Abstract
Heat shock protein 90 (HSP90) is involved in protein folding and functions as a chaperone for numerous client proteins, many of which are important in non-small cell lung cancer (NSCLC) pathogenesis. We sought to define preclinical effects of the HSP90 inhibitor NVP-AUY922 and identify predictors of response. We assessed in vitro effects of NVP-AUY922 on proliferation and protein expression in NSCLC cell lines. We evaluated gene expression changes induced by NVP-AUY922 exposure. Xenograft models were evaluated for tumor control and biological effects. NVP-AUY922 potently inhibited in vitro growth in all 41 NSCLC cell lines evaluated with IC50 < 100 nM. IC100 (complete inhibition of proliferation) < 40 nM was seen in 36 of 41 lines. Consistent gene expression changes after NVP-AUY922 exposure involved a wide range of cellular functions, including consistently decreased dihydrofolate reductase (DHFR) after exposure. NVP-AUY922 slowed growth of A549 (KRAS mutant) xenografts, and achieved tumor stability and decreased epidermal growth factor receptor (EGFR) protein expression in H1975 xenografts, a model harboring a sensitizing and a resistance mutation for EGFR tyrosine kinase inhibitors in the EGFR gene. This data will help inform the evaluation of correlative data from a recently completed phase II NSCLC trial and a planned phase IB trial of NVP-AUY922 in combination with pemetrexed in NSCLC.
Keywords: NVP-AUY922, HSP90, Lung Cancer
Introduction
Heat shock proteins (including HSP90) serve as molecular chaperones required for stability, post-translation modification, and function of multiple client proteins.(1) Expression of heat shock proteins (HSP) is increased at times of physiologic stress, and these effects are believed to support cell survival. Preclinical data indicates that malignant cells have increased levels of active HSP90, and mutated oncogenic proteins are more reliant on HSP90 function.(1) Increased HSP90 expression has been linked to worse prognosis in NSCLC patients.(2) Based on these data, as well as a long list of client proteins important in malignancy,(3) HSP90 has emerged a promising therapeutic target.(1)
HSP90 operates in an ATP-dependent manner with several co-chaperones. HSP90 inhibitors developed to date block the N terminal ATP binding pocket, altering normal HSP90 function and leading to release of client proteins with eventual destruction via the ubiquitin-proteosome pathway.(1) Inhibition of HSP90 releases heat shock factor 1 (HSF1), a transcription factor which controls HSP expression.(4) This response increases expression of other family members, including HSP70 and HSP72, limiting the biologic effects of HSP90 inhibition.(4) Induction of this response is a pharmacodynamic endpoint to evaluate target inhibition by HSP90 inhibitors. Based on the wide range of client proteins, HSP90 inhibitors are expected to have diverse molecular effects.
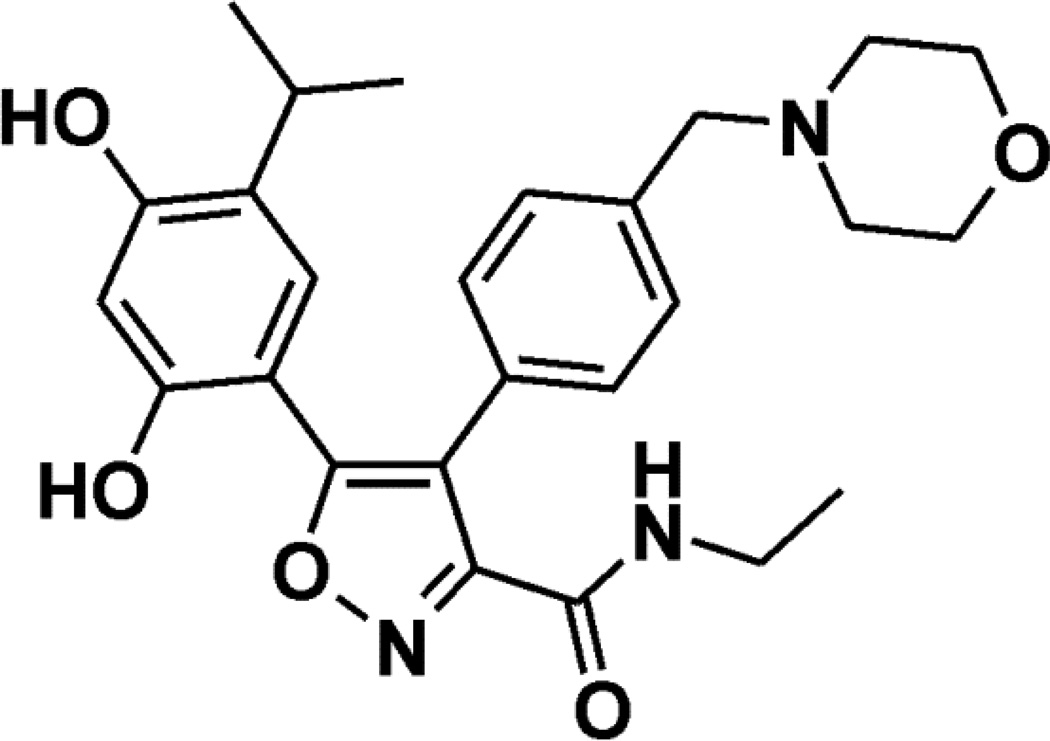
Naturally occurring HSP90 inhibitors include the geldanamycin class, of which 17-AAG has been most extensively evaluated. Phase I and II trials evaluating 17-AAG demonstrate evidence of activity.(5–9) Other synthetic inhibitors have been developed, including NVP-AUY922, a novel isoxazole-based inhibitor (Figure 1).(10) The compound inhibits HSP90α and β subunits at low nM concentrations, whereas the other two HSP90 family members Grp94 and Trap-1 are inhibited at 25–40 fold higher concentrations.(11, 12) A phase I trial of NVP-AUY922 demonstrated tolerability with prolonged disease stabilization in some patients.(13)
Figure 1. Chemical structure of NVP-AUY922.
The chemical structure of NVP-AUY922 is shown
It is now recognized that NSCLC arises as a result of several driver mutations.(14) Epidermal growth factor receptor (EGFR) mutations, particularly deletions in exon 19 or a point mutation in exon 21 (L858R), are seen in approximately 10% of NSCLC and are associated with a favorable prognosis(15–18) and radiographic response to EGFR tyrosine kinase inhibitors (TKIs).(19–21) Cell lines with activating EGFR mutations are reliant on HSP90 function for protein stabilization and undergo growth inhibition when HSP90 function is inhibited.(22, 23) HSP90 inhibition can overcome the T790M secondary mutation which confers resistance to EGFR TKIs in preclinical models.(23, 24) Other important proteins considered relevant in EGFR mutant NSCLC are HSP90 clients.(25, 26) c-RAF is an HSP90 client protein, and this has been hypothesized as an explanation of sensitivity among ras mutant NSCLC models.(27, 28) ALK gene-rearrangements have emerged as an important target in NSCLC.(29) This abnormality has been successfully targeted by the ALK inhibitor crizotinib.(30) Two studies of HSP90 inhibitors (IPI-504 and ganetespib) have shown clinical responses among patients with ALK gene-rearrangements.(31, 32) Phase II studies using NVP-AUY922 are now underway, and have shown radiographic responses in EGFR mutant NSCLC, ALK gene-rearranged NSCLC as well as patients with neither of these molecular abnormalities. (33, 34)
Materials and Methods
Cell lines, cell cultures and reagents
NVP-AUY922 was studied in 41 human NSCLC cell lines in vitro: A427, A549, Calu-1, Calu-3, Calu-6, H23, H226, H358, H460, H520, H522, H596, H810, H838, H1155, H1299, H1385, H1435, H1437, H1563, H1568, H1581, H1651, H1703, H1755, H1793, H1836, H1944, H1975, H2030, H2073, H2106, H2110, H2135, H2172, H2228, H2286, H2342, H2405, HCC827, and SHP77, all of which were obtained from ATCC and identity was confirmed by evaluating single tandem repeats using Cell ID System, Part#TM074 (Promega, Madison, Wisconsin), with comparison to the ATCC database within 3 months of the described experiments.
A549 was cultured in HAM’s F12 (ATCC) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% penicillin G-streptomycin-fungizone solution (PSF, Irvine Scientific, Santa Ana, CA). Calu-3 was grown in EMEM (ATCC) supplemented with 10% heat-inactivated FBS and 1% PSF. H1155, H1435, H1581 (10% FBS), and H2286 and H2405 (2% BSA) were grown in ACL-4. H1793, H1836, H2342, and H810 were grown in HITES supplemented with 5% heat-inactivated FBS and PSF. The remaining lines were cultured in RPMI 1640 (Cellgro, Manassas, VA) supplemented with 10% heat-inactivated FBS, 2 mmol/L glutamine (Invitrogen, Carlsbad, CA), and PSF.
Microarray data preparation
Agilent microarray analyses were used to assess baseline gene expression for 34 of the cell lines (data was unavailable for the remaining seven lines).(35, 36) Briefly, cells were grown to log phase. RNA was extracted using the RNeasy Kit (Qiagen, Germantown, MD). Purified RNA was eluted in 30–60 µl DEPC water, and the quantity of RNA was measured by spectral analysis using the Nanodrop Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). RNA separation via capillary electrophoresis using the Agilent 2000 Bioanalyzer was performed to determine RNA quality.
Baseline microarrays were performed on Agilent Human 1A V2 chips. Individual cell lines were characterized by comparison to a reference pool (consisting of equal amounts of RNA from 45 NSCLC cell lines) on a single slide in which the mixed pool RNA was labeled with cyanine-3 and the individual cell lines with cyanine-5.(37)
A subset of eight cell lines representative of different subtypes of NSCLC (A549, H1155, H1435, H1793, H1975, H2172, H23, and HCC827) were evaluated at baseline and after exposure (1, 24 and in some cases 48 hours) to NVP-AUY922 at different concentrations (10, 50 and 100 nM). In each case, RNA from a cell line prior to treatment was labeled with cyanine-3 while the corresponding post-treatment sample was labeled with cyanine-5 using Agilent whole human genome 4×44 K chips.
Microarray slides were read using an Agilent Scanner. Calculation of gene expression values was performed using Agilent Feature Extraction software version 7.5. Extracted data was imported into Rosetta Resolver 5.1 to create expression profiles for each individual cell line experiment. Cluster analysis was performed in Resolver, and cell line profiles were exported to Excel (Microsoft) for additional analysis of the distribution of gene expression changes across the various subtypes and the individual cell line response data. Baseline (GSE 43567) and treatment (GSE 43568) array data has been submitted and accepted in the GEO repository.
Statistical Methods
Baseline gene expression analyses: Cell lines were separated into “sensitive” and “resistant” groups based on IC100 values. Only probes with greater than two fold change in expression and a p-value <0.01 in at least one experiment were analyzed. Multiple probes identifying a single gene are reported as a single gene. For correlations between expression of protein levels and NVP-AUY922 sensitivity, Pearson’s χ2 test was performed.
Pre vs. post-treatment expression analysis (two types of analyses were performed):
To determine effects of NVP-AUY922 on global gene expression, a direct comparison was performed by hybridizing RNA from exposed and unexposed lines to the same array. Changes in gene expression were considered significant if the fold change was greater than 2 with a multitest corrected (Benjamini-Hochberg) p-value < 0.01.
To identify genes for which mean expression intensity differed between two conditions on separate arrays (e.g. differences between sensitive and resistant cell lines at 24 hours), we performed a one-way Analysis of Variance (ANOVA) with multitest correction using the Benjamini-Hochberg FDR with a corrected p-value cutoff of 0.05.
Proliferation assays
Cells were seeded in duplicate at 5,000–10,000 cells per well. The day after plating (day 1), NVP-AUY922 was added at 1 µM and 2-fold dilutions over 12 concentrations. Data were compared to untreated controls. Cells were counted on the day drug was added and five days later and these two counts were compared. Cells were harvested by trypsinization and counted immediately using a Coulter Z2 particle counter (Beckman Coulter Inc, Fullerton, CA). Percent growth inhibition, defined as 100 × (1 − [generations in treated wells/generations in untreated controls]) was determined. Experiments were performed in duplicate. Error bars represent standard error for each experiment. Non-linear curve fitting was performed by fitting curves to data points using the Proc NLIN function in SAS for Windows version 9.2 (SAS Institute, Inc., Cary, NC) using a basic four-parameter sigmoid model. IC50 (concentration needed to prevent 50% of cell population doublings) and IC100 (complete inhibition of proliferation) were the summary outcome measures interpolated from the resulting curves.
Western blots
The same subset of 8 cell lines evaluated for pre- and post-exposure gene expression (A549, H23, H1435, H2172, HCC827, H1155, H1793, and H1975) growing in log-phase were exposed to media with or without 50 and 100 nM NVP-AUY922 for 30 minutes, 18 and 24 hours prior to cell lysis (the only exception being dihydrofolate reductase (DHFR), which was exposed to 50 nM NVP-AUY922 for 1, 24 and 48 hours to evaluate the timepoints used in the microarray experiment). Cells were washed in ice cold PBS and lysed at 4°C in lysis buffer. Insoluble material was cleared by centrifugation at 10,000g for 10 min. Protein was quantitated using BCA (Pierce Biochemicals, Rockford, IL, USA), resolved by SDS-PAGE, and transferred to nitrocellulose membranes (Invitrogen, Carlsbad, CA, USA) with 1–2 cell lines per blot. Total ERK expression was detected by the monoclonal antibody p44/42 map Kinase antibody tERK (Cell Signaling, Beverly, MA, USA). Phospho-ERK expression was detected by the monoclonal anti-phospho-ERK antibody phospho-44/42 map Kinase (Thr 202/Tyr 204) antibody pERK (Cell Signaling). Total AKT expression was detected by the monoclonal antibody Total AKT antibody #9272 (Cell Signaling). Expression of phosphorylated AKT at serine 308 and serine 473 were detected by the monoclonal antibodies Phospho AKT (Ser308) and phospho AKT (Ser 473) respectively (Cell Signalling). HSP70 expression was detected by the monoclonal antibody #SPA-810 Hsp70 (Assay Designs, Ann Arbor, MI). DHFR expression was detected by monoclonal antibody #ab124814 (Abcam, Cambridge, MA). Quantification for DHFR was performed by comparing each timepoint to control normalized to tubulin using Alpha Innotech AlphaView® Software version 3.0.3.0 (Santa Clara, CA). Tubulin expression was detected by α-Tubulin antibody #2144 (Cell Signaling). In H1975 xenograft tumors, Phospho-AKT levels were detected with a phospho-Akt (Ser473) antibody (#9271; Cell Signaling Technology, Inc.), total AKT levels were detetcted with a total Akt antibody (#9271; Cell Signaling Technology, Inc.), EGFR expression was detected using an anti-HER1 antibody (Cell Signalling #2232) and β-actin was detected using a monoclonal antibody (Chemicon # MAB1501).
Tumor Xenografts
To establish tumor xenografts, A549 (107) or H1975 (5 × 106) cells were injected in 200 µl HBSS subcutaneously in the right flank. Fragments from established H1975 tumors (app. 30 mm3) were serially transplanted subcutaneously onto recipient mice which were used for efficacy studies. When tumors reached a size of 100–200 mm3, animals were randomized into treatment groups (n=8) and intravenous NVP-AUY922 was initiated weekly (qw) or thrice weekly (3qw). Tumor volumes (measured manually with calipers) and body weights were measured 2–3 times/week and presented as means for each group. Error bars represent the standard error of the mean. Tumors volume was estimated using the formula: (L × W2 × π/6), where width (W) and length (L) are the two largest diameters. Tumor and body weight data were analyzed by one-way ANOVA with post hoc Dunnet’s test for comparison of treatment vs. control group. The post hoc Tukey test was used for intragroup comparison. Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software, CA, USA). As a measure of efficacy, the %T/C value is calculated at the end of the experiment according to: (Δtumor volumetreated/Δtumor volumecontrol) × 100; where Δtumor volumes represent the mean tumor volume on the evaluation day minus the mean tumor volume at the start of the experiment. Invasive procedures were performed under Forene anesthesia. All experiments were performed using female Harlan HsdNpa: Athymic Nude-nu mice which were obtained from Novartis internal breeding stocks (Laboratory Animal Services, Novartis Pharma AG, Basel, Switzerland). Animals were kept under optimized hygienic conditions (OHC) with a 12 hour dark, 12 hour light cycle. Animals were fed food and water ad libitum. All animal experiments were performed in adherence to the Swiss law for animal protection. The experimental protocols were approved by the Swiss Kantonal Veterinary Office of Basel Stadt, Basel, Switzerland.
Results
NVP-AUY922 potently inhibits NSCLC growth in vitro
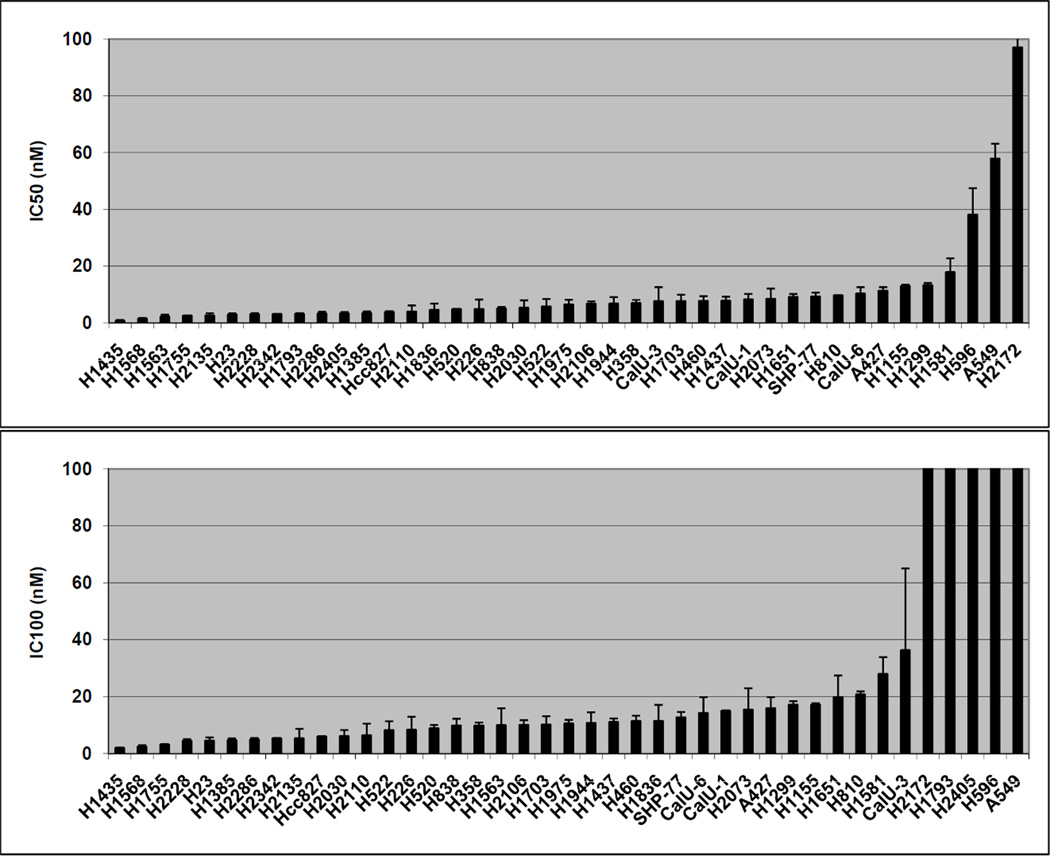
All 41 cell lines were sensitive to the anti-proliferative effects of NVP-AUY922, with IC50 < 100 nM (Figure 2A). In contrast to near-uniformity in sensitivity, relative differences were seen with regard to IC100 (complete inhibition of proliferation) among cell lines (Figure 2B). 36 cell lines had an IC100 < 40 nM while 5 had an IC100 > 200 nM.
Figure 2. In vitro sensitivity to NVP-AUY922.
41 cell lines with IC50 represented in nM (A) and IC100 represented in nM (B). Error bars represent SE based on multiple experiments
Western Blot of NSCLC cell lines demonstrates that differences in protein expression correlate with in vitro sensitivity to NVP-AUY922
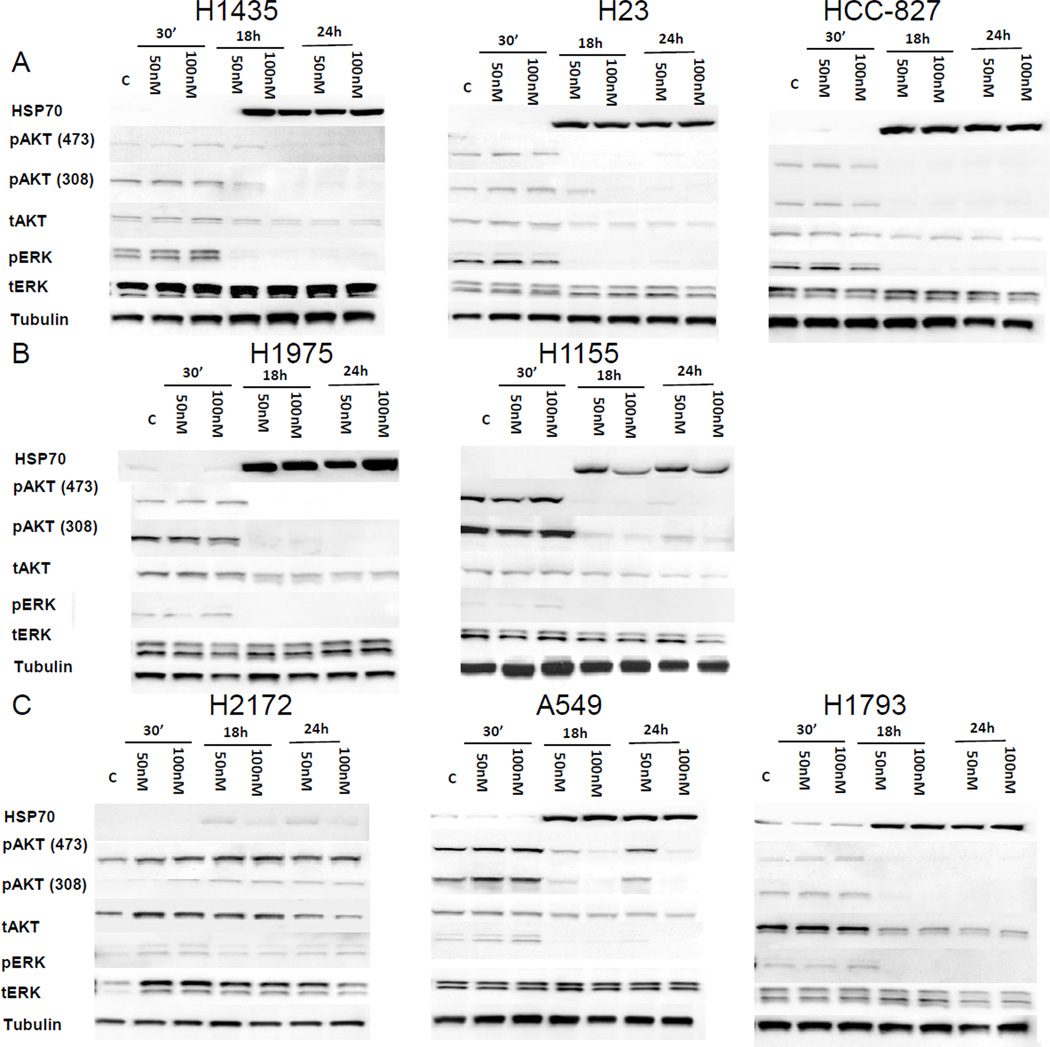
To assess the biochemical effects of NVP-AUY922 and predictors of in vitro sensitivity, Western blots were performed to assess HSP70, total and phospho ERK (pERK) and total and phospho AKT (pAKT: serine 308 or 473) at baseline and after 50 and 100 nM NVP-AUY922 exposure for 30 minutes, 18 and 24 hours (Figure 3). 8 cell lines were selected for this analysis based on NVP-AUY922 sensitivity and molecular characteristics. Three harbored KRAS mutations (A549, H23, and H1155) and two harbored EGFR mutations, HCC827 which is sensitive to EGFR TKIs and H1975 which is resistant. Evaluated lines included 3 with IC50 < 5 nM and IC100 < 40 nM (Figure 3A), 2 with IC50 > 5 nM and IC100 < 40 nM (Figure 3B) and 3 with IC100 > 200 nM (Figure 3C). Little change in protein levels was seen 30 minutes after exposure, but HSP70 was reliably increased in all evaluated cell lines after 18 hours. After NVP-AUY922 exposure, pERK and pAKT were inhibited to a high degree in cell lines with IC100 < 40 nM (Figure 3A, 3B). Among cell lines with IC100 > 200 nM, pAKT and pERK inhibition was more variable (Figure 3C).
Figure 3. Immunoblot evaluation evaluating effects of NVP-AUY922 in vitro.
Immunoblot evaluation for cell lines with IC50 < 5 nM and IC100 < 40 nM (A), IC50 > 5 nM and IC100 < 40 nM (B) and IC100 > 40 nM (C) are shown. Measured proteins include HSP70, pERK 1/2 (Thr202/Tyr204), total ERK1/ERK2, pAKT (Ser473/Ser308), total AKT, and Tubulin at baseline and after 30 minutes, 18 and 24 hours of treatment with 50 nM or 100 nM of NVP-AUY922 as indicated.
NVP-AUY922 induces stereotyped changes in gene expression
Changes in gene expression after NVP-AUY922 exposure were evaluated in the 8 cell lines described in the preceding section and over a range of exposure times (1, 24 and in some lines 48 hours) and concentrations (10, 50 and 100 nM). For each cell line, exposed and unexposed samples were compared on a single microarray with the unexposed sample serving as the control.
Time dependent gene expression changes were evaluated. 86 genes were more than two-fold changed in expression with unadjusted p-value < 0.01 in at least one experiment after one hour of 100 nM NVP-AUY922 exposure (data not shown), including increases in HSP family members HSPA1A (HSP70 protein A1) and HSPA6 (HSP70B). However, these results did not meet the cut-off for statistical significance (FDR < 0.01). More significant changes in gene expression occurred by 24 hours, and changes at 24 hours correlated well with 48 hours with slightly more pronounced changes after longer exposure (data not shown).
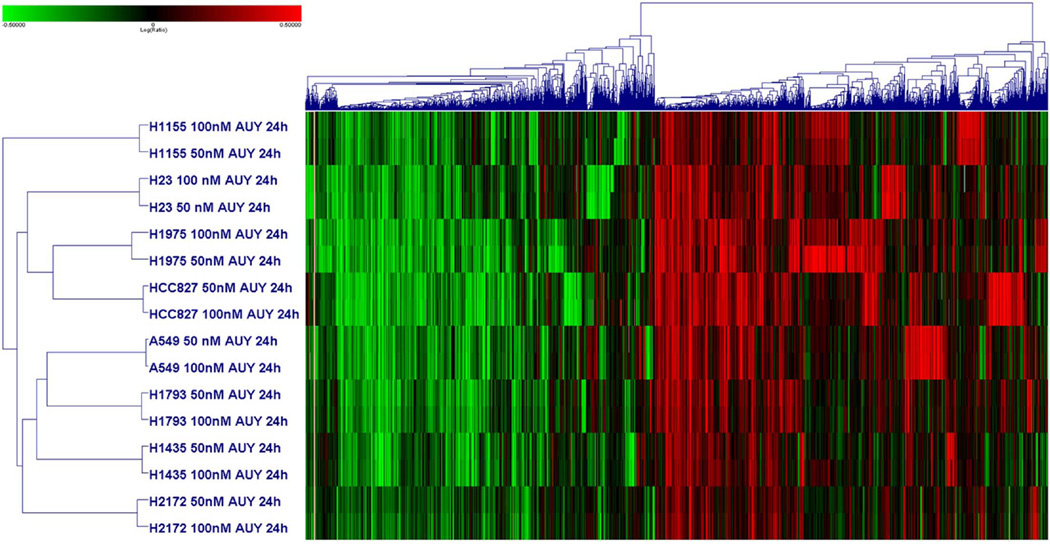
Concentration dependent gene expression changes were less pronounced. Changes in response to 10 nM of NVP-AUY922 generally mirrored those at higher concentrations but were less pronounced (data not shown). Changes in response to 50 nM and 100 nM NVP-AUY922 were very similar (correlation > 0.9 for each cell line). Thus, data from 50 nM and 100 nM experiments were pooled for analysis. After a 24 hour exposure to 50 or 100 nM NVP-AUY922, 7078 genes demonstrated at least twofold change in expression and a p-value < 0.01 in at least one cell line without FDR (Figure 4). Expression of 11 genes were significantly changed in all 16 cell lines evaluated and included the up-regulated HSPA1A (HSP70), ENST00000330775, THC1872885, C1orf63 and BX428745 genes and the down-regulated IER2, MCM7, EGR1, TNFRSF12A, C15orf39 and VDR genes. We identified genes whose expression significantly changed in at least 13 of the 16 experiment to account for outliers and false positives, yielding 69 genes including dihydrofolate reductase (DHFR), which decreased in response to therapy (Supplemental Figure 1).
Figure 4. Heat maps from microarray analyses of gene expression changes after exposure to NVP-AUY922.
Heatmap showing hierarchical clustering of 8 cell lines after 24 hour exposure to either 50 or 100 nM of NVP-AUY922 using 7078 genes that demonstrated greater than twofold expression change at a p-value < 0.01 in at one experiment.
Using PANTHER (Protein ANalysis THrough Evolutionary Relationships) gene ontology,(38) pathways with the greatest change in expression in response to NVP-AUY922 were the angiogenesis (p = 1.65 × 10−5), TGF-β signaling (p = 6.92 × 10−5) and EGFR (p = 9.88 × 10−5) pathways. The most significant effects with respect to molecular function were catalytic activity (p = 1.16 × 10−18), binding (p = 1.36 × 10−18) and protein binding (p = 2.56 × 10−16).
Western Blot confirms decreased DHFR expression in many cell lines after exposure to NVP-AUY922, particularly at 24 hours
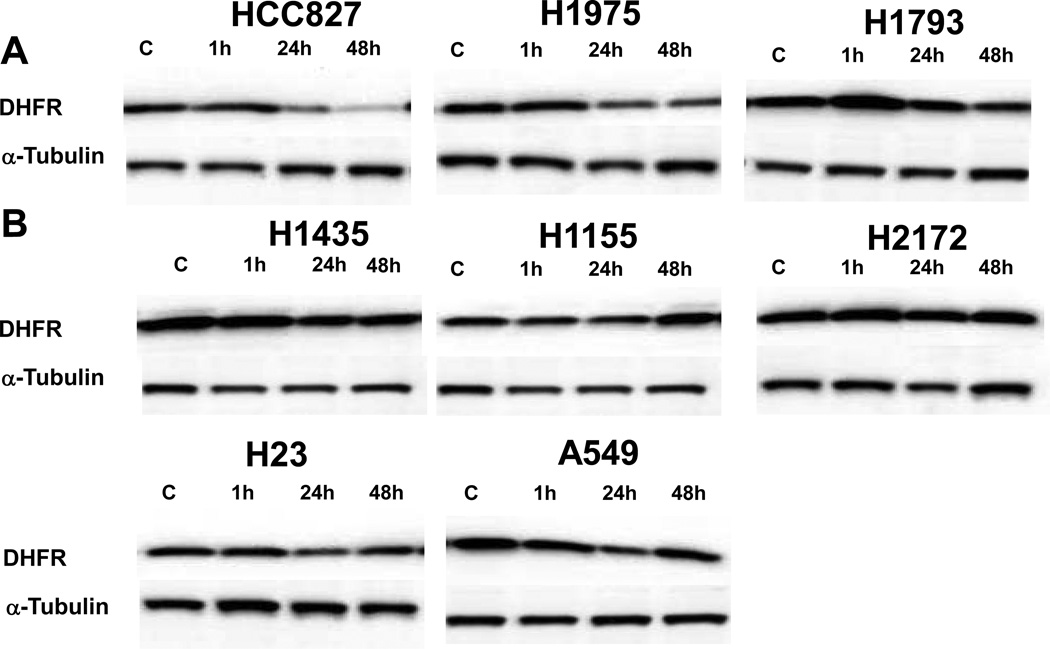
To evaluate whether the decreased DHFR expression seen in the microarray experiments led to decreased DHFR protein levels, the eight cell lines evaluated in the microarray experiments were exposed to 50 nM of NVP-AUY922 at the same time points (1 hour, 24 hour and 48 hour) as the microarray experiment. Western blots were performed to assess DHFR compared to an α-tubulin control (Figure 5). Three of the cell lines showed levels of DHFR less than 40% of baseline when compared to a tubulin control after 48 hours of NVP-AUY922 exposure (Figure 5A). The other five cell lines did not show as significant reductions in DHFR levels after NVP-AUY922 exposure (Figure 5B). DHFR levels did not predict sensitivity among the cell lines.
Figure 5. Immunoblot evaluation of dihydrofolate reductase (DHFR) in response to NVP-AUY922 exposure.
Immunoblot evaluation for DHFR as compared to α-tubulin after exposure to 50 nM of NVP-AUY922 for 1, 24 and 48 hours as compared to controls (C). The eight cell lines evaluated by microarray experiments are shown, including three cell lines with DHFR levels less than 40% of baseline when compared to a tubulin control at 48 hours (A) and five cell lines with less or no decrease in DHFR expression after exposure (B).
Baseline gene expression predicts NVP-AUY922 sensitivity
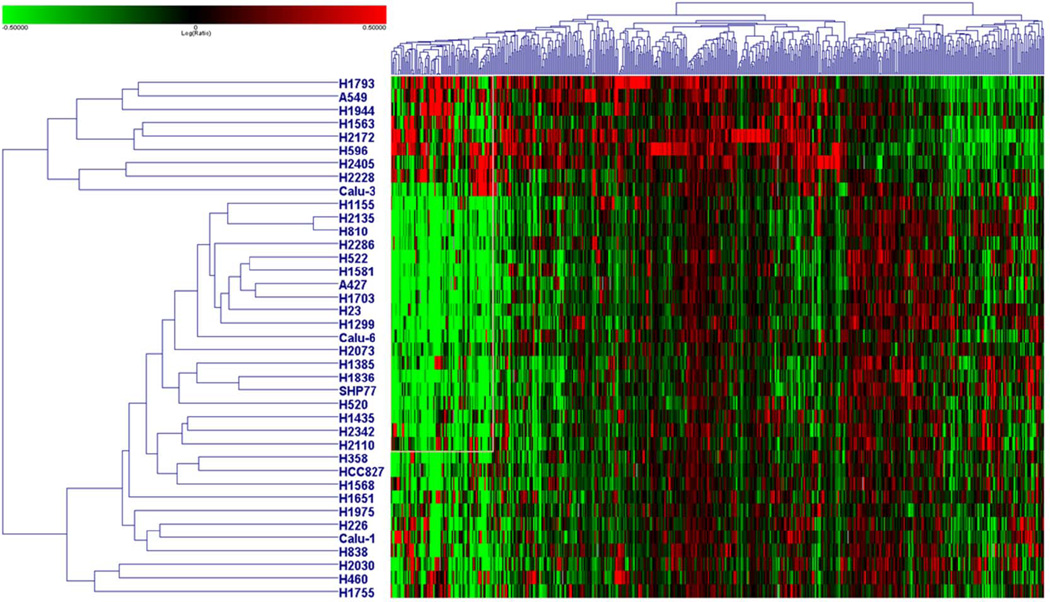
To identify genes associated with NVP-AUY922 sensitivity, baseline gene expression was compared between all 5 resistant lines (IC100 > 200 nM) and the 29 sensitive lines (IC100 < 40 nM) for which baseline gene expression data was available. For each baseline array, cell line specific RNA was compared to a control reference mixture of pooled NSCLC cell line RNA. Probes with a greater than two fold deviation from control and p < 0.01 were considered significant and used in the ANOVA analysis comparing sensitive and resistant cell lines. 521 genes were identified (p < 0.01 without FDR) (Figure 6). When the same analysis was repeated using FDR multi-test correction, 4 probes retained significance (increased expression of KIAA1324L and decreased expression of THC1923177, DSG3 and PI3 in sensitive lines as compared to resistant lines), but relevance to sensitivity after NVP-AUY922 exposure is not clear.
Figure 6. Heatmap showing baseline gene expression differences based on NVP-AUY922 sensitivity.
Heatmap showing hierarchical clustering of 34 cell lines using 521 genes associated with NVP-AUY922 sensitivity. Genes were identified using ANOVA comparing baseline arrays of 5 resistant (IC100 > 200 nM; listed on the top of the heatmap) to 29 sensitive cell lines (IC100 < 40 nM).
Gene expression changes in response to NVP-AUY922 correlate with sensitivity
To assess the difference in gene expression after NVP-AUY922 exposure between sensitive and resistant cell lines, the eight cell lines with post-treatment gene expression data were evaluated using ANOVA comparing the five sensitive and three resistant cell lines (with and without FDR correction). After one hour of 100 nM NVP-AUY922 exposure, four probes were differentially expressed based on sensitivity prior to FDR adjustment. In the sensitive lines, A_32_P150802 and PRAM1 expression increased with drug exposure, and LIN37, CLCA3 expression decreased compared to resistant lines (data not shown). After 24 hours of 50 or 100 nM NVP-AUY922 exposure, 1982 genes were associated with sensitivity of which 127 remained significant after FDR correction (Supplemental Figure 2). Genes identified among these 127 were AHSA1, the gene coding the HSP90 co-chaperone AHA1 as well as HSP10 (HSPE1) and HSP60 (HSPD1). All three genes increased in response to NVP-AUY922 exposure but had significantly higher expression in resistant lines.
In vivo effects of NVP-AUY922
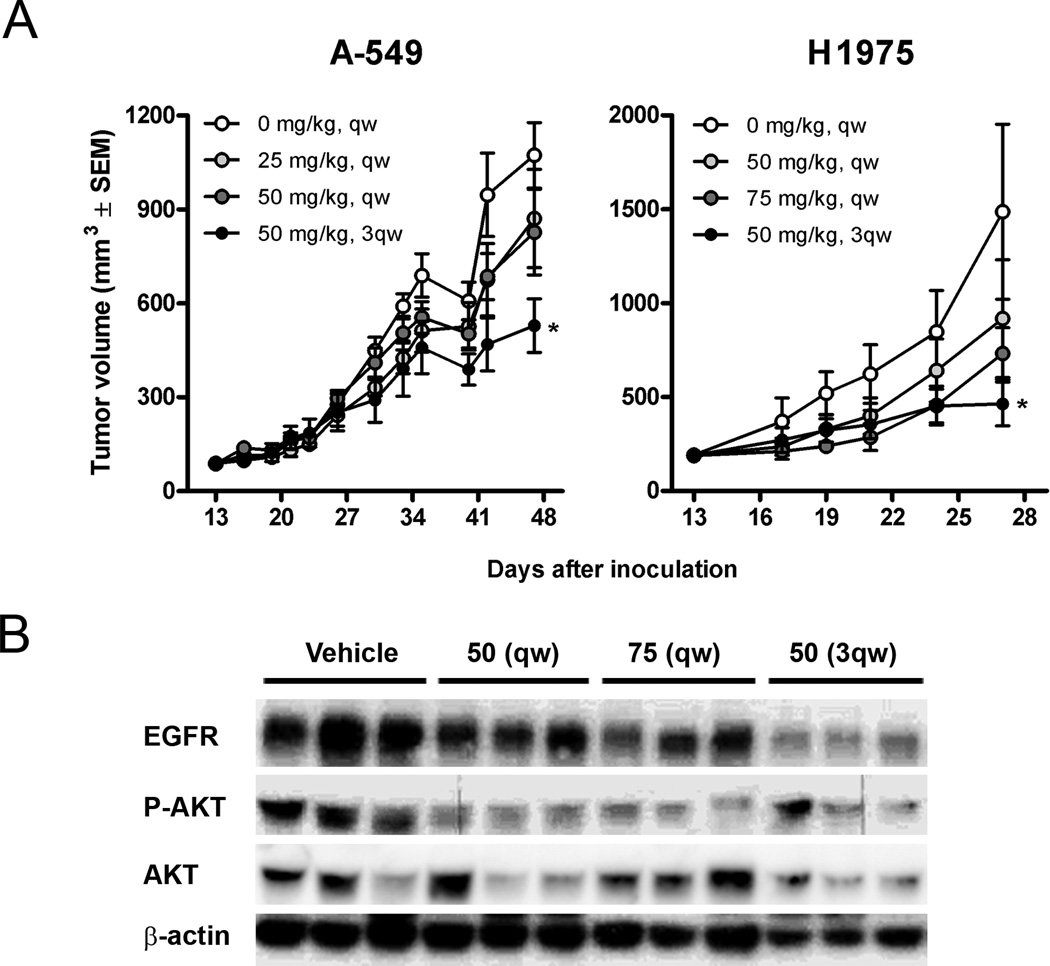
Effects of NVP-AUY922 were assessed in xenograft models. The A549 and H1975 cell lines were injected subcutaneously in nude mice. For each line, 32 mice were randomized to one of four arms, three active treatment arms and a control arm. The A549 cell line, which is relatively resistant in vitro, demonstrated a dose dependent reduction in tumor growth rates at doses of 25 mg/kg weekly, 50 mg/kg weekly and 50 mg/kg thrice weekly (Figure 7A). All three dosing schemes were tolerable, but the thrice weekly schedule led to a reduction in animal weight (Supplementary Table 1). The H1975 cell line, which harbors an activating and resistance EGFR mutation for EGFR TKIs and was relatively sensitive to NVP-AUY922 in vitro, grew more quickly than the A549 xenografts, and the animals needed to be sacrificed after 27 days, as opposed to 48 days in A549 xenografts (Figure 7A). In H1975 xenografts, all dosing schemes were tolerable, without reduction in body weight at 50 mg/kg weekly, 75 mg/kg weekly and 50 mg/kg thrice weekly. Tumor growth inhibition was again dose dependent, with essential stabilization of tumor size in the 50 mg/kg thrice weekly group (Supplementary Table 1). These results are in contrast to the A549 xenografts in which no difference was seen between tumors in treated and control mice after 27 days. To evaluate effects of HSP90 inhibition on the pathway driving H1975 proliferation, EGFR and pAKT levels were evaluated six hours after the final dose (Fig 7B) compared to tAKT and tubulin controls. EGFR levels decreased in a dose and dose-frequency dependent manner whereas pAKT levels were highly suppressed with all schedules.
Figure 7. In vivo effects of NVP-AUY922 in two xenograft models.
(A) 32 mice carrying subcutaneous A549 or H1975 xenograft tumors were randomized into 4 groups (n=8) 13 days after inoculation to receive intravenous doses of vehicle or NVP-AUY922 at 25–75 mg/kg weekly (qw) or 50 mg/kg thrice weekly (3qw). Tumor volumes are shown as means ± SEM. Significantly different antitumor effect compared to vehicle controls (p<0.05, one-way ANOVA post hoc Dunnet’s) are noted with asterisks. (B) Western blot analysis demonstrating effect of HSP90 inhibition on EGFR and phospho-AKT (P-AKT) in H1975 tumor xenografts dissected 6 hours after last treatment.
Discussion
HSP90 has emerged as an attractive target for therapy in a broad range of malignancies. We evaluated a novel synthetic diarylisoxazol resorcinol HSP90 inhibitor, NVP-AUY922, across a broad range of NSCLC models. We demonstrated potent growth inhibition across a broad sample of NSCLC cell lines with maximum IC50 < 100 nM. Evaluated cell lines harbored KRAS mutations (H23, H1944, A549, H1155, H1385, H2122, H358, SHP77, SKLU1), EGFR TKI sensitizing and resistant EGFR mutations (HCC827 and H1975 respectively), HER2 amplification (Calu-3), BRAF mutations (H1755), PI3KCA mutations (H1975, H1563), NRAS mutations (H1299, H2135), and PTEN loss (H1155). In general, these findings are similar to those seen in vitro by others.(39) However, there are some differences in the observed results, and these may be secondary to our proliferation assay, in which calculations are based on changes in generations over time rather than cell counts remaining at the end of the experiment. Our results also differ somewhat compared to other HSP90 inhibitors,(40) but as differences in clinical activity have also been seen, (33, 34) it is very possible that the spectrum of activity differs among compounds.
Changes we describe in gene expression following NVP-AUY922 exposure are consistent with those observed by others, without alteration of known HSP90 client proteins.(41, 42) Our evaluation confirms increased HSP family member expression, which we see as early as one hour after NVP-AUY922 exposure. Additional changes were seen across a broad range of cellular activities, reflecting the diverse action of HSP90. Many of the genes for which expression changed in all or most exposed cell lines were heat shock proteins, including HSP70 which is known to increase in response to HSP90 inhibition, but also HSP90, potentially in an effort to maintain effective levels in the setting of functional inhibition. Changes in genes involved in cell cycle could correspond with the profound inhibitory effect of NVP-AUY922.
In our study, many changes seen, such as those seen in pERK after NVP-AUY922 exposure, were felt to be downstream effects of pathways disrupted by HSP90 inhibition. Decreased DHFR expression post-NVP-AUY922 exposure was seen in some sensitive cell lines, and this observation has potential implications for treatment with pemetrexed, a standard chemotherapeutic used in NSCLC. DHFR is inhibited by pemetrexed, and reduction in DHFR expression may serve as a marker for pemetrexed responsiveness,(43) while increased DHFR has been associated with development of pemetrexed resistance.(44) Therefore, combination of NVP-AUY922 with pemetrexed based therapy is a potential area for future investigation, and a clinical trial in that setting is planned. At this time, we are exploring the dynamics of the changes in DHFR as will as evaluating potential sequencing of the two agents to support the planned clinical trial.
To identify baseline molecular characteristics of cell lines that were most sensitive to NVP-AUY922, cell lines were separated based on IC100 into a sensitive (IC100 < 40 nM) and resistant group (IC100 > 200 nM). Genes differentiating these cell lines at baseline represent diverse molecular pathways. Based on the relatively smaller number of resistant cell lines, the relevance of this gene set is considered exploratory and will require validation in ongoing trials. Although four probes were differentially expressed based on sensitivity to NVP-AUY922 after one hour of exposure, one does not code a known gene, while the other three code proteins for which testing is not commonly performed, making them poor candidates for biomarkers. In nearly 2000 genes, expression differed after 24 hours of NVP-AUY922 exposure in 5 sensitive and 3 resistant cell lines (based on IC100). These genes again represent a wide range of pathways, and many changes were likely related to greater growth inhibition in sensitive cell lines. One intriguing gene identified was AHSA1, activator of heat shock 90-KD protein ATPase, also called AHA1. AHA1 binds HSP90 and stimulates the activity of it ATPase.(45) AHA1 is upregulated by exposure to the HSP90 inhibitor 17-AAG, and it was one of the genes upregulated in our treatment experiments.(46) siRNA knockdown of AHA1 induced increased sensitivity to HSP90 inhibition with 17-AAG.(46) HSP10 and HSP60 also increased disproportionately after NVP-AUY922 exposure in resistant cell lines. These two genes are located head to head, separated by a bidirectional promoter,(47) and increased expression of these genes represents another potential resistance mechanism. We are planning further experiments to evaluate these genes as potential resistance mechanisms.
Our in vivo findings corroborate the in vitro findings although they were in general less profound, with the H1975 cell line inhibited with thrice weekly NVP-AUY922 and relative resistance for the A549 line. The H1975 xenograft model mimics a common clinical scenario of patients with known EGFR mutations who develop a secondary mutation and experience clinical progression with EGFR TKI therapy. Although pAKT was inhibited at low doses in tumors, the clinical stabilization of disease correlated with EGFR inhibition with thrice weekly dosing as demonstrated by Western blot. Clinical data with NVP-AUY922 now demonstrates a 20% response rate in patients with EGFR mutant tumors after EGFR TKI progression.(33) Based on this laboratory and clinical data, secondary resistance in patients with EGFR mutations will be evaluated in an expansion cohort of an ongoing clinical trial, and in some patients, biopsies during treatment will be obtained to correlate EGFR mutations and expression with clinical response to NVP-AUY922.(33) In patients harboring an EGFR mutation, a study of NVP-AUY922 vs. chemotherapy after progression on an EGFR TKI is planned, and an ongoing study is evaluating NVP-AUY922 along with erlotinib.(48, 49)
In summary, our evaluation demonstrates that NVP-AUY922 is an extremely potent inhibitor of NSCLC cell lines. Although consistent patterns of gene expression changes are seen in response to exposure, these effects span a large number of pathways. The finding of a decrease in DHFR after NVP-AUY922 exposure in some cell lines has implications for therapy with pemetrexed, and a study of pemetrexed plus NVP-AUY922 is planned. As a result of the high degree of sensitivity and the diverse set of pathways involved, clinical development of NVP-AUY922 initially has been in a broad subset of NSCLC patients. The data in the setting of EGFR TKI resistant EGFR mutant NSCLC justifies a particular focus in that setting.
Supplementary Material
Acknowledgements
Funding came from 1K23CA149079, the Wolfen Family Clinical/Translational Lung Cancer Research program, The One Ball Matt Memorial Golf Tournament and NIH CTSI UL1TR000124.
Grant Support
1K23CA149079 from the National Cancer Institute funds E.B. Garon
CTSI UL1TR000124 from the National Institute of Health funds S.M. Dubinett
Novartis funded laboratory research in the laboratory of D.J. Slamon.
The Wolfen Family Clinical/Translational Lung Cancer Research Program gives funding for the laboratory of D.J. Slamon
The One Ball Matt Memorial Golf Tournament funds research by E.B. Garon.
Footnotes
Conflicts of Interest: EBG received research funding from Novartis, and SI, EA, MRJ, CQ and ML are full time employees of Novartis
References
- 1.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5:761–772. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 2.Gallegos Ruiz MI, Floor K, Roepman P, Rodriguez JA, Meijer GA, Mooi WJ, et al. Integration of gene dosage and gene expression in non-small cell lung cancer, identification of HSP90 as potential target. PLoS One. 2008;3:e0001722. doi: 10.1371/journal.pone.0001722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solit DB, Basso AD, Olshen AB, Scher HI, Rosen N. Inhibition of heat shock protein 90 function down-regulates Akt kinase and sensitizes tumors to Taxol. Cancer Res. 2003;63:2139–2144. [PubMed] [Google Scholar]
- 4.Powers MV, Clarke PA, Workman P. Death by chaperone: HSP90, HSP70 or both? Cell Cycle. 2009;8:518–526. doi: 10.4161/cc.8.4.7583. [DOI] [PubMed] [Google Scholar]
- 5.Ramalingam SS, Egorin MJ, Ramanathan RK, Remick SC, Sikorski RP, Lagattuta TF, et al. A phase I study of 17-allylamino-17-demethoxygeldanamycin combined with paclitaxel in patients with advanced solid malignancies. Clin Cancer Res. 2008;14:3456–3461. doi: 10.1158/1078-0432.CCR-07-5088. [DOI] [PubMed] [Google Scholar]
- 6.Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, et al. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res. 2008;14:8302–8307. doi: 10.1158/1078-0432.CCR-08-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner AJMJ, Chugh R, et al. Inhibition of heat shock protein 90 (Hsp90) with the novel agent IPI-504 in metastatic GIST following failure of tyrosine kinase inhibitors (TKI) or other sarcomas: clinical results from a phase I trial. J Clin Oncol. 2008;26 Abs 10503. [Google Scholar]
- 8.Modi S, Stopeck AT, Gordon MS, Mendelson D, Solit DB, Bagatell R, et al. Combination of trastuzumab and tanespimycin (17-AAG, KOS-953) is safe and active in trastuzumab-refractory HER-2 overexpressing breast cancer: a phase I dose-escalation study. J Clin Oncol. 2007;25:5410–5417. doi: 10.1200/JCO.2007.11.7960. [DOI] [PubMed] [Google Scholar]
- 9.Banerji U, O'Donnell A, Scurr M, Pacey S, Stapleton S, Asad Y, et al. Phase I pharmacokinetic and pharmacodynamic study of 17-allylamino, 17-demethoxygeldanamycin in patients with advanced malignancies. J Clin Oncol. 2005;23:4152–4161. doi: 10.1200/JCO.2005.00.612. [DOI] [PubMed] [Google Scholar]
- 10.Jensen MR, Schoepfer J, Radimerski T, Massey A, Guy CT, Brueggen J, et al. NVP-AUY922: a small molecule HSP90 inhibitor with potent antitumor activity in preclinical breast cancer models. Breast Cancer Res. 2008;10:R33. doi: 10.1186/bcr1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eccles SA, Massey A, Raynaud FI, Sharp SY, Box G, Valenti M, et al. NVP-AUY922: a novel heat shock protein 90 inhibitor active against xenograft tumor growth, angiogenesis, and metastasis. Cancer Res. 2008;68:2850–2860. doi: 10.1158/0008-5472.CAN-07-5256. [DOI] [PubMed] [Google Scholar]
- 12.Massey AJ, Schoepfer J, Brough PA, Brueggen J, Chene P, Drysdale MJ, et al. Preclinical antitumor activity of the orally available heat shock protein 90 inhibitor NVP-BEP800. Mol Cancer Ther. 2010;9:906–919. doi: 10.1158/1535-7163.MCT-10-0055. [DOI] [PubMed] [Google Scholar]
- 13.Samuel TA, Sessa C, Britten C, Milligan KS, Mita MM, Banerji U, et al. AUY922, a novel HSP90 inhibitor: Final results of a first-in-human study in patients with advanced solid malignancies. J Clin Oncol. 2010;28(15s) (suppl; abstr 2528) J Clin Oncol. 2009;27:Abstract 3532. [Google Scholar]
- 14.Kris MGJB, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Aronson SL, Engelman JA, Shyr Y, Khuri FR, Rudin CM, Garon EB, Pao W, Schiller JH, Haura EB, Shirai K, Giaccone G, Berry LD, Kugler K, Minna JD, Bunn PA. Identification of driver mutations in tumor specimens from 1,000 patients with lung adenocarcinoma: The NCI’s Lung Cancer Mutation Consortium (LCMC) J Clin Oncol. 2011;29(suppl) abstr CRA7506. [Google Scholar]
- 15.Janne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol. 2005;23:3227–3234. doi: 10.1200/JCO.2005.09.985. [DOI] [PubMed] [Google Scholar]
- 16.Taron M, Ichinose Y, Rosell R, Mok T, Massuti B, Zamora L, et al. Activating mutations in the tyrosine kinase domain of the epidermal growth factor receptor are associated with improved survival in gefitinib-treated chemorefractory lung adenocarcinomas. Clin Cancer Res. 2005;11:5878–5885. doi: 10.1158/1078-0432.CCR-04-2618. [DOI] [PubMed] [Google Scholar]
- 17.Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst. 2005;97:339–346. doi: 10.1093/jnci/dji055. [DOI] [PubMed] [Google Scholar]
- 18.Takano T, Ohe Y, Sakamoto H, Tsuta K, Matsuno Y, Tateishi U, et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non-small-cell lung cancer. J Clin Oncol. 2005;23:6829–6837. doi: 10.1200/JCO.2005.01.0793. [DOI] [PubMed] [Google Scholar]
- 19.Ono M, Hirata A, Kometani T, Miyagawa M, Ueda S, Kinoshita H, et al. Sensitivity to gefitinib (Iressa, ZD1839) in non-small cell lung cancer cell lines correlates with dependence on the epidermal growth factor (EGF) receptor/extracellular signal-regulated kinase 1/2 and EGF receptor/Akt pathway for proliferation. Mol Cancer Ther. 2004;3:465–472. [PubMed] [Google Scholar]
- 20.Amann J, Kalyankrishna S, Massion PP, Ohm JE, Girard L, Shigematsu H, et al. Aberrant epidermal growth factor receptor signaling and enhanced sensitivity to EGFR inhibitors in lung cancer. Cancer Res. 2005;65:226–235. [PubMed] [Google Scholar]
- 21.Benz MR, Herrmann K, Walter F, Garon EB, Reckamp KL, Figlin R, et al. 18F-FDG PET/CT for Monitoring Treatment Responses to the Epidermal Growth Factor Receptor Inhibitor Erlotinib. J Nucl Med. 2011;52:1684–1689. doi: 10.2967/jnumed.111.095257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimamura T, Lowell AM, Engelman JA, Shapiro GI. Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res. 2005;65:6401–6408. doi: 10.1158/0008-5472.CAN-05-0933. [DOI] [PubMed] [Google Scholar]
- 23.Kobayashi N, Toyooka S, Soh J, Yamamoto H, Dote H, Kawasaki K, et al. The anti-proliferative effect of heat shock protein 90 inhibitor, 17-DMAG, on non-small-cell lung cancers being resistant to EGFR tyrosine kinase inhibitor. Lung Cancer. 2012 Feb;75(2):161–166. doi: 10.1016/j.lungcan.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 24.Shimamura T, Li D, Ji H, Haringsma HJ, Liniker E, Borgman CL, et al. Hsp90 inhibition suppresses mutant EGFR-T790M signaling and overcomes kinase inhibitor resistance. Cancer Res. 2008;68:5827–5838. doi: 10.1158/0008-5472.CAN-07-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem. 2002;277:39858–39866. doi: 10.1074/jbc.M206322200. [DOI] [PubMed] [Google Scholar]
- 26.Maulik G, Kijima T, Ma PC, Ghosh SK, Lin J, Shapiro GI, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res. 2002;8:620–627. [PubMed] [Google Scholar]
- 27.Blasco RB, Francoz S, Santamaria D, Canamero M, Dubus P, Charron J, et al. c-Raf, but not B-Raf, is essential for development of K-Ras oncogene-driven non-small cell lung carcinoma. Cancer Cell. 2011;19:652–663. doi: 10.1016/j.ccr.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dogan T, Harms GS, Hekman M, Karreman C, Oberoi TK, Alnemri ES, et al. X-linked and cellular IAPs modulate the stability of C-RAF kinase and cell motility. Nat Cell Biol. 2008;10:1447–1455. doi: 10.1038/ncb1804. [DOI] [PubMed] [Google Scholar]
- 29.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 30.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sequist LV, Gettinger S, Senzer NN, Martins RG, Janne PA, Lilenbaum R, et al. Activity of IPI-504, a novel heat-shock protein 90 inhibitor, in patients with molecularly defined non-small-cell lung cancer. J Clin Oncol. 2010;28:4953–4960. doi: 10.1200/JCO.2010.30.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong KKM, Goldman JW, Paschold EH, Horn L, Lufkin JM, Blackman RK, Teofilovici F, Shapiro G, Socinski MA. An open-label phase II study of the Hsp90 inhibitor ganetespib (STA-9090) as monotherapy in patients with advanced non-small cell lung cancer (NSCLC) J Clin Oncol. 2011;29(suppl) abstr 7500. [Google Scholar]
- 33.Garon EBMT, Barlesi F, Gandhi L, Sequist LV, Kim SW, Groen HJM, Besse B, Smit EF, Kim DW, Akimov M, Avsar E, Bailey S, Felip E. Phase II study of the HSP90 inhibitor AUY922 in patients with previously treated, advanced non-small cell lung cancer (NSCLC) American Society of Clinical Oncology. 2012;2012 [Google Scholar]
- 34.Felip ECE, Barlesi F, Gandhi L, Sequist L, Kim SW, Groen HJM, Besse B, Kim DW, Smit E, Akimov M, Avsar E, Pain S, Ofosu-Appiah W, Garon EB. Phase II activity of the HSP90 inhibitor AUY922 in patients with ALK-rearranged (ALK+) or EGFR-mutated advanced non-small cell lung cancer (NSCLC) Proceedings of the European Society of Medical Oncology Meeting. 2012 [Google Scholar]
- 35.Wilson CADJ, Bernardo G, Rong HM, Ginther C, Ferdman R, Cook AM, Finn RS, Slamon DJ. Cell differentiation and dominant signaling pathway signatures in the molecular classification of human breast cancer cell lines. Breast Cancer Res. 2005;7(Suppl 2) S 4.25. [Google Scholar]
- 36.Finn RS, Dering J, Ginther C, Wilson CA, Glaspy P, Tchekmedyian N, et al. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/"triple-negative" breast cancer cell lines growing in vitro. Breast Cancer Res Treat. 2007;105:319–326. doi: 10.1007/s10549-006-9463-x. [DOI] [PubMed] [Google Scholar]
- 37.Garon EB, Finn RS, Hosmer W, Dering J, Ginther C, Adhami S, et al. Identification of common predictive markers of in vitro response to the Mek inhibitor selumetinib (AZD6244; ARRY-142886) in human breast cancer and non-small cell lung cancer cell lines. Mol Cancer Ther. 2010;9:1985–1994. doi: 10.1158/1535-7163.MCT-10-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomas PD, Kejariwal A, Campbell MJ, Mi H, Diemer K, Guo N, et al. PANTHER: a browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. doi: 10.1093/nar/gkg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ueno T, Tsukuda K, Toyooka S, Ando M, Takaoka M, Soh J, et al. Strong anti-tumor effect of NVP-AUY922, a novel Hsp90 inhibitor, on non-small cell lung cancer. Lung cancer. 2012;76:26–31. doi: 10.1016/j.lungcan.2011.09.011. [DOI] [PubMed] [Google Scholar]
- 40.Shimamura T, Perera SA, Foley KP, Sang J, Rodig SJ, Inoue T, et al. Ganetespib (STA-9090), a Nongeldanamycin HSP90 Inhibitor, Has Potent Antitumor Activity in In Vitro and In Vivo Models of Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2012;18:4973–4985. doi: 10.1158/1078-0432.CCR-11-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clarke PA, Hostein I, Banerji U, Stefano FD, Maloney A, Walton M, et al. Gene expression profiling of human colon cancer cells following inhibition of signal transduction by 17-allylamino-17-demethoxygeldanamycin, an inhibitor of the hsp90 molecular chaperone. Oncogene. 2000;19:4125–4133. doi: 10.1038/sj.onc.1203753. [DOI] [PubMed] [Google Scholar]
- 42.Maloney A, Clarke PA, Naaby-Hansen S, Stein R, Koopman JO, Akpan A, et al. Gene and protein expression profiling of human ovarian cancer cells treated with the heat shock protein 90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2007;67:3239–3253. doi: 10.1158/0008-5472.CAN-06-2968. [DOI] [PubMed] [Google Scholar]
- 43.Wu MF, Hsiao YM, Huang CF, Huang YH, Yang WJ, Chan HW, et al. Genetic determinants of pemetrexed responsiveness and nonresponsiveness in non-small cell lung cancer cells. J Thorac Oncol. 2010;5:1143–1151. doi: 10.1097/JTO.0b013e3181e0b954. [DOI] [PubMed] [Google Scholar]
- 44.Zhang D, Ochi N, Takigawa N, Tanimoto Y, Chen Y, Ichihara E, et al. Establishment of pemetrexed-resistant non-small cell lung cancer cell lines. Cancer Lett. 2011;309:228–235. doi: 10.1016/j.canlet.2011.06.006. [DOI] [PubMed] [Google Scholar]
- 45.Lotz GP, Lin H, Harst A, Obermann WM. Aha1 binds to the middle domain of Hsp90, contributes to client protein activation, and stimulates the ATPase activity of the molecular chaperone. J Biol Chem. 2003;278:17228–17235. doi: 10.1074/jbc.M212761200. [DOI] [PubMed] [Google Scholar]
- 46.Holmes JL, Sharp SY, Hobbs S, Workman P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008;68:1188–1197. doi: 10.1158/0008-5472.CAN-07-3268. [DOI] [PubMed] [Google Scholar]
- 47.Hansen JJ, Bross P, Westergaard M, Nielsen MN, Eiberg H, Borglum AD, et al. Genomic structure of the human mitochondrial chaperonin genes: HSP60 and HSP10 are localised head to head on chromosome 2 separated by a bidirectional promoter. Hum Genet. 2003;112:71–77. doi: 10.1007/s00439-002-0837-9. [DOI] [PubMed] [Google Scholar]
- 48.Johnson MLYH, Hart E, Worden R, Rademaker F, Miller C, Patel JD, Kris MG, Miller VA, Riely GJ. A phase I dose-escalation study of the HSP90 inhibitor AUY922 and erlotinib for patients with EGFR mutant lung cancer with acquired resistance (AR) to EGFR tyrosine kinase inhibitors (EGFR TKIs). 12th Annual Targeted Therapies of the Treatment of Lung Cancer Meeting; 2012; Santa Monica CA. 2012. [Google Scholar]
- 49.Johnson MLYH, Hart EM, Worden R, Rademaker F, Gupta R, Miller C, Patel JD, Kris MG, Miller VA, Riely GJ. A phase I dose-escalation study of the HSP90 inhibitor AUY922 and erlotinib for patients with EGFR mutant lung cancer with acquired resistance (AR) to EGFR tyrosine kinase inhibitors (EGFR TKIs) American Society of Clinical Oncology. 2012;2012 doi: 10.1200/JCO.2014.59.7328. abstr 3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.