Abstract
Paclitaxel is a standard chemotherapeutic agent for ovarian cancer. PEA-15 (phosphoprotein enriched in astrocytes-15 kDa) regulates cell proliferation, autophagy, apoptosis, and glucose metabolism and also mediates AKT-dependent chemoresistance in breast cancer. PEA-15's functions are tightly regulated by its phosphorylation status at Ser104 and Ser116. However, the effect of PEA-15 phosphorylation status on chemosensitivity of cancer cells remains unknown. Here, we tested the hypothesis that PEA-15 phosphorylated at both Ser104 and Ser116 (pPEA-15) sensitizes ovarian cancer cells to paclitaxel. We first found that knockdown of PEA-15 in PEA-15-high-expressing HEY and OVTOKO ovarian cancer cells resulted in paclitaxel resistance, whereas re-expression of PEA-15 in these cells led to paclitaxel sensitization. We next found that SKOV3.ip1-DD cells (expressing phosphomimetic PEA-15) were more sensitive to paclitaxel than SKOV3.ip1-AA cells (expressing nonphosphorylatable PEA-15). Compared to SKOV3.ip1-vector and SKOV3.ip1-AA cells, SKOV3.ip1-DD cells displayed reduced cell viability, inhibited anchorage-independent growth, and augmented apoptosis when treated with paclitaxel. Furthermore, HEY and OVTOKO cells displayed enhanced paclitaxel sensitivity when transiently overexpressing phosphomimetic PEA-15 and reduced paclitaxel sensitivity when transiently overexpressing nonphosphorylatable PEA-15. These results indicate that pPEA-15 sensitizes ovarian cancer cells to paclitaxel. cDNA microarray analysis suggested that SCLIP (SCG10-like protein), a microtubule (MT)-destabilizing protein, is involved in pPEA-15-mediated chemosensitization. We found that reduced expression and possibly posttranslational modification of SCLIP following paclitaxel treatment impaired SCLIP's MT-destabilizing effect, thereby promoting induction of mitotic arrest and apoptosis by paclitaxel. Our findings highlight the importance of pPEA-15 as a promising target for improving the efficacy of paclitaxel-based therapy in ovarian cancer.
Keywords: PEA-15, SCLIP, paclitaxel, chemosensitivity, ovarian cancer
Introduction
Paclitaxel is a chemotherapeutic drug commonly used for the treatment of ovarian, breast, and other malignancies (1, 2). Paclitaxel kills tumor cells by increasing microtubule (MT) polymerization via binding to the β-subunit in preformed MTs (3) and by stabilizing the MT network via direct interaction with MTs (4). Thus, paclitaxel interferes with MT function and disrupts the mitotic spindle, leading to G2/M arrest and eventually programmed cell death (5, 6). Paclitaxel has limited effectiveness in the treatment of ovarian cancer because of intrinsic or acquired resistance of cancer cells to the drug (7-12). Identifying molecular determinants of sensitivity to paclitaxel may lead to improvement of the therapeutic efficacy of the drug in ovarian cancer.
Previous studies showed that high expression level of PEA-15 (phosphoprotein enriched in astrocytes-15 kDa) in cancer cells correlated with longer survival of patients with ovarian cancer (13) and lower proliferation and less invasiveness of breast cancer cells (14, 15). PEA-15 is a multifunctional protein regulating cell proliferation, apoptosis, autophagy, and glucose metabolism (16). PEA-15 controls cell survival by interfering with the apoptotic or autophagic pathways (13, 14) and controls cell proliferation by interfering with the ERK pathway (17, 18). PEA-15's functions are highly dependent on its phosphorylation status (16). PEA-15 is phosphorylated at Ser104 by protein kinase C and at Ser116 by calmodulin-dependent protein kinase II and AKT/PKB (19, 20). Previously, PEA-15 was shown to mediate paclitaxel resistance in breast cancer cells and protect these cells from paclitaxel-induced apoptosis through inhibition of the stress kinase pathway (21). However, the effect of PEA-15 phosphorylation status on chemosensitivity of cancer cells remains unknown.
SCLIP (SCG10-like protein), also known as STMN3, belongs to the stathmin/OP18 family, which also includes stathmin, SCG10 (superior cervical ganglion-10 protein), and RB3 (stathmin-like protein B3) (22). Like other members of the stathmin/OP18 family, SCLIP functions as a regulator of MT dynamics (22). The stathmin/OP18 family proteins prevent MT polymerization by sequestering soluble tubulin heterodimers (22, 23) and promote MT depolymerization by increasing MT catastrophe rate (22, 24-26). Stathmin has been shown to regulate resistance of cancer cells to MT-targeting drugs, including paclitaxel, by affecting their binding to MTs and delaying G2/M transition (27). To date, there is no direct evidence implicating SCLIP in chemosensitivity of cancer cells. Here, we showed for the first time that SCLIP is involved in PEA-15-mediated chemosensitization in ovarian cancer cells.
In the present study, we investigated the effect of PEA-15 phosphorylation status on the sensitivity of ovarian cancer cells to paclitaxel. We demonstrated for the first time that PEA-15 phosphorylated at both Ser104 and Ser116 (pPEA-15) sensitized ovarian cancer cells to paclitaxel by impairing SCLIP-mediated MT destabilization, thereby enhancing induction of mitotic arrest and apoptotic cell death by paclitaxel. Our findings provide a rationale for future clinical studies using pPEA-15 as a biomarker to improve the efficacy of paclitaxel-based therapy in ovarian cancer.
Materials and Methods
Cell culture
OVCAR3 ovarian cancer cells were obtained from American Type Culture Collection. KOC-7c ovarian cancer cells were provided by Dr. Toru Sugiyama (Kurume University, Kurume, Japan); OVTOKO ovarian cancer cells by Dr. Hiroshi Minaguchi (Yokohama City University, Yokohama, Japan); HEY and OVCA 432 ovarian cancer cells by Dr. Robert Bast (The University of Texas MD Anderson Cancer Center, Houston, TX); and SKOV3.ip1 ovarian cancer cells by Dr. Mien-Chie Hung (The University of Texas MD Anderson Cancer Center, Houston, TX). SKOV3.ip1-vector cells stably transfected with pcDNA3.1 empty vector, SKOV3.ip1-AA cells stably expressing nonphosphorylatable PEA-15 (AA; Ser104 and Ser116 of PEA-15 substituted with alanine), and SKOV3.ip1-DD cells stably expressing phosphomimetic PEA-15 (DD; Ser104 and Ser116 of PEA-15 substituted with aspartic acid) were generated previously in our laboratory (28). All cell lines used in this study except for KOC-7c were validated using a short tandem repeat method based on primer extension to detect single base deviations in February 2013 by the Characterized Cell Line Core Facility at MD Anderson Cancer Center (29). We are unable to provide the exact dates on which we obtained these cell lines. HEY, OVCA 432, and SKOV3.ip1 cells were maintained in DMEM/F12 (Invitrogen), and OVCAR3, KOC-7c, and OVTOKO cells in RPMI 1640 (Invitrogen).
siRNA transfection
Cells were seeded in 6-well plates and transfected the next day with an ON-TARGET SMART siPEA-15 pool (a mixture of 4 designed siRNAs; Dharmacon), an ON-TARGET SMART siSCLIP pool (a mixture of 4 designed siRNAs; Dharmacon), or a scrambled control siRNA pool (a mixture of 4 siControl nontargeting siRNAs; Dharmacon) at a final siRNA concentration of 100 nM using Oligofectamine (Invitrogen) as described previously (14). The cells were used for chemosensitivity analyses 24 hours after transfection.
Plasmid transfection
Cells were transfected with plasmids of interest using the Neon Transfection System (Invitrogen) according to the manufacturer's instructions. Plasmids used in this study included pCMV6-Entry vector (Origene), pCMV6-Entry vector carrying SCLIP (Origene), pcDNA3.1 empty vector, and pcDNA3.1 vector carrying PEA-15, nonphosphorylatable PEA-15 (AA; Ser104 and Ser116 of PEA-15 substituted with alanine), or phosphomimetic PEA-15 (DD; Ser104 and Ser116 of PEA-15 substituted with aspartic acid). All the pcDNA3.1 vectors were generous gifts from Dr. Mark H. Ginsberg (17) (University of California, San Diego, CA). Cells were used for chemosensitivity analysis 24 hours after transfection.
Western blot analysis
Western blotting was done as described previously (14). Proteins of interest were probed using the following primary antibodies (1:1000 dilution) purchased from Cell Signaling Technology or other suppliers as indicated: anti-PEA-15 (Santa Cruz Biotechnology), anti-pPEA-15 at Ser104, anti-pPEA-15 at Ser116 (Invitrogen), anti-SCLIP (Proteintech Group), anti-detyrosinated α-tubulin (Millipore), anti-acetylated α-tubulin (Sigma-Aldrich), anti-α-tubulin, anti-β-actin (Sigma-Aldrich), anti-caspase 3, anti-caspase 8, anti-caspase 9, and anti-PARP. The secondary antibodies used were horseradish peroxidase-conjugated IgG (Invitrogen) for chemiluminescent signal detection and Alexa Fluor-conjugated IgG (Invitrogen) for fluorescence signal detection. Expression levels of target proteins were quantified using National Institutes of Health Image J software (http://rsb.info.nih.gov/ij/) and then normalized to the β-actin protein level.
Growth inhibition assay
Cell growth inhibition by paclitaxel (Supplementary Fig S1; App Pharmaceuticals) was determined using the WST-1 cell proliferation assay or trypan blue exclusion assay as described previously (28, 30). For the WST-1 assay, cells were seeded in 96-well plates and treated the next day with paclitaxel (0-10 μM). After 72 hours of paclitaxel treatment, optical density (OD) at 450 nm was determined, and median growth inhibition (IC50), which corresponds to the concentration of paclitaxel that results in 50% inhibition of net cell growth, was calculated. For the trypan blue exclusion assay, cells were seeded in 6-well plates and treated the next day with paclitaxel. After 72 hours of paclitaxel treatment, cell viability was determined using Vi-CELL series cell viability analyzers (Beckman Coulter) according to the manufacturer's instructions.
Anchorage-independent growth assay
Anchorage-independent growth assay was performed as described previously (14). Briefly, SKOV3.ip1 stable cells were resuspended in 0.4% agarose growth medium with or without paclitaxel. The cell suspensions were then plated in 6-well plates containing solidified 0.8% agarose in growth medium. Three weeks later, colonies greater than 80 μm in diameter were counted using the GelCount system (Oxford Optronix).
Cell cycle analysis
Cells were grown overnight and then treated with paclitaxel for 24 or 36 hours. The cells were then harvested, fixed, treated with RNase/propidium iodide (PI), and subjected to cell cycle analysis by flow cytometry as described previously (31).
cDNA microarray analysis
Total RNA was extracted from SKOV3.ip1-S116A cells (stably expressing nonphosphorylatable PEA-15 at Ser116, which was substituted with alanine) and SKOV3.ip1-S116D cells (stably expressing phosphomimetic PEA-15 at Ser116, which was substituted with aspartic acid) using RNeasy mini kit (Qiagen) according to the manufacturer's instructions. The integrity of the purified RNA was assessed using an Agilent 2100 BioAnalyzer (Agilent Technologies). The Affymetrix HGU133 plus platform was used for hybridization, staining, and imaging of the arrays by following the manufacturer's instructions. Data obtained from the microarrays were normalized by the robust multiarray average method (32). Analysis-of-variance P values and fold changes for gene expression were calculated using R statistical software version 2.12.2. A threshold cutoff was set to false discovery rate less than 0.01 and at least a 2-fold geometric change in gene-level expression between SKOV3.ip1-S116A and SKOV3.ip1-S116D cells. The microarray data have been deposited into the Gene Expression Omnibus database under the accession number GSE37934.
Quantitative RT-PCR
Total RNA was extracted from SKOV3.ip1 stable cells using an RNA prep kit (Invitrogen) according to the manufacturer's instructions. First-strand cDNAs were reverse-transcribed using the ImProm-II reverse transcriptase system kit (Promega) by following the manufacturer's protocol. The quantitative PCR reactions were performed using the SYBR green qPCR kit (Bio-Rad) with a pair of SCLIP primers: 5′-GGAGCTGCAAAAGCGGCTGG-3′ (forward) and 5′-CTGCTTCAGCACCTGCGCCT-3′ (reverse). Primers for human β-actin mRNA control were 5′-GCG GGAAATCGT GCGTGACATT-3′ (forward) and 5′-AGACAGTCTCCACTCACCCAGGAAG-3′ (reverse). Human β-actin mRNA was used as a normalization control. The mRNA levels of SCLIP in SKOV3.ip1 stable cells were first normalized to the mRNA levels of the housekeeping gene β-actin, and then the fold induction of SCLIP mRNA was calculated on the basis of the SCLIP mRNA level in SKOV3.ip1-vector cells.
Mitotic index determination
SKOV3.ip1 stable cells were grown overnight and then treated with paclitaxel for 12 hours. The cells were harvested, fixed in ice-cold 70% ethanol, and permeabilized with 0.25% Triton X-100. The cells were then incubated with anti-phosphohistone H3 antibody (Cell Signaling) and subsequently with FITC-conjugated secondary antibody (Millipore). The cells were treated with RNase/PI and then analyzed for mitotic index by flow cytometry as described previously (33).
Immunofluorescence staining of MTs
SKOV3.ip1 stable cells grown in culture chamber slides were treated with paclitaxel for 6 or 12 hours. The cells were fixed with ice-cold methanol, permeabilized with 0.2% Triton X-100, and blocked with 3% bovine serum albumin in PBS. The cells were then incubated with the following primary antibodies: anti-α-tubulin (Cell Signaling), anti-phosphohistone H3 (Cell Signaling), anti-acetylated α-tubulin (Sigma-Aldrich), or anti-detyrosinated α-tubulin (Millipore), followed by incubation with FITC-conjugated secondary antibodies (Invitrogen). The slides were mounted with mounting solution containing DAPI (Invitrogen). The MT network and mitotic spindles were photographed under 400X magnification using an Eclipse 80i fluorescence microscope (Nikon). Fluorescence intensity of the MT network was quantified using NIS-Elements BR3.1 software (Nikon).
Separation of soluble and polymerized tubulin
SKOV3.ip1 stable cells were grown overnight and then treated with paclitaxel for 12 hours. Soluble and polymerized tubulin were separated from the cultured cells as described previously (34) and then analyzed by western blotting.
Statistical analysis
Each experiment was performed at least in duplicate with three repeats, and data were expressed as means ± standard deviation. Statistical analyses were performed using SAS 9.3 software (SAS Institute). Analysis of variance was used to evaluate the significance of differences in means among different groups. P values less than 0.05 were considered significant.
Results
PEA-15 silencing decreased sensitivity of ovarian cancer cells to paclitaxel
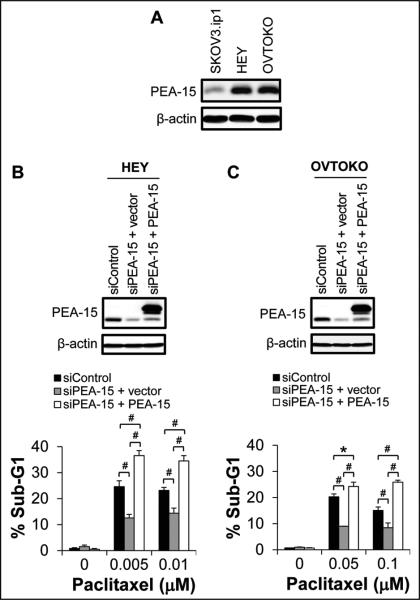
To assess whether PEA-15 mediates chemosensitization in ovarian cancer cells, we silenced PEA-15 expression in PEA-15-high-expressing HEY and OVTOKO ovarian cancer cells using PEA-15-targeting siRNA (Fig. 1) and then tested sensitivity by cell cycle analysis. PEA-15 silencing resulted in a 12% reduction in the sub-G1 fraction in HEY cells (0.005 μM paclitaxel; P < 0.001; Fig. 1B) and an 11.2% reduction in the sub-G1 fraction in OVTOKO cells (0.05 μM paclitaxel; P < 0.001; Fig. 1C) compared to the sub-G1 fraction in control cells treated with scrambled siRNA after paclitaxel treatment, indicating that PEA-15 silencing reduces paclitaxel sensitivity in ovarian cancer cells. To verify that loss of PEA-15 expression was responsible for the reduced paclitaxel sensitivity, we re-expressed PEA-15 in siRNA PEA-15-treated cells and observed a 24% increase in the sub-G1 fraction in HEY cells (0.005 μM paclitaxel; P < 0.001; Fig. 1B) and a 15.2% increase in the sub-G1 fraction in OVTOKO cells (0.05 μM paclitaxel; P < 0.001; Fig. 1C) compared to the sub-G1 fraction in control cells transfected with vector after paclitaxel treatment. These results demonstrate that PEA-15 mediates paclitaxel sensitization in ovarian cancer cells.
Figure 1.
PEA-15 mediated paclitaxel sensitization in HEY and OVTOKO cells. A, Expression of PEA-15 in SKOV3.ip1, HEY, and OVTOKO cells was determined by western blotting. β-actin served as a loading control. HEY (B) and OVTOKO (C) cells were treated with PEA-15-targeting siRNA and then transiently transfected 48 hours later with the pcDNA3.1 vector or the pcDNA3.1 vector carrying PEA-15. Expression of PEA-15 was determined by western blotting 48 hours after transfection (upper panels). β-actin served as a loading control. Following PEA-15 silencing and then re-expression, the sensitivities of HEY and OVTOKO cells to paclitaxel were determined by trypan blue exclusion assay 72 hours after paclitaxel treatment (lower panels). *, P < 0.05; #, P < 0.001.
PEA-15-mediated paclitaxel sensitization was dependent on its phosphorylation status
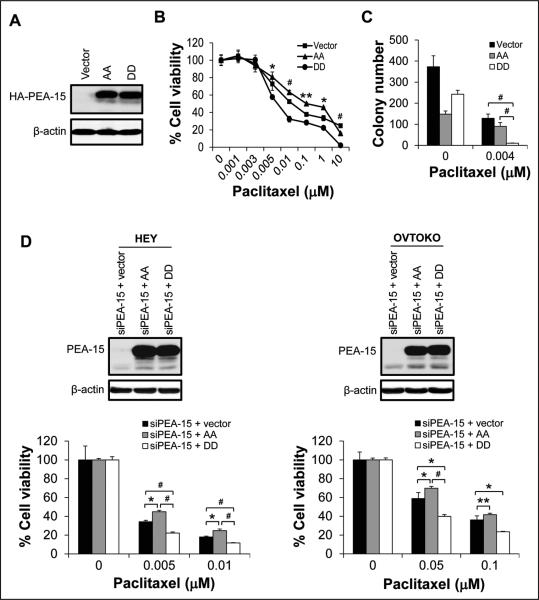
PEA-15 is known to mediate paclitaxel resistance in breast cancer (21). PEA-15's functions are phosphorylation dependent (16). To date, the effect of PEA-15 phosphorylation status on chemosensitivity of cancer cells remains unclear. We hypothesized that PEA-15 phosphorylated at both Ser104 and Ser116 sensitizes ovarian cancer cells to paclitaxel. To test this hypothesis, we used SKOV3.ip1-vector cells, SKOV3.ip1-AA cells stably expressing nonphosphorylatable PEA-15, and SKOV3.ip1-DD cells stably expressing phosphomimetic PEA-15 (Fig. 2A). SKOV3.ip1-DD cells (IC50 = 0.01 μM) were more sensitive to paclitaxel than were SKOV3.ip1-vector cells (IC50 = 0.02 μM) and SKOV3.ip1-AA cells (IC50 = 0.11 μM) (Fig. 2B). Furthermore, SKOV3.ip1-DD cells displayed a 92.2% reduction in colony formation compared to SKOV3.ip1-vector cells (P < 0.001) and an 88.9% reduction in colony formation compared to SKOV3.ip1-AA cells (P < 0.001) after paclitaxel treatment (Fig. 2C). To confirm that PEA-15-mediated paclitaxel sensitization was dependent on its phosphorylation status, we silenced PEA-15 expression using PEA-15-targeting siRNA and then transiently overexpressed AA and DD in HEY and OVTOKO ovarian cancer cells (Fig. 2D) and assessed paclitaxel sensitivity using trypan blue exclusion assay. Compared to cells transfected with vector, HEY cells transfected with DD showed a 12% reduction (P < 0.001) in viability, whereas HEY cells transfected with AA showed a 10.6% increase (P < 0.05) in viability after treatment with paclitaxel (0.005 μM; Fig. 2D). Similarly, compared to cells transfected with vector, OVTOKO cells transfected with DD showed a 19% reduction (P < 0.001) in viability, whereas OVTOKO cells transfected with AA showed an 11% increase (P < 0.05) in viability after treatment with paclitaxel (0.05 μM; Fig. 2D). These results indicate that PEA-15-mediated paclitaxel sensitization depends on PEA-15 phosphorylation status.
Figure 2.
Overexpression of PEA-15 phosphorylated at both Ser104 and Ser116 enhanced the sensitivity of ovarian cancer cells to paclitaxel. A, Expression of HA (hemagglutinin)-tagged PEA-15 was determined by western blotting in SKOV3.ip1 stable cells. β-actin served as a loading control. The sensitivity of SKOV3.ip1 stable cells to paclitaxel was determined by (B) WST-1 assay 72 hours after paclitaxel treatment and (C) anchorage-independent growth assay 3 weeks after paclitaxel treatment. In A-C, vector, SKOV3.ip1-vector; AA, SKOV3.ip1-AA (stably expressing nonphosphorylatable PEA-15 at Ser104 and Ser116, which were substituted with alanine); DD, SKOV3.ip1-DD (stably expressing phosphomimetic PEA-15 at Ser104 and Ser116, which were substituted with aspartic acid). D, PEA-15 expression in HEY and OVTOKO cells was silenced using PEA-15-targeting siRNA, and then cells were transiently transfected with the pcDNA3.1 vector, the pcDNA3.1 vector carrying nonphosphorylatable PEA-15 (AA; Ser104 and Ser116 of PEA-15 substituted with alanine), or the pcDNA3.1 vector carrying phosphomimetic PEA-15 (DD; Ser104 and Ser116 of PEA-15 substituted with aspartic acid). Expression of PEA-15 was determined by western blotting 48 hours after transfection (upper panels). β-actin served as a loading control. Following transfection, the sensitivity of HEY and OVTOKO cells to paclitaxel was determined by trypan blue exclusion assay 72 hours after paclitaxel treatment (lower panels). *, P < 0.05; **, P < 0.01; #, P < 0.001.
pPEA-15-mediated paclitaxel sensitization correlated with enhancement of mitotic arrest and apoptosis
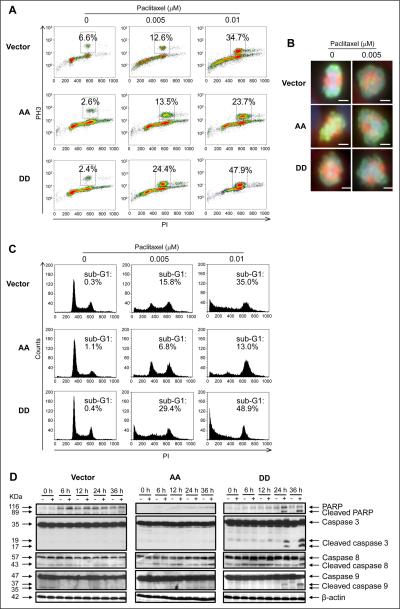
Paclitaxel induces G2/M arrest and apoptotic cell death (5, 6, 8). Thus, we hypothesized that pPEA-15 increases paclitaxel sensitivity in ovarian cancer cells by enhancing mitotic arrest and apoptosis. As expected, flow cytometry analysis showed that paclitaxel exposure led to a robust mitotic arrest in a dose-dependent manner (data not shown). After exposure to paclitaxel (0.01 μM), SKOV3.ip1-DD cells displayed a 13.2% increase in mitotic accumulation, whereas SKOV3.ip1-AA cells displayed a 10.9% decrease in mitotic accumulation, compared to SKOV3.ip1-vector control cells (Fig. 3A). This result suggests that pPEA-15 increases paclitaxel cytotoxicity by enhancing paclitaxel-induced mitotic arrest.
Figure 3.
Overexpression of PEA-15 phosphorylated at both Ser104 and Ser116 promoted induction of mitotic arrest and apoptosis by paclitaxel. SKOV3.ip1 stable cells were treated with paclitaxel and then subjected to immunofluorescence staining with anti-phosphohistone H3 antibody and analyses by (A) flow cytometry 12 hours after paclitaxel treatment and (B) fluorescence microscopy 6 hours after paclitaxel treatment. In A, the x-axis indicates the intensity of propidium iodide (PI) staining, and the y-axis indicates the intensity of phosphohistone H3 (PH3) staining. In B, scale bar, 100 μm. C, SKOV3.ip1 stable cells were treated with paclitaxel for 36 hours and then subjected to cell cycle analysis using PI staining and flow cytometry. The x-axis indicates the intensity of PI staining, and the y-axis (counts) shows the cell number. D, SKOV3.ip1 stable cells were treated with paclitaxel for 36 hours and then subjected to western blotting to verify cleavage of PARP and caspases 3, 8, and 9. β-actin served as a loading control. Vector, SKOV3.ip1-vector; AA, SKOV3.ip1-AA (stably expressing nonphosphorylatable PEA-15 at Ser104 and Ser116, which were substituted with alanine); DD, SKOV3.ip1-DD (stably expressing phosphomimetic PEA-15 at Ser104 and Ser116, which were substituted with aspartic acid).
It is known that paclitaxel-induced mitotic arrest is a result of inhibition of spindle organization (35). We observed that in the absence of paclitaxel, in dividing SKOV3.ip1-vector and SKOV3.ip1-AA cells, mitotic spindles were normal, whereas in dividing SKOV3.ip1-DD cells, the majority of mitotic spindles were abnormal and monopolar and contained ball-shaped aggregations of condensed chromosomes (Fig. 3B). After paclitaxel treatment, cells that were blocked at mitosis had abnormal spindles, and the majority of metaphase spindles displayed either monopolar or bipolar organization with chromosomes scattered throughout the spindles (Fig. 3B). However, no significant differences in spindle organization were observed among SKOV3.ip1-vector, SKOV3.ip1-AA, and SKOV3.ip1-DD cells after paclitaxel treatment. These findings demonstrate that when PEA-15 is phosphorylated at both Ser104 and Ser116, it promotes paclitaxel's ability to block cell cycle progression at mitosis.
We next investigated the effect of PEA-15 phosphorylation status on apoptosis induction by paclitaxel by cell cycle analysis. After exposure to paclitaxel (0.01 μM), SKOV3.ip1-DD cells showed a 14% increase in the sub-G1 fraction, whereas SKOV3.ip1-AA cells showed a 12% decrease in the sub-G1 fraction, compared to SKOV3.ip1-vector control cells (Fig. 3C). Similarly, when siRNA PEA-15-treated HEY and OVTOKO ovarian cancer cells (Fig. 2D) and KOC-7c ovarian cancer cells were transiently transfected with DD, these cells displayed 10-15% increases in the sub-G1 fraction compared to the cells transfected with vector after paclitaxel treatment (Supplementary Fig. S2A-C). We further confirmed the enhanced apoptosis in SKOV3.ip1-DD cells after paclitaxel treatment by Annexin-V and 7-AAD staining. Compared to SKOV3.ip1-vector control cells, SKOV3.ip1-DD cells showed 19.8% and 30.1% increases in apoptosis when treated with 0.005 and 0.01 μM paclitaxel, respectively (Supplementary Fig. S2D). These results indicate that when PEA-15 is phosphorylated at both Ser104 and Ser116, it enhances paclitaxel-induced apoptosis. Moreover, after paclitaxel treatment, we observed significant cleavage of PARP and caspases 3, 8, and 9 in SKOV3.ip1-DD cells compared to SKOV3.ip1-vector and SKOV3.ip1-AA cells (Fig. 3D). The addition of caspase 8 and 9 inhibitors reduced paclitaxel-induced apoptosis by 28.5% and 4.8%, respectively, in SKOV3.ip1-AA cells and by 56.6% and 49.5%, respectively, in SKOV3.ip1-DD cells (Supplementary Fig. S3). These results indicate that the enhanced apoptosis resulting from PEA-15 phosphorylation is a consequence of cleavage of both caspases 8 and 9, suggesting that paclitaxel induces apoptosis at least in part through activation of both the death effector and mitochondrial pathways.
SCLIP was involved in pPEA-15-mediated paclitaxel sensitization
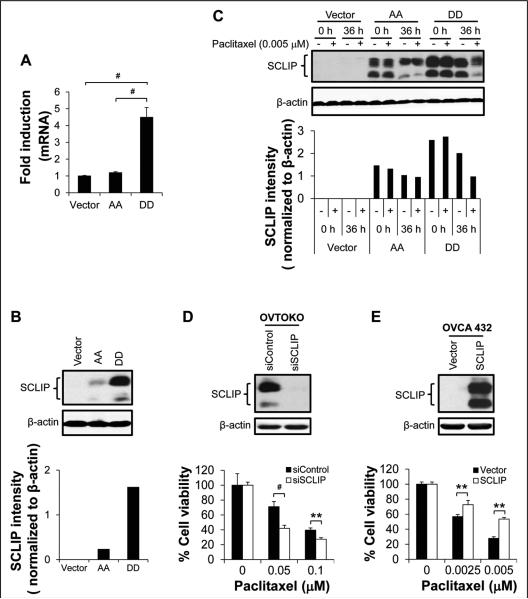
To dissect the molecular mechanisms of PEA-15-mediated chemosensitization of ovarian cancer cells, we analyzed our cDNA microarray data obtained previously using SKOV3.ip1-S116A cells (Ser116 of PEA-15 substituted with alanine) and SKOV3.ip1-S116D cells (Ser116 of PEA-15 substituted with aspartic acid). We found that SCLIP was highly expressed in SKOV3.ip1-S116D cells. On the basis of this finding, we assessed SCLIP expression in SKOV3.ip1 stable cells by RT-PCR and western blotting. The expression of SCLIP was 4.5-fold higher at the mRNA level (P < 0.001; Fig. 4A and Supplementary Fig. S4) and 1.6-fold higher at the protein level in SKOV3.ip1-DD cells than in SKOV3.ip1-vector control cells (Fig. 4B).
Figure 4.
SCLIP was involved in pPEA-15-mediated paclitaxel sensitization. In paclitaxel-untreated SKOV3.ip1 stable cells, SCLIP (A) mRNA levels were determined by RT-PCR and (B) protein levels by western blotting (upper panel) and densitometric quantification (lower panel). C, SKOV3.ip1 stable cells were treated with paclitaxel, and 36 hours later, SCLIP expression was determined by western blotting (upper panel) and densitometric quantification (lower panel). β-actin served as a loading control. Vector, SKOV3.ip1-vector; AA, SKOV3.ip1-AA (stably expressing nonphosphorylatable PEA-15 at Ser104 and Ser116, which were substituted with alanine); DD, SKOV3.ip1-DD (stably expressing phosphomimetic PEA-15 at Ser104 and Ser116, which were substituted with aspartic acid). D, OVTOKO cells were treated with SCLIP-targeting siRNA, and SCLIP expression was determined by western blotting 72 hours later (upper panel). β-actin served as a loading control. Following SCLIP silencing, the sensitivity of OVTOKO cells to paclitaxel was determined by trypan blue exclusion assay 72 hours after paclitaxel treatment (lower panel). E, OVCA 432 were transiently transfected with pCMV6-Entry vector or pCMV6-Entry vector carrying SCLIP, and SCLIP expression was determined by western blotting 48 hours later (upper panel). β-actin served as a loading control. Following SCLIP overexpression, the sensitivity of OVCA 432 cells to paclitaxel was determined by trypan blue exclusion assay 72 hours after paclitaxel treatment (lower panel). *, P < 0.05; **, P < 0.01; #, P < 0.001.
Because SCLIP regulates cellular functions by modulating MT dynamics (23, 26) and stathmin, a protein in the same family as SCLIP, has been implicated in paclitaxel resistance (27), we hypothesized that SCLIP is involved in pPEA-15-mediated chemosensitization. We next investigated whether the high expression level of SCLIP was associated with the enhanced chemosensitivity in SKOV3.ip1-DD cells. We assessed SCLIP expression at both the mRNA and protein levels in SKOV3.ip1 stable cells after paclitaxel treatment. No changes were observed in SCLIP expression at the mRNA level after paclitaxel treatment (data not shown). Interestingly, the protein level of SCLIP in paclitaxel-treated SKOV3.ip1-DD cells was only 51.2% of that in untreated SKOV3.ip1-DD cells, whereas SCLIP expression in paclitaxel-treated SKOV3.ip1-AA cells was not substantially different from that in untreated SKOV3.ip1-AA cells (Fig. 4C). Furthermore, SCLIP silencing in SKOV3.ip1-DD cells using SCLIP-targeting siRNA resulted in a 9% increase (P < 0.001) in the sub-G1 fraction compared to the sub-G1 fractions in the control cells treated with scrambled siRNA after treatment with paclitaxel (0.005 μM) for 24 hours (Supplementary Fig. S5A). In contrast, transient re-expression of SCLIP in siRNA SCLIP-treated SKOV3.ip1-DD cells led to a 21.9% reduction (P < 0.001) in the sub-G1 fraction compared to cells transfected with vector after treatment with paclitaxel (0.005 μM) for 24 hours (Supplementary Fig. S5A). Similar results were obtained when cells were treated with paclitaxel for 36 hours (Supplementary Fig. S5A). These results indicate that reduced SCLIP expression following paclitaxel exposure correlates with increased paclitaxel sensitivity of SKOV3.ip1-DD cells.
To elucidate whether SCLIP regulates paclitaxel sensitization in ovarian cancer cells, we first silenced SCLIP expression in SCLIP-high-expressing OVTOKO and HEY cells and then analyzed cell viability by trypan blue exclusion assy. Compared to cells treated with scrambled siRNA, OVTOKO cells with SCLIP silencing displayed a 29.1% reduction in cell viability (0.05 μM paclitaxel; P < 0.001; Fig. 4D) and HEY cells with SCLIP silencing displayed an 18.6% reduction in cell viability (0.005 μM paclitaxel; P < 0.01; Supplementary Fig. S5B) after 72 hours of treatment with paclitaxel. In contrast, when SCLIP was overexpressed in SCLIP-low-expressing OVCA 432 cells and OVCAR3 cells, we observed a 16% increase in cell viability in OVCA 432 cells (0.0025 μM paclitaxel; P < 0.01; Fig. 4E) and a 17.7% increase in cell viability in OVCAR3 cells (0.0025 μM paclitaxel; P < 0.05; Supplementary Fig. S5C) compared to cells transfected with vector after 72 hours of paclitaxel treatment. These results demonstrate that SCLIP promotes paclitaxel resistance in ovarian cancer cells.
Interestingly, a shift in SCLIP molecular weight was noted in SKOV3.ip1-DD cells but not in SKOV3.ip1-AA cells following paclitaxel treatment (Fig. 4C, upper panel). Paclitaxel has been reported to induce serine phosphorylation of the 66-kDa Shc isoform (36). Thus, the shift in SCLIP molecular weight may be a result of phosphorylation of SCLIP in paclitaxel-treated SKOV3.ip1-DD cells. However, phosphatase treatment of cell lysates did not revise the paclitaxel-induced shift of SCLIP molecular weight but only partially reduced SCLIP intensity (data not shown). These results suggest that reduced expression and possibly posttranslational modification of SCLIP following paclitaxel treatment impair SCLIP's inhibitory effect on paclitaxel, which in turn enhances the sensitivity of SKOV3.ip1-DD cells to paclitaxel.
pPEA-15 overexpression enhanced the MT-polymerizing effect of paclitaxel
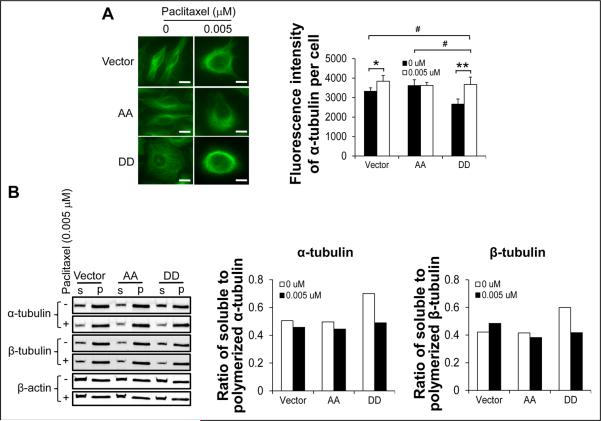
Paclitaxel exerts its antitumor activity by inducing cell cycle arrest at mitosis through enhancement of MT polymerization (35). In contrast, SCLIP sequesters soluble tubulin heterodimers and destabilizes MT polymers (22, 23). Since both paclitaxel and SCLIP modulate MT dynamics, we analyzed MT organization in SKOV3.ip1 stable cells by immunofluorescence staining following paclitaxel exposure. In the absence of paclitaxel, SKOV3.ip1-vector and SKOV3.ip1-AA cells contained long MT polymers, whereas SKOV3.ip1-DD cells had short and diffuse MT polymers that formed a meshwork of MTs (Fig. 5A, left panel). After a 12-hour exposure to paclitaxel, the MT network in SKOV3.ip1-AA cells bundled with parallel MTs radiating out from the nucleus, whereas MTs were more condensed and oriented in parallel fashion and reorganized into thick ring-like bundles near the nucleus in SKOV3.ip1-vector and SKOV3.ip-DD cells (Fig. 5A, left panel). Similar phenomena were observed when cells were treated with paclitaxel for 3 or 6 hours (data not shown). Furthermore, the fluorescence intensity of α-tubulin in untreated SKOV3.ip1-DD cells was significantly lower than that in untreated SKOV3.ip1-vector and SKOV3.ip1-AA cells (P < 0.01; Fig. 5A, right panel). Following paclitaxel treatment, α-tubulin intensity in SKOV3.ip1-DD cells dramatically increased and was similar to that in SKOV3.ip1-vector and SKOV3.ip1-AA cells.
Figure 5.
Examination of MT network and polymerization in SKOV3.ip1 stable cells in the presence or absence of paclitaxel. A, MT network was examined by immunofluorescence staining (left panel) and fluorescence intensity quantification (right panel) in SKOV3.ip1 stable cells using anti-α-tubulin antibody after a 12-hour incubation in the absence or presence of paclitaxel. Scale bar, 100 μm. B, Soluble (S) and polymerized (P) α- and β-tubulin subunits were isolated from SKOV3.ip1 stable cells after a 12-hour incubation with paclitaxel and then analyzed by western blotting (left panel) and densitometric quantification (middle and right panels). β-actin served as a loading control. Vector, SKOV3.ip1-vector; AA, SKOV3.ip1-AA (stably expressing nonphosphorylatable PEA-15 at Ser104 and Ser116, which were substituted with alanine); DD, SKOV3.ip1-DD (stably expressing phosphomimetic PEA-15 at Ser104 and Ser116, which were substituted with aspartic acid). *, P < 0.05; **, P < 0.01; #, P < 0.001.
To confirm immunofluorescence staining results, we further quantified soluble and polymerized α- and β-tubulin from cells after 12 hours of paclitaxel exposure. In the absence of paclitaxel, SKOV3.ip1-DD cells had more soluble α- and β-tubulin subunits than did SKOV3.ip1-vector and SKOV3.ip1-AA cells (Fig. 5B), which might be a result of SCLIP-dependent MT destabilization in SKOV3.ip1-DD cells. However, paclitaxel exposure reduced the content of both soluble α-tubulin and soluble β-tubulin subunits, indicating enhanced polymerization of tubulin into MTs in SKOV3.ip1-DD cells after paclitaxel exposure (Fig. 5B). In contrast, no significant changes were observed in the ratio of soluble to polymerized tubulin in SKOV3.ip1-vector and SKOV3.ip1-AA cells following paclitaxel exposure (Fig. 5B). These results suggest that increased paclitaxel sensitivity of SKOV3.ip1-DD cells may be a result of enhanced MT polymerization.
pPEA-15 overexpression promoted the MT-stabilizing effect of paclitaxel
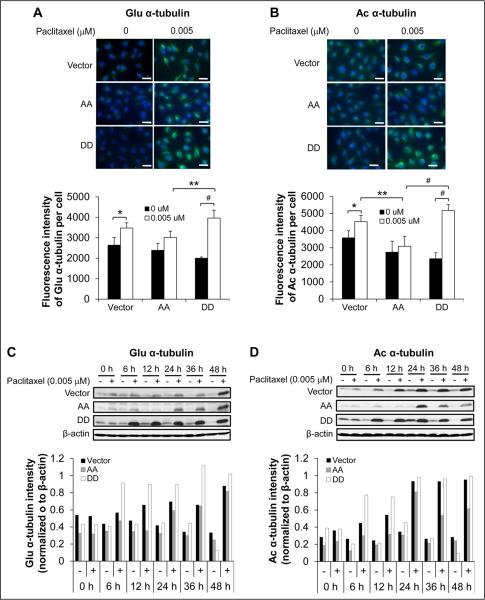
Posttranslational modifications of MTs are known to stabilize individual MTs in response to various stimuli. These modifications include α-tubulin detyrosination at the C-terminus (37) and acetylation at Lys40 (38), which occur primarily on tubulin assembled into MTs and have been correlated with MT stability (39, 40). To address whether increased paclitaxel sensitivity of SKOV3.ip1-DD cells correlates with enhanced MT stability, we assessed the assembled MTs by immunofluorescence staining for detyrosinated (Glu) and acetylated (Ac) α-tubulin subunits. In the absence of paclitaxel, all SKOV3.ip1 stable cells showed weak staining for both Glu and Ac α-tubulin (Fig. 6A and 6B, upper panels). SKOV3.ip1-DD cells had less intense staining for both Glu and Ac α-tubulin than did SKOV3.ip1-vector and SKOV3.ip1-AA cells (Fig. 6A and 6B, lower panels), indicating that tubulin is less polymerized in untreated SKOV3.ip1-DD cells. As expected, a 1-h paclitaxel exposure resulted in an increase in the intensity of both Glu and Ac α-tubulin staining in each cell type (Fig. 6A and 6B). Paclitaxel-treated SKOV3.ip1-DD cells displayed more intense staining for both Glu and Ac α-tubulin than did paclitaxel-treated SKOV3.ip1-vector and SKOV3.ip1-AA cells (Fig. 6A and 6B).
Figure 6.
Examination of detyrosinated (Glu) and acetylated (Ac) α-tubulin expression in SKOV3.ip1 stable cells in the presence or absence of paclitaxel. Expression of (A) Glu and (B) Ac α-tubulin was examined by immunofluorescence staining (upper panels) and fluorescence intensity qualification (lower panels) in SKOV3.ip1 stable cells after a 1-hour incubation in the absence or presence of paclitaxel. Scale bar, 10 μm. Expression of (C) Glu and (D) Ac α-tubulin was examined by western blotting (upper panels) and densitometric quantification (lower panels) in SKOV3.ip1 stable cells after a 48-hour incubation with paclitaxel. Vector, SKOV3.ip1-vector; AA, SKOV3.ip1-AA (stably expressing nonphosphorylatable PEA-15 at Ser104 and Ser116, which were substituted with alanine); DD, SKOV3.ip1-DD (stably expressing phosphomimetic PEA-15 at Ser104 and Ser116, which were substituted with aspartic acid). *, P < 0.05; **, P < 0.01; #, P < 0.001.
Immunofluorescence staining results were further confirmed by western blot analysis. The expression levels of both Glu (Fig. 6C) and Ac (Fig. 6D) α-tubulin were low in all untreated cells. In SKOV3.ip1-DD cells, the levels of both Glu (Fig. 6C) and Ac (Fig. 6D) α-tubulin dramatically increased 6 hours after paclitaxel exposure and remained high up to 48 hours. In SKOV3.ip1-vector and SKOV3.ip1-AA cells, the levels of Glu α-tubulin increased slightly at 24 hours after paclitaxel exposure and were similar to the levels in SKOV3.ip1-DD cells at 48 hours (Fig. 6C), whereas the levels of Ac α-tubulin peaked at 24 hours in both cell lines and then remained high up to 48 hours in SKOV3.ip1-vector cells but decreased in SKOV3.ip1-AA cells (Fig. 6D). The elevated acetylation and detyrosination of α-tubulin in paclitaxel-treated SKOV3.ip1-DD cells reflected the enhancement of MT polymerization and stability. These results clearly demonstrate that pPEA-15 overexpression enhances paclitaxel-induced MT polymerization and stability, resulting in increased sensitivity of SKOV3.ip1-DD cells to paclitaxel.
Discussion
Our results demonstrate that when PEA-15 is phosphorylated at both Ser104 and Ser116, it increases the sensitivity of ovarian cancer cells to paclitaxel by impairing SCLIP-mediated MT destabilization, which is a result of reduced expression and possibly posttranslational modification of SCLIP following paclitaxel treatment. Impairment of SCLIP-mediated MT destabilization promotes induction of MT polymerization and stabilization by paclitaxel, leading to enhanced mitotic arrest and apoptotic cell death.
Our results here show that pPEA-15 mediates paclitaxel sensitization in ovarian cancer cells by activating both the death effector and the mitochondrial pathways. In contrast to our findings, Stassi and colleagues reported that PEA-15 mediated resistance to paclitaxel-induced apoptosis in breast cancer cells (21). This discrepancy may be due to differences in the phosphorylation status of PEA-15 as well as differences in the cell types, i.e., breast cancer cells vs. ovarian cancer cells. PEA-15's functions are tightly regulated by its phosphorylation status. Studies have shown that phosphorylation at Ser104 inhibits binding of PEA-15 to ERK, whereas phosphorylation at Ser116 promotes binding of PEA-15 to FADD (41). Furthermore, PEA-15 phosphorylation at Ser116 is required for its antiapoptotic activity in both 293 cells and U373MG glioma cells (20), whereas monophosphorylation at either Ser104 or Ser116 is essential for inhibition of Fas-mediated but not TNFα-mediated apoptosis in NIH-3T3 cells (41). These studies clearly indicate that phosphorylation status of PEA-15 plays a crucial role in determining its biological functions. In addition, our finding that pPEA-15 sensitizes ovarian cancer cells to paclitaxel strongly suggests that PEA-15 phosphorylation status is also critical for the responsiveness of cancer cells to chemotherapy.
MT dynamics have been considered a key factor in determining the chemosensitivity of cancer cells (5). Subtle suppression of MT dynamics by paclitaxel inhibits the assembly and functioning of the mitotic spindle, leading to mitotic arrest and apoptotic cell death (5, 6). Thus, cellular molecules that enhance paclitaxel's effect on MT polymerization and stabilization may improve its cytotoxicity (42). Our results here showed that overexpression of PEA-15 that is phosphorylated at both Ser104 and Ser116 markedly increased paclitaxel's ability to induce mitotic arrest and apoptosis in ovarian cancer cells. Furthermore, dramatic enhancement in MT polymerization and in tubulin acetylation and detyrosination was observed in pPEA-15-overexpressing SKOV3.ip1-DD cells as early as 6 hours following paclitaxel exposure. It has been reported that paclitaxel's effect on MT dynamics is at least partially due to its effect on tubulin acetylation and detyrosination, established markers of MT stability (39, 40). This information in combination with our results indicates that the mechanism underlying the high sensitivity of SKOV3.ip1-DD cells to paclitaxel is the enhanced MT polymerization and stabilization, which lead to mitotic arrest and apoptosis.
In the present study, we demonstrated that overexpression of PEA-15 phosphorylated at both Ser104 and Ser116 induced high-level expression of SCLIP, whereas paclitaxel exposure resulted in a dramatic reduction in SCLIP expression at the protein level. SCLIP is able to inhibit MT polymerization by sequestering soluble tubulin (43) and promote MT depolymerization by increasing the MT catastrophe rate (24, 25). Stathmin has been implicated in modulating the sensitivity of cancer cells to MT-targeting drugs by interfering with interaction of the drugs with MTs (31, 44). However, up to now, the biological functions of SCLIP and its role in chemosensitization have been largely elusive, and these functions were only inferred, in part, by extrapolation from what is known about other stathmin/OP18 family members. Our study here provides direct evidence that SCLIP has a functional effect on chemosensitivity of paclitaxel through a MT-dependent mechanism. The reduced SCLIP expression following paclitaxel treatment impaired SCLIP's MT-destabilizing effect as supported by high-level expression of Glu and Ac tubulin, thereby facilitating paclitaxel's promotion of MT assembly and apoptosis.
In addition to reduced SCLIP expression, a shift in SCLIP molecular weight was noted after paclitaxel exposure, which may have resulted from enhanced phosphorylation of SCLIP. Phosphorylation of stathmin/OP18 family proteins at serine residues has been reported to impair their effects on MT dynamics and halt their antagonistic activity against MT-targeting drugs (22). Since there are no antibodies available to detect the phosphorylation status of SCLIP, we performed phosphatase treatment of cell lysates to verify whether the molecular weight shift of SCLIP was a result of phosphorylation. However, phosphatase treatment did not revise the shift of SCLIP molecular weight but only partially reduced the intensity of SCLIP (data not shown), indicating that phosphorylation is not the sole reason for SCLIP molecular weight change. Thus, other unknown modifications, possibly glycosylation, may also be responsible for the change in SCLIP molecular weight and influence SCLIP's effect on MT instability. Further studies are needed to address the mechanisms that underlie the effect of pPEA-15 on SCLIP expression and the effect of paclitaxel on SCLIP functioning.
Our study defines an important role for pPEA-15 in regulating chemosensitization of paclitaxel in ovarian cancer cells: pPEA-15 impairs the MT-destabilizing activity of SCLIP, thereby enhancing the effects of paclitaxel on MT assembly, mitosis, and apoptosis. In addition, our results suggest that pPEA-15 could be used as a biomarker to predict the responsiveness of ovarian tumors to paclitaxel treatment and that patients with ovarian tumors expressing high levels of pPEA-15 would be more likely to benefit from paclitaxel treatment than other patients. In our future studies, we will examine the association between PEA-15 phosphorylation status and the pathological grade of ovarian tumors and evaluate the in vivo impact of PEA-15 phosphorylation status on the effectiveness of paclitaxel in a human ovarian carcinoma xenograft.
Supplementary Material
Acknowledgments
We acknowledge Stephanie P. Deming of the Department of Scientific Publications for her editorial assistance, the Flow Cytometry and Cellular Imaging Core Facility at MD Anderson Cancer Center for flow cytometry analyses, the Genomics Core Facility at MD Anderson Cancer Center for cDNA microarray analysis, and Tamer S. Kaoud (The University of Texas at Austin, TX) for providing paclitaxel structure.
Grant Support: This work was supported by NIH/NCI grant (R01CA127562 03) awarded to N. T. Ueno and NIH/NCI Cancer Center Support Grant (CA016672) awarded to The University of Texas MD Anderson Cancer Center.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Mekhail TM, Markman M. Paclitaxel in cancer therapy. Expert Opin Pharmacother. 2002;3:755–66. doi: 10.1517/14656566.3.6.755. [DOI] [PubMed] [Google Scholar]
- 2.Rua F, Sadeghi SJ, Castrignano S, Di Nardo G, Gilardi G. Engineering Macaca fascicularis cytochrome P450 2C20 to reduce animal testing for new drugs. J Inorg Biochem. 2012;117:277–84. doi: 10.1016/j.jinorgbio.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Xiao H, Verdier-Pinard P, Fernandez-Fuentes N, Burd B, Angeletti R, Fiser A, et al. Insights into the mechanism of microtubule stabilization by Taxol. Proc Natl Acad Sci U S A. 2006;103:10166–73. doi: 10.1073/pnas.0603704103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parness J, Horwitz SB. Taxol binds to polymerized tubulin in vitro. J Cell Biol. 1981;91:479–87. doi: 10.1083/jcb.91.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nat Rev Cancer. 2004;4:253–65. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 6.Yvon AM, Wadsworth P, Jordan MA. Taxol suppresses dynamics of individual microtubules in living human tumor cells. Mol Biol Cell. 1999;10:947–59. doi: 10.1091/mbc.10.4.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matei D. Novel agents in ovarian cancer. Expert Opin Investig Drugs. 2007;16:1227–39. doi: 10.1517/13543784.16.8.1227. [DOI] [PubMed] [Google Scholar]
- 8.Goncalves A, Braguer D, Kamath K, Martello L, Briand C, Horwitz S, et al. Resistance to Taxol in lung cancer cells associated with increased microtubule dynamics. Proc Natl Acad Sci U S A. 2001;98:11737–42. doi: 10.1073/pnas.191388598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peer D, Dekel Y, Melikhov D, Margalit R. Fluoxetine inhibits multidrug resistance extrusion pumps and enhances responses to chemotherapy in syngeneic and in human xenograft mouse tumor models. Cancer Res. 2004;64:7562–9. doi: 10.1158/0008-5472.CAN-03-4046. [DOI] [PubMed] [Google Scholar]
- 10.Tan M, Jing T, Lan KH, Neal CL, Li P, Lee S, et al. Phosphorylation on tyrosine-15 of p34(Cdc2) by ErbB2 inhibits p34(Cdc2) activation and is involved in resistance to taxol-induced apoptosis. Mol Cell. 2002;9:993–1004. doi: 10.1016/s1097-2765(02)00510-5. [DOI] [PubMed] [Google Scholar]
- 11.Giannakakou P, Sackett DL, Kang YK, Zhan Z, Buters JT, Fojo T, et al. Paclitaxel-resistant human ovarian cancer cells have mutant beta-tubulins that exhibit impaired paclitaxel-driven polymerization. J Biol Chem. 1997;272:17118–25. doi: 10.1074/jbc.272.27.17118. [DOI] [PubMed] [Google Scholar]
- 12.Di Michele M, Marcone S, Cicchillitti L, Della Corte A, Ferlini C, Scambia G, et al. Glycoproteomics of paclitaxel resistance in human epithelial ovarian cancer cell lines: towards the identification of putative biomarkers. J Proteomics. 2010;73:879–98. doi: 10.1016/j.jprot.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 13.Bartholomeusz C, Rosen D, Wei C, Kazansky A, Yamasaki F, Takahashi T, et al. PEA-15 induces autophagy in human ovarian cancer cells and is associated with prolonged overall survival. Cancer Res. 2008;68:9302–10. doi: 10.1158/0008-5472.CAN-08-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bartholomeusz C, Gonzalez-Angulo AM, Kazansky A, Krishnamurthy S, Liu P, Yuan LX, et al. PEA-15 inhibits tumorigenesis in an MDA-MB-468 triple-negative breast cancer xenograft model through increased cytoplasmic localization of activated extracellular signal-regulated kinase. Clin Cancer Res. 2010;16:1802–11. doi: 10.1158/1078-0432.CCR-09-1456. [DOI] [PubMed] [Google Scholar]
- 15.Glading A, Koziol JA, Krueger J, Ginsberg MH. PEA-15 inhibits tumor cell invasion by binding to extracellular signal-regulated kinase 1/2. Cancer Res. 2007;67:1536–44. doi: 10.1158/0008-5472.CAN-06-1378. [DOI] [PubMed] [Google Scholar]
- 16.Fiory F, Formisano P, Perruolo G, Beguinot F. Frontiers: PED/PEA-15, a multifunctional protein controlling cell survival and glucose metabolism. Am J Physiol Endocrinol Metab. 2009;297:E592–601. doi: 10.1152/ajpendo.00228.2009. [DOI] [PubMed] [Google Scholar]
- 17.Krueger J, Chou FL, Glading A, Schaefer E, Ginsberg MH. Phosphorylation of phosphoprotein enriched in astrocytes (PEA-15) regulates extracellular signal-regulated kinase-dependent transcription and cell proliferation. Mol Biol Cell. 2005;16:3552–61. doi: 10.1091/mbc.E04-11-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sulzmaier FJ, Valmiki MK, Nelson DA, Caliva MJ, Geerts D, Matter ML, et al. PEA-15 potentiates H-Ras-mediated epithelial cell transformation through phospholipase D. Oncogene. 2011 doi: 10.1038/onc.2011.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kubes M, Cordier J, Glowinski J, Girault JA, Chneiweiss H. Endothelin induces a calcium-dependent phosphorylation of PEA-15 in intact astrocytes: identification of Ser104 and Ser116 phosphorylated, respectively, by protein kinase C and calcium/calmodulin kinase II in vitro. J Neurochem. 1998;71:1307–14. doi: 10.1046/j.1471-4159.1998.71031307.x. [DOI] [PubMed] [Google Scholar]
- 20.Trencia A, Perfetti A, Cassese A, Vigliotta G, Miele C, Oriente F, et al. Protein kinase B/Akt binds and phosphorylates PED/PEA-15, stabilizing its antiapoptotic action. Mol Cell Biol. 2003;23:4511–21. doi: 10.1128/MCB.23.13.4511-4521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stassi G, Garofalo M, Zerilli M, Ricci-Vitiani L, Zanca C, Todaro M, et al. PED mediates AKT-dependent chemoresistance in human breast cancer cells. Cancer Res. 2005;65:6668–75. doi: 10.1158/0008-5472.CAN-04-4009. [DOI] [PubMed] [Google Scholar]
- 22.Cassimeris L. The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol. 2002;14:18–24. doi: 10.1016/s0955-0674(01)00289-7. [DOI] [PubMed] [Google Scholar]
- 23.Charbaut E, Curmi PA, Ozon S, Lachkar S, Redeker V, Sobel A. Stathmin family proteins display specific molecular and tubulin binding properties. J Biol Chem. 2001;276:16146–54. doi: 10.1074/jbc.M010637200. [DOI] [PubMed] [Google Scholar]
- 24.Belmont LD, Mitchison TJ. Identification of a protein that interacts with tubulin dimers and increases the catastrophe rate of microtubules. Cell. 1996;84:623–31. doi: 10.1016/s0092-8674(00)81037-5. [DOI] [PubMed] [Google Scholar]
- 25.Manna T, Thrower D, Miller HP, Curmi P, Wilson L. Stathmin strongly increases the minus end catastrophe frequency and induces rapid treadmilling of bovine brain microtubules at steady state in vitro. J Biol Chem. 2006;281:2071–8. doi: 10.1074/jbc.M510661200. [DOI] [PubMed] [Google Scholar]
- 26.Gavet O, Ozon S, Manceau V, Lawler S, Curmi P, Sobel A. The stathmin phosphoprotein family: intracellular localization and effects on the microtubule network. J Cell Sci. 1998;111(Pt 22):3333–46. doi: 10.1242/jcs.111.22.3333. [DOI] [PubMed] [Google Scholar]
- 27.Alli E, Bash-Babula J, Yang JM, Hait WN. Effect of stathmin on the sensitivity to antimicrotubule drugs in human breast cancer. Cancer Res. 2002;62:6864–9. [PubMed] [Google Scholar]
- 28.Lee J, Bartholomeusz C, Krishnamurthy S, Liu P, Saso H, LaFortune T, et al. PEA-15 unphosphorylated at both serine 104 and serine 116 inhibits ovarian cancer cell tumorigenicity and progression through blocking β-catenin. Oncogenesis. 2012:1. doi: 10.1038/oncsis.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lacroix M. Persistent use of “false” cell lines. International Journal of Cancer. 2008;122:1–4. doi: 10.1002/ijc.23233. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Chen B, Chen J, Cai X, Xia G, Liu R, et al. Synthesis and antitumor efficacy of daunorubicin-loaded magnetic nanoparticles. Int J Nanomedicine. 2011;6:203–11. doi: 10.2147/IJN.S16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alli E, Yang JM, Ford JM, Hait WN. Reversal of stathmin-mediated resistance to paclitaxel and vinblastine in human breast carcinoma cells. Mol Pharmacol. 2007;71:1233–40. doi: 10.1124/mol.106.029702. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.Gillespie DA, Walker M. Mitotic index determination by flow cytometry. Subcell Biochem. 2006;40:355–8. doi: 10.1007/978-1-4020-4896-8_25. [DOI] [PubMed] [Google Scholar]
- 34.Iancu C, Mistry SJ, Arkin S, Wallenstein S, Atweh GF. Effects of stathmin inhibition on the mitotic spindle. J Cell Sci. 2001;114:909–16. doi: 10.1242/jcs.114.5.909. [DOI] [PubMed] [Google Scholar]
- 35.Jordan MA, Toso RJ, Thrower D, Wilson L. Mechanism of mitotic block and inhibition of cell proliferation by taxol at low concentrations. Proc Natl Acad Sci U S A. 1993;90:9552–6. doi: 10.1073/pnas.90.20.9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang CP, Horwitz SB. Taxol mediates serine phosphorylation of the 66-kDa Shc isoform. Cancer Res. 2000;60:5171–8. [PubMed] [Google Scholar]
- 37.Arce CA, Hallak ME, Rodriguez JA, Barra HS, Caputto R. Capability of tubulin and microtubules to incorporate and to release tyrosine and phenylalanine and the effect of the incorporation of these amino acids on tubulin assembly. J Neurochem. 1978;31:205–10. doi: 10.1111/j.1471-4159.1978.tb12449.x. [DOI] [PubMed] [Google Scholar]
- 38.LeDizet M, Piperno G. Identification of an acetylation site of Chlamydomonas alpha-tubulin. Proc Natl Acad Sci U S A. 1987;84:5720–4. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Black MM, Baas PW, Humphries S. Dynamics of alpha-tubulin deacetylation in intact neurons. J Neurosci. 1989;9:358–68. doi: 10.1523/JNEUROSCI.09-01-00358.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gundersen GG, Khawaja S, Bulinski JC. Postpolymerization detyrosination of alpha-tubulin: a mechanism for subcellular differentiation of microtubules. J Cell Biol. 1987;105:251–64. doi: 10.1083/jcb.105.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Estelles A, Charlton CA, Blau HM. The phosphoprotein protein PEA-15 inhibits Fas- but increases TNF-R1-mediated caspase-8 activity and apoptosis. Dev Biol. 1999;216:16–28. doi: 10.1006/dbio.1999.9510. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed AA, Wang X, Lu Z, Goldsmith J, Le XF, Grandjean G, et al. Modulating microtubule stability enhances the cytotoxic response of cancer cells to Paclitaxel. Cancer Res. 2011;71:5806–17. doi: 10.1158/0008-5472.CAN-11-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jourdain L, Curmi P, Sobel A, Pantaloni D, Carlier MF. Stathmin: a tubulin-sequestering protein which forms a ternary T2S complex with two tubulin molecules. Biochemistry. 1997;36:10817–21. doi: 10.1021/bi971491b. [DOI] [PubMed] [Google Scholar]
- 44.Balachandran R, Welsh MJ, Day BW. Altered levels and regulation of stathmin in paclitaxel-resistant ovarian cancer cells. Oncogene. 2003;22:8924–30. doi: 10.1038/sj.onc.1207060. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.