Abstract
Objective
Hexanucleotide repeat expansions in C9ORF72 underlie a significant fraction of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS). This study investigates the frequency of C9ORF72 repeat expansions in clinically diagnosed late-onset Alzheimer’s disease (AD).
Design, setting and patients
This case-control study genotyped the C9ORF72 repeat expansion in 872 unrelated familial AD cases and 888 controls recruited as part of the NIA-LOAD cohort, a multi-site collaboration studying 1000 families with two or more individuals clinically diagnosed with late-onset-AD.
Main Outcome Measure
We determined the presence or absence of the C9ORF72 repeat expansion by repeat-primed PCR, the length of the longest non-expanded allele, segregation of the genotype with disease, and clinical features of repeat expansion carriers.
Results
Three families showed large C9ORF72 hexanucleotide repeat expansions. Two additional families carried more than 30 repeats. Segregation with disease could be demonstrated in 3 families. One affected expansion carrier had neuropathology compatible with AD. In the NIA-LOAD series, the C9ORF72 repeat expansions constituted the second most common pathogenic mutation, just behind the PSEN1 A79V mutation, highlighting the heterogeneity of clinical presentations associated with repeat expansions.
Interpretation
C9ORF72 repeat expansions explain a small proportion of patients with a clinical presentation indistinguishable from AD, and highlight the necessity of screening “FTD genes” in clinical AD cases with strong family history.
Introduction
Alzheimer’s Disease (AD) constitutes the most common cause of dementia in clinical practice1. AD manifests clinically with progressive cognitive dysfunction with prominent memory loss, while its neuropathology is characterized by the presence of extracellular neuritic plaques and intracellular neurofibrillary tangles1. Familial AD can be caused by mutations in amyloid precursor protein (APP), presenilin 1 (PSEN1), and presenilin 2 (PSEN2)2–6. Interestingly, mutations in genes typically associated with frontotemporal dementia (FTD), including microtubule associated protein tau (MAPT) and granulin (GRN), have also been reported in clinical AD cases6–13, highlighting the overlapping phenotypes of AD and amnestic-FTD.
Recently, an intronic hexanucleotide repeat (GGGGCC) in the C9ORF72 gene was identified as a frequent cause of sporadic and familial FTD and amyotrophic lateral sclerosis (ALS)14, 15. Because some individuals with clinical AD carry mutations in FTD genes7–13, 16–18, two studies investigated the role of C9ORF72 expansions in AD, with conflicting results. The first report identified C9ORF72 repeat expansions in 3 of 342 cases with familial AD and 6 of 711 cases with sporadic AD. Interestingly, reanalysis of autopsy material from two expansion carriers showed FTD pathology19, suggesting that both patients had amnestic presentations of FTD rather than AD. The second study identified no repeat expansions in 568 patients with probable AD by clinical criteria and concluded that C9ORF72 repeats are specific for FTD20.
To clarify the role of C9ORF72 hexanucleotide repeat expansions in familial late-onset AD, this study screens 872 unrelated AD cases and 888 unrelated controls and investigates whether expanded repeats are associated with age at dementia onset. Furthermore, we address whether a larger, but non-expanded, GGGGCC allele is associated with risk for AD or age at onset.
Material and Methods
Patients
Individuals from 872 unrelated families with at least two individuals affected by AD and 888 unrelated unaffected controls from the National Institute of Aging Late Onset Alzheimer Disease Family Study (NIA-LOAD Family Study) were included. All AD cases had been diagnosed with dementia of the Alzheimer's type using criteria equivalent to the National Institute of Neurological and Communication Disorders and Stroke-Alzheimer's Disease and Related Disorders Association (NINCDS-ADRDA) for probable AD21, 22. All patients had a family history of Alzheimer's disease, but not other types of dementia or other neurodegenerative disease. Probands were required to have a diagnosis of definite or probable late-onset AD (onset >60) and a sibling with definite, probable or possible late-onset AD with a similar age at onset. A third biologically-related family member (first, second or third degree) was also required, regardless of affected status. If unaffected, this individual had to be ≥60 years of age, but ≥50 years of age if diagnosed with LOAD or mild cognitive impairment23. Within each pedigree, we selected a single individual to screen by identifying the youngest affected family member with the most definitive diagnosis (i.e. individuals with autopsy confirmation were chosen over those with clinical diagnosis only). A summary of the demographics of all subjects is shown in Table 1. Written informed consent was obtained from all participants, and the study was approved by local IRB committees.
Table 1.
Cohort Demographics
| AD Cases | Controls | |
|---|---|---|
| Number | 872 | 888 |
| Age (years)a | 75.8 ± 8.9 | 71.41 ± 6.7 |
| Age range (years) | 42–101 | 48–98 |
| Male (%) | 33.9 | 39.7 |
| APOE 4+ (%)b | 76.2 | 30.6 |
| No. C9ORF72 (%) | 5 (0.57) | 1 (0.11) |
For AD cases, age indicates onset of symptoms but refers to age at last assessment for controls.
APOE 4+ refers to those carrying at least 1 APOE 4 allele.
Genetic analysis
Repeat-primed PCR
The presence of the expanded hexanucleotide repeat and the number of repeat units in the longest allele was determined using previously reported methods for repeat-primed PCR and fluorescence-based fragment size analysis14. Briefly, repeat-primed PCR was performed in a total reaction volume of 28 µl containing 100 ng genomic DNA, 1× FastStart PCR Master Mix (Roche Applied Science, Indianapolis, IN, USA), 3.5% DMSO, 1× Q solution (Quiagen, Valencia, CA) and 0.18 mM of deazaGTP (NEB, Ipswich, MA). Primer concentrations and sequences (chr9:27563580F and chr9:27563465R) were the same as previously reported14. PCR products were run on an ABI® 3130×l Genetic Analyzer (Applied Biosystems) and analyzed using GeneMapper®. Consistent with standards used in prior studies, a sample was considered to have a repeat expansion when assay replicates demonstrated >30 peaks and a decrementing saw-tooth pattern with 6 base-pair periodicity (Supplementary Figure 1). A normal repeat alleles was defined as 30 or fewer peaks14, 15, 19, 20.
Cross-repeat PCR
In all cases, repeat expansions identified with repeat-primed PCR were confirmed by attempting to PCR across the GGGGCC repeat using fluorescence-based fragment size analysis (“cross-repeat PCR”) as reported with minor modifications14. Briefly, using previously reported primers with fluorescent labels, PCR was performed on 100 ng genomic DNA with 1× Phusion High-Fidelity DNA Polymerase Master mix (Thermo Fisher Scientific, Lafayette, CO), 5% DMSO, 1.25M betaine, and 0.2 mM deazaGTP. Products were run on a 2% agarose gel for visual inspection and then analyzed for fragment size determination as described for repeat-primed PCR. Based on our analysis, cross-repeat PCR can amplify alleles with ≤35 GGGGCC repeats but not the 700–1600 repeats reported for pathological expansions14. To investigate repeat expansion segregation, we also genotyped all family members (n=491) from every pedigree with ≥10 hexanucleotide repeats.
Southern Blot
Southern blot hybridization analysis was conducted using previously described methods and probe sequences14 to estimate the number of repeat units in expansion carriers.
Risk Haplotype Genotyping
We also analyzed all repeat expansion carriers for the 24 SNP “at-risk” haplotype that has been associated with pathologically expanded C9ORF72 repeats24. All samples were genotyped with the Illumina Human 610 Beadchip with direct genotyping of all analyzed SNPs. Stringent quality control criteria was applied to remove low-quality SNPs23. We used the entire NIA-LOAD GWAS dataset and the HapMap CEU population as reference populations. MACH25 software was used to phase the 24 SNPs.
Statistical and Bioinformatic Analyses
A linear regression model (SAS) was used to test whether the number of GGGGCC repeats was associated with risk for AD, including age, gender and APOE genotype as covariates. Association with age at onset (AAO) was carried out using the Kaplan-Meier method and tested for significant differences using a Cox proportional hazards model (proc PHREG, SAS) that included gender and APOE.
RESULTS
C9ORF72 repeat expansion frequency and segregation in AD families
We screened 872 unrelated familial AD cases and 888 unrelated controls for expansions of the hexanucleotide repeat in C9ORF72. Five Caucasian individuals with a clinical diagnosis of AD (0.57%) and 1 normal control (age 73) showed an abnormal expansion with repeat-primed PCR, defined as more than 30 repeats with 6 base-pair periodicity and the expected decrementing saw-tooth pattern (Supplementary Figure 1). To better clarify the number of repeats in these patients, we attempted to PCR across the GGGGCC repeat. However, in all 6 individuals, only a single peak representing the normal size allele was obtained. Southern-blot of AD probands showed expansions of 1200–1300 GGGGCC repeats for Families 1, 2 and 3, but only 35–100 repeat units in Families 4 and 5 (data not shown). Regardless of repeat size, all 5 AD cases were found to carry the “at-risk” haplotype associated with pathological repeat expansions in all FTD and ALS patients reported to date (Supplementary Table 1)24. All 5 of these patients had also tested negative for the most common mutations in APP, PSEN1, PSEN2, GRN and MAPT16.
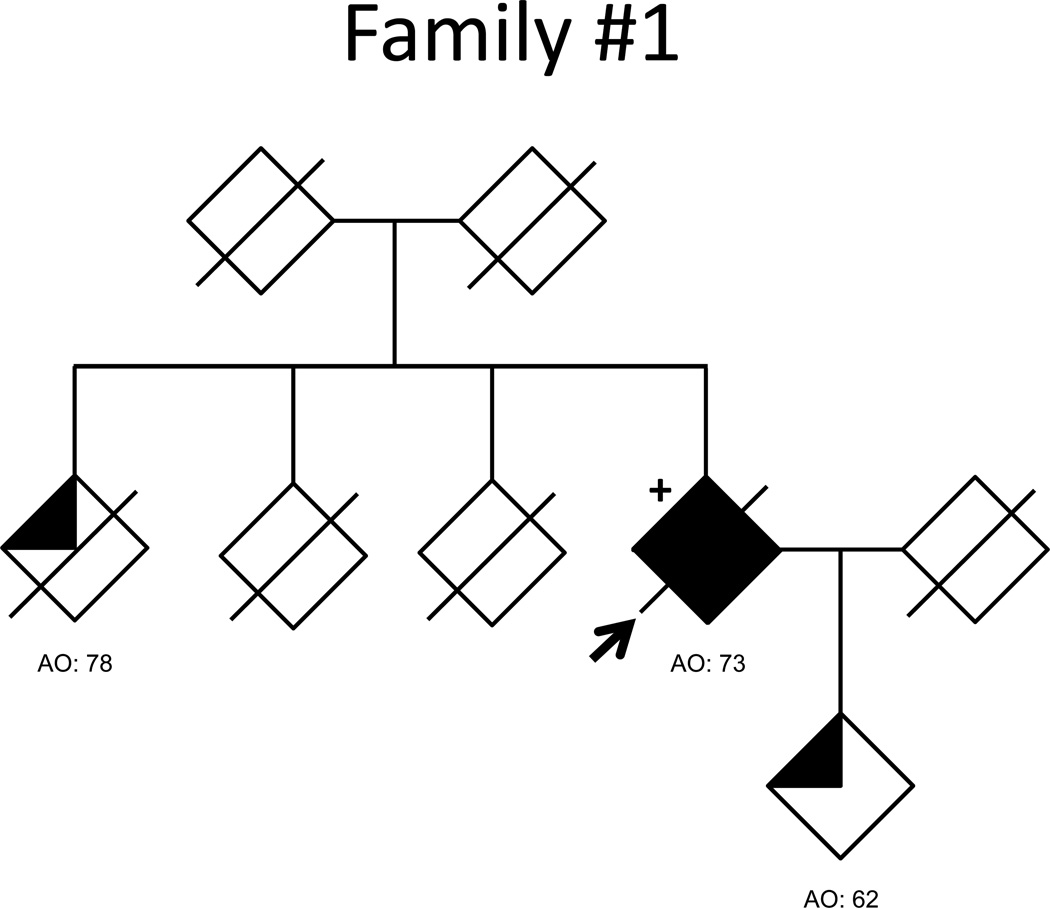
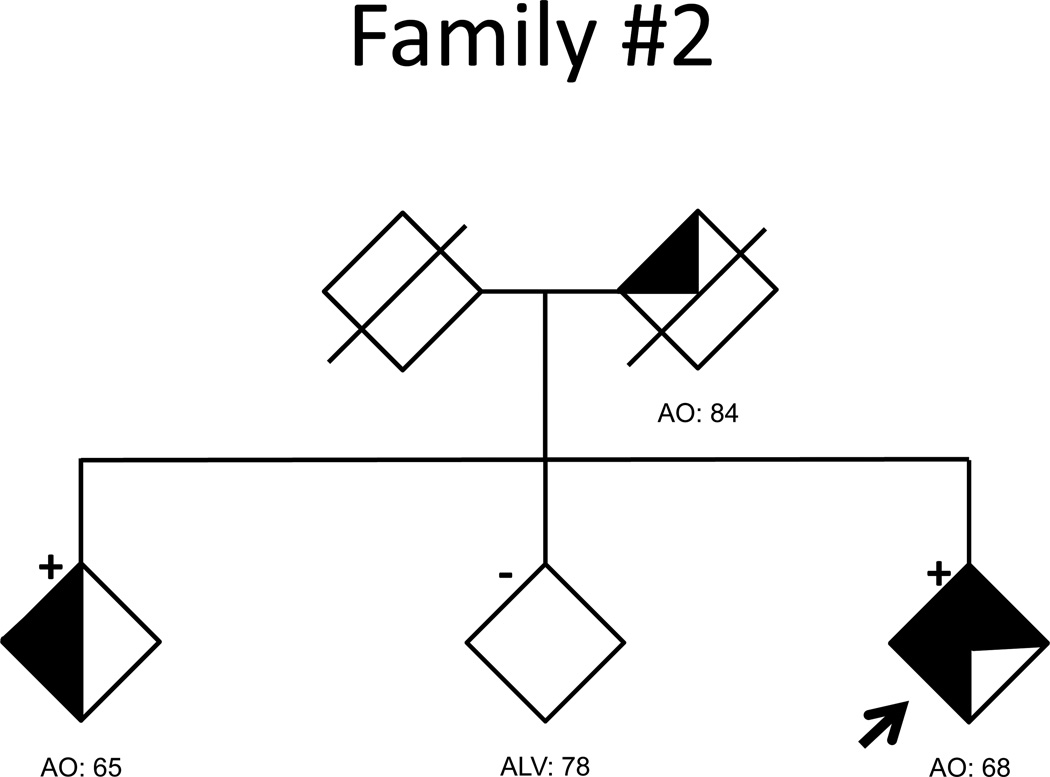
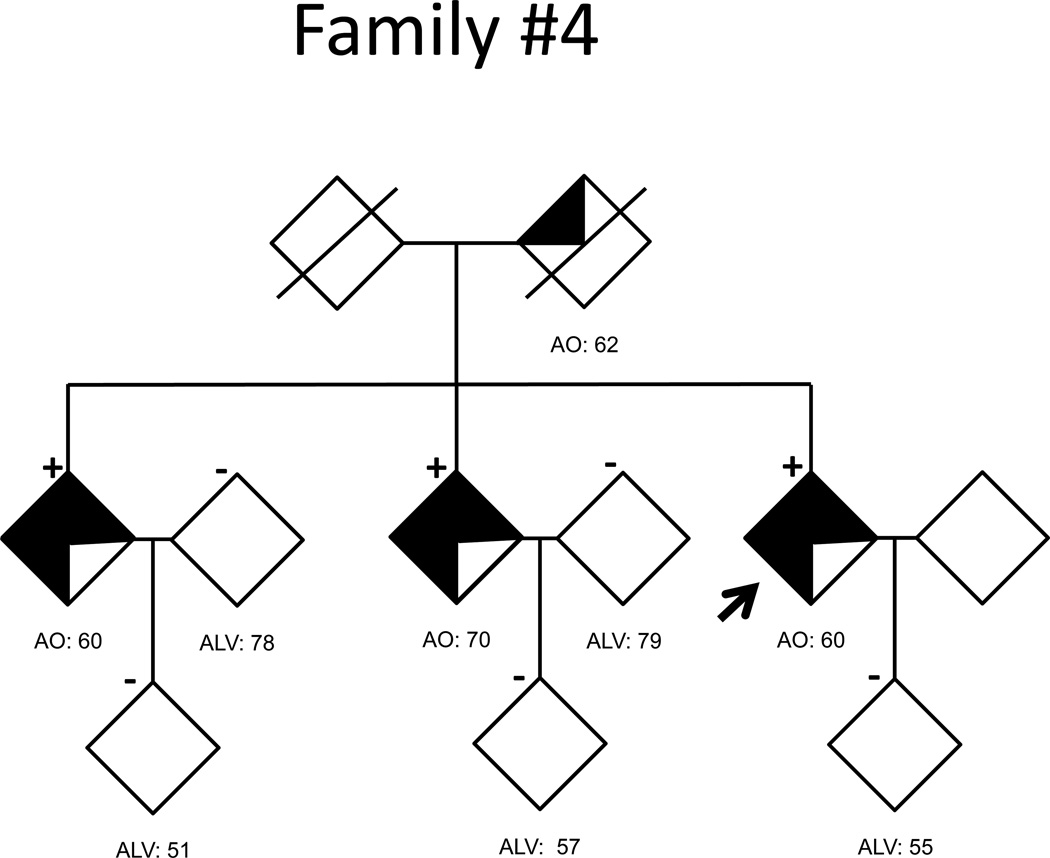
Pedigrees for the 5 families with abnormal (>30) C9ORF72 repeats are shown in the Figure and clinically summarized in Table 2. All additional affected individuals had been diagnosed with probable AD based on the NINCDS-ADRDA criteria. DNA for additional family members was available for all but Family 1. Family 2 showed complete segregation of the large repeat expansion and risk haplotype with disease status (Figure, Supplementary Table 2). Family 3, in which all the affected individuals are APOE 3/3 homozygous, showed segregation of the expanded repeat and risk haplotype except for a single individual who developed dementia with only 7 repeat units. Additionally, this individual did not carry the risk haplotype and had dementia onset at a later age than relatives with repeat expansions. In Families 4 and 5 (repeat expansions between 35 and 100 repeats), the characteristic repeat-primed chromatogram pattern and the risk haplotype segregated perfectly with disease status. Overall, in these five families, 10 of 11 affected individuals for whom DNA was available carried abnormal GGGGCC expansions and the risk haplotype. It is important to note that all the individuals with more than 30 repeats also carried the risk haplotype.
Figure 1. AD pedigrees carrying abnormal C9ORF72 repeat expansions.
Fully shaded symbols indicate autopsy-confirmed AD. Clinical diagnoses were made using NINCDS-ADRDA criteria, with three-quarters and one-half shading indicating probable and possible AD, respectively. Repeat expansion genotypes are indicated by ‘+’ when present, and ‘−‘ when absent. The absence of a genotype symbol indicates that DNA was not available for analysis. An arrow marks the proband of each pedigree.
Table 2.
Segregation analysis of pedigrees with repeat expansions
| Affected with AD | Unaffected | ||||
|---|---|---|---|---|---|
| C9ORF72 repeat expansion | + | − | + | − | |
| Overall | # of Individuals | 10 | 1 | 0 | 8 |
| Mean Age ± SDa | 65.3 ± 4.9 | 65 | 60 | 69.2 ± 11.3 | |
| Age range | 60–73 | 55–79 | |||
| Family 1 | # of Individuals | 1 | 0 | 0 | 0 |
| Ages | 73 | - | - | - | |
| Family 2 | # of Individuals | 2 | 0 | 0 | 1 |
| Ages | 65,68 | - | - | 78 | |
| Family 3 | # of Individuals | 2 | 1 | 0 | 1 |
| Ages | 60 | 65 | - | 72 | |
| Family 4 | # of Individuals | 3 | 0 | 0 | 5 |
| Ages | 60,60,70 | - | - | 55–79 | |
| Family 5 | # of Individuals | 2 | 0 | 0 | 1 |
| Ages | 67,70 | - | - | 72 | |
For AD patients, age refers to age at symptom onset, while for unaffected individuals it refers to the age at last assessment.
The age at onset for individuals with more than 30 repeats was earlier than cases with normal repeat alleles (65.6 ± 5.5 vs 71.4 ± 6.8; p=0.04). This association retained statistical significance even after APOE genotypes were included in the model (p=0.03).
Clinical case and neuropathology descriptions
The proband of Family 1 (1200–1300 repeats), an APOE 2/3 carrier, was diagnosed with dementia, Alzheimer type, at age 73. Formal neurocognitive testing was not available, but prominent memory loss with repetitive questioning and wandering behavior had developed by age 78. Upon death 12 years after diagnosis, a brain autopsy was carried out by the Brain Bank at McLean Hospital (Belmont, MA) in 1999. Microscopic examination found extensive plaque and tangle pathology. Neurofibrillary tangles and neuritic plaques (>40 plaques per 100× field) were present throughout the neocortex, hippocampus, amygdala and nucleus basalis of Meynert. Scattered neocortical Lewy bodies were seen. Severe neuronal loss in the substantia nigra pars compacta was accompanied by a few Lewy bodies. This case would be diagnosed as having NIA-Reagan high likelihood criteria for AD and coexisting neocortical-predominant LBD using current criteria26, 27. Tissue was no longer available to carry out additional relevant staining (e.g. tau, ubiquitin, TDP-43, or p62), preventing an assessment for possible FTD pathology.
Aside from age at symptom onset and meeting clinical criteria for probable AD, limited clinical information was available for the remaining index cases. The proband of Family 2 (1200–1300 repeats) was diagnosed clinically with probable AD at the age of 71, three years after disorientation and memory loss began. By the time of study inclusion at age 74, the patient was non-verbal and too impaired to participate in cognitive testing. The proband of Family 3 (1200–1300 repeats) was diagnosed with probable AD at the age of 60. Three years later, the Clinical Dementia Rating (CDR) was 0.5, which progressed over three years to CDR=1.0. Review of limited caregiver reports suggested difficulties with disinhibition and anxiety were out-of-proportion to memory impairment. No additional clinical information was available for members of Families 4 and 5 (35–100 repeats in each).
Associations of C9ORF72 repeat length in AD
Previous studies in ALS and FTD have clearly demonstrated that C9ORF72 repeat expansions are causative for disease14. However, the minimum repeat number required for disease has not been established. Furthermore, it is not known if higher repeat numbers (but still within the normal range) are associated with risk for ALS or FTD. To address this question in AD, we compared the longest non-expanded allele in cases and controls. The average longest allele was not statistically different from controls (6.5± 4.1 repeat units vs 4.48 ± 3.7, p=0.10), and the distribution of longest allele lengths were similar. Thus within the normal range, higher repeat numbers do not appear to be a risk factor for AD in this population. Furthermore, we found no association between the length of the longest non-expanded allele and age at onset (p=0.52), nor evidence for an interaction with APOE genotype.
Comparison of the C9ORF72 repeat expansion frequency with other pathogenic gene mutations within the same cohort
In a previous study, we sequenced APP, PSEN1, PSEN2, MAPT and GRN genes in a discovery series comprised of 439 cases included in this study16. The most common pathogenic mutation identified by sequencing in the discovery series (PSEN1, A79V), was then genotyped in the entire cohort (follow-up series). Overall the A79V mutation was found in 4 of the 872 cases (0.48%)16 compared to the 5 pedigrees where abnormal C9ORF72 repeat expansions were found. Furthermore, we analyzed the overall frequency of “AD gene” mutations (APP, PSEN1, and PSEN2), vs. FTD gene mutations (MAPT, GRN and C9ORF72), and found that 1.82% of probands carry a pathogenic, or very likely pathogenic, mutation in APP, PSEN1 and PSEN2, while a slightly large number (1.94%) have mutations in MAPT, GRN, or C9ORF72.
DISCUSSION
This study assessed C9ORF72 hexanucleotide expansions in familial late-onset AD cases and normal controls, identifying 5 AD families carrying abnormal C9ORF72 hexanucleotide repeat expansions. This frequency is very similar to that found in an independent AD series15, but significantly lower than in FTD or ALS.
Three families with clinical AD (0.34%) were found to have repeat expansions in the range reported for FTD and ALS (>1000). In a previous report documenting expansions in clinically diagnosed AD, re-evaluation of autopsy material demonstrated FTD pathology and suggested that AD cases with C9ORF72 repeat expansions represent amnestic variants of FTD9, 14, 15, 28, 29. We were unable to perform an equivalent analysis because autopsies had not been performed or tissue was no longer available. Therefore, even in the proband from Family 1, where the neuropathology showed coexisting AD and LBD pathology, we cannot rule out the possibility that the families identified in this study also represent amnestic presentations of FTD rather than AD. It is notable that several C9ORF72 FTD cases reported, several have shown concurrent AD pathology9, 14, 15, 28, 29 and an additional expansion carrier with FTD+ALS demonstrated enough plaque and tangle pathology to meet diagnostic criteria for AD28. These cases suggest that some individuals with C9ORF72 repeat expansions could present with clinical symptoms of AD and have a high enough burden of AD neuropathology that biomarker analysis (cerebrospinal tau or Aβ, and/or PIB-PET neuroimaging) would also support an AD diagnosis. This hypothesis is supported three recently reported individuals with early onset AD, CSF profiles typical of AD30, who were found to carry C9ORF72 repeat expansions. In this setting, the correct diagnosis (amnestic FTD) would presumably only be reached by neuropathologic studies or genetic testing. Our cases and previous studies reinforce the heterogeneous clinical and neuropathological presentations of C9ORF72 repeat expansions (ALS, FTLD, FTLD-ALS, and clinical AD).
We also identified two families carrying smaller, but abnormal repeats (>35, but smaller than 100 units). Despite segregating with disease status, it remains unclear whether these smaller repeat expansions cause disease, increase risk for dementia, or are incidental. Future studies correlating quantified repeat sizes with disease status will be required to answer this question.
Although the frequency of large C9ORF72 repeat expansions is low in our cohort, it is the second most common pathogenic mutation (3/872), just behind PSEN1 A79V (4/872). In addition, mutations in “FTD genes” were as common as mutations in “AD genes” (1.94% compared to 1.87%). Our results confirm that the clinical phenotype of mutations in “FTD genes”, including GRN, MAPT and C9ORF72, can be clinically indistinguishable from typical AD. This fact has important implications for clinicians, who should consider both “FTD” and “AD” genes when evaluating families with strong histories of AD.
Supplementary Material
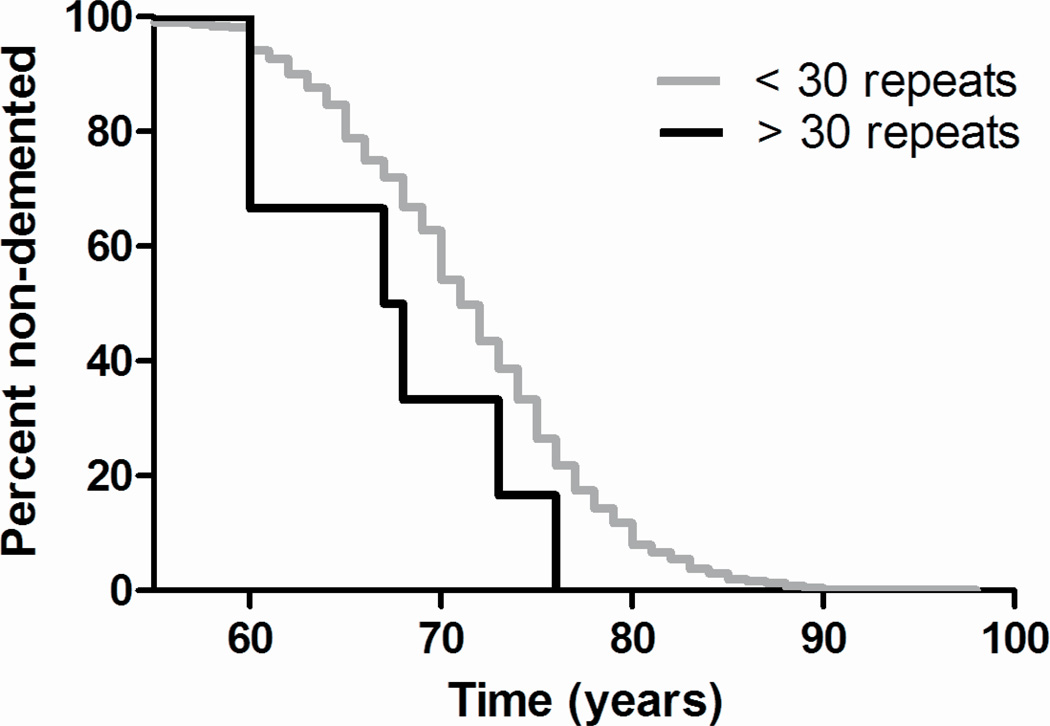
Figure 2. C9ORF72 repeat expansions correlate with an earlier age at onset.
Age at onset (AAO) was analyzed for association with repeat expansion genotypes by the Kaplan-Meier method and tested for significant differences using a proportional hazards model (proc PHREG, SAS). Repeat expansion carriers show an earlier AAO than non-carriers (68.25 ± 5.8 vs 71.4 ± 6.8; p=0.04).
Acknowledgements
This work was supported by grants from the National Institutes of Health (P30-NS069329-01, R01-AG035083, and K08-NS075094). Samples from the National Cell Repository for Alzheimer's Disease (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) were used in this study. We thank contributors, including the Alzheimer's Disease Centers and the Washington University Neuromuscular Genetics Project who collected samples used in this study, as well as patients and their families, whose help and participation made this work possible.
REFERENCES
- 1.Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. Science translational medicine. 2011 Apr 6;3(77):77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rogaeva EA, Fafel KC, Song YQ, et al. Screening for PS1 mutations in a referral-based series of AD cases: 21 novel mutations. Neurology. 2001 Aug 28;57(4):621–625. doi: 10.1212/wnl.57.4.621. [DOI] [PubMed] [Google Scholar]
- 3.Brickell KL, Leverenz JB, Steinbart EJ, et al. Clinicopathological concordance and discordance in three monozygotic twin pairs with familial Alzheimer's disease. J Neurol Neurosurg Psychiatry. 2007 Oct;78(10):1050–1055. doi: 10.1136/jnnp.2006.113803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athan ES, Williamson J, Ciappa A, et al. A founder mutation in presenilin 1 causing early-onset Alzheimer disease in unrelated Caribbean Hispanic families. Jama. 2001 Nov 14;286(18):2257–2263. doi: 10.1001/jama.286.18.2257. [DOI] [PubMed] [Google Scholar]
- 5.Goate A, Chartier-Harlin MC, Mullan M, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer's disease. Nature. 1991 Feb 21;349(6311):704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 6.Cruchaga C, Chakraverty S, Mayo K, et al. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer's disease families. PLoS One. 2012;7(2):e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cortini F, Fenoglio C, Guidi I, et al. Novel exon 1 progranulin gene variant in Alzheimer's disease. Eur J Neurol. 2008 Oct;15(10):1111–1117. doi: 10.1111/j.1468-1331.2008.02266.x. [DOI] [PubMed] [Google Scholar]
- 8.Rademakers R, Baker M, Gass J, et al. Phenotypic variability associated with progranulin haploinsufficiency in patients with the common 1477C-->T (Arg493X) mutation: an international initiative. Lancet Neurol. 2007 Oct;6(10):857–868. doi: 10.1016/S1474-4422(07)70221-1. [DOI] [PubMed] [Google Scholar]
- 9.Brouwers N, Nuytemans K, van der Zee J, et al. Alzheimer and Parkinson diagnoses in progranulin null mutation carriers in an extended founder family. Arch Neurol. 2007 Oct;64(10):1436–1446. doi: 10.1001/archneur.64.10.1436. [DOI] [PubMed] [Google Scholar]
- 10.Lindquist SG, Holm IE, Schwartz M, et al. Alzheimer disease-like clinical phenotype in a family with FTDP-17 caused by a MAPT R406W mutation. European journal of neurology : the official journal of the European Federation of Neurological Societies. 2008 Apr;15(4):377–385. doi: 10.1111/j.1468-1331.2008.02069.x. [DOI] [PubMed] [Google Scholar]
- 11.Rademakers R, Dermaut B, Peeters K, et al. Tau (MAPT) mutation Arg406Trp presenting clinically with Alzheimer disease does not share a common founder in Western Europe. Human mutation. 2003 Nov;22(5):409–411. doi: 10.1002/humu.10269. [DOI] [PubMed] [Google Scholar]
- 12.Momeni P, Pittman A, Lashley T, et al. Clinical and pathological features of an Alzheimer's disease patient with the MAPT Delta K280 mutation. Neurobiology of aging. 2009 Mar;30(3):388–393. doi: 10.1016/j.neurobiolaging.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ludolph AC, Kassubek J, Landwehrmeyer BG, et al. Tauopathies with parkinsonism: clinical spectrum, neuropathologic basis, biological markers, and treatment options. Eur J Neurol. 2009 Mar;16(3):297–309. doi: 10.1111/j.1468-1331.2008.02513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dejesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron. 2011 Oct 20;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renton AE, Majounie E, Waite A, et al. A Hexanucleotide Repeat Expansion in C9ORF72 Is the Cause of Chromosome 9p21-Linked ALS-FTD. Neuron. 2011 Oct 20;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruchaga C, Chakraventy S, Mayo K, Vallania FL, Mitra A, Faber KM. Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s Disease families. PLoS One. 2012 doi: 10.1371/journal.pone.0031039. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kelley BJ, Haidar W, Boeve BF, et al. Alzheimer disease-like phenotype associated with the c.154delA mutation in progranulin. Arch Neurol. 2010 Feb;67(2):171–177. doi: 10.1001/archneurol.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reed LA, Grabowski TJ, Schmidt ML, et al. Autosomal dominant dementia with widespread neurofibrillary tangles. Ann Neurol. 1997 Oct;42(4):564–572. doi: 10.1002/ana.410420406. [DOI] [PubMed] [Google Scholar]
- 19.Majounie E, Abramzon Y, Renton AE, et al. Repeat expansion in C9ORF72 in Alzheimer's disease. N Engl J Med. 2012 Jan 19;366(3):283–284. doi: 10.1056/NEJMc1113592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rollinson S, Halliwell N, Young K, et al. Analysis of the hexanucleotide repeat in C9ORF72 in Alzheimer's disease. Neurobiol Aging. 2012 Mar 10; doi: 10.1016/j.neurobiolaging.2012.01.109. [DOI] [PubMed] [Google Scholar]
- 21.Berg L, McKeel DW, Jr, Miller JP, et al. Clinicopathologic studies in cognitively healthy aging and Alzheimer's disease: relation of histologic markers to dementia severity, age, sex, and apolipoprotein E genotype. Arch Neurol. 1998 Mar;55(3):326–335. doi: 10.1001/archneur.55.3.326. [DOI] [PubMed] [Google Scholar]
- 22.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984 Jul;34(7):939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 23.Wijsman EM, Pankratz ND, Choi Y, et al. Genome-wide association of familial late-onset Alzheimer's disease replicates BIN1 and CLU and nominates CUGBP2 in interaction with APOE. PLoS genetics. 2011 Feb;7(2):e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mok K, Traynor BJ, Schymick J, et al. Chromosome 9 ALS and FTD locus is probably derived from a single founder. Neurobiol Aging. 2012 Jan;33(1):209, e203–e208. doi: 10.1016/j.neurobiolaging.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Y, Abecasis GR. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am J Hum Genet. 2006;S79:2290. [Google Scholar]
- 26.Hansen L, Salmon D, Galasko D, et al. The Lewy body variant of Alzheimer's disease: a clinical and pathologic entity. Neurology. 1990 Jan;40(1):1–8. doi: 10.1212/wnl.40.1.1. [DOI] [PubMed] [Google Scholar]
- 27.Forstl H, Burns A, Luthert P, Cairns N, Levy R. The Lewy-body variant of Alzheimer's disease. Clinical and pathological findings. Br J Psychiatry. 1993 Mar;162:385–392. doi: 10.1192/bjp.162.3.385. [DOI] [PubMed] [Google Scholar]
- 28.Murray ME, DeJesus-Hernandez M, Rutherford NJ, et al. Clinical and neuropathologic heterogeneity of c9FTD/ALS associated with hexanucleotide repeat expansion in C9ORF72. Acta Neuropathol. 2011 Dec;122(6):673–690. doi: 10.1007/s00401-011-0907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boeve BF, Boylan KB, Graff-Radford NR, et al. Characterization of frontotemporal dementia and/or amyotrophic lateral sclerosis associated with the GGGGCC repeat expansion in C9ORF72. Brain. 2012 Mar;135(Pt 3):765–783. doi: 10.1093/brain/aws004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallon D, Rovelet-Lecrux A, Deramecourt V, et al. Definite Behavioral Variant of Frontotemporal Dementia with C9ORF72 Expansions Despite Positive Alzheimer's Disease Cerebrospinal Fluid Biomarkers. J Alzheimers Dis. 2012 Jul 5; doi: 10.3233/JAD-2012-120877. [DOI] [PubMed] [Google Scholar]
- 31.Tolboom N, Koedam EL, Schott JM, et al. Dementia mimicking Alzheimer's disease Owing to a tau mutation: CSF and PET findings. Alzheimer Dis Assoc Disord. 2010 Jul-Sep;24(3):303–307. doi: 10.1097/WAD.0b013e3181cf35ec. [DOI] [PubMed] [Google Scholar]
- 32.Lindquist SG, Schwartz M, Batbayli M, Waldemar G, Nielsen JE. Genetic testing in familial AD and FTD: mutation and phenotype spectrum in a Danish cohort. Clin Genet. 2009 Aug;76(2):205–209. doi: 10.1111/j.1399-0004.2009.01191.x. [DOI] [PubMed] [Google Scholar]
- 33.Carecchio M, Fenoglio C, De Riz M, et al. Progranulin plasma levels as potential biomarker for the identification of GRN deletion carriers. A case with atypical onset as clinical amnestic Mild Cognitive Impairment converted to Alzheimer's disease. J Neurol Sci. 2009 Dec 15;287(1–2):291–293. doi: 10.1016/j.jns.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 34.Brouwers N, Sleegers K, Engelborghs S, et al. Genetic variability in progranulin contributes to risk for clinically diagnosed Alzheimer disease. Neurology. 2008 Aug 26;71(9):656–664. doi: 10.1212/01.wnl.0000319688.89790.7a. [DOI] [PubMed] [Google Scholar]
- 35.Ostojic J, Elfgren C, Passant U, et al. The tau R406W mutation causes progressive presenile dementia with bitemporal atrophy. Dement Geriatr Cogn Disord. 2004;17(4):298–301. doi: 10.1159/000077158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.