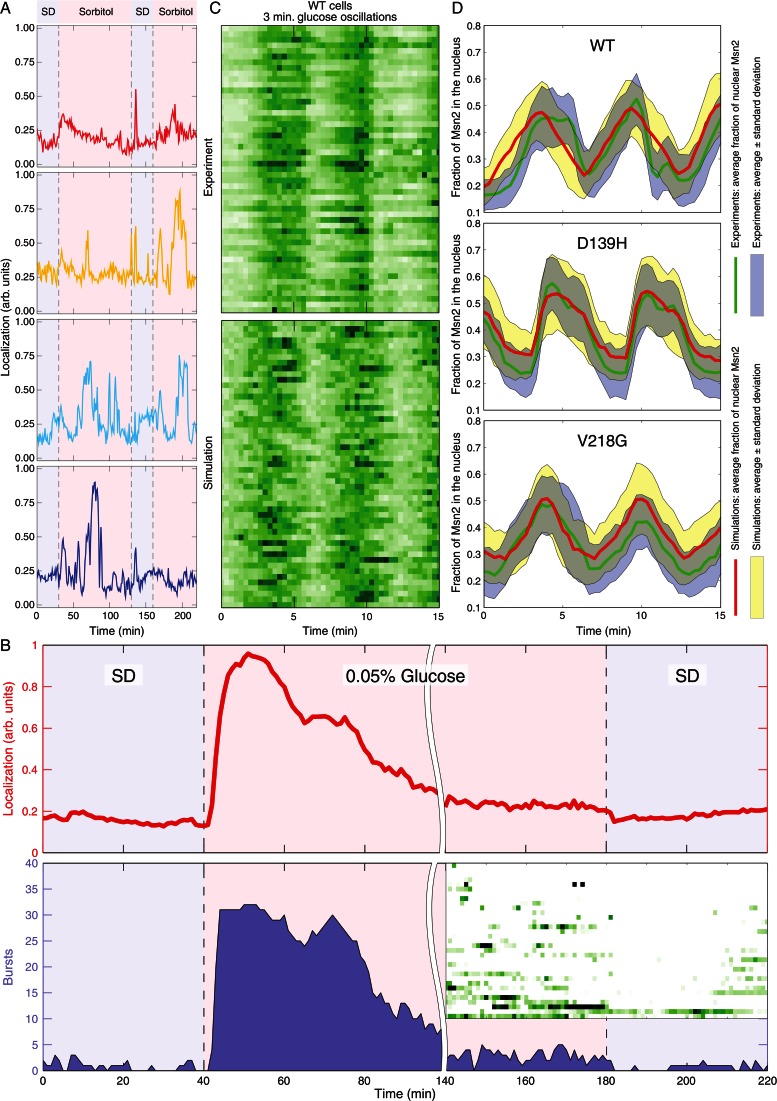
A noteworthy aspect of Msn2 relocalization in response to stress is that different cells behave differently. As noted in Figure 7A, this individual behavior is not due to differences among cells but instead is an idiosyncratic behavior exhibited by each individual cell. We subjected cells to an initial stress, returned them to the unstressed condition, and after a fixed interval subjected them to the same stress again. As is evident from tracings of Msn2 localization in four individual cells (Figure 7A), some cells respond robustly to the initial stress and then either respond robustly to the second stress or do not. Similarly, some cells might not respond to the initial stress and then either respond or not respond to the secondary stress. Thus there was no consistent correlation in the behavior of a cell to the initial input with that of the response to the second input. As noted, no correlation existed with the stage of the cell cycle either. From these observations, we conclude that the response of cells to stress is predominantly stochastic.
FIGURE 7:
Nuclear localization of Msn2 in response to stress is stochastic. (A) Y3630 (MSN2-GFP msn4∆ prototroph) cells immobilized in a microfluidic chamber were imaged while perfused with SD for 30 min, then subjected to 1 M sorbitol stress for 100 min, allowed to recover in SD for 30 min, and again stressed by SD + 1 M sorbitol for the duration of the time course. Shown are traces of Msn2-GFP nuclear localization in four different cells. (B) Y3630 (MSN2-GFP msn4∆ prototroph) cells immobilized in a microfluidic chamber were imaged while perfused with SD for 40 min, shifted to glucose-limited medium for 140 min, and then returned to SD. The top graph plots the average Msn2 nuclear localization over all cells, and the shaded area in the bottom graph shows the total number of cells with nuclear localization above a threshold of background fluorescence as a function of time. The inset shows a kymograph of individual cell behavior for the last 80 min of the experiment, spanning the transition from limited glucose to SD. Data were collected at 1-min intervals. (C) Top, kymograph of nuclear localization of Msn2 in Y3630 cells subjected to 3-min cycle of glucose addition and withdrawal. Data were collected at 20-s intervals. Bottom, simulated kymograph of wild-type cells subjected to 3-min cycles of glucose addition and withdrawal, obtained by Gillespie simulations of the model depicted in Figure 5. (D) Msn2-GFP nuclear localization was monitored in WT and two of the pka-wimp strains subjected to 3-min cycles of glucose removal and addition. The green line in each plot shows the averaged nuclear occupancy for all cells over the experiment, and the blue shading is the average ± SD. Gillespie simulations were performed as described in the Supplemental Methods, based on the model in Figure 5. The red line is averaged nuclear localization from these simulations, and the yellow shading indicates ± SD. See also Supplemental Figure S5.
A second component of the Msn2 response to stress is an increase in excited or bursting behavior after application of stress. As noted in Figure 7B, cells grown in SD show little bursting activity. Subjecting cells to a glucose downshift yields an increase in nuclear localization of Msn2 followed by adaptation in which the average nuclear localization returns to essentially prestressed levels. However, even in this adaptation period, the bursting behavior is much higher than observed under prestressed conditions and, even more strikingly, much higher than observed immediately after returning cells to the initial glucose levels. Thus stress induces an increase in noisy behavior of Msn2 even after cells adapt to the new conditions.
We find that signaling components contribute to the level of noise in the response of Msn2 to stress. Whereas strains attenuated for PKA activity continue to exhibit bursting activity, suggesting that the PKA pathway has little influence on the stochastic behavior of Msn2 (Supplemental Figure S5, A–F), deletion of SNF1 increases the frequency of bursting behavior both after application of stress and in the absence of stress (Supplemental Figure S4). From the data shown in Supplemental Figure S4, we calculate the coefficient of variation for unstimulated cells, proportional to the noise level and averaged over all times, to be 7.4% in snf1Δ cells, as opposed to 5.5% in SNF1 cells (Supplemental Figure S4C). Similarly, the coefficient of variation for stimulated cells is 12.7% in snf1Δ cells versus 9.0% in SNF1 cells (Supplemental Figure S4F). Thus SNF1 but not PKA appears to buffer the cells from fluctuations in Msn2 nuclear localization.
We attempted to identify the source of the erratic bursting behavior of Msn2 through stochastic modeling of the signaling circuit for Msn2 localization depicted in Figure 5. Using Gillespie simulations of this model of Msn2 regulation, employing the absolute levels of contributing components previously proposed (Gonze et al., 2008), we can achieve reasonably faithful recreation of the pattern of Msn2 localization both in wild-type cells and cells attenuated for PKA signaling (Figure 7, C and D). Furthermore, computationally altering the levels of components in the PKA pathway had little effect on the predicted behavior of Msn2, consistent with our experimental results described earlier suggesting that PKA does not contribute to the bursting behavior of Msn2. Instead, the most significant contributing factor in our simulations to the noisy behavior of Msn2 was the absolute levels of Msn2 in the cells. Simulating increased levels of Msn2 resulted in reduced bursting behavior, whereas decreased levels lead to increased bursting behavior. Consistent with this modeling, the level of noise in an MSN2-GFP/MSN2-GFP diploid during the bursting phase after a glucose downshift is less than that in an MSN2-GFP/msn2Δ diploid (Supplemental Figure S5, G–I). Moreover, even two distinguishable pools of Msn2 within the same cell do not respond in complete synchrony (Supplemental Figure S5, J–L). These results suggest that, although, as noted earlier, the signaling pathways themselves are sources of noise, that noise is amplified by the small number of transcription factors that reside downstream of the signaling networks.
DISCUSSION
What are the roles of the different signaling pathways in Msn2 nuclear localization?
Msn2 responds to a wide variety of stresses by relocalization from the cytoplasm to the nucleus (Görner et al., 1998). Our results address how these different stresses converge on Msn2. PKA-dependent phosphorylation of sites within the nuclear import domain of Msn2 blocks migration of Msn2 into the nucleus, and PKA-dependent phosphorylation of a site within the nuclear export domain promotes Msn2 exodus from the nucleus (Görner et al., 2002). Consistent with this regulation, we find that simply inactivating PKA rapidly drives Msn2 into the nucleus and reactivation rapidly drives Msn2 out of the nucleus. Given this result and the fact that PKA activity tightly correlates with external glucose levels (Zaman et al., 2008), nuclear localization of Msn2 in response to a glucose downshift might have been explained solely by signaling through PKA, without having to invoke any other glucose-responsive regulatory components.
Two observations, however, document that PKA is not solely responsible for the glucose effects on Msn2 localization. First, the pathway bandwidth for Msn2 localization in response to cycles of PKA inhibition and activation is less than that for glucose addition and withdrawal. That is, while step functions of PKA inhibition/activation of 2- or 3-min duration elicit distinct and corresponding waves of Msn2 nuclear localization, this response is blurred for stimuli of 1-min cycles (Supplemental Figure S3C). However, 1-min cycles of glucose addition and withdrawal elicit distinct waves of Msn2 nuclear localization (Supplemental Figure S3A). Thus glucose engages processes in addition to PKA that enhance signal fidelity and resolution. We note that this transmission fidelity is comparable to, but somewhat higher than, that for signaling through the HOG MAP kinase pathway (McClean et al., 2011). Second, cycles of glucose removal and addition yield cycles of Msn2 nuclear localization even in strains in which PKA is unresponsive to glucose levels (Figure 3I and Supplemental Figure S3, K and N). Thus the glucose effect on Msn2 localization can proceed through a PKA-independent signaling network. Prior results as well as our modeling and experimental data suggest that this additional input proceeds through SNF1 and, to a lesser extent, PP1 (De Wever et al., 2005). In particular, we propose that PP1 promotes glucose-dependent Msn2 entry into the nucleus on a fast time scale and SNF1 stimulates exit at a slower rate. Moreover, our results with different tpk-wimp strains indicate that SNF1 activity toward Msn2 is inversely correlated with PKA levels: higher PKA levels yield lower SNF1 activity and, consequently, slower nuclear Msn2 exit. These observations underpin the model for Msn2 regulation in response to glucose depicted in Figure 5 and yield robust modeling of the behavior of Msn2 in wild-type and mutant strains we examined.
Nitrogen withdrawal also stimulates Msn2 migration into the nucleus. Previous work suggested that the TORC1 network mediates perception of nitrogen availability. However, we find that inhibition of TORC1 by rapamycin addition, which should mimic nitrogen withdrawal were that model correct (Beck and Hall, 1999; Görner et al., 2002), has no effect on Msn2 localization (Figure 4B). Instead, Msn2 nuclear localization in response to nitrogen withdrawal is almost completely dependent on signaling through PKA. Thus, whereas inhibition of TORC1 may sensitize Msn2 localization to other stresses, immediate perception of stress from nitrogen downshift proceeds through PKA rather than TORC1. This suggests that the pathways for carbon and nitrogen signaling may be less segregated than previously appreciated.
Increased osmolarity, whether sorbitol or salt, affects Msn2 localization in a complex manner. Addition of an osmolite induces rapid nuclear relocalization, followed by nuclear exit and then a second wave of nuclear localization, the timing and duration of which are dose dependent. We note that this differs from the behavior of Msn2 in response to salt reported previously by Hao and O'Shea (2011). Our results indicate that this complex behavior results from a complex interplay of signaling networks. The initial wave of nuclear occupancy depends predominantly on PKA signaling with additional input through SNF1. The second wave depends absolutely on signaling through the Hog1 pathway and partially on SNF1 but is the consequence of posttranslational events stimulated by these signaling pathways. That is, some product(s) induced by osmolite-stimulated Hog1 signaling promotes Msn2 localization, although the identity of that product is unclear. This observation helps to explain the previously noted interplay between Msn2 and the Hog pathway on stress response (Rep et al., 2000; Capaldi et al., 2008).
Is localization sufficient for transcriptional activation?
Using an Msn2-dependent reporter construct, we examined the correlation on a cell-by-cell basis between nuclear localization of Msn2 and subsequent transcriptional activation. Although this correlation is reasonably high, it is by no means absolute. In fact, some cells show no correlation, and some exhibit anticorrelation. This indicates that nuclear localization alone is not sufficient for transcriptional activation, suggesting that at least one additional event, such as specific DNA binding, modification of the activation domain, or recruitment of additional factors, must occur for productive transcription (Durchschlag et al., 2004). The high correlation between nuclear occupancy and transcription suggests that this second event is also stimulated by the signaling process that drives Msn2 into the nucleus, perhaps through phosphorylation of the transcriptional activation domain of Msn2 or of some other component of the transcriptional machinery, such as Mediator. Finally, we note that some cells yield transcriptional activation even in the absence of any noticeable Msn2 nuclear accumulation, supporting the notion of a low level of Msn2 nuclear occupancy even under resting conditions.
We determined the time window over which the integrated Msn2 nuclear occupancy best correlates with subsequent reporter response within cells. Surprisingly, this window did not encompass the initial burst of Msn2 nuclear localization after the application of stress but instead implicated the subsequent period of erratic bursting behavior as the main contributor to productive transcriptional activation. This conclusion is consistent with previous observations suggesting that periods of bursting promote transcription better than sustained nuclear localization (Hao and O'Shea, 2011). Moreover, this observation suggests that the initial wave of nuclear localization may not be accompanied by the secondary event necessary for productive transcription. This raises the possibility that signaling processes with different time scales are responsible for the stress-induced transcription.
What causes the stochastic bursting behavior of Msn2?
We attempted to address how random bursts of Msn2 arise after application of stress. Garmendia-Torres et al. (2007) proposed that random fluctuations in Msn2 nuclear occupancy resulted from an oscillatory pattern of cAMP levels arising from a feedback loop through PKA that promotes cAMP decay. Although this might be a contributing factor, we observe bursting behavior even in strains in which PKA is not responsive to cAMP levels. Thus PKA oscillatory cycles cannot be the major driver for the observed bursting behavior. Instead, our results suggest that noise within the signaling pathways and stochastic events amplified by the small number of target molecules are the major contributors to bursting behavior.
Bursting of Msn2 nuclear occupancy occurs erratically, although the extent of bursting is increased by the application of stress. Randomness, or noise, in cellular processes such as transcription can arise from extrinsic factors, such as differences in size and cell cycle position, or from intrinsic factors, such as stochastic assembly of transcription complexes or remodeling of local chromatin structure (Thattai and van Oudenaarden, 2001; Elowitz et al., 2002; Raser and O'Shea, 2004; Xu et al., 2006). Stewart-Ornstein et al. (2012) demonstrated that a measurable level of noise arises within the signaling network upstream of transcriptional output, sufficient to provide a noise signature to all of those genes responsive to a common signaling pathway. We find little evidence for factors such as cell size or cell position contributing to the randomness of Msn2 localization. Instead, most of the noise arises from noise in the signaling and receiver elements of the stress pathways. Deletion of SNF1 significantly enhanced the bursting behavior of Msn2, even under unstressed conditions, suggesting that SNF1 activity contributes to noise suppression within the signaling network. Moreover, the extent of bursting immediately diminishes after removal of a stress signal, suggesting that the active signaling pathway per se enhances noise. However, a major source of the noise arises from the small number of Msn2 within the cell, ∼100 molecules/cell (Ghaemmaghami et al., 2003). First, nuclear localization of separately marked versions of Msn2 is not fully correlated (Supplemental Figure S5, J–L), indicating random behavior downstream of the signaling networks. Moreover, stochastic modeling of our hypothesized stress-responsive network predicts bursting behavior whose extent and level of intensity are relatively insensitive to changes in the levels of signaling components but are quite sensitive to changes in the number of Msn2 molecules in the cell. Consistent with those calculations, we find that bursting after adaptation is more extensive in an MSN2-GFP/msn2∆ than in an MSN2-GFP/MSN2-GFP diploid. Thus the random bursting behavior of Msn2 appears to involve the small number of Msn2 molecules in the cell. It is worth noting that this mechanism would contribute to the noise signature in transcriptional output identified by Stewart-Ornstein et al. (2012) and explain the unusually high cell-to-cell variation in the levels of stress-responsive proteins (Newman et al., 2006).
Individual response of cells to stress
This study highlights the large variation of responses of genetically identical cells to the same stress, both in the behavior of Msn2 and in the extent of transcriptional output. We suggest that this variation is not simply a consequence of a theoretical limit to the precision of signaling pathways in a microorganism, that is, that cells simply cannot do a better job of signal fidelity (e.g., see Yu et al., 2008), but instead reflects the evolutionary advantage of identical cells being able to mount distinct responses to a single input. Mounting a robust stress response protects cells from a sustained stressful environment but precludes cells from a rapid resumption of growth after removal of the stress. Cells subjected to a stress cannot predict whether the stress will be of short duration, for which the optimum strategy would be not to mount a stress response, or of long duration, for which the optimum strategy would be to mount a robust response. Thus being able to mount different responses to the same stress ensures that some cells within the population will have selected the optimum strategy, regardless of what the future holds. This ability for cells to “hedge their bets” has been observed for other aspects of the yeast stress response and for many other microorganisms, suggesting that this ability is a pervasive strategy (Kussell and Leibler, 2005; Nachman et al., 2007; Pelet et al., 2011).
MATERIALS AND METHODS
Strain growth and construction
Standard methods were used to grow, maintain, and construct strains (Burke et al., 2005). Strains used in this study were prototrophic and derived from W303 and are listed in Supplemental Table S1. Nitrogen-free medium consisted of 2% glucose and 1.7g/l Yeast Nitrogen Base without amino acids and without ammonium sulfate (Difco, Franklin Lakes, NJ). 1NM-PP1 (kindly provided by Kevan Shokat, University of California, San Francisco, San Francisco, CA) was dissolved in MeOH; rapamycin and cycloheximide were dissolved in dimethyl sulfoxide (DMSO). All drugs were added in volumes ≤0.03% of the total medium volume to minimize the stress effects of MeOH or DMSO.
The MSN2-GFP construct described in Görner et al. (1998) and provided by Stephen Garrett (University of Medicine & Dentistry of New Jersey, Newark, NJ) was used for integration into the endogenous MSN2 locus. The 5′ end of genomic MSN2 was replaced with URA3 and then the MSN2-GFP PCR fragment inserted in place of URA3 via 5-fluoroorotic acid counterselection. MSN2-mCherry in the STRE(4)-PLEU2-VENUS strain was similarly constructed. STRE(4)-PLEU2-LacZ and STRE(4)-PLEU2-VENUS strains were derived from CTT1-18 (Marchler et al., 1993), kindly provided by Claudio De Virgilio (University of Fribourg, Switzerland), and integrated into the URA3 locus of our strains. PKA analogue–sensitive and attenuated strains were previously described (Wang et al., 2004; Zaman et al., 2009).
Microfluidics
Two-channel flow cells were fabricated from polydimethylsiloxane as described (Hersen et al., 2008). The flow cell was pretreated for 10–15 min with 2 mg/ml concanavalin A dissolved in a solution of 5 mM CaCl2 and 5 mM MnCl2, pH 6–7. Cells (3 ml) grown at 25°C to an OD600 of 0.2 were concentrated by centrifugation, loaded into the flow chamber, and incubated for 4 min. Medium was connected to the two inflows with polyethylene tubing (Intramedic, inner diameter, 0.86 mm; outer diameter, 1.27 mm; BD, Franklin Lakes, NJ), and the outflow was similarly connected to a waste tube. The flow was maintained by gravity. The inflows could be closed by manually clipping the tubing, and media could be changed almost instantaneously by opening one inlet and closing the other.
Time-lapse microscopy
Cultures in the flow cells were imaged on a wide-field inverted microscope (DeltaVision; Applied Precision, Issaquah, WA) with a charge-coupled device camera (CoolSNAP HQ; Roper Scientific, Tucson, AZ), using a 100× oil-immersion objective at 25°C. The percentage transmittance for all channels was set at 10%, and the exposure times were 200 ms for GFP, 250 ms for mCherry, 150 ms for Venus, and 100 ms for CFP. Four 1-μm z-stacks were taken of each focal plane using the DeltaVision softWoRx software. Live imaging was also performed with a custom-built two-photon scanning microscope constructed around an upright Olympus BX51 (Olympus, Tokyo, Japan). Fluorescence photons were collected through a numerical aperture 0.9 Olympus water immersion objective X60 LUMPlanFl/IR and detected with high–quantum efficiency, hand-peaked GaAsP photomultipliers (H10770PA-40; Hamamatsu, Hamamatsu, Japan). The microscope was run through the MATLAB-written software ScanImage (MathWorks, Natick, MA), modified to control a piezo-objective (PI, Auburn, MA). Images were taken with an excitation wavelength of 920 nm. The laser intensity was set to 8.8 mW, measured at the level of the sample. Six 1-μm z-stacks were recorded every 20 s.
Image processing
The z-stacks were projected into one image by the maximum pixel intensity method. Images were segmented using a custom Hough transform algorithm in MATLAB. Fluorescence intensities in the nucleus and cytoplasm, after removal of background noise and illumination correction, were used to estimate the nuclear localization of Msn2 over time. The average nuclear fluorescence intensity was computed using the mean of the top 20% brightest pixels within each cell. For images that contained information in multiple color channels (e.g., Msn-GFP and Venus-mCherry), similar image analysis was done for all channels, involving background correction, image segmentation, and cell tracking. Subsequent data analysis used standard routines in MATLAB, which are available in the following toolboxes: Image Processing Toolbox, Curve Fitting Toolbox, Global Optimization Toolbox, and SimBiology. We developed several graphical user interfaces that helped us process a large number of experiments in a consistent manner. Details are provided in the Supplemental Methods. Deconvolution and detailed manual analysis of several experiments using CellProfiler image analysis software (www.cellprofiler.org) allowed us to estimate the amount of nuclear Msn2-GFP in unstressed cells (∼5–10%), as well as in maximally stressed cells (∼90–95%).
Real-time quantitative PCR
Cell cultures were grown at 25°C in control medium, vacuum filtered onto 0.45-μm nitrocellulose filters (Millipore, Billerica, MA), and washed into experimental medium. Samples were then collected by brief centrifugation and flash frozen in liquid nitrogen. RNA was extracted using Tri Reagent, treated with DNAse I (RNAse-Free DNAse Set; Qiagen, Valencia, CA), and converted to cDNA using the SuperScript First-Strand Synthesis System for real time-PCR (Invitrogen, Carlsbad, CA). The real-time PCR mixture was assembled with Power SYBR Green (Applied Biosystems, Foster City, CA) into 96-well Optical Reaction Plates (Applied Biosystems) and cycled in the ABI 7900 HT thermocycler. Results for LacZ and Venus RNAs were normalized to ACT1, ARP4, and TFB3 RNAs.
Simulations
Methods, equations, and parameter selections for simulations are described in the Supplemental Methods and in Supplemental Tables S2 and S3. MATLAB implementations of our deterministic and stochastic models are available as supplemental files.
Supplementary Material
Acknowledgments
We are grateful to Kevan Shokat, Stephen Garrett, and Claudio De Virgilio for reagents and plasmids. This research was supported by National Institutes of Health Grants GM076562 to J.R.B. and HG004708 to A.V.M., and Center for Quantitative Biology/National Institutes of Health Grant P50 GM071508.
Abbreviations used:
- AMP
adenosine monophosphate
- GFP
green fluorescent protein
- PKA
protein kinase A
- PP1
protein phosphatase 1
- TORC1
target of rapamycin complex 1
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-12-0870) on April 24, 2013.
*Present address: Department of Biochemistry and Molecular Biology, Penn State College of Medicine, Hershey, PA 17033.
REFERENCES
- Beck T, Hall MN. The TOR signalling pathway controls nuclear localization of nutrient- regulated transcription factors. Nature. 1999;402:689–692. doi: 10.1038/45287. [DOI] [PubMed] [Google Scholar]
- Berry DB, Gasch AP. Stress-activated genomic expression changes serve a preparative role for impending stress in yeast. Mol Biol Cell. 2008;19:4580–4587. doi: 10.1091/mbc.E07-07-0680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry DB, Guan Q, Hose J, Haroon S, Gebbia M, Heisler LE, Nislow C, Giaever G, Gasch AP. Multiple means to the same end: the genetic basis of acquired stress resistance in yeast. PLoS Genet. 2011;7:e1002353. doi: 10.1371/journal.pgen.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke DJ, Amberg DC, Strathern JN. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi AP, Kaplan T, Liu Y, Habib N, Regev A, Friedman N, O'Shea EK. Structure and function of a transcriptional network activated by the MAPK Hog1. Nat Genet. 2008;40:1300–1306. doi: 10.1038/ng.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Ammerer G, Posas F. Controlling gene expression in response to stress. Nat Rev Genet. 2011;12:833–845. doi: 10.1038/nrg3055. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Posas F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 2010;29:4–13. doi: 10.1038/emboj.2009.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C, Loewith R. The TOR signalling network from yeast to man. Int J Biochem Cell Biol. 2006;38:1476–1481. doi: 10.1016/j.biocel.2006.02.013. [DOI] [PubMed] [Google Scholar]
- De Wever V, Reiter W, Ballarini A, Ammerer G, Brocard C. A dual role for PP1 in shaping the Msn2-dependent transcriptional response to glucose starvation. EMBO J. 2005;24:4115–4123. doi: 10.1038/sj.emboj.7600871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durchschlag E, Reiter W, Ammerer G, Schüller C. Nuclear localization destabilizes the stress-regulated transcription factor Msn2. J Biol Chem. 2004;279:55425–55432. doi: 10.1074/jbc.M407264200. [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Garmendia-Torres C, Goldbeter A, Jacquet M. Nucleocytoplasmic oscillations of the yeast transcription factor Msn2: evidence for periodic PKA activation. Curr Biol. 2007;17:1044–1049. doi: 10.1016/j.cub.2007.05.032. [DOI] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W, Bower K, Howson R, Belle A, Dephoure N, O'Shea E, Weissman J. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Giaever G, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Gonze D, Jacquet M, Goldbeter A. Stochastic modelling of nucleocytoplasmic oscillations of the transcription factor Msn2 in yeast. J Royal Soc Int. 2008;5(suppl 1):S95–S109. doi: 10.1098/rsif.2008.0141.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Martinez-Pastor MT, Estruch F, Ammerer G, Hamilton B, Ruis H, Schüller C. Nuclear localization of the C2H2 zinc finger protein Msn2p is regulated by stress and protein kinase A activity. Genes Dev. 1998;12:586–597. doi: 10.1101/gad.12.4.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görner W, Durchschlag E, Wolf J, Brown EL, Ammerer G, Ruis H, Schüller C. Acute glucose starvation activates the nuclear localization signal of a stress-specific yeast transcription factor. EMBO J. 2002;21:135–144. doi: 10.1093/emboj/21.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao N, O'Shea EK. Signal-dependent dynamics of transcription factor translocation controls gene expression. Nat Struct Mol Biol. 2011;19:31–39. doi: 10.1038/nsmb.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersen P, McClean MN, Mahadevan L, Ramanathan S. Signal processing by the HOG MAP kinase pathway. Proc Nat Acad Sci USA. 2008;105:7165–7170. doi: 10.1073/pnas.0710770105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquet M, Renault G, Lallet S, Mey JD, Goldbeter A. Oscillatory nucleocytoplasmic shuttling of the general stress response transcriptional activators Msn2 and Msn4 in Saccharomyces cerevisiae. J Cell Biol. 2003;161:497–505. doi: 10.1083/jcb.200303030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klosinska MM, Crutchfield CA, Bradley PH, Rabinowitz JD, Broach JR. Yeast cells can access distinct quiescent states. Genes Dev. 2011;25:336–349. doi: 10.1101/gad.2011311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kussell E, Leibler S. Phenotypic diversity, population growth, and information in fluctuating environments. Science. 2005;309:2075–2078. doi: 10.1126/science.1114383. [DOI] [PubMed] [Google Scholar]
- Marchler G, Schüller C, Adam G, Ruis H. A Saccharomyces cerevisiae UAS element controlled by protein kinase A activates transcription in response to a variety of stress conditions. EMBO J. 1993;12:1997–2003. doi: 10.1002/j.1460-2075.1993.tb05849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayordomo I, Estruch F, Sanz P. Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (Stress Response Element)-regulated genes. J Biol Chem. 2002;277:35650–35656. doi: 10.1074/jbc.M204198200. [DOI] [PubMed] [Google Scholar]
- McClean MN, Hersen P, Ramanathan S. Measuring in vivo signaling kinetics in a mitogen-activated kinase pathway using dynamic input stimulation. Methods Mol Biol. 2011;734:101–119. doi: 10.1007/978-1-61779-086-7_6. [DOI] [PubMed] [Google Scholar]
- Nachman I, Regev A, Ramanathan S. Dissecting timing variability in yeast meiosis. Cell. 2007;131:544–556. doi: 10.1016/j.cell.2007.09.044. [DOI] [PubMed] [Google Scholar]
- Newman JRS, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, DeRisi JL, Weissman JS. Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature. 2006;441:840–846. doi: 10.1038/nature04785. [DOI] [PubMed] [Google Scholar]
- Pelet S, Rudolf F, Nadal-Ribelles M, Nadal Ed, Posas F, Peter M. Transient activation of the HOG MAPK pathway regulates bimodal gene expression. Science. 2011;332:732–735. doi: 10.1126/science.1198851. [DOI] [PubMed] [Google Scholar]
- Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Santhanam A, Hartley A, Düvel K, Broach JR, Garrett S. PP2A phosphatase activity is required for stress and Tor kinase regulation of yeast stress response factor Msn2p. Eukaryotic Cell. 2004;3:1261–1271. doi: 10.1128/EC.3.5.1261-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Ornstein J, Weissman JS, El-Samad H. Cellular noise regulons underlie fluctuations in Saccharomyces cerevisiae. Mol Cell. 2012;45:483–493. doi: 10.1016/j.molcel.2011.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thattai M, van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci USA. 2001;98:8614–8619. doi: 10.1073/pnas.151588598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Pierce M, Schneper L, Gu¨ldal CG, Zhang X, Tavazoie S, Broach JR. Ras and Gpa2 mediate one branch of a redundant glucose signaling pathway in yeast. PLoS Biol. 2004;2:e128. doi: 10.1371/journal.pbio.0020128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu EY, Zawadzki KA, Broach JR. Single-cell observations reveal intermediate transcriptional silencing states. Mol Cell. 2006;23:219–229. doi: 10.1016/j.molcel.2006.05.035. [DOI] [PubMed] [Google Scholar]
- Yu RC, Pesce CG, Colman-Lerner A, Lok L, Pincus D, Serra E, Holl M, Benjamin K, Gordon A, Brent R. Negative feedback that improves information transmission in yeast signalling. Nature. 2008;456:755–761. doi: 10.1038/nature07513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Schneper L, Slonim N, Broach JR. Glucose regulates transcription in yeast through a network of signaling pathways. Mol Syst Biol. 2009;5:245. doi: 10.1038/msb.2009.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S, Lippman SI, Zhao X, Broach JR. How Saccharomyces responds to nutrients. Annu Rev Genet. 2008;42:27–81. doi: 10.1146/annurev.genet.41.110306.130206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.