Abstract
Import-Karyopherin or Importin proteins bind nuclear localization signals (NLSs) to mediate the import of proteins into the cell nucleus. Karyopherin β2 or Kapβ2, also known as Transportin, is a member of this transporter family responsible for the import of numerous RNA binding proteins. Kapβ2 recognizes a targeting signal termed the PY-NLS that lies within its cargos to target them through the nuclear pore complex. The recognition of PY-NLS by Kapβ2 is conserved throughout eukaryotes. Kap104, the Kapβ2 homolog in Saccharomyces cerevisiae, recognizes PY-NLSs in cargos Nab2, Hrp1, and Tfg2. We have determined the crystal structure of Kapβ2 bound to the PY-NLS of the mRNA processing protein Nab2 at 3.05-Å resolution. A seven-residue segment of the PY-NLS of Nab2 is observed to bind Kapβ2 in an extended conformation and occupies the same PY-NLS binding site observed in other Kapβ2•PY-NLS structures.
Keywords: Karyopherin, Importin, Nuclear Import, Nuclear Localization Signal, Nucleocytoplasmic Transport, Nuclear pore, Nab2
Introduction
Karyopherin β proteins (Kaps; also known as Importins and Exportins) are responsible for the majority of nucleocytoplasmic transport in eukaryotic cells. At least 20 members of the Kap family have been identified in humans, whereas 14 Kaps are known in S. cerevisiae. Each Kap binds a unique set of cargos and targets them to the nuclear pore complex. Kaps bind nuclear localization signals (NLSs) or nuclear export signals (NESs) in cargo proteins to direct them in and out of the nucleus, respectively. Kap-cargo interactions and transport directionality are in turn regulated by the Ran GTPase nucleotide cycle.1
The Karyopherin β2 (Kapβ2; also known as Transportin) importin recognizes a class of NLS in its cargos termed the PY-NLS.2-4 These 15- to 100-residue long sequences are diverse and cannot be sufficiently described by a traditional consensus sequence. PY-NLSs are instead described by a collection of physical rules that include the requirements for intrinsic structural disorder, overall basic character, and a set of sequence motifs. PY-NLS motifs consist of an N-terminal hydrophobic or basic motif and a C-terminal RX2-5PY motif.4-6
The shuttling heterogeneous nuclear ribonucleoprotein Nab2 is essential for mRNA export in Saccharomyces cerevisiae. Nab2 recognizes poly(A) RNA and binds to the nuclear pore-associated protein myosin-like protein 1 (Mlp1), which functions in both mRNA export and quality control.7 Nab2 contains a PY-NLS, which is recognized by the yeast homolog of Kapβ2, Kap104, for import into the nucleus (Fig 1.A).8 The binding of cytosolic Kap104 to the PY-NLS of Nab2 has been implicated in the release of DEAD-box RNA helicase Dbp5 from mRNA to allow for translation.9,10 Following Nab2 release, the Kap104-Nab2 complex is translocated through the nuclear pore complex into the nucleus, where RanGTP and mRNA act cooperatively to dissociate Nab2 from Kap104.
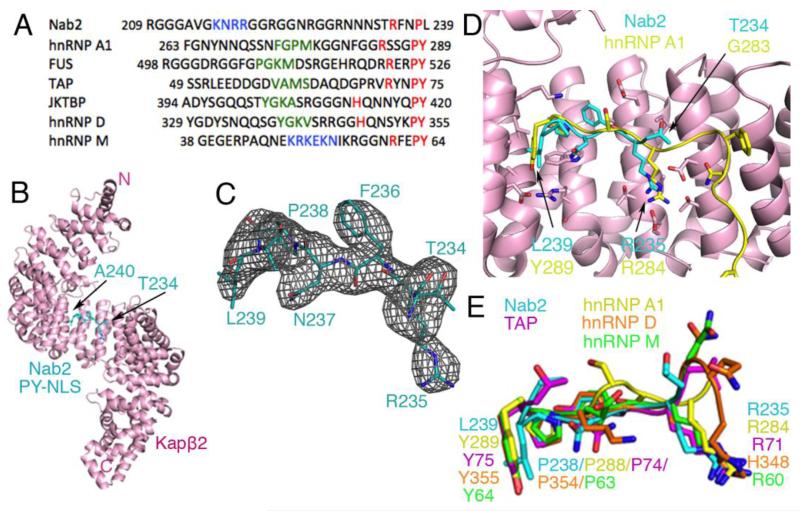
Fig. 1. Structure and comparative analysis of Nab2PY-NLS.
(A) Sequence alignment of several PY-NLSs. This signal consists of an N-terminal hydrophobic or basic motif (green and blue, respectively) and a C-terminal RX2-5PY motif (red). (B) Overall structure of the Kapβ2•Nab2PY-NLS complex. The karyopherin is in pink and the PY-NLS cyan. (C) mFo-DFc difference map, contoured at 2.0σ (grey), at the PY-NLS binding site of Kapβ2 before the Nab2PY-NLS residues were included in the model. The final refined model of Nab2PY-NLS residues Thr234 to Ala240 are in cyan. (D) Superposition of Kapβ2 residues 301-634 for Kapβ2s bound to PY-NLSs from cargos Nab2 and hnRNP A1 (PDB ID 2H4M; Cα rmsd 0.35 Å). Kapβ2 of the Nab2 complex is shown in pink. The Nab2 PY-NLS is in cyan and the hnRNP A1 PY-NLS is in yellow. (E) Similar superpositions of Kapβ2 as in D, to compare PY-NLSs from Nab2 (cyan), hnRNP A1 (yellow), hnRNP M (green), hnRNP D (orange), and TAP/NXF1 (purple).
All previous structures of PY-NLSs bound to Kapβ2 are of signals that contain the canonical PY dipeptide motif. The PY-NLS of Nab2 is unusual in that it contains a homologous PL dipeptide motif at its C-terminus. Mutagenesis studies suggest that some PY dipeptides in PY-NLSs can be replaced by Pφ dipeptides where φ is any hydrophobic residue without losing binding affinity for the Kap.8 Here we report the 3.05-Å-crystal structure of Kapβ2 bound to the PY-NLS of Nab2, which shows for the first time the homologous PL dipeptide motif. The structure explains how an aliphatic hydrophobic residue is able to substitute structurally for the conserved tyrosine in a PY-NLS.
Materials and Methods
Protein Expression, Purification, and Complex Assembly
Human Kapβ2 (Uniprot ID U72069) was expressed in pGEX-TEV vector [pGEX-4T3 (GE Healthcare) with a TEV cleavage site] as a GST fusion protein and purified as previously described.4 Kapβ2 with a truncated loop, which does not interfere with NLS binding, was used for crystallization (residues 337-367 of Kapβ2 were replaced with a GGSGGSG linker).4 Residues 205-242 of S. cerevisiae Nab2 (Nab2PY-NLS; Nab2, Uniprot ID P32505) were also expressed as a GST fusion protein.8 GST-Nab2PY-NLS was purified by affinity and ion exchange chromatography. GST-Nab2PY-NLS and Kapβ2 were mixed at molar ratio of 5:1. The GST tag was removed with TEV protease and the complex further purified by gel filtration in buffer composed of 20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM DTT, 2 mM magnesium acetate, and 1 mM EGTA with 20% (v/v) glycerol. The complex was concentrated to 13 mg/mL for crystallization.
Crystallization and structure determination of the Kapβ2•Nab2PY-NLS complex
Purified Kapβ2•Nab2PY-NLS complex was screened against MCSG1-4 (Microlytic North America, USA) and ProComplex (Qiagen, USA) crystallization screens via sitting drop vapor diffusion at 20°C (0.4 μL protein + 0.4 μL reservoir solution) using a Phoenix (Art Robins, USA) liquid handling system. Crystals with well-formed morphology were obtained in several conditions. Many crystals did not yield useful diffraction, but crystals grown with crystallization condition MCSG3-H11 (700 mM sodium citrate tribasic and 100 mM Bis-trispropane pH 7.0) diffracted to 3.05-Å resolution. Crystals were cryo-protected by addition of ~20% (v/v) glycerol, and flash-cooled by immersion in liquid nitrogen. Diffraction data, recorded at the X29A beamline at the Brookhaven National Laboratory at a wavelength of 1.0705 Å, were processed using HKL3000.12 The structure was determined by molecular replacement using PHASER13 with a search model of human Kapβ2 from the PDB Code 2QMR (A chain).11 Several rounds of refinement using REFMAC514 and manual model building with COOT15, were performed. The high resolution structure of Kapβ2 bound to the PY-NLS of Fused in Sarcoma protein (PDB code 4FDD)6 was used to guide manual model building. Residues 234-240 of Nab2 were built into the electron density maps at the last stages of the refinement [Fig 1.A]. The final model of the Kapβ2•Nab2PY-NLS complex shows excellent stereochemical parameters (Table I). Illustrations were prepared with PyMol (http://www.pymol.org).
Table I. Crystallographic statistics for HsKapβ2•ScNab2PY-NLS complex.
| Crystal Parameters | |
| Space group | P21212 |
| Cell dimensions: | |
| a, b, c (Å) | 132.2, 172.4, 68.4 |
| α, β, γ (°) | 90, 90, 90 |
| Matthew’s coefficient (Å3/Da) | 4.14 |
| Solvent content (%) | 70 |
| Data Collection | |
| Wavelength (Å) | 1.0750 |
| Resolution (Å) | 50.00-3.05 (3.29-3.05)a |
| Rsym(%) | 12.3 |
| Number of unique reflections | 30,914 (1536) a |
| Number of reflections in Rfree set | 1538 |
| Mean Redudancy | 8.8 (9.0) a |
| Overall Completeness (%) | 100.0 (100.0) |
| Mean I/σ | 16.6 (1.18) a |
| Refinement Residuals | |
| Rfree (%) | 23.7 |
| Rwork (%) | 18.4 |
| Completeness (%) | 99.5 (94.6) |
| Model Quality | |
| MRSD bond lengths (Å) | 0.0109 |
| RMSD bond angles (°) | 1.5380 |
| Molprobity Ramachandran: | |
| Favored region (%) | 94.8 |
| Allowed region (%) | 99.9 |
| Mean overall B-factor | |
| HsKapβ2 (Å2) | 104.7 |
| ScNAB2-NLS (Å2) | 101.7 |
| Model Contents | |
| Protomers in ASU | |
| HsKapβ2 | 1 |
| No. of HsKapβ2 residues | 840 |
| No. of HsKapβ2 atoms | 6676 |
| ScNAB2-NLS | 1 |
| No. of ScNAB2-NLS residues | 7 |
| No. of ScNAB2-NLS atoms | 57 |
| No. of water atoms | 0 |
| PDB accession code | 4H1K |
Values in parenthesis correspond to the highest-resolution shell
Results and Discussion
Structure of the Kapβ2•Nab2PY-NLS complex
We have determined the 3.05 Å crystal structure of human Kapβ2 bound to the PY-NLS segment of S. cerevisiae Nab2 that spans residues 205-242 (Fig 1.B). Kapβ2, as shown previously11,17, is a superhelical protein composed of 20 α-helical HEAT repeats. Each HEAT repeat is composed of two antiparallel α-helices, A and B, each lining the convex and concave sides of the superhelix, respectively. Seven residues of the Nab2PY-NLS (residues 234-240) are modeled and shown to bind the previously described PY-NLS binding site in the C-terminal arch of Kapβ2.4-6,17 Residues 234-240 of the Nab2PY-NLS peptide bind in extended conformation, tracing a path along the concave surface of Kapβ2 similar to other structurally characterized PY-NLSs, such as that from hnRNP A1 (Fig 1.C, D).4 Residues 204-233 and 241-242 of the bound Nab2PY-NLS peptide were not modeled due to weak electron density.
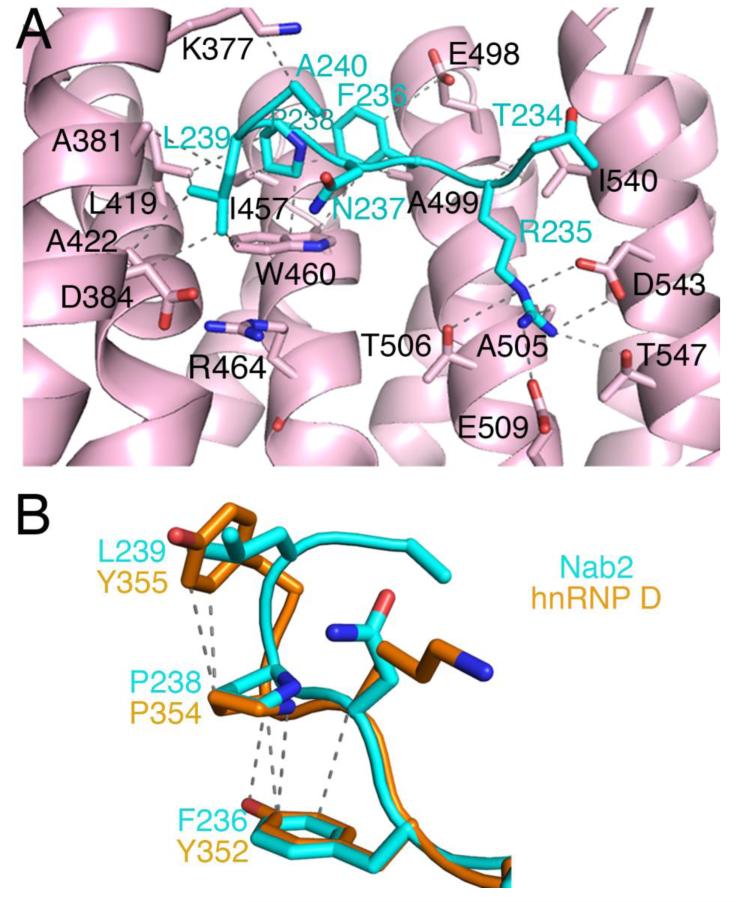
Residues 234TRFNPL240 of Nab2 occupy the same binding site on Kapβ2 as the 284RX2-5PY289 motifs of other PY-NLSs such as those from cargos hnRNP A1, hnRNP D, hnRNP M, and TAP/NXF1 (Fig 1.E).4,5,17 All previous structures of PY-NLSs are of peptides that contain the canonical PY dipeptide. This Nab2 PY-NLS structure shows for the first time the homologous PL dipeptide motif. Like PY motifs, the PL motif of the Nab2PY-NLS also makes numerous contacts with hydrophobic residues of Kapβ2 (Fig 2.A). Pro-238 of Nab2 interacts primarily through hydrophobic interactions with the sidechains Leu-419, Ile-457, and Trp-460 of Kapβ2. Leu-239 of the Nab2 PL motif makes hydrophobic interactions with Leu-419, Ala-381, Ala-422, and Trp-460 of Kapβ2. Previous mutational analysis showed that mutation of the PL motif (wildtype Nab2PY-NLS binds Kapβ2 with KD=37 nM) in Nab2 to PY improved binding affinity to Kap104 by about three-fold (KD=13 nM for the PY mutant of Nab2).8 This increase in binding energy may be due to the aromatic ring of tyrosine making additional hydrophobic contacts with Kapβ2 side chains and/or the tyrosine making polar interactions with Arg-464 of Kapβ2 as seen in other Kapβ2-PY-NLS structures (PDB ID: hnRNP A1, 2H4M; hnRNP M, 2OT8; TAP, 2Z5K; hnRNP D, 2Z5N).
Fig. 2. The Kapβ2•Nab2PY-NLS Interface.
(A) The Nab2PY-NLS (cyan) makes numerous hydrophobic contacts with Kapβ2 (pink). In addition, Arg-235 of the Nab2PY-NLS makes multiple salt bridges and hydrogen bonds with Kapβ2. Interacting residues on Kapβ2 are labeled in black and contacts (4.0 Å or less) are indicated by dashed lines. (B) Comparison of the PY motifs of PY-NLSs from Nab2 (cyan) and hnRNP D (orange). Intramolecular contacts (4.0 Å or less) in hnRNP D are shown with dashed lines.
Further N-terminus, Phe-236 also makes hydrophobic interactions with Ala-499, Glu-498 and Trp-460 sidechains of Kapβ2. The Phe236 sidechain is also positioned close to Pro-238 (within 4.5 Å). Similar conformations were observed for the equivalent position in PY-NLSs of hnRNP D, hnRNP M and TAP/NXF1 where a phenylalanine is also present (Fig 2.B).4,5,17 Previous biochemical studies have shown that Phe-236 contributes significantly to Kap104-Nab2 interactions and that a hydrophobic residue at this position is important.8 Phe-236 likely contributes entropically through intramolecular interactions that preorganize the PL motif for Kapβ2 binding. The neighboring Arg-235, which is the N-terminal arginine of the RX2-5PY/L motif, makes multiple salt bridges and hydrogen bonds with Asp-543, Thr-506, Glu-509, and Thr-547 of Kapβ2 (Fig 2.A). Beyond residue Thr-234, electron density is too weak to model the N-terminal portion of the Nab2 PY-NLS.
Protein Data Bank Codes
Atomic coordinates and structure factors of the Kapβ2•Nab2PY-NLS complex were deposited to the PDB on 10 Sepetember 2012 with accession codes 4H1K. The NYSGRC target identifiers for the PSI:Biology targets Kapβ2 and Nab2PY-NLS are “NYSGRC-020458” and “NYSGRC-020441”, respectively.
Acknowledgements
We thank Z. Zhang for advice on protein purification and M. Rout for discussions. This work is funded by the National Institutes of Health U01 GM98256-01 (YMC, ASC), U54 GM094662 (SCA) and R01-GM069909 (YMC), Welch Foundation (I-1532; YMC), Leukemia and Lymphoma Society Scholar award (YMC), CPRIT (RP120352; YMC) and UT Southwestern Endowed Scholars Program (YMC). This publication was made possible by the Center for Synchrotron Biosciences grant, P30-EB-009998, from the National Institute of Biomedical Imaging and Bioengineering (NIBIB). Use of the National Synchrotron Light Source, Brookhaven National Laboratory, was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-98CH10886. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Basic Energy Sciences. Access to the LRL-CAT beam line facilities at Sector 31 of the APS was provided by Eli Lilly, which operates the facility
References
- 1.Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- 2.Chook Y, Suel K. Nuclear import by karyopherin-βs: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu D, Farmer A, Chook Y. Recognition of nuclear targeting signals by Karyopherin-β proteins. Curr Opin Struc Bio. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee B, Cansizoglu A, Suel K, Louis T, Zhang Z, Chook Y. Rules for nuclear localization sequence recognition by Karyopherinβ2. Cell. 2006;126:544–558. doi: 10.1016/j.cell.2006.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cansizoglu A, Lee B, Zhang Z, Fontoura B, Chook Y. Structure-based design of a pathway-specific nuclear import inhibitor. Nat. Struc. Mol. Bio. 2007;14:452–454. doi: 10.1038/nsmb1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang ZC, Chook YM. Structural and energetic basis of ALS-causing mutations in the atypical proline-tyrosine nuclear localization signal of the Fused in Sarcoma protein (FUS) Proc Natl Acad Sci (USA) 2012;109:12017–12021. doi: 10.1073/pnas.1207247109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fasken M, Stewart M, Corbett A. Functional significance of the interaction between the mRNA-binding protein, Nab2, and the nuclear pore-associated Protein, Mlp1, in mRNA export. J Bio Chem. 2008;283:27130–27143. doi: 10.1074/jbc.M803649200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suel K, Gu H, Chook Y. Modular organization and combinatorial energetics of prolinetyrosine nuclear localizations signals. PLoS Biol. 2008;6:1253–1267. doi: 10.1371/journal.pbio.0060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee D, Aitchison J. Kap104-mediated nuclear import. Nuclear localization signals in mRNA-binding proteins and the role of Ran and RNA. J Bio Chem. 2009;274:29031–29037. doi: 10.1074/jbc.274.41.29031. [DOI] [PubMed] [Google Scholar]
- 10.Tran E, Zhou Y, Corbett A, Wente S. The DEAD-box protein Dbp5 controls mRNA export by triggering specific RNA:protein remodeling events. Mol. Cell. 2007;28:850–859. doi: 10.1016/j.molcel.2007.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Cansizoglu AE, Chook YM. Conformational heterogeneity of Karyopherin β2 is segmental. Structure. 2007;15:1431–1441. doi: 10.1016/j.str.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta crystallographica. Section D, Biological crystallography. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 13.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. PHASER crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collaborative Computing Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 15.Emsley P, Cowtan K. COOT: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 16.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucl Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. http://molprobity.biochem.duke.edu/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Imasaki T, Shimizu T, Hashimoto H, Hidaka Y, Kose S, Imamoto N, Yamada M, Sato M. Structural basis for substrate recognition and dissociation by human tranportin 1. Mol Cell. 2007;28:57–67. doi: 10.1016/j.molcel.2007.08.006. [DOI] [PubMed] [Google Scholar]