Abstract
In addition to their natural role in eukaryotic genome evolution, transposons can be powerful tools for functional genomics in diverse taxa. The piggyBac transposon has been applied as such in eukaryotic parasites, both protozoa and helminths, and in several important vector mosquitoes. piggyBac is advantageous for functional genomics because of its ability to transduce a wide range of taxa, its capacity to integrate large DNA ‘cargoes’ relative to other mobile genetic elements, its propensity to target transcriptional units and its ability to re-mobilize without leaving a pattern of non-excised sequences or ‘footprint’ in the genome. We recently demonstrated that piggyBac can integrate transgenes into the genome of the parasitic nematode Strongyloides ratti, an important model for parasitic nematode biology and a close relative of the significant human pathogen S. stercoralis. Unlike transgenes encoded in conventional plasmid vectors, which we assume are assembled into multi-copy episomal arrays as they are in Caenorhabditis elegans, transgenes integrated via piggyBac are not only stably inherited in S. ratti, they are also continuously expressed. This has allowed derivation of the first stable transgene expressing lines in any parasitic nematode, a significant advance in the development of functional genomic tools for these important pathogens.
Keywords: transposon, piggyBac, transgenesis, parasitic nematode, Strongyloides, chromosomal integration, gene silencing
Introduction
Parasitism by nematodes is often regarded as peripheral in human medicine, but, in fact, these infections account for an enormous burden of disease and disability when viewed globally. In the aggregate, parasitic nematodes infect roughly a quarter of the world’s population and cause acute illness, chronic debilitation, disfigurement, blindness and failure to thrive in children.1-3 The burden of disease due to nematode parasitism rests disproportionately on developing nations, but some of these agents are endemic in economically stressed communities in the USA.4 Parasitic nematodes in the genus Strongyloides infect a wide range of animal species including pigs, ruminants, horses and humans. S. stercoralis is a significant human pathogen, infecting approximately 200 million people. It is unique among nematodes parasitizing humans in its ability to autoinfect and thus self-replicate in its host. This can result in potentially fatal disseminated hyperinfection in patients immunocompromised by HTLV-1 infection or undergoing immunosuppressive drug treatment.5 There are no practical vaccines against parasitic nematode infection, and the armamentarium of drugs effective against these parasites is small and threatened by resistance. Modern approaches to discovery of new drug or vaccine targets in parasitic nematodes are hampered by the lack of functional genomic tools for these organisms. Futhermore, there are many interesting basic biological questions, among them ones pertaining to the genetic bases for the evolution of parasitism among nematodes6,7 and to the extreme longevity of parasitic nematodes compared with their free-living counterparts,8 which might be addressed at the functional level if more robust molecular genetic tools were in hand. Strongyloides and related genera would be particularly well suited to basic studies of the kind just described because their life cycles include alternative pathways of development leading to relatively short-lived free-living adults or relatively long-lived parasitic ones (Fig. 1).5 Methods for transgenesis and for disruption or silencing of specific genes in parasitic nematodes, which could facilitate both basic studies and the discovery of novel drug and vaccine targets, have been very slow to develop owing to the very complex and protracted life histories of these parasites and to the fact that none of them can be propagated in vitro.
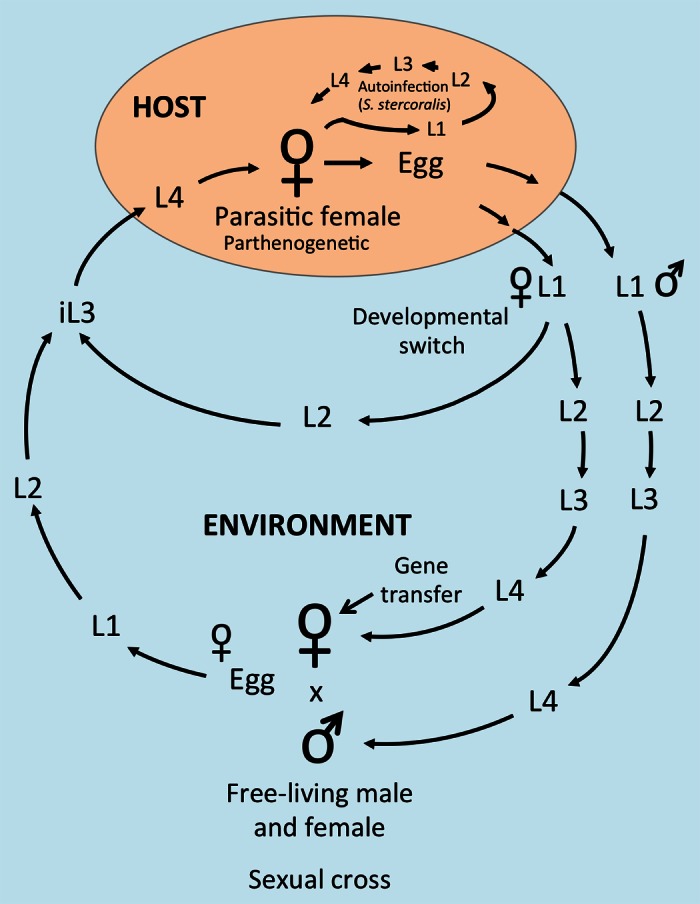
Figure 1. Schematic of the life cycle of Strongyloides spp For the genus Strongyloides, only parasitic females have been observed, and these reproduce by mitotic parthenogenesis. Depending on the species, either eggs or first stage larvae (L1), hatched within the host intestine, are passed in the host feces. Resulting post parasitic larvae are male or female. Female larvae have alternative developmental pathways leading either directly to infective third-stage larvae (L3i) or to a generation of free-living females. All male L1 develop to free-living males. Free-living adults mate and produce a generation of progeny, which in S. stercoralis and S. ratti are uniformly fated to develop to L3i. L3i invade the host by skin penetration and develop via the L4 to parasitic females. Strongyloides stercoralis is unique in its ability to develop precociously to the L3i within the primary host and thereby initiate repetitive autoinfective cycles that lead to geometrically expanding parasite burdens with serious, potentially fatal consequences for the host. Because of their anatomical similarity to hermaphrodites of Caenorhabditis elegans, free-living females of Strongyloides are readily transfected with DNA constructs by standard techniques for gonadal microinjection.
To address this methodological gap, our laboratory has pursued techniques for transgenesis in Strongyloides, which, because of some unique aspects of its life cycle, to be discussed below, is a more tractable model for parasitic nematode biology. Methods for gene transfer into S. stercoralis, involving only slight modifications of standard techniques for gonadal microinjection in C. elegans,9,10 were developed a decade ago11 and promoter-regulated, tissue-specific transgene expression was achieved in 2006.12 Despite these early successes, lines of Strongyloides that stably inherit and express transgene sequences were not achieved until 2012, when Shao et al. reported integrative transformation and establishment of stable transgenic lines in S. ratti.13 This significant result hinged upon chromosomal integration of transgene sequences, and this was achieved using elements of the piggyBac transposon system. This paper is the subject of this Commentary.
Mobile genetic elements (MGEs) occur naturally in the genomes of parasitic nematodes and some likely originated via horizontal transfer from the host
MGEs are important drivers of genome evolution in eukaryotic organisms, and genomic study of nematodes (including parasitic species) reveals that this is no less true for these worms.14 Both long-terminal repeat retrotransposons (LTRR) and Non-LTRR have been have been discovered in Ascaris lumbricoides,15,16 and a family of non-LTRR, dubbed the Dingo retrotransposons, was recently reported from the genome of the hookworm Ancylostoma caninum.17 In addition to retrotransposons, Class II MGEs, which are DNA transposons, have been discovered in C. elegans (active transposons Tc1-Tc5)18 and a number of parasitic nematodes, particularly within phylogenetic Clade V.19 These parasites include the trichostrongyles, which are important parasites of ruminant livestock, and hookworms, which are important pathogens of both humans and domestic animals. The Class II MGEs discovered in Clade V parasites include Tc1-like transposons such as hctcl from the trichostrongyle Hemonchus contortus and Mariner-like elements, including mle-1 from the trichostrongyle Trichostrongylus colubriformis and Bandit from the hookworm A. caninum.20 We have also discovered SMART from the threadworm Strongyloides stercoralis, a Clade IV parasite, only distantly related to the hookworms and trichostrongyles (Lok et al. unpublished). A defining characteristic of DNA transposons is their propensity for horizontal transfer between species. Phylogenetic analysis revealed that, at the time of its discovery, the closest known relative of the Bandit transposon from A. caninum was the Mariner-like element Hsmar-1 from humans,20 strongly suggesting that Bandit originated from horizontal transfer of a Mariner-like element to hookworms from their human hosts. Our own phylogenetic study of SMART in S. stercoralis (unpublished) reveals, similarly, that its closest relatives are Bandit from A. caninum and Hsmar-1 suggesting that this transposon was also acquired by S. stercoralis via horizontal transfer from its human host. It is interesting to note that retrotransposons have evidently been involved in horizontal transfer of other sequences to nematodes from other species. One prominent example of this is the apparent transfer of genes of insect origin, such as those encoding Diapausins, to the nematode Pristionchus pacificus, which lives in necromenic association with beetles.21
DNA transposons have practical applications in functional genomic study of parasitic nematodes and other parasitic helminths
The ability of MGEs to integrate into eukaryotic genomes makes them potential vehicles for experimental transformation of parasitic nematodes and other modifications of their genomes. Recent papers from Brindley and collegues demonstrate the capabilities of pseudotyped retroviruses to integrate transgenes into the genomes of parasitic flatworms, the blood flukes Schistosoma mansoni22,23 and S. japonicum.24 The Brindley lab has also achieved integrative transformation of S. mansoni using a modified DNA transposon, piggyBac, by transducing larval stages (schistosomules) with plasmid vectors encoding a reporter transgene flanked by truncated piggyBac inverted terminal repeats (ITRs) and capped mRNA encoding the piggyBac transposase.25 Demonstrating the feasibility of using MGEs as vectors for integrative transgenesis in schistosomes proved to be crucial in the development of methods for stable transgenesis in parasitic nematodes.
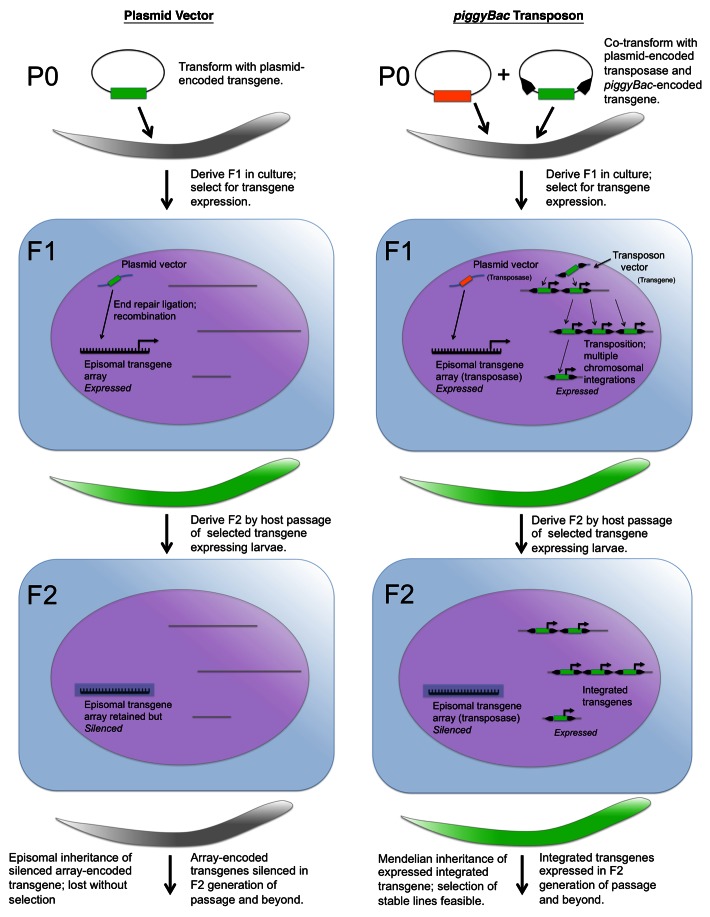
As previously indicated, our overall approach to transgenesis in parasitic nematodes was to select a subject species, preferably one of medical significance, that was most amenable to adaptation of standard methods for gene transfer in C. elegans, which involve infusing solutions of vector DNA into the gonadal syncytium by microinjection.9,10,26. The unique life cycles of Strongyloides spp, comprising both free-living and parasitic generations, provide access to free-living female worms of a similar size and body plan to C. elegans hermaphrodites, including a conspicuous gonadal syncytium that can be easily microinjected with nucleic acid solutions.11 Once it was determined that endogenous 3′ and 5′ regulatory sequences were necessary for promoter-regulated expression of transgenes in S. stercoralis,12 it became feasible to isolate transformed progeny of microinjected free-living females, rear them to infective stage larvae in vitro and to propagate these F1 transgenics through repeated rounds of alternating host and culture passage.27 As indicated, host passage is required for establishment of transgenic lines of any obligately parasitic helminth, as none can be cultured continuously in vitro. Strongyloides offers the advantage of one or more generations of free-living development, enhancing its utility as a subject for initial attempts at transgenesis. However, even in these most tractable of parasitic nematodes, free-living cycles must alternate with parasitic ones in deriving and maintaining transgenic lines. This notwithstanding, our first attempt at host passage of F1 transgenic S. stercoralis revealed that a significant proportion of the progeny of parasitic females transformed with plasmid vectors by gonadal microinjection retained transgene sequences through at least three generations of host and culture passage, indicating that germline transformation had occurred. F1 transgenic S. stercoralis larvae generated by this technique express transgenes in anatomical patterns that are consistent with their C. elegans orthologs. However, parasites transformed in this manner show no evidence of transgene expression in the F2 generation of passage and beyond.27 We hypothesize that this represents active silencing of transgenes in the F2 and subsequent generations of transgenic S. stercoralis (Fig. 2).
Figure 2. Observed expression patterns and hypothesized inheritance modes of plasmid- and transposon-encoded transgenes in germline cells of Strongyloides following transformation of parental free-living females by gonadal microinjection. Cartoon depicts assembly of plasmid-encoded transgenes into multi-copy episomal arrays, which are expressed in the F1 generation of passage and silenced in subsequent generations. Also depicted is chromosomal integration of piggyBac transposon-encoded transgenes, catalyzed by a plasmid-encoded and therefore episomally expressed transposase enzyme. Note that silencing of arrays encoding the transposase in worms transformed with piggyBac vectors will prevent inadvertent re-mobilization of the integrated transgene copies in the F2 generation and beyond. Green and gray worm cartoons indicate parasites that express or do not express the transgene, respectively. Green and orange inserts in plasmid and transposon cartoons denote transgene and transposase coding sequences, respectively. Black arrowheads flanking the transgene insert in the transposon vector cartoon denote the piggyBac ITRs. Cartoon design is based on the concept of Mello and Fire26 in their depiction of hypothetical processing and inheritance of plasmid-encoded transgenes delivered by microinjection into oocyte nuclei of C. elegans.
In assessing possible explanations for transgene silencing in S. stercoralis, we assumed that like C. elegans, these parasites assemble the preponderance of microinjected plasmid-based vectors into tandem, multi-copy arrays located in episomes (Fig. 2).9 Based upon this assumption, we hypothesized that these sequences are silenced by S. stercoralis (and, as subsequent unpublished studies showed, by S. ratti as well) because of their highly repetitive nature and/or their episomal location.13 We resolved to test this hypothesis by integrating a reporter transgene of proven functionality into the genome of S. stercoralis or S. ratti and searching for evidence of expression in F2 transgenic worms. Because of its demonstrated capability to integrate transgenes into the chromosomes of S. mansoni,25 we selected the piggyBac transposon as a system for transgene integration. As an additional precaution against epigenetic silencing, we also tested insulator sequences from another MGE, the gypsy retrovirus, that are known to resist positional silencing of transgenes in Drosophila.28
The piggyBac transposon, originally discovered in the cabbage looper moth Trichoplusia ni,29 has a number of advantages that enhance its utility as a tool for transgenesis or other genome modifications in a broad range of eukaryotic taxa. Its high frequency of transposition and its capacity for transposing DNA inserts or ‘cargoes’ that are large (up to 18 kb) relative to other MGEs30-32 make it an efficient vector system for transgenesis. Moreover, piggyBac has a propensity to integrate into coding sequences,33,34 and this makes it an attractive agent for forward genetics using insertional mutagenesis30,33 and for ‘trapping’ of genes and regulatory elements such as promoters and enhancers.35 Finally, piggyBac may be remobilized from the genomes of transposed organisms without leaving a ‘footprint’ or characteristic pattern of unexcised nucleotides as seen with other transposons.31,32 These advantages, coupled with its ability to transpose in a wide variety taxa, have stimulated interest in piggyBac as a vector system for functional genomic study of numerous divergent parasites and their arthropod vectors. In addition to schistosomes25 and Strongyloides,13,36 piggyBac has become the vector of choice for transgenesis and mutagenesis in the malaria parasites Plasmodium falciparum37 and P. berghei38 and in Anopheles39,40 and Aedes41-43 mosquitoes. Its tendency to transpose into transcriptional units has also enabled its use in enhancer trapping studies in Anopheles gambiae.35
Whereas efforts to achieve stable, heritable expression of transgenes encoded in conventional plasmid vectors were uniformly unsuccessful, attempts to accomplish this using integrative transformation by piggyBac were immediately successful in S. stercoralis and S. ratti. Parental free-living female S. stercoralis or S. ratti were co-transformed by microinjection with a donor plasmid encoding a GFP reporter transgene flanked by the piggyBac inverted terminal repeats and the gypsy insulator sequences and a helper plasmid encoding the piggyBac transposase. Host passage of F1 co-transformed parasites resulted in the first F2 individuals observed to date that expressed the reporter transgene (Fig. 2).13,36 Subsequent experiments in S. ratti, which has the advantage over S. stercoralis of a well-adapted rodent host in which to undertake serial passage, demonstrated that these transgene-expressing F2 worms could be used to establish stable lines that uniformly transmit and express the reporter.13 These experiments also demonstrated that the gypsy insulators were not necessary to sustain transgene expression per se and that integration into the chromosome is the essential key to avoiding the active silencing of transgenes. It is noteworthy that silencing of arrays in the F2 generation and beyond actually works to advantage in the piggyBac system by preventing inadvertent re-mobilization of stably integrated transgenes in subsequent generations (Fig. 2). Beyond this, our studies in S. ratti demonstrated that integrations occurred at a frequency of 30–50 per genome at TTAA sites typical of the piggyBac transposon and that a significant proportion of these integrations were in the coding regions of genes, also typical of piggyBac. Thus, the piggyBac system has the potential to be a robust method not only for transgenesis in S. ratti, but also for identification of regulatory elements and for unbiased forward genetics.
Stable transgenesis in S. ratti mediated by piggyBac could provide an entré into other functional genomic methods for parasitic nematodes. For example it could facilitate development of gene silencing by RNAi as a functional genomic method in this parasite. Like other animal parasitic nematodes, Strongyloides has proven largely refractory to post-transcriptional silencing of genes by administration of sequence-specific double-stranded (ds) RNA. While most components of the RNAi pathway are conserved in animal parasitic nematodes, deficiencies in dsRNA transporters homologous to C. elegans sid-1 and sid-2 have been noted in some,44 and this has been confirmed by genomic and transcriptomic study in Strongyloides (Lok et al. unpublished). It is possible that stable heterologous expression of one or both C. elegans dsRNA transporters could result in stable lines of transgenic parasite with enhanced sensitivity to RNAi and therefore greater utility as subjects for functional genomics.
Optimization of integrative transgenesis in Strongyloides will also require solution of some problems highlighted in our study. One in particular is the large number of transgene copies integrated by the piggyBac system as presently configured. This could result in nonspecific phenotypes, including loss of fitness in transgenic lines due to overexpression of transgenes or insertional mutagenesis. A clever technique, termed MosSCI (Mos induced single copy insertion), exists for single-copy transgene integration in C. elegans, which involves introducing a unique Mos1 transposon site into the genome and then creating episomal arrays containing a transgene encoding both the functional sequence of interest and a positive selection marker, another transgene encoding the Mos1 transposase under an inducible promoter and negative selection markers to subsequently eliminate the extrachromosomal array.45,46 The challenge in C. elegans is to distinguish between worms expressing the transgene of interest from the episomal array and worms with a transgene integration. This is accomplished through the visual and negative selection markers encoded in the array. The technical challenges of creating strains of Strongyloides with unique transposon sites as integration targets are daunting but not insurmountable. Moreover, the selectable markers that facilitate single-copy integration in C. elegans are not currently available in Strongyloides, but alternatives exist in the form of fluorescent markers that are expressed only in integrants of the F2 generation and beyond and can be selected manually or via fluorescence activated sorting.47 More positively, the fact that Strongyloides appears to silence episomal transgene arrays, should eliminate the confounding factor of transgene expression from arrays that pertains in C. elegans and obviate the measures necessary to eliminate array-carrying worms. It is also noteworthy that the Mos1 transposon has been used in methods for gene deletion (MosDEL)48 and gene conversion (MosTIC)49 in C. elegans, and these systems might also be used to advantage in Strongyloides.
Ultimately, it will be highly desirable to apply the piggyBac transposon system to transgenesis in a wider range of animal parasitic nematodes. The chief hurdle in these studies will not be the mechanism of transgene integration but rather the physical mode of delivering DNA into the parasite. Among parasitic nematodes, availability of free-living females with similar gonadal morphology to C. elegans is limited to Strongyloides and its relatives. It is unlikely, therefore, that gonadal microinjection will be broadly applicable for gene transfer into other medically important species of parasitic nematode. More likely is a system in which piggyBac or other integrating MGE is introduced via chemically mediated gene transfer, as was recently accomplished for the filaria Brugia malayi.50 This system does not employ microinjection for gene delivery, but rather involves culturing worms through one or more molt cycles in the presence of DNA-calcium co-precipitates. Consequently it could be applicable to the wide range of parasitic nematodes that can be induced to develop for short intervals either in vitro or in vivo in the presence of these co-precipitates. Coupled with an efficient system for chromosomal integration of transgenes such as the piggyBac transposon, this method could make the powerful technique of transgenesis available, and thus greatly facilitate functional genomic study for a wide variety of parasitic nematodes.
Conclusion
MGEs, including DNA transposons and pseudotyped retroviruses have proven to be efficient vehicles for integration of transgenes into the chromosomes of parasitic helminths. The piggyBac transposon has numerous advantages as a tool for transgenesis including the capacity for integrating large DNA inserts and a high frequency of transposition in a wide range of taxa. Among eukaryotic pathogens these include helminths such as Schistosoma mansoni, parasitic protozoa such as Plasmodium falciparum and several species of important vector mosquitoes. Recently, we have used the piggyBac transposon to integrate transgenes into the genome of the parasitic nematode Strongyloides ratti. This finding is highly significant because chromosomal integration sufficed to overcome active silencing of transgenes observed in previous attempts at stable transgenesis using conventional plasmid vectors that are presumably assembled by the parasite into repetitive episomal arrays. This has allowed derivation of the first lines of a parasitic nematode that stably transmit and express transgenes. This system will facilitate functional genomic study in parasitic nematodes and could hasten the identification of new drug and vaccine targets that are crucial to the control and elimination of these important pathogens.
Acknowledgments
I am grateful for advice and support from Paul Brindley on use of the piggyBac transposon system. I also thank Adrian Streit, Hongguang Shao and Jonathan Stoltzfus for critical comments on the manuscript. This work received support from US National Institutes of Health grants AI82548 to James Lok and Edward Pearce, AI50668 and AI22662 to James Lok and RR02512 to Mark Haskins.
Glossary
Abbreviations:
- MGE
mobile genetic element
- LTRR
long terminal repeat retrotransposon
- ITR
inverted terminal repeat
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/24417
References
- 1.Awasthi S, Bundy DA, Savioli L. Helminthic infections. BMJ. 2003;327:431–3. doi: 10.1136/bmj.327.7412.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan MS. The global burden of intestinal nematode infections--fifty years on. Parasitol Today. 1997;13:438–43. doi: 10.1016/S0169-4758(97)01144-7. [DOI] [PubMed] [Google Scholar]
- 3.Hotez PJ, Fenwick A, Savioli L, Molyneux DH. Rescuing the bottom billion through control of neglected tropical diseases. Lancet. 2009;373:1570–5. doi: 10.1016/S0140-6736(09)60233-6. [DOI] [PubMed] [Google Scholar]
- 4.Hotez PJ. Neglected infections of poverty in the United States of America. PLoS Negl Trop Dis. 2008;2:e256. doi: 10.1371/journal.pntd.0000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viney ME, Lok JB. Strongyloides spp. In: The C. elegans Research Community, ed. WormBook, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dieterich C, Sommer RJ. How to become a parasite - lessons from the genomes of nematodes. Trends Genet. 2009;25:203–9. doi: 10.1016/j.tig.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Thompson FJ, Barker GL, Hughes L, Viney ME. Genes important in the parasitic life of the nematode Strongyloides ratti. Mol Biochem Parasitol. 2008;158:112–9. doi: 10.1016/j.molbiopara.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 8.Gardner MP, Gems D, Viney ME. Extraordinary plasticity in aging in Strongyloides ratti implies a gene-regulatory mechanism of lifespan evolution. Aging Cell. 2006;5:315–23. doi: 10.1111/j.1474-9726.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- 9.Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–70. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans TC. Transformation and microinjection. In: The C. elegans Research Community, ed. WormBook, 2006. [Google Scholar]
- 11.Lok JB, Massey HC., Jr. Transgene expression in Strongyloides stercoralis following gonadal microinjection of DNA constructs. Mol Biochem Parasitol. 2002;119:279–84. doi: 10.1016/S0166-6851(01)00414-5. [DOI] [PubMed] [Google Scholar]
- 12.Li X, Massey HC, Jr., Nolan TJ, Schad GA, Kraus K, Sundaram M, et al. Successful transgenesis of the parasitic nematode Strongyloides stercoralis requires endogenous non-coding control elements. Int J Parasitol. 2006;36:671–9. doi: 10.1016/j.ijpara.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Shao H, Li X, Nolan TJ, Massey HC, Jr., Pearce EJ, Lok JB. Transposon-mediated chromosomal integration of transgenes in the parasitic nematode Strongyloides ratti and establishment of stable transgenic lines. PLoS Pathog. 2012;8:e1002871. doi: 10.1371/journal.ppat.1002871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brindley PJ, Laha T, McManus DP, Loukas A. Mobile genetic elements colonizing the genomes of metazoan parasites. Trends Parasitol. 2003;19:79–87. doi: 10.1016/S1471-4922(02)00061-2. [DOI] [PubMed] [Google Scholar]
- 15.Aeby P, Spicher A, de Chastonay Y, Müller F, Tobler H. Structure and genomic organization of proretrovirus-like elements partially eliminated from the somatic genome of Ascaris lumbricoides. EMBO J. 1986;5:3353–60. doi: 10.1002/j.1460-2075.1986.tb04650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Felder H, Herzceg A, de Chastonay Y, Aeby P, Tobler H, Müller F. Tas, a retrotransposon from the parasitic nematode Ascaris lumbricoides. Gene. 1994;149:219–25. doi: 10.1016/0378-1119(94)90153-8. [DOI] [PubMed] [Google Scholar]
- 17.Laha T, Kewgrai N, Loukas A, Brindley PJ. The dingo non-long terminal repeat retrotransposons from the genome of the hookworm, Ancylostoma caninum. Exp Parasitol. 2006;113:142–53. doi: 10.1016/j.exppara.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Bessereau JL. Transposons in C. elegans. In: The C. elegans Research Community, ed. WormBook, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blaxter ML, De Ley P, Garey JR, Liu LX, Scheldeman P, Vierstraete A, et al. A molecular evolutionary framework for the phylum Nematoda. Nature. 1998;392:71–5. doi: 10.1038/32160. [DOI] [PubMed] [Google Scholar]
- 20.Laha T, Loukas A, Wattanasatitarpa S, Somprakhon J, Kewgrai N, Sithithaworn P, et al. The bandit, a new DNA transposon from a hookworm-possible horizontal genetic transfer between host and parasite. PLoS Negl Trop Dis. 2007;1:e35. doi: 10.1371/journal.pntd.0000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rödelsperger C, Sommer RJ. Computational archaeology of the Pristionchus pacificus genome reveals evidence of horizontal gene transfers from insects. BMC Evol Biol. 2011;11:239. doi: 10.1186/1471-2148-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kines KJ, Mann VH, Morales ME, Shelby BD, Kalinna BH, Gobert GN, et al. Transduction of Schistosoma mansoni by vesicular stomatitis virus glycoprotein-pseudotyped Moloney murine leukemia retrovirus. Exp Parasitol. 2006;112:209–20. doi: 10.1016/j.exppara.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 23.Rinaldi G, Eckert SE, Tsai IJ, Suttiprapa S, Kines KJ, Tort JF, et al. Germline transgenesis and insertional mutagenesis in Schistosoma mansoni mediated by murine leukemia virus. PLoS Pathog. 2012;8:e1002820. doi: 10.1371/journal.ppat.1002820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang S, Brindley PJ, Zeng Q, Li Y, Zhou J, Liu Y, et al. Transduction of Schistosoma japonicum schistosomules with vesicular stomatitis virus glycoprotein pseudotyped murine leukemia retrovirus and expression of reporter human telomerase reverse transcriptase in the transgenic schistosomes. Mol Biochem Parasitol. 2010;174:109–16. doi: 10.1016/j.molbiopara.2010.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales ME, Mann VH, Kines KJ, Gobert GN, Fraser MJ, Jr., Kalinna BH, et al. piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. FASEB J. 2007;21:3479–89. doi: 10.1096/fj.07-8726com. [DOI] [PubMed] [Google Scholar]
- 26.Mello C, Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–82. doi: 10.1016/S0091-679X(08)61399-0. [DOI] [PubMed] [Google Scholar]
- 27.Junio AB, Li X, Massey HC, Jr., Nolan TJ, Todd Lamitina S, Sundaram MV, et al. Strongyloides stercoralis: cell- and tissue-specific transgene expression and co-transformation with vector constructs incorporating a common multifunctional 3′ UTR. Exp Parasitol. 2008;118:253–65. doi: 10.1016/j.exppara.2007.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markstein M, Pitsouli C, Villalta C, Celniker SE, Perrimon N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat Genet. 2008;40:476–83. doi: 10.1038/ng.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cary LC, Goebel M, Corsaro BG, Wang HG, Rosen E, Fraser MJ. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–69. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- 30.Nakazawa Y, Huye LE, Dotti G, Foster AE, Vera JF, Manuri PR, et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–36. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding S, Wu X, Li G, Han M, Zhuang Y, Xu T. Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 2005;122:473–83. doi: 10.1016/j.cell.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Li MA, Turner DJ, Ning Z, Yusa K, Liang Q, Eckert S, et al. Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 2011;39:e148. doi: 10.1093/nar/gkr764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balu B, Adams JH. Functional genomics of Plasmodium falciparum through transposon-mediated mutagenesis. Cell Microbiol. 2006;8:1529–36. doi: 10.1111/j.1462-5822.2006.00776.x. [DOI] [PubMed] [Google Scholar]
- 34.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–7. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 35.O’Brochta DA, Alford RT, Pilitt KL, Aluvihare CU, Harrell RA., 2nd piggyBac transposon remobilization and enhancer detection in Anopheles mosquitoes. Proc Natl Acad Sci U S A. 2011;108:16339–44. doi: 10.1073/pnas.1110628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lok JB. Nucleic acid transfection and transgenesis in parasitic nematodes. Parasitology. 2012;139:574–88. doi: 10.1017/S0031182011001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balu B, Shoue DA, Fraser MJ, Jr., Adams JH. High-efficiency transformation of Plasmodium falciparum by the lepidopteran transposable element piggyBac. Proc Natl Acad Sci U S A. 2005;102:16391–6. doi: 10.1073/pnas.0504679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fonager J, Franke-Fayard BM, Adams JH, Ramesar J, Klop O, Khan SM, et al. Development of the piggyBac transposable system for Plasmodium berghei and its application for random mutagenesis in malaria parasites. BMC Genomics. 2011;12:155. doi: 10.1186/1471-2164-12-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, Benedict MQ. Germline transformation of the malaria vector, Anopheles gambiae, with the piggyBac transposable element. Insect Mol Biol. 2001;10:597–604. doi: 10.1046/j.0962-1075.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 40.Perera OP, Harrell RA, II, Handler AM. Germ-line transformation of the South American malaria vector, Anopheles albimanus, with a piggyBac/EGFP transposon vector is routine and highly efficient. Insect Mol Biol. 2002;11:291–7. doi: 10.1046/j.1365-2583.2002.00336.x. [DOI] [PubMed] [Google Scholar]
- 41.Kokoza V, Ahmed A, Wimmer EA, Raikhel AS. Efficient transformation of the yellow fever mosquito Aedes aegypti using the piggyBac transposable element vector pBac[3xP3-EGFP afm] Insect Biochem Mol Biol. 2001;31:1137–43. doi: 10.1016/S0965-1748(01)00120-5. [3xP3-EGFP afm] [DOI] [PubMed] [Google Scholar]
- 42.Labbé GM, Nimmo DD, Alphey L. piggybac- and PhiC31-mediated genetic transformation of the Asian tiger mosquito, Aedes albopictus (Skuse) PLoS Negl Trop Dis. 2010;4:e788. doi: 10.1371/journal.pntd.0000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodrigues FG, Oliveira SB, Rocha BC, Moreira LA. Germline transformation of Aedes fluviatilis (Diptera:Culicidae) with the piggyBac transposable element. Mem Inst Oswaldo Cruz. 2006;101:755–7. doi: 10.1590/S0074-02762006000700008. [DOI] [PubMed] [Google Scholar]
- 44.Dalzell JJ, McVeigh P, Warnock ND, Mitreva M, Bird DM, Abad P, et al. RNAi effector diversity in nematodes. PLoS Negl Trop Dis. 2011;5:e1176. doi: 10.1371/journal.pntd.0001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frøkjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, et al. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–83. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frøkjær-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–8. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lok JB, Artis D. Transgenesis and neuronal ablation in parasitic nematodes: revolutionary new tools to dissect host-parasite interactions. Parasite Immunol. 2008;30:203–14. doi: 10.1111/j.1365-3024.2008.01006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frøkjaer-Jensen C, Davis MW, Hollopeter G, Taylor J, Harris TW, Nix P, et al. Targeted gene deletions in C. elegans using transposon excision. Nat Methods. 2010;7:451–3. doi: 10.1038/nmeth.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Robert V, Bessereau JL. Targeted engineering of the Caenorhabditis elegans genome following Mos1-triggered chromosomal breaks. EMBO J. 2007;26:170–83. doi: 10.1038/sj.emboj.7601463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu S, Liu C, Tzertzinis G, Ghedin E, Evans CC, Kaplan R, et al. In vivo transfection of developmentally competent Brugia malayi infective larvae. Int J Parasitol. 2011;41:355–62. doi: 10.1016/j.ijpara.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]