Abstract
We recently reported that acute stress causes a substantial upregulation of the epigenetic mark, Histone H3 Lysine 9 Trimethyl (H3K9me3) in the rat hippocampus within an hour of acute stress exposure. To determine the function of this change we used ChIP-sequencing to determine where this silencing mark was being localized. We found that it showed a strong bias toward localization at more active classes of retrotransposable elements and away from genes. Further, we showed that the change was functional in that it reduced transcription of some of these elements (notably the endogenous retrovirus IAP and the B2 SINE). In this commentary we examine these results, which appear to describe a selective genomic stress response and relate it to human health and disease, particularly stress related maladies such as Post-traumatic Stress Disorder, which have recently been shown to have both epigenetic elements in their causation as well as differences in epigenetic marking of retrotransposons in human patients.
Keywords: epigenetic, hippocampus, post-traumatic stress disorder, corticosteroid, heterochromatin, chromatin
Introduction
Stress is a common life event, which has substantial, often long-term, effects upon the brain and behavior. Stress can induce structural changes in the brain and contribute to a variety of chronic diseases from post-traumatic stress disorder to depression to diabetes.1,2 Much fruitful work has come from the study of stress since the discipline was pioneered by Selye decades ago.3 The ability of stress to cause persistent changes has long been of interest to neurobiologists and clinicians, as has the apparent variability in resilience to stress within populations, even those which are relatively genetically similar such as inbred strains of laboratory animals.4 As a consequence, epigenetic explanations for the effects of stress upon the brain and behavior have been sought and, with increasing frequency, found.5,6
Our own work established that large, rapid (less than 2 h) and regionally selective increases in the heterochromatin mark Histone 3 Lysine 9 trimethyl (H3K9me3) within the rat hippocampus as a consequence of acute restraint stress,7 a standard rodent model of moderate acute stress. This change persisted for at least 24 h but had begun to habituate by 7 d of repeated stress and was absent after 3 weeks. Whether this represents learning or adaptation to a predictable stressor is, as yet, uncertain. Most observations of epigenetic changes in as a consequence of stress involve relatively constrained regions of the genome, such as a single promoter CpG island or transcriptional start site.5 As the change we observed was of a large magnitude and seemed to involve a rapid and metabolically expensive increase in heterochromatin we wondered if it might involve control of potentially destabilizing mobile genetic elements in the genomes of hippocampal neurons.
Decades before the complete sequencing of a mammalian genome it was known that much, if not most of the genome did not consist of genes. However, little interest was shown in this genomic terra incognita as it had been pronounced “junk” by eminences such as Francis Crick and Susumu Ohno.8,9 It was assumed that most of this DNA was silent and irrelevant to function or phenotype. More recently, due in large part to the results obtained by the ENCODE project, it has become clear that not only is most of this DNA transcriptionally active10,11 but many have functional roles as well.12
While some have argued that transposable elements are purely parasitic,9 their discoverer, McClintock, felt they had adaptive use under conditions of stress.13 As is often the case in biology, both perspectives contain some truth. These elements have been implicated in processes as diverse as transcriptional regulation, A to I editing, RNA splicing and subcellular localization.14,15 In the context of the central nervous system, it has recently been established that transposition of LINE elements plays a regular part in neural development and neurogenesis,16 perhaps playing a role in generating neuronal diversity similar to that played by the V(D)J system in generating diversity in the immune system.17,18 However they are also implicated in a number of disorders, such as cancer.19 With regard to the nervous system the Alu SINEs have been shown to have altered expression in neurodegenerative diseases such as Creutzfeldt-Jakob and Alzheimer disease20 and as a principle player in the pathogenesis of age related macular degeneration of the retina.21 The latter paper is particularly interesting in that the mechanism of Alu RNA induced cell death and degeneration was via inflammasome activation, suggesting that excess transposon expression, can, like stress, contribute to “brain weathering” and accelerated aging.22
Stress Induced Epigenetic Silencing of Transposons in the Hippocampus
The neurons of the mammalian hippocampus are unique in a number of ways. They are responsible for the formation of long-term memories and are one of only a few populations of adult neurons with a resident stem cell population and active neurogenesis. Further, they are exquisitely sensitive to the effects of environmental stress and glucocorticoid stress hormones.6,23,24 Stress can lower the levels of neurogenesis and cause profound structural changes in the dendritic arbors of hippocampal pyramidal cells.23,24 Given their terminally differentiated status, sensitivity to stress and role in memory formation, we reasoned that they may require an active mechanism to control unstable genomic elements, like retrotransposons, which would endanger their ability to maintain their function and specialized phenotype across a lifetime. We suspected that the rapid increase in H3K9me3 we observed was a part of such a genomic stability mechanism as it would rapidly silence large regions of the genome and reduce the likelihood of stress causing transpositions or transcriptional disruptions.
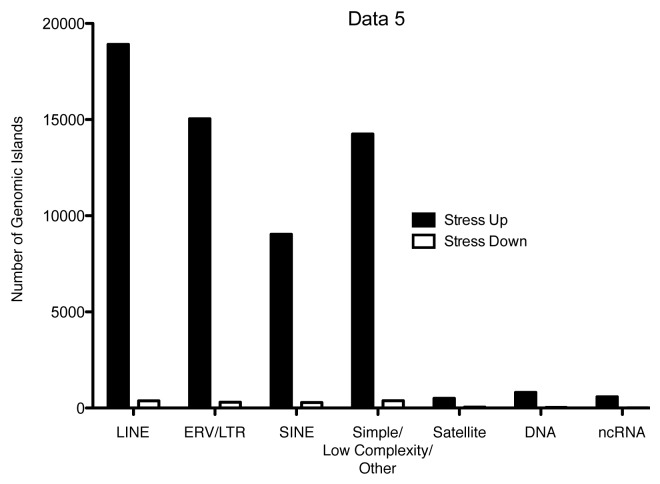
When acute stress induced H3K9me3 levels in the rat hippocampus were examined using ChIP Sequencing, we observed little change in the levels of the mark within gene bodies or promoters. Instead, as we had predicted, H3K9me3 reads associated with transposons and repetitive elements increased by 32% from control levels. When the number of transposable or repetitive element loci showing increases in H3K9me3 after stress was compared with those showing decreases the trend was even more marked, more than 59,000 loci showed an increase with stress while only 1,400 or so showed a decrease.25 While other groups have shown changes in brain H3K9 methylation in association with environmental influences such as treatment with drugs of abuse, e.g.,26,27 they have not observed the same directionality of effect, suggesting a unique function for the effects we have observed in the hippocampus. Phylogenetic analysis of the types of elements showing the largest increases in H3K9me3 revealed an interesting pattern, the more active classes of retrotransposable elements, LINEs, SINEs and ERV/LTR showed higher levels of the silencing mark after stress, while more stable elements like simple repeats and DNA transposons were more likely either to not change or show a reduction in the levels of the mark (Fig. 1).
Figure 1. Phylogenetic breakdown of numbers of 5 kb mobile or repetitive element genomic islands showing increased H3Kme3 levels (Stress Up) vs. reduced levels (Stress Down). Adapted from reference 25.
In line with the generally repressive role for H3K9me3 we found that acute stress was associated with the suppression of transcription of the RNAs for some of these elements, notably the rodent SINE, B2 and the rodent ERV IAP elements. In keeping with our observations of H3K9me3 levels after stress, this suppression was not observed in the other brain regions tested (frontal cortex and cerebellum) or in peripheral tissues like liver, heart or skeletal muscle.
In an attempt to begin to elucidate the mechanism by which the increase in H3K9me3 in the hippocampus might be mediated, we examined the expression of the histone H3 lysine 9 specific methyltransferases G9a, GLP, Suv39h1 and h2 with RT-PCR and in situ hybridization. We found no change in G9a, GLP or Suv39h1 with either method, though we did see an increase in Suv39h2 expression within 30 min of stress onset (Fig. 2). Using ChIP Sequencing targeted at the glucocorticoid receptor (GR), we demonstrated that acute corticosterone treatment induces increased binding of GR in the region of the Suv39h2 gene,25 suggesting that the increase in H3K9me3 is mediated by the genomic action of the GR. Thus the apparent specificity of the effect to the rodent hippocampus may be due to its high levels of GR expression relative to other brain regions. In humans, where GR is more highly expressed in cortex in addition to hippocampus,28-30 this effect may therefore be more widespread in our own species.
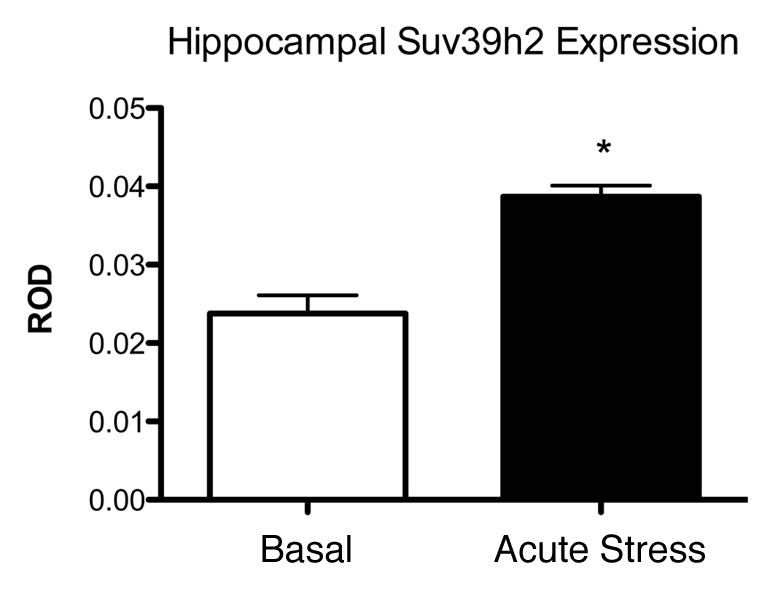
Figure 2. Levels of the H3K9 specific methyltransferases Suv39h2 mRNA in rat hippocampus under basal conditions and 1 h after acute stress. Adapted from reference 25.
A Genomic Stress Response
Taken in sum these findings are suggestive that, at least in certain stress sensitive tissues like the hippocampus, there is an active regulatory mechanism governing the expression of a large number of mobile genetic elements and that it is subject to regulation by activated GR. As hippocampal neurons are vital to long-term memory formation and their loss or instability would reduce the ability to encode new memory, the hypothesis that this response is a genomic stability mechanism is an appealing one. An alternate, but not mutually exclusive hypothesis derives from the contrast of our findings with regard to the downregulation of Alu like SINE elements by restraint stress with the findings of the Goodrich lab (see above) with regard to heat shock. They found that heat shock increased the expression of B2 and Alu RNA and that the RNA bound RNA polII and blocked transcription of a number of genes31,32 and a similar rise in B2 elements has been observed in models of stroke.33 This sort of transcriptional repression would be a useful means of preventing the accumulation of mis-folded proteins during hyperthermic episodes or severe brain injury. On the other hand, suppression of protein synthesis, upon which long-term memory formation depends,34,35 during a stressful and potentially life threatening (but not physically damaging) event would obviate one of the major adaptive benefits of having memory in the first place: remembering how to avoid stress and death Thus the actions of transposable elements in the brain seem to be encoded to the type and severity of the threat to the organ and the organism. Of particular interest in this regard is the recent observation that Alu and LINE elements are differentially methylated in combat veterans who have acquired post-traumatic stress disorder vs. combat veterans who do not.36 Given the transgenerational epigenetic nature of PTSD,37,38 and our own work, one could hypothesize that the disorder might involve a failure of the sort of genomic stress response we describe here. Together these results suggests that the expression of at least some transposable elements may be as exquisitely regulated and environmentally tuned as any gene. Whether this is the case for the majority of these elements remains to be seen.
What occurs when this (at this point speculative) system fails to function properly is another question. Stress and corticosteroid action have been linked to a variety of disorders, but rarely in a consistent one to one fashion, such as might be seen with a rabies infection, where the mere presence of live virus in a patient is necessary and sufficient for the disease. Stress related diseases show highly variable levels of resilience within both human populations and laboratory animals. While the reasons for some of this variance have been successfully explicated, much remains unexplained.39 It is certainly tempting to imagine that some of this variation might arise from inter-individual diversity of transposable elements a phenomenon that is now well established in humans.40 A related and perhaps more obvious question is: does stress increase the level of somatic transposition in the brain or elsewhere? If so it seems likely that this could have far reaching consequences for human health and disease.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/24555
References
- 1.McEwen BS, Gianaros PJ. Central role of the brain in stress and adaptation: links to socioeconomic status, health, and disease. Ann N Y Acad Sci. 2010;1186:190–222. doi: 10.1111/j.1749-6632.2009.05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McEwen BS. Stress, sex, and neural adaptation to a changing environment: mechanisms of neuronal remodeling. Ann N Y Acad Sci. 2010;1204(Suppl):E38–59. doi: 10.1111/j.1749-6632.2010.05568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Selye H. A Syndrome Produced by Diverse Nocuous Agents. Nature. 1936;138:32. doi: 10.1038/138032a0. [DOI] [PubMed] [Google Scholar]
- 4.Karatsoreos IN, McEwen BS. Psychobiological allostasis: resistance, resilience and vulnerability. Trends Cogn Sci. 2011;15:576–84. doi: 10.1016/j.tics.2011.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Hunter RG. Epigenetic effects of stress and corticosteroids in the brain. Front Cell Neurosci. 2012;6:18. doi: 10.3389/fncel.2012.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McEwen BS, Eiland L, Hunter RG, Miller MM. Stress and anxiety: structural plasticity and epigenetic regulation as a consequence of stress. Neuropharmacology. 2012;62:3–12. doi: 10.1016/j.neuropharm.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–7. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohno S. So much “junk” DNA in our genome. Brookhaven Symp Biol. 1972;23:366–70. [PubMed] [Google Scholar]
- 9.Orgel LE, Crick FH. Selfish DNA: the ultimate parasite. Nature. 1980;284:604–7. doi: 10.1038/284604a0. [DOI] [PubMed] [Google Scholar]
- 10.Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, et al. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–9. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- 11.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–8. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 12.Maurano MT, Humbert R, Rynes E, Thurman RE, Haugen E, Wang H, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 14.Kugel JF, Goodrich JA. Non-coding RNAs: key regulators of mammalian transcription. Trends Biochem Sci. 2012;37:144–51. doi: 10.1016/j.tibs.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walters RD, Kugel JF, Goodrich JA. InvAluable junk: the cellular impact and function of Alu and B2 RNAs. IUBMB Life. 2009;61:831–7. doi: 10.1002/iub.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 17.Muotri AR, Marchetto MC, Coufal NG, Gage FH. The necessary junk: new functions for transposable elements. Hum Mol Genet. 2007;16 Spec No. 2:R159–67. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- 18.Jones JM, Gellert M. The taming of a transposon: V(D)J recombination and the immune system. Immunol Rev. 2004;200:233–48. doi: 10.1111/j.0105-2896.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 19.Kozeretska IA, Demydov SV, Ostapchenko LI. Mobile genetic elements and cancer. From mutations to gene therapy. Exp Oncol. 2011;33:198–205. [PubMed] [Google Scholar]
- 20.Kiesel P, Gibson TJ, Ciesielczyk B, Bodemer M, Kaup FJ, Bodemer W, et al. Transcription of Alu DNA elements in blood cells of sporadic Creutzfeldt-Jakob disease (sCJD) Prion. 2010;4:87–93. doi: 10.4161/pri.4.2.11965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–59. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McEwen BS. Physiology and neurobiology of stress and adaptation: central role of the brain. Physiol Rev. 2007;87:873–904. doi: 10.1152/physrev.00041.2006. [DOI] [PubMed] [Google Scholar]
- 23.McEwen BS. Plasticity of the hippocampus: adaptation to chronic stress and allostatic load. Ann N Y Acad Sci. 2001;933:265–77. doi: 10.1111/j.1749-6632.2001.tb05830.x. [DOI] [PubMed] [Google Scholar]
- 24.McEwen BS, Gianaros PJ. Stress- and allostasis-induced brain plasticity. Annu Rev Med. 2011;62:431–45. doi: 10.1146/annurev-med-052209-100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter RG, Murakami G, Dewell S, Seligsohn M, Baker ME, Datson NA, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci U S A. 2012;109:17657–62. doi: 10.1073/pnas.1215810109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maze I, Feng J, Wilkinson MB, Sun H, Shen L, Nestler EJ. Cocaine dynamically regulates heterochromatin and repetitive element unsilencing in nucleus accumbens. Proc Natl Acad Sci U S A. 2011;108:3035–40. doi: 10.1073/pnas.1015483108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perlman WR, Webster MJ, Herman MM, Kleinman JE, Weickert CS. Age-related differences in glucocorticoid receptor mRNA levels in the human brain. Neurobiol Aging. 2007;28:447–58. doi: 10.1016/j.neurobiolaging.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 29.Watzka M, Bidlingmaier F, Beyenburg S, Henke RT, Clusmann H, Elger CE, et al. Corticosteroid receptor mRNA expression in the brains of patients with epilepsy. Steroids. 2000;65:895–901. doi: 10.1016/S0039-128X(00)00205-1. [DOI] [PubMed] [Google Scholar]
- 30.Perlman WR, Webster MJ, Kleinman JE, Weickert CS. Reduced glucocorticoid and estrogen receptor alpha messenger ribonucleic acid levels in the amygdala of patients with major mental illness. Biol Psychiatry. 2004;56:844–52. doi: 10.1016/j.biopsych.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 31.Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–9. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- 32.Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, et al. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Kalkkila JP, Sharp FR, Kärkkäinen I, Reilly M, Lu A, Solway K, et al. Cloning and expression of short interspersed elements B1 and B2 in ischemic brain. Eur J Neurosci. 2004;19:1199–206. doi: 10.1111/j.1460-9568.2004.03233.x. [DOI] [PubMed] [Google Scholar]
- 34.Flexner JB, Flexner LB, Stellar E, De La Haba G, Roberts RB. Inhibition of protein synthesis in brain and learning and memory following puromycin. J Neurochem. 1962;9:595–605. doi: 10.1111/j.1471-4159.1962.tb04216.x. [DOI] [PubMed] [Google Scholar]
- 35.McGaugh JL. Memory--a century of consolidation. Science. 2000;287:248–51. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 36.Rusiecki JA, Chen L, Srikantan V, Zhang L, Yan L, Polin ML, et al. DNA methylation in repetitive elements and post-traumatic stress disorder: a case-control study of US military service members. Epigenomics. 2012;4:29–40. doi: 10.2217/epi.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yehuda R, Bell A, Bierer LM, Schmeidler J. Maternal, not paternal, PTSD is related to increased risk for PTSD in offspring of Holocaust survivors. J Psychiatr Res. 2008;42:1104–11. doi: 10.1016/j.jpsychires.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yehuda R, Bierer LM. Transgenerational transmission of cortisol and PTSD risk. Prog Brain Res. 2008;167:121–35. doi: 10.1016/S0079-6123(07)67009-5. [DOI] [PubMed] [Google Scholar]
- 39.Russo SJ, Murrough JW, Han MH, Charney DS, Nestler EJ. Neurobiology of resilience. Nat Neurosci. 2012;15:1475–84. doi: 10.1038/nn.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]