Abstract
Background
The D typing strategies in several European countries protect carriers of D category VI (DVI) from anti-D immunization but not carriers of other partial D. Besides DVI, one of the clinically most important partial D is D category IV (DIV). A detailed description and direct comparison of the different DIV types was missing.
Study design and methods
RHD nucleotide sequences were determined from genomic DNA. D epitope patterns were established with commercial monoclonal anti-D panels.
Results
DIV comprises several variants of the D antigen with distinct serology, molecular structures, evolutionary origins and ethnic prevalences. The DIV phenotype is determined by 350H shared by all, but not limited to, DIV variants which are further divided into DIVa and DIVb. The DIVa phenotype is expressed by DIV type 1.0 harboring 350H and the dispersed amino acids 62F, 137V and 152T. The DIVb phenotype is expressed by DIV type 3 to type 5 representing RHD-CE-D hybrids. 4 of the 6 postulated DIV variants were encountered among 23 DIV samples analyzed. Of 12 DIV carriers with anti-D, 10 were female and 7 likely immunized by pregnancy. 2 DIV related alleles are newly described: DWN which differs from DIV type 4 by 350D and epitope pattern. DNT carries 152T, known to cause a large D antigen density.
Conclusion
DIV alleles arose from at least 2 independent evolutionary events. DIV type 1.0 with DIVa phenotype belongs to the oldest extant human RHD alleles. DIV type 2 to type 5 with DIVb phenotype arose from more recent gene conversions. Anti-D immunization, especially dreaded in pregnancies, will be avoided not only in carriers of DVI but also in carriers of other D variants like DIV, if our proposed D typing strategy is adopted.
Introduction
D is the clinically most important protein antigen on red blood cells (RBC) and the leading cause of alloimmunization in D negative individuals. In 1953, allo-anti-D was also detected in D positive individuals.1 This ostensible contradiction was explained by partial defects of the D antigen. Individuals lacking part of the D antigen may develop an antibody against the missing part following transfusion, transplant or pregnancy. The term “partial D” for these D variants was introduced in 1984;2,3 however, D variants missing different epitopes had been recognized much earlier.
In 1959, the D antigen was divided into RhA, RhB, RhC 4,5 and RhD 6,7 and summarized as the 4 “blood factors”.8 Independent of this earlier work,4-7,9 D categories I to VI were defined in 1962,10 of which D category I was retracted11,12 while D category VII was added later.12,13 The D category I samples of 1977 represented a heterogeneous set of weak D phenotypes from Caucasians,11 difficult to characterize by serology because of the weak expression of the D antigen,11 while the heterogeneity of the weak D types likely involved was unknown at the time. The original nomenclature comprised of the “blood factors”4,5 was abandoned for the D category classification,10 which is still in use.14
The current terminology for D categories comprises DII to DVII.15 Several subtypes of D categories14 have since been detected as well as many partial D, whose serological appearance and molecular basis did not match any defined D category. Today, D categories represent only a fraction of all partial D alleles. At present, 85 partial D are listed in the internet-based registry of RHD alleles (The RhesusBase),16 of which only 26 belong to D categories and their subtypes.
DII is very rare; only 3 individuals expressing DII are known in 2 pedigrees.14,15 DVII is the most prevalent D category in Caucasians with a phenotype frequency of 1 in 900 in Germany.17 DVI, not to be confused with DIV, is much less frequent with 1 in 6214,18 but is the clinically most relevant partial D in Europeans with respect to immunization by normal D.19 Therefore, in several European countries a D typing strategy involving the use of two monoclonal anti-D antibodies that do not recognize DVI is mandatory for recipients.20 While this strategy prevents anti-D formation in DVI individuals, there are other partial D that are typed as D positive, like DIV, R0Har, DNB, DVII, DIII, weak D type 4.2 and DV.21 These partial D may remain undetected and anti-D immunizations may occur with the consequence of transfusion incompatibility and complications during pregnancy.22,23
The RhD protein has 12 transmembraneous segments and forms 6 extracellular loops. DII, DHK (identical to DV type 5) and DVII type 1 are caused by single amino acid substitutions in the extracellular loops 6, 4 and 2, respectively. These substitutions are not related to the RhCE amino acid sequence and are, hence, not caused by gene conversions.24,25 In contrast, DIII, DIV, DV and DVI harbor amino acid substitutions which may be explained by gene conversions because they are found in the RhCE protein.
DIV has been divided serologically into DIVa and DIVb using polyclonal anti-Goa, an antibody defining the low-prevalence RhD antigen RH30 (Goa). All DIVa are Goa positive and DIVb are considered Goa negative.11 Similarly, DIVa but not DIVb carry the high-prevalence D epitope 4 (epD4) detectable with monoclonal antibodies (9 epitope model).26 Molecular characterization suggested different phylogenetic origins for DIVa and DIVb.27-29 Several molecular subtypes have been reported for DIVb,30-35 while DIVa is less diverse at the molecular level.
A complete record of the DIV types and their phylogeny was missing. We provide a detailed and systematic description of DIV alleles and propose an easy and comprehensive nomenclature. A large case collection of DIV carriers with anti-D is presented. We also propose an improved routine typing strategy, which would avoid anti-D immunization in pregnancies and transfusions of individuals carrying the DIV phenotype or some other partial D.
Materials and Methods
Immunohematology
Serologic testing of IgG and IgM monoclonal antibodies for agglutination was done by a gel matrix test (LISS-Coombs 37°C with polyspecific rabbit anti-IgG and monoclonal anti-C3d, clone C139-9, ID Micro Typing System; DiaMed; Cressier sur Morat, Switzerland).36 Monoclonal IgM anti-D approved for routine use in Germany were BS226 and BS232 (Seraclone Anti-D (RH1); Biotest, Dreieich, Germany), RUM-1 (immuClone Anti-D rapid; Immucor, Rödermark, Germany) and D175-2 (immuClone Anti-D fast; Immucor). Several monoclonal anti-D from RhD typing kits were used as detailed in the Results (D Screen; Diagast, Loos, France; and Advanced Partial RhD Typing Kit; Alba Bioscience, Edinburgh, UK). Two monoclonal IgG anti-D LOR17-6C7 and LHM76/58 came from the International Workshop on Monoclonal Antibodies against Human Red Blood Cells and Related Antigens in Nantes 1996.37 The sources of further antisera are detailed in Table 3.
Table 3.
Serologic reactivity with panels of anti-D
| Monoclonal anti-D |
Reactivity* |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone | Isotype | epD† | DIV type 1 ccDee | DIV type 3 CcDee | DIV type 4 CcDee | DIV type 5 ccDEe | DWN ccDee | DNU ccDEe | DNB ccDEe | DIIIc CCDee | DIII type 4 CcD.ee | DNT CcDee |
| BS226‡ | IgM | 6.4 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| BS232‡ | IgM | 6.4 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ND | ++++ |
| RUM-1‡ | IgM | 6.1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ND | ++++ |
| D175-2‡ | IgM | 6.1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | +++ | ++++ |
| HM10‡ | IgM | 6.6 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| HM16§ | IgG | 6.4 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| P3X61§ | IgM | 6.1 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| P3X35§ | IgG | 5.4 | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | +++ | ++++ | ++++ | ++++ |
| P3X212 11 F1§ | IgM | 8.2 | ++++ | +++ | +++ | ++++ | +++ | +++ | +++ | ++++ | ++++ | +++ |
| P3X212 23 B10§ | IgM | 9.1 | - | - | - | - | - | - | ++ | ++++ | ++++ | ++++ |
| P3X241§ | IgG | 5.4 | ++++ | ++++ | ++++ | +++ | +++ | +++ | +++ | ++++ | ++++ | ++++ |
| P3X249§ | IgG | 2.1 | - | - | - | - | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| P3X290§ | IgG | 3.1 | - | - | - | - | ++++ | +++ | +++ | ++++ | ++++ | ++++ |
| LHM76/58∥ | IgG1λ | ND | ++++ | - | - | - | ++++ | +++ | +++ | ++++ | ++++ | ++++ |
| LHM76/59∥ | IgG1 | ND | - | - | - | - | ++++ | +++ | +++ | ++++ | +++ | ++++ |
| LHM174/102∥ | IgG3κ | 1.2 | - | - | - | - | +++ | +++ | +++ | ++++ | ++++ | +++ |
| LHM50/2B∥ | IgG1λ | 6.3 | ++++ | - | ++ | (+) | ++++ | ++++ | +++ | ++++ | ++++ | ++++ |
| LHM169/81∥ | IgG3κ | 1.1 | - | - | - | - | +++ | +++ | +++ | ++++ | ++++ | ++++ |
| ESD1∥ | IgG1κ | ND | - | - | - | - | ++++ | +++ | ++ | ++++ | ND | ++++ |
| LHM76/55∥¶ | IgG1κ | 3.1 | - | - | - | - | ++++ | +++ | +++ | ++++ | ++++ | ++++ |
| LHM77/64∥¶ | IgG1κ | 9.1 | - | - | - | - | - | - | +++ | ++++ | +++ | ++++ |
| LHM70/45∥¶ | IgG1λ | 1.2 | - | - | - | - | + | +++ | +++ | ++++ | ++++ | +++ |
| LHM59/19∥¶ | IgG3κ | 8.1 | +++ | +++ | +++ | ++++ | +++ | +++ | +++ | ++++ | +++ | ++++ |
| LHM169/80∥¶ | IgG3λ | 6.3 | ++++ | ++++ | +++ | ++++ | ++++ | +++ | +++ | +++ | ++++ | ++++ |
| LHM57/17∥ | IgG1λ | 6.3 | ++ | ++ | + | +++ | +++ | ++ | + | +++ | ND | +++ |
| LDM-1¶ | IgM | ND | +++ | + | + | +++ | ++ | ++ | + | +++ | ND | +++ |
| BS221** | IgG | 6.3 | +++ | ++ | +++ | ++ | +++ | +++ | +++ | ++++ | ND | ++++ |
| BS227** | IgG | 2.2 | - | - | - | - | + | +++ | +++ | ++++ | ++++ | ++++ |
| BS228** | IgG | 6.3 | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | +++ | ++++ | ND | ++++ |
| BS229** | IgG | 5.4 | ++++ | ++++ | ++++ | ++++ | ++++ | ++++ | +++ | ++++ | ND | +++ |
| BS231** | IgG | 5.4 | ++++ | ++++ | ++++ | +++ | ++++ | ++++ | +++ | ++++ | ND | ++++ |
| H41** | IgG | 3.1 | - | - | - | - | +++ | ++ | +++ | ++++ | ++++ | +++ |
| LOR17-6C7 | IgG | 4.1 | ++++ | - | - | - | - | ++++ | - | ++++ | ++++ | ++++ |
Gel matrix tests with antiglobulin.
epD patterns as described previously by Scott.26 ND - not determined. NA – not applicable.
Monoclonal anti-D approved for routine use in Germany.
D-Screen; Diagast.
Advanced Partial RhD Typing Kit; Alba Bioscience. The results obtained with LHM76/58 and with monoclonal antibody no. 74 documented as LHM76/58 in the Nantes workshop37 differed. The reactivity of ESD1 differed from that of “ESD1M?” (monoclonal anti-D no. 1-56) documented in the Paris workshop where specificities for epD4.1 and epD9.1 were listed.26
ID-Partial Rh D-Typing Set; DiaMed.
Monoclonal anti-D Panel; Biotest.
Commercial monoclonal IgM anti-C were MS24 (Ortho BioClone; Ortho, Neckargemünd, Germany and Gamma-clone; Immucor, Norcross, USA) and MS273 (immuClone 2; Immucor, Rödermark, Germany). The anti-Goa sera A1149 and DL34678B were kindly provided by M.K. Moulds and showed identical results with all partial D and weak D types except weak D type 4.0. The 3 weak D type 4.0 samples tested did not react with the A1149 serum, were noted as Goa negative in the figure, but reacted with the DL34678B serum possibly due to an additional specificity in the DL34678B serum.
The mean antigen density was determined by flow cytometry according to the protocol described previously38 with monoclonal IgG anti-D BS221, BS227, BS228, BS229, BS231 and H4111B7 (Biotest). The secondary antibody was goat anti-human IgG, Fab-fragment, FITC-conjugated (Jackson ImmunoResearch Laboratories, West Grove, USA).
Molecular methods
RHD and RHCE nucleotide sequencing from genomic DNA for exons 1 through 10 including adjacent flanking intron regions and RHD exon specific PCR was performed as described.31,39-41 All DIV type 1.0 samples were sequenced in full length as well as DWN, DNT and at least one example of each reported DIV allele; further samples were identified by RHD exon specific PCR and nucleotide sequencing of relevant RHD exons. The presence of the ceTI typical nucleotide substitution 1025C>T was determined by sequencing of RHCE exon 7.42
Serological population screen
From February until September 1995, we examined 78,156 blood donations in South-Western Germany, of whom 60,965 were D positive. An autoanalyzer (Olympus PK7100; Olympus, Hamburg, Germany) was used to test for reactivity with 7 IgM anti-D including BS226 and BS232 (Biotest), P3X212 11 F1 and P3X212 23 B10 (Diagast) and RUM-1 (Immucor).17,43 P3X212 23 B10 binds to epD9,15 also published as epD23,26,44 which is absent from DIVa and DIVb. P3X212 11 F1 binds to epD8,15 also published as epD22,26,44 which is absent e.g. from DVII, DVI, R0Har and DHMI. The antibodies were used in a dilution of 1:1,000 in phosphate buffered 0.9% NaCl (Immusol; Dade Behring, Liederbach, Germany) with 0.22% bovine serum albumin (Ortho, Neckargemünd, Germany) as described previously.43 25 μl of an 1.6% RBC suspension prepared in Immusol with 0.1% bromeline (Roth, Karlsruhe, Germany) was mixed with 25 μl antibody solution. After 1 h at 25 °C the incubation mixture was read with the autoanalyzer. D positive donors typing negative with P3X212 23 B10 or P3X212 11 F1 were further tested with panels of monoclonal anti-D. All DIV were also characterized by molecular methods. Several publications have covered samples from this 9 months’ blood donor survey in 1995,17,19,24,29,31,32,38,45-48 which commenced the molecular D variant analysis that took place in Ulm since.
Rhesus Immunization Registry
Samples from D positive patients with suspected anti-D were collated in the Rhesus Immunization Registry (RIR) from 1998 to 2009.21 A possible immunization event, such as a transfusion or pregnancy, had been documented in all patients.
Nomenclature
We followed the common practice to abbreviate the names of the D categories; for example, we used the abbreviation DIV for D category IV.
All names for alleles and other genetic structures are italicized; all names for phenotypes and protein structures are in regular font.
DIII type 4 and DIII type 5 are related to DIV type 1.0 and presented for comparison. DIII type 5 was first characterized in 2002;28 according to recent findings the nucleotide sequence of DIIIa is in fact identical to DIII type 5,49,50 while the sequence originally assigned to DIIIa in 1997 was incomplete.51 These findings imply that the phenotypes previously described by DIII type 5 28 and DIIIa 51 are identical. We are aware of the internet-based allele terminology that had been drafted by an ISBT subcommittee and proposed at the time of manuscript submission.52 Because 15 years of literature refers to a putative nucleotide structure dubbed DIIIa,51 now known to be incorrect,50 we propose discontinuing the use of DIIIa as an allele designation. We advocate reserving the designation DIIIa for the description of the serologic phenotype,12 which represents its original definition, and using DIII type 5 for the allele and DIII type 5 for the protein,28 which makes the DIIIa allele and DIIIa protein designations51 obsolete.
The allele DWN was named after the two characteristic amino acid substitutions tryptophane (W) and asparagine (N) and has been observed before without attribution of a name.53 The designation DNT was derived from the amino acid substitution asparagine (N) to threonine (T).
Results
RHD alleles
We collected and characterized 23 samples with DIV phenotype (Table 1), 16,27,28,31-35,54-58 among which 4 molecular structures were encountered. The 3 DIV type 1.0 samples differed from the DIV type 1.1 described in 199530 by the presence of the additional amino acid substitution A137V28 (Table 1). The partial D DNT carried the amino acid substitution N152T in isolated form; which is one of the four amino acid substitutions characteristic of DIV type 1.0. The partial D DWN is similar to DIV type 4 except for amino acid position 350, which is variant in all DIV types but normal in DWN.
Table 1.
Molecular bases of DIV types and the 2 partial D DWN and DNT
|
RHD allele |
Predicted membrane localization† | Probands of this study |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Proposed name* | Previous designation16, 54, 55 | Nucleotide substitution in RHD gene† | Effect on protein sequence | Exon involved | probable Haplotype | Phenotype | n | References | |
| DIV type 1.0 ‡ | DIVa-2 | dispersed 186G>T 410C>T 455A>C 1048G>C |
dispersed L62F A137V N152T D350H |
2 3 3 7 |
TM TM TM EF |
cDe | ccDee | 3 | 28 |
| DIV type 1.1 | DIVa type 1 54 | dispersed 186G>T 455A>C 1048G>C |
dispersed L62F N152T D350H |
2 3 7 |
TM TM EF |
NA§ | NA | 0 | 30, 55; original decription30 shown to be incorrect56 |
| DIV type 2 | DIVb type 2 | gene conversion 1048C to 1193T |
gene conversion 350H to 398V |
part of 7 to 9 | EF, TM, IC | NA | NA | 0 | 30, 55 |
| DIV type 3 ‡∥ | DIVb type 3 | gene conversion 916A to 1193T |
gene conversion 306I to 398V |
6 to 9 | TM, IC, EF | CDe | CcDee CCDee CcDEe |
5 3 1 |
31, 32, 55 |
| DIV type 4 ‡ | DIVb type 4 | gene conversion 1048C to 1061A |
gene conversion 350H to 354N |
part of 7 | EF | CDe | CcDee | 8 | 33, 34, 55 |
| DIV type 5 ‡ | DIVb(J) | gene conversion 941T to 1193T |
gene conversion 314V to 398V |
7 to 9 | TM, IC, EF | cDE | ccDEe | 3 | 54 |
| DWN ‡ | NA | gene conversion 1053T to 1061A |
gene conversion 353W to 354N |
part of 7 | EF | cDe | ccDee | 1 | this study |
| DNT ‡ | NA | 455A>C | N152T | 3 | TM | CDe | CcDee | 1 | this study57, 58 |
This name is also proposed for the D variant protein, for which it should be used in regular font (no italics).
Numbering refers to cDNA; all gene conversions from RHCE. TM = transmembraneous; IC = intracellular; EF = exofacial.
The nucleic acid sequence data were deposited in EMBL under accession numbers HF549086 (DIV type 1.0), HF549087 (DIV type 3), HF549085 (DIV type 4), HF549083 (DWN), HF549084 (DNT). Hyodo et al.35 have deposited AB037270 (cDNA of DIV type 5).
NA = not available.
The 3’ breakpoint was confirmed (n = 6) as described previously.31
Anti-D immunization
In an internet-based survey from 1998 to 2009 (Rhesus Immunization Registry), we collated cases of anti-D immunizations occurring in D positive individuals.21 124 samples were submitted and analyzed at the molecular level. 12 were DIV, among which DIV type 4 and type 3 were the most frequent (Tables 1 and 2). Strong anti-D antibodies were observed in DIV type 1.0 and type 4 carriers. Also DWN and DNT were recognized because their carriers had produced anti-D.
Table 2.
Allo-anti-D in patients with DIV, DWN and DNT between 1998 and 2008
| Possible immunization events |
||||||||
|---|---|---|---|---|---|---|---|---|
| RHD allele | Case* | Sex | Phenotype | Anti-D titer† | RBC unit transfusion | Pregnancies | Date | Ethnicity |
| DIV type 1.0 | RIR-24‡ RIR-118 |
female female |
ccDee ccDee |
256 16 |
none none |
2∥ 2 live births 1 miscarriage∥ |
before 1997 2006 |
Mulatto (Dominican Republic) African (Togo) |
| DIV type 3 | RIR-23 RIR-26 RIR-94 |
female female female |
CcDee CcDee CcDee |
1 2 8 |
none unknown 2 D positive∥ |
2∥ at least 1 unknown |
before 1997 before 2000 1996 |
Swiss German German |
| DIV type 4 | RIR-18 RIR-29 RIR-80 RIR-85 RIR-105 |
female male female female male |
CcDee CcDee CcDee CcDee CcDee |
4 256 1 128 8 |
none 1 D positive∥ unknown none 10 D positive∥ |
4∥ NA§ 7∥ 2live births 3miscarriages∥ NA |
before 1990 2000 before 1997 before 1997 2006 |
German German Yugoslavian German Turkish |
| DIV type 5 | RIR-87 RIR-119 |
female female |
ccDEe ccDEe |
2 8 |
unknown several∥ |
9∥ 1 |
before 1997 1988 |
German German |
| DWN ¶ | RIR-112 | female | ccDee | 16 | several | at least 1∥ | before 2000 | African (USA) |
| DNT | RIR-84 | female | CcDee | 16 | unknown | 2∥ | 2001 | German |
Rhesus Immunization Registry (RIR) entries, data available online.21
Auto-anti-D were excluded by negative direct antiglobulin tests in 8 of 14 samples. RIR-80, RIR-85, RIR-94, RIR-105, RIR-118 and RIR-119 samples had a positive direct antiglobulin test but antibody elution was negative; autologous absorptions were generally not performed due to lack of an appropriate amount of RBC. Titers were done in gel matrix test.36 Additional antibodies were anti-E (RIR-23); anti-E (RIR-29); anti-Fya (RIR-84); anti-Fyb, anti-S and anti-M (RIR-85); anti-E and anti-K (RIR-112); and anti-C (RIR-119).
RIR-24 compound heterozygous for DIV type 1.0/RHDΨ.
NA = not applicable.
Likely immunization cause.
There was one additional DWN sample with anti-D titer 32 and anti-E, carrying the DWN/RHDΨ genotype.
Ethnicity
At least 2 of the 3 DIV type 1.0 carriers were of African origin; the ethnicity of the 3rd carrier is unknown. All 20 DIV type 3 to DIV type 5 carriers were Caucasians; 17 were donors or patients from Germany and 1 patient each was of Turkish, Yugoslavian and Swiss origin. The DNT carrier was a Caucasian from Germany while the DWN carrier was an African American.
Immunohematology
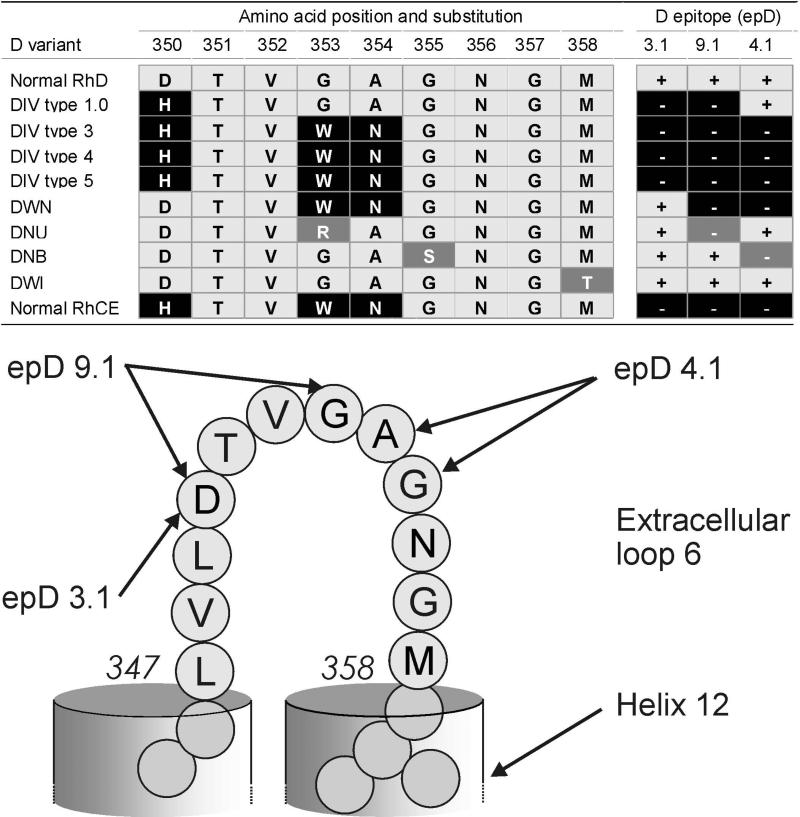
DIV types and related D variants were tested with monoclonal anti-D to establish the epitope patterns (Table 3).26,37 Samples of DIIIc,32 DIII type 4,32 DNU,59 DNB59 and DWI60 were tested for comparison. Characteristic of DIV were the lack of reactivity with anti-D clones specific for epD 1.1, 1.2, 2.1, 2.2, 3.1 and 9.1. An antibody directed against epD4.1 (clone no. LOR17-6C7) recognized DIV type 1.0 but not DIV type 3 to type 5, DWN and DNB. Only DIV type 1.0 samples were Goa positive (Fig. 1).
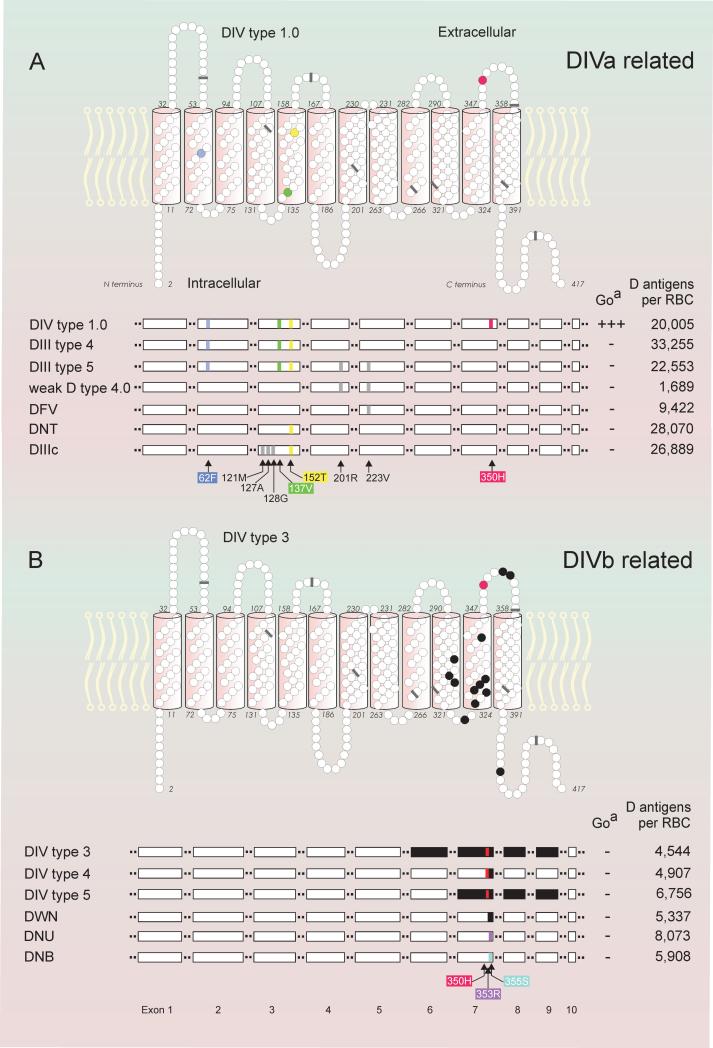
Figure 1.
Molecular structure, antigen density and anti-Goa reactivity of DIV variants. DIVa-related variants (panels A) and DIVb-related variants (panels B) are shown with models of the 2-dimensional structure (upper panels of A and B) and the linear exon structure (lower panels of A and B). In the 2-dimensional models of the RhD protein, amino acid substitutions are indicated (colored circles) together with the positions of the 9 exon boundaries (bars) as reflected in the RHD cDNA. DIV type 1.0 exemplifies the typical structure of DIVa-related variants with dispersed amino acid substitutions and DIV type 3 represents a typical structure of DIVb variants resulting from gene conversions (black circles). The linear exon structures of several related D variants are compared (white horizontal bars, lower panels of A and B); typical amino acid substitutions corresponding to nucleotide substitutions (colored and gray vertical bars) as well as RHCE-like gene conversions (black bars) are indicated. Antigen densities of DIII type 4 and DIIIc,32 of weak D type 4.0 and DFV29 and of DIV type III, DNU and DNB59 were published previously.
Antigen densities
The number of D antigens per RBC were large in DIV type 1.0, DIII type 4 and type 5 as well as DNT, while moderately reduced in DIV type 3 to type 4, DWN, DNU and DNB (Fig.1).
Population frequencies
Among 78,156 blood donations,17 we detected 8 samples with DIV phenotype by applying a high throughput serologic screening procedure followed by further serological and molecular characterization. 4 DIV type 3, 3 DIV type 4 and 1 DIV type 5 donors were identified (Table 4), who are among the 23 DIV samples listed in Table 1. Furthermore, we detected 80 samples with DVII phenotype and 1 sample each of DNU and DFW.
Table 4.
Phenotype frequencies of D variants in the Southwest German population
| Phenotype frequency |
|||
|---|---|---|---|
| partial D | Estimate | 95% Confidence interval* | Donations† (n) |
| DVII | 1:977 | 1:788 - 1:1,264 | 80 |
| all DIV | 1:9,770 | 1:5,238 - 1:23,792 | 8 |
| DIV type 1.0 | NA | 1:26,087 - NA | 0 |
| DIV type 3 | 1:19,539 | 1:8,143 - 1:57,215 | 4 |
| DIV type 4 | 1:26,052 | 1:9,647 - 1:95,545 | 3 |
| DIV type 5 | 1:78,156 | 1:14,683 - 1:1,532,470 | 1 |
| DNU | 1:78,156 | 1:14,683 - 1:1,532,470 | 1 |
| DFW | 1:78,156 | 1:14,683 - 1:1,532,470 | 1 |
95% confidence interval of the frequency estimate according to Poisson distribution.
D variants among 78,156 blood donations, of which 60,965 were D positive. All DIV samples came from different donors. The 80 donations with DVII were from 71 donors.
DIV and ceTI
Our 3 DIV type 1.0 samples were serologically C negative with 3 monoclonal IgM anti-C including Gamma-clone, which is known to detect weak C expression in rare samples with the DIVa(C) phenotype.42,61 They all carried the RHCE allele ceTI 42 as evident from the presence of a heterozygous T/C at nucleotide position 1,025. Samples of DIV type 3 (n = 3), DIV type 4 (n = 2), DIV type 5 (n = 3), DWN (n = 1), DIII type 4 (n = 1), DIII type 5 (n = 1) and DNT (n = 1) did not carry the ceTI allele.
Discussion
A comprehensive characterization is available for most of the clinically important partial D but was missing for the group of DIV alleles. The current D typing strategy applied in Germany and several European countries protects individuals with DVI from immunization by normal D. However, patients with DIV and other partial D are typed D positive and hence can get immunized by transfusion of D positive blood. An improved typing strategy will be particularly relevant in women in childbearing age and girls. To avoid adverse effects of anti-D immunization, detailed knowledge is required for the involved alleles and the susceptibility to become immunized by normal D. This objective prompted us to address the molecular basis and to determine the clinical importance of the various DIV alleles.
What was initially defined as DIV, was later recognized to comprise a group of different phenotypes and alleles. DIV and its subgroups DIVa and DIVb are serologically defined; DIV lacks epD 1, 2, 3 and 9 or parts thereof;24,26,60,62,63 DIVa carries epD4 and the low prevalence antigen Goa (RH30), which are absent from DIVb (Table 5).11,26,64 The differences between DIVa and DIVb at the serological level are mirrored by distinct molecular structures, evolutionary origins and ethnic prevalences (Fig. 1 and Table 5). 350H is the only variant amino acid residue that is shared by all DIV types.
Table 5.
Differences between DIVa and DIVb
| DIVa* DIV type 1.0† |
DIVb* DIV type 2 to type 5† |
|
|---|---|---|
| Epitopes | ||
| Goa | positive | negative |
| epD4 | positive | negative |
| D antigen expression | high | moderately reduced |
| RHD nucleotide substitutions | dispersed | RHD-CE-D gene conversions |
| RHCE | associatied with ceTI | regular exon 7 |
| Haplotype | cDe or (C)De‡ | CDe or cDE |
| Phylogeny | old extant allele, primordial allele of the DIVa cluster in the African cluster | gene conversions in the Eurasian cluster |
| Typical ethnic origin | African | Caucasian |
Defined serologically
Defined at the molecular level
A minor fraction of DIVa samples expresses a weak C antigen.64
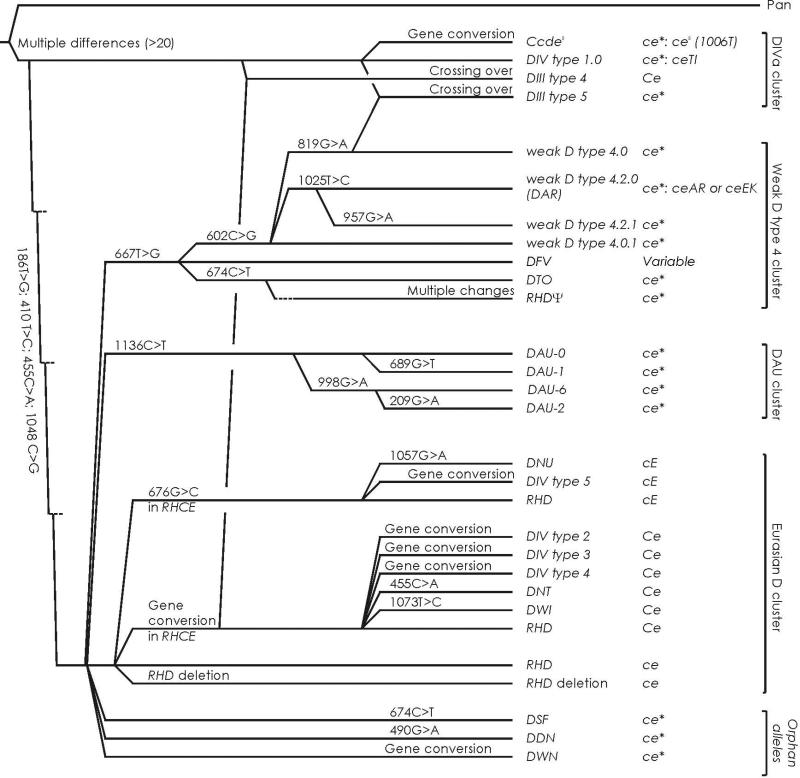
The DIVa phenotype is expressed by DIV type 1.0. The original publication of DIV type 1.130 has been incorrect;56 if DIV type 1.1 is ever found, the DIVa phenotype will need to be tested in that sample. We observed DIV type 1.0 only (Table 1), which is the primordial allele of the DIVa cluster (Fig. 2).28 The DIVa cluster is one of the 3 African clusters and encompasses also the DIII type 4 and DIII type 5 alleles as well as the (C)cdes haplotype. All these DIVa-related variants (Fig. 1A) share the amino acids 62F, 137V and 152T, which are likely remnants of evolutionary old alleles.28,32 62F and 137V are highly conserved in the animal kingdom and occur in the Rh-related proteins RhAG, RhBG and RhCG. 152T is an RhCE typical amino acid present in several mammal species and is regarded as the specific amino acid substitution of the DIVa cluster relative to the normal Eurasian RHD allele.28 We observed the corresponding D variant with N152T as the sole difference to the normal RhD protein. This allele, dubbed DNT, was evolutionary unrelated to the DIVa cluster and may belong to the Eurasian D cluster because it differed in haplotype. Furthermore, the DNT sample was found in a Caucasian, while DIV type 1.0 typically occurs in Africans.
Figure 2.
Phylogeny of DIV. The phylogenetic tree is based on a previous tree for RHD29,39 with 4 main branches representing allele clusters. The three African D clusters have characteristic primordial amino acids and typically occur in a cDe haplotype while the Eurasian D cluster has normal RHD as the primordial structure in which the Ce and cE alleles of RHCE evolved. The DIVa cluster is one of the 3 African D clusters; D variants of this cluster share the ancestral amino acid substitutions 62F, 137V and152T relative to the consensus RhD. In contrast, DIV variants with DIVb phenotype evolved in the Eurasian D cluster by gene conversions. The typically associated RHCE alleles are shown.
The DIVb phenotype is expressed by DIV type 2 to type 5, which are structurally distinct from DIV type 1 because they are encoded by RHD-CE-D hybrid alleles of the RHD gene (Table 1, Fig. 1B and Table 5). They arose as members of the Eurasian D cluster (Fig. 2). While DIV type 1 typically occurs in Africans, all our samples of DIV type 3 to type 5 were from Caucasians (Table 5). DIV type 4 represents the RHD-CE-D gene conversion with the shortest RHCE segment encoding only 3 amino acids (Table 1). All DIVb-related variants share the structural motif of DIV type 4 but harbor further stretches of RhCE-like amino acids which are located in the transmembraneous or intracellular section of the protein (Fig. 1B).
D350H, the sole amino acid substitution shared by all DIV variants, is part of extracellular loop 6 of the RhD protein (Fig. 1 and Fig. 3).65 This RhCE typical substitution causes a change in charge and hydrophobicity likely affecting anti-D binding. The associated antigenic loss defines the DIV phenotype. The D variant DWN was similar to DIV type 4 but lacked the critical D350H substitution and was instrumental in determining the influence of 350D for D antigen expression. The comparison of epitopes expressed by several D variants with substitutions at loop 6 (Table 3) corroborated previous reports24,60,62,63 on the allocation of epD 3, 4 and 9 to distinct amino acids of loop 6 (Fig. 3).
Figure 3.
Amino acids at extracellular loop 6 critical for D epitope (epD) expression. epD were allocated on the basis of reactivities with monoclonal anti-D P3X290 and LHM56/55 (epD 3.1), with LOR17-6C7 (epD 4.1) as well as P3X212 23 B10 and LHM77/64 (epD 9.1). Further splits of these epD were previously described.24,60 DNAK is another partial D65 with a single amino acid substitution in loop 6 (G357D, not shown).
In 1977, DIV could be divided into DIVa and DIVb by serologic criteria.11,26 DIVa is Goa positive and occurs in African populations, while DIVb lacks Goa and occurs in Caucasians. The first 2 alleles expressing these 2 DIV phenotypes were characterized on a molecular level in 1995 and named DIVa and DIVb.30 Neither allele was observed among our 24 DIV samples or has been confirmed since the original description. 4 additional DIV alleles have been described since 1995 and confirmed in this study.
We propose to reserve the designations DIVa and DIVb for the description of the serologic phenotypes and to use the designations DIV type 1 to type 5 for the alleles characterized molecularly (Table 1). The 2 initially described alleles 30 were named DIV type 1.1 and DIV type 2 (Table 1).55DIV type 1.028 has been confirmed independently,56,66 while DIV type 2 has not been confirmed and may attract further scrutiny. Alleles encoding the DIVb phenotype are labeled DIV type 2 to type 5 (Table 1). DIV type 2 cannot be frequent,30,55 if extant at all, and DIV types 3 and 4 kept their original names.32-34 The allele DIVb(J), first described in Japan,35 was designated DIV type 5.
A small fraction of DIVa samples express C antigen very weakly.61 The initial presumption42 that this C expression may be caused by the RHCE variant ceTI has not been confirmed;67 recently, this weak C expression in DIVa(C) was attributed to a hybrid allele at the RHCE locus.64 Our 3 DIV type 1.0 samples, lacking C expression, occurred in a genotype heterozygous for ceTI, which is in accordance with independent observations.42,67 The ceTI allele has not been found associated with the DIVb phenotype, at least not in our sample collection.
All DIV variants appear D positive by routine serology, because their densities are larger than 4,500 D antigens per RBC. DIVa-related D variants, which carry the amino acid substitution N152T, had a density of larger than 20,000 D antigens per RBC (Fig. 1A), while DIVb-related D variants had a moderately reduced density of lower than 8,000 D antigens per RBC (Fig. 1B). Typical antigen densities68 are 22,750 for CDe/CDe (R1R1)32 and 24,500 for CDe/cDE (R1R2).32 N152T may facilitate the integration of the RhD protein into the RBC membrane.31 Accordingly, the D variant DNT with the single amino acid substitution N152T had an unusual large density of 28,000 D antigens per RBC.57 Similarly, increased antigen densities were observed in other D variants harboring the N152T substitution such as DIV type 1, DIII type 4 and DVI type III.19,31 The relatively large antigen density of DIII type 5 as compared to weak D type 4.0 may be due to the presence of 152T as well. DIII type 5 arose from a recombination between alleles of the DIVa and the weak D type 4 clusters (Fig. 2).28,69 The substitution T201R likely caused the low antigen density of weak D type 4.0 (Fig. 1A).
During the years 1998 - 2009, cases of D positive patients with anti-D were collated in the Rhesus Immunization Registry.21 Among 124 cases was 1 patient with anti-LWa, the others had an allo- or an auto-anti-D. Allo-anti-D was confirmed in 70 cases comprising 12 DIV, 12 DVI, 11 R0Har, 9 DNB, 6 DVII, 5 DIII, 2 DV, 2 weak D type 4.2/DAR; and 1 each of DAU-3, DAU-4, DDE, DFL, DHMI, DMI, DNT, DOL, DWN, weak D type 11 and weak D type 15. The number of registered cases did not correlate with the population frequency of the involved D variants: the prevalence of D variants in the population of Southern Germany are 1 in 977 for DVII, 1 in 6214 for DVI,18 and 1 in 9770 for DIV (Table 4). Individuals with DVII are, therefore, much less prone to anti-D immunization than individuals with DVI and DIV. Among the 12 cases with DIV and anti-D were only 2 males. 7 immunizations were suspected or confirmed to be caused by pregnancy (Table 2). 79% of all patients with partial D and anti-D were females and pregnancy was the most frequent immunization cause. Therefore, women of childbearing age and girls would especially benefit from measures to prevent further anti-D immunization. In a study conducted in the USA, among 21 cases of anti-D immunizations in D-positive individuals one third were cases with DIV70 including 6 DIVa and 1 DIVb; 6 cases involved weak D type 4.2.
The present typing strategy for the D antigen in recipients, introduced 1996 in Germany, aims at preventing anti-D immunization in patients with DVI (1:6,214),18 the clinically most relevant partial D in Caucasians.19,71 DNB (1:292 to 1:1644)59 and DVII are the most frequent known partial D in Caucasians but of lesser clinical relevance. Currently 2 monoclonal anti-D that react with epD6 26 and do not recognize DVI are used for typing of recipients. Consequently, recipients with DVI are typed and transfused D negative. However, recipients with DIV are not protected by this strategy developed in Europe18,71 and adapted to Caucasians. Electing reagents for typing of the D antigen is posing an ongoing challenge.72
It would be valuable to also protect carriers of other clinically relevant partial D, including DIV, from becoming immunized by the D antigen. To this end, an adaptation of the current serological D typing strategy for recipients would be necessary and can be implemented at the routine level. A set of 2 monoclonal anti-D antibodies could be used with one not recognizing DVI, and another not recognizing other clinically important D variants. For instance, an antibody binding epD9.1, like P3X212 23 B10 and LHM77/64 (Table 3), would neither recognize any DIV type (Table 3) nor R0Har 73 or give only weakly positive results with several other partial D. Other useful target epitope and monoclonal anti-D may be epD3.1 and P3X290 or ESD1 (Table 3). For clinical purposes, appropriate monoclonal anti-D might be selected according to the prevalence of partial D in the population served.74 Discrepant results with the 2 monoclonal antibodies should prompt identification of the allele by molecular methods or evaluation by panels of monoclonal anti-D. Pregnant women and their children would especially benefit from this modified typing strategy.
Acknowledgements
The authors dedicate this paper to the memory of John James Moulds, MT(ASCP)SBB, who died on June 13, 2011 at the age of 67. With his lifetime contributions to blood group serology, he was a key person for our field and contributed profoundly to our understanding of blood group antigens and to establishing the foundation, on which all molecular immunohematology is built.
We thank all contributors of the Rhesus Imunization Registry framework21 for procuring blood samples and clinical data of D positive patients with anti-D carrying the D variants DIV, DIII, or DNB. Such samples were provided by J. Burkhart, München, W. Endres, Stuttgart, M. Heuberger, Kassel, B. Just, Hagen, A. Mildner, Reutlingen, E. Piro-Biehler, Weiden, P. Pirzer, Amberg, and A. Seltsam, Springe, all from Germany; S. Kilga-Nogler, Innsbruck, Austria; the late N. Eicher, Bern, Switzerland; J. Jorgensen, Aarhus, Denmark; M.R. Combs, Durham, NC, S. Frandson, San Diego, CA, and M.K. Moulds, Shreveport, LA, United States. DWI was a gift from G. F. Körmöczi, Vienna, Austria. One additional sample of DWN/RHDpsi came from S. Kuhn, Würzburg, Germany. The authors acknowledge the serology advice by Marylin K. Moulds and the technical assistance by Donna M. Baker and David Hanna in Shreveport, LA, and by Marianne Lotsch, Anita Link and Hedwig Erne in Ulm. This work was supported by the Intramural Research Programs of the DRK-Blutspendedienst Baden-Württemberg – Hessen, Mannheim and the NIH Clinical Center.
Footnotes
Statement of Disclaimer: The views expressed do not necessarily represent the view of the National Institutes of Health, the Department of Health and Human Services, or the U.S. Federal Government.
Conflict of Interest: FFW and WAF receive royalties for RHD genotyping patents. WAF holds intellectual property rights for RHD genotyping and serves on the Scientific Advisory Board of Immucor (non-remunerated). JMM serves as principal investigator for an investigator agreement with BioArray Solutions, Immucor. IvZ declares no conflict of interest.
Reference List
- 1.Argall CI, Ball JM, Trentelman E. Presence of anti-D antibody in the serum of a Du patient. J.Lab.Clin.Med. 1953;41:895–8. [PubMed] [Google Scholar]
- 2.Salmon C, Cartron JP, Rouger P. The Human Blood Groups. Masson; New York: 1984. [Google Scholar]
- 3.Issitt PD, Telen MJ. Introduction of the term “partial D”. Transfusion. 1996;36(8):761–2. doi: 10.1046/j.1537-2995.1996.36296181917.x. [DOI] [PubMed] [Google Scholar]
- 4.Unger LJ, Wiener AS. Some observations on blood factors RhA, RhB and RhC of the Rh-Hr blood group system. Blood. 1959;14:522–34. [PubMed] [Google Scholar]
- 5.Unger LJ, Wiener AS, Katz L. Studies on blood factors RhA, RhB, and RhC. J.Exp.Med. 1959;110:495–510. doi: 10.1084/jem.110.4.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sacks MS, Wiener AS, Jahn EF, Spurling CL, Unger LJ. Isosensitization to a new blood factor, RhD, with special reference to its clinical importance. Ann.Int.Med. 1959;51:740–7. doi: 10.7326/0003-4819-51-4-740. [DOI] [PubMed] [Google Scholar]
- 7.Unger LJ, Wiener AS, Katz L. Studies of the quantitative antiglobulin test with serums of specificities anti-Rh O, anti-Rh A, anti-Rh B, anti-Rh C, and anti-Rh D. Am.J.Clin.Pathol. 1959 Dec;32:499–506. doi: 10.1093/ajcp/32.6.499. [DOI] [PubMed] [Google Scholar]
- 8.Wiener AS, Unger LJ. Further observations on the blood factors Rh-A, Rh-B, Rh-C and Rh-D. Transfusion. 1962 Jul;2:230–3. doi: 10.1111/j.1537-2995.1962.tb00230.x. [DOI] [PubMed] [Google Scholar]
- 9.Wiener AS, Unger LJ. Rh factors related to the Rh0 factor as a source of clinical problems. J.Amer.Med.Assoc. 1959;169:696–9. doi: 10.1001/jama.1959.03000240034008. [DOI] [PubMed] [Google Scholar]
- 10.Tippett P, Sanger R. Observations on subdivisions of Rh antigen D. Vox Sang. 1962;7:9–13. doi: 10.1111/j.1423-0410.1962.tb03223.x. [DOI] [PubMed] [Google Scholar]
- 11.Tippett P, Sanger R. Further observations on subdivisions of the Rh antigen D. Ärztl.Lab. 1977;23:476–80. doi: 10.1111/j.1423-0410.1962.tb03223.x. [DOI] [PubMed] [Google Scholar]
- 12.Tippett P, Lomas-Francis C, Wallace M. The Rh antigen D: partial D antigens and associated low incidence antigens. Vox Sang. 1996;70:123–31. doi: 10.1111/j.1423-0410.1996.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 13.Lomas C, Bruce M, Watt A, Gabra GS, Mueller S, Tippett P. TAR+ individuals with anti-D, a new category DVII. Transfusion. 1986;26(6):560. Ref Type: Abstract. [Google Scholar]
- 14.Daniels GL. Human Blood Groups. 2 ed. Blackwell Science; Oxford: 2002. [Google Scholar]
- 15.Lomas C, McColl K, Tippett P. Further complexities of the Rh antigen D disclosed by testing category DII cells with monoclonal anti-D. Transfus.Med. 1993;3:67–9. doi: 10.1111/j.1365-3148.1993.tb00106.x. [DOI] [PubMed] [Google Scholar]
- 16.Wagner FF. The RhesusBase. DRK-Blutspendedienst Baden-Württemberg - Hessen; Ulm: 1998. [Google Scholar]
- 17.Flegel WA, Wagner FF. The frequency of RHD protein variants in Caucasians. Transfusion Clinique et Biologique. 1996;3:10s. Ref Type: Abstract. [Google Scholar]
- 18.Wagner FF, Kasulke D, Kerowgan M, Flegel WA. Frequencies of the blood groups ABO, Rhesus, D category VI, Kell, and of clinically relevant high-frequency antigens in South-Western Germany. Infusionsther.Transfusionsmed. 1995;22(5):285–90. doi: 10.1159/000223144. [DOI] [PubMed] [Google Scholar]
- 19.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Three molecular structures cause Rhesus D category VI phenotypes with distinct immunohematologic features. Blood. 1998;91(6):2157–68. [PubMed] [Google Scholar]
- 20.Flegel WA, Khull S, Wagner FF. Primary anti-D immunization by weak D type 2 RBC. Transfusion. 2000;40(4):428–34. doi: 10.1046/j.1537-2995.2000.40040428.x. [DOI] [PubMed] [Google Scholar]
- 21.Flegel WA. The Rhesus Immunization Surveillance, 1998 - 2009. DRK-Blutspendedienst Baden-Württemberg - Hessen; Ulm: 1998. [Google Scholar]
- 22.Lurie S, Eliezer E, Piper I, Woliovitch I. Is antibody screening in Rh (D)-positive pregnant women necessary? J.Matern.Fetal Neonatal.Med. 2003 Dec;14(6):404–6. doi: 10.1080/14767050412331312260. [DOI] [PubMed] [Google Scholar]
- 23.Prasad MR, Krugh D, Rossi KQ, O'Shaughnessy RW. Anti-D in Rh positive pregnancies. Am.J.Obstet.Gynecol. 2006 Oct;195(4):1158–62. doi: 10.1016/j.ajog.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 24.Avent ND, Jones JW, Liu W, Scott ML, Voak D, Flegel WA, Wagner FF, Green C. Molecular basis of the D variant phenotypes DNU and DII allows localization of critical amino acids required for expression of Rh D epitopes epD3, 4 and 9 to the sixth external domain of the Rh D protein. Br.J.Haematol. 1997;97:366–71. doi: 10.1046/j.1365-2141.1997.632710.x. [DOI] [PubMed] [Google Scholar]
- 25.Rouillac C, Le Van Kim C, Beolet M, Cartron JP, Colin Y. Leu110Pro substitution in the RhD polypeptide is responsible for the DVII category blood group phenotype. Am.J.Hematol. 1995;49:87–8. doi: 10.1002/ajh.2830490115. [DOI] [PubMed] [Google Scholar]
- 26.Scott M. Section 1A: Rh serology. Coordinator's report. Transfus.Clin.Biol. 2002 Jan;9(1):23–9. doi: 10.1016/s1246-7820(01)00211-7. [DOI] [PubMed] [Google Scholar]
- 27.Rouillac C, Gane P, Cartron JP, Le Pennec PY, Colin Y. Molecular basis of the altered antigenic expression of RhD in weak D (Du) and RhC/e in RN phenotypes. Blood. 1996 Jun 1;87:4853–61. [PubMed] [Google Scholar]
- 28.Wagner FF, Ladewig B, Angert KS, Heymann GA, Eicher NI, Flegel WA. The DAU allele cluster of the RHD gene. Blood. 2002 Jul 1;100(1):306–11. doi: 10.1182/blood-2002-01-0320. [DOI] [PubMed] [Google Scholar]
- 29.Flegel WA, von Zabern I, Doescher A, Wagner FF, Strathmann KP, Geisen C, Palfi M, Pisacka M, Poole J, Polin H, et al. D variants at the RhD vestibule in the weak D type 4 and Eurasian D clusters. Transfusion. 2009 Oct 6;49(6):1059–69. doi: 10.1111/j.1537-2995.2009.02102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rouillac C, Colin Y, Hughes-Jones NC, Beolet M, D'Ambrosio A-M, Cartron JP, Le Van Kim C. Transcript analysis of D category phenotypes predicts hybrid Rh D-CE-D proteins associated with alteration of D epitopes. Blood. 1995;85:2937–44. [PubMed] [Google Scholar]
- 31.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood. 1999;93(1):385–93. [PubMed] [Google Scholar]
- 32.Wagner FF, Frohmajer A, Ladewig B, Eicher NI, Lonicer CB, Müller TH, Siegel MH, Flegel WA. Weak D alleles express distinct phenotypes. Blood. 2000;95(8):2699–708. [PubMed] [Google Scholar]
- 33.Wagner FF, Gassner C, Eicher NI, Lonicer C, Flegel WA. Characterization of D category IV type IV, DFW, and DNB. Transfusion. 1998;38(S1):63S. Ref Type: Abstract. [Google Scholar]
- 34.Körmöczi GF, Förstemann E, Gabriel C, Mayr WR, Schönitzer D, Gassner C. Novel weak D types 31 and 32: adsorption-elution-supported D antigen analysis and comparison to prevalent weak D types. Transfusion. 2005;45(10):1574–80. doi: 10.1111/j.1537-2995.2005.00580.x. [DOI] [PubMed] [Google Scholar]
- 35.Hyodo H, Ishikawa Y, Kashiwase K, Ogawa A, Watanabe Y, Tsuneyama H, Toyoda C, Uchikawa M, Akaza T, Fujii T, et al. Polymorphisms of RhDVa and a new RhDVa-like variant found in Japanese individuals. Vox Sang. 2000;78(2):122–5. doi: 10.1159/000031162. [DOI] [PubMed] [Google Scholar]
- 36.Flegel WA, Eicher NI, Doescher A, Hustinx H, Gowland P, Monsouri Taleghani B, Petershofen EK, Bauerfeind U, Ernst M, von Zabern I, et al. In-frame triplet deletions in RHD alter the D antigen phenotype. Transfusion. 2006;46(12):2156–61. doi: 10.1111/j.1537-2995.2006.01046.x. [DOI] [PubMed] [Google Scholar]
- 37.Rouger P, Muller J-Y. Third International Workshop and Symposium on Monoclonal Antibodies against Human Red Cells and Related Antigens: Section RH. Transfus.Clin.Biol. 1996;3(6):329–541. [PubMed] [Google Scholar]
- 38.Flegel WA, Curin-Serbec V, Delamaire M, Donvito B, Ikeda H, Jorgensen J, Kumpel BM, Le Pennec P-Y, Pisacka M, Tani Y, et al. Section 1B: Rh flow cytometry. Coordinator's report. Rhesus index and antigen density: an analysis of the reproducibility of flow cytometric determination. Transfus.Clin.Biol. 2002;9:33–44. doi: 10.1016/s1246-7820(01)00213-0. [DOI] [PubMed] [Google Scholar]
- 39.Wagner FF, Ladewig B, Flegel WA. The RHCE allele ceRT: D epitope 6 expression does not require D-specific amino acids. Transfusion. 2003 Sep;43(9):1248–54. doi: 10.1046/j.1537-2995.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- 40.Chen Q, Flegel WA. Random survey for RHD alleles among D+ European persons. Transfusion. 2005;45(7):1183–91. doi: 10.1111/j.1537-2995.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- 41.Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet. 2001;2(1):10. doi: 10.1186/1471-2156-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vege S, Meyer W, Copeland T, Westhoff CM. A new RHce allele, RHCE*ceTI, is associated with C typing discrepancies and is linked to RHD*DVIa. Transfusion. 2007;47(S3):159A. Ref Type: Abstract. [Google Scholar]
- 43.Flegel WA, Kaucher M, Schoty S, Hochgeladen E, Northoff H. Automated testing for Rhesus-antigen with monoclonal antibodies. In: Chester MA, Johnson U, Lundblad A, et al., editors. Proceedings of the Second International Workshop and Symposium on Monoclonal Antibodies against Human Red Blood Cells and Related Antigens. 1 ed. Symposium Secretariat; Lund: 1990. p. 151. [Google Scholar]
- 44.Scott M. Rh serology - coordinator's report. Transfus.Clin.Biol. 1996;3:333–7. doi: 10.1016/s1246-7820(96)80040-1. [DOI] [PubMed] [Google Scholar]
- 45.Flegel WA, Hillesheim B, Wagner FF. RHD category VII is caused by a single molecular event [Abstract]. Transfusion Clinique et Biologique. 1996;3:33s. Ref Type: Abstract. [Google Scholar]
- 46.Flegel WA, Wagner FF. RHD antigen density and agglutination in RHD variant red cells. Transfus.Clin.Biol. 1996;3:385–6. doi: 10.1016/s1246-7820(96)80049-8. [DOI] [PubMed] [Google Scholar]
- 47.Gassner C, Schmarda A, Kilga-Nogler S, Jenny-Feldkircher B, Rainer E, Müller TH, Wagner FF, Flegel WA, Schönitzer D. RHD/CE typing by polymerase chain reaction using sequence-specific primers. Transfusion. 1997;37(10):1020–6. doi: 10.1046/j.1537-2995.1997.371098016439.x. [DOI] [PubMed] [Google Scholar]
- 48.Müller TH, Wagner FF, Trockenbacher A, Eicher NI, Flegel WA, Schönitzer D, Schunter F, Gassner C. PCR screening for common weak D types shows different distributions in three Central European populations. Transfusion. 2001;41(1):45–52. doi: 10.1046/j.1537-2995.2001.41010045.x. [DOI] [PubMed] [Google Scholar]
- 49.Westhoff CM, Vege S, Hue-Roye K, Huang C, Reid ME. DIIIa and DIII type 5 partial D phenotypes are encoded by the same RHD variant allele. Transfusion. 2007;47(S3):158A. Ref Type: Abstract. [Google Scholar]
- 50.Westhoff CM, Vege S, Halter-Hipsky C, Whorley T, Hue-Roye K, Lomas-Francis C, Reid ME. DIIIa and DIII Type 5 are encoded by the same allele and are associated with altered RHCE*ce alleles: clinical implications. Transfusion. 2010 Jun;50(6):1303–11. doi: 10.1111/j.1537-2995.2009.02573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang CH, Chen Y, Reid M. Human DIIIa erythrocytes: RhD protein is associated with multiple dispersed amino acid variations. Am.J.Hematol. 1997;55:139–45. doi: 10.1002/(sici)1096-8652(199707)55:3<139::aid-ajh4>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 52.Storry JR, Castilho L, Daniels G, Flegel WA, Garratty G, Francis CL, Moulds JM, Moulds JJ, Olsson ML, Poole J, et al. International Society of Blood Transfusion Working Party on red cell immunogenetics and blood group terminology: Berlin report. Vox Sang. 2011 Jul;101(1):77–82. doi: 10.1111/j.1423-0410.2010.01462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fichou Y, Le MC, Bryckaert L, Guerry C, Benech C, Dupont I, Jamet D, Ferec C, Chen JM. Variant screening of the RHD gene in a large cohort of subjects with D phenotype ambiguity: report of 17 novel rare alleles. Transfusion. 2012;52(4):759–64. doi: 10.1111/j.1537-2995.2011.03350.x. [DOI] [PubMed] [Google Scholar]
- 54.Reid ME, Lomas-Francis C. The Blood Group Antigen Facts Book. 2 ed. Academic Press; San Diego: 2003. [Google Scholar]
- 55.Avent ND, Reid ME. The Rh blood group system: a review. Blood. 2000;95(2):375–87. [PubMed] [Google Scholar]
- 56.Reid ME, Ripaux M, Auxerre C, Reussel M, Nataf J, Rouger P, Le Pennec P-Y, Peyrard T. DIVa and DIVa-2 are encoded by the same RHD allele. Transfusion. 2012;52(Suppl. S3):34A. Ref Type: Abstract. [Google Scholar]
- 57.von Zabern I, Wagner FF, Hanna D, Moulds JM, Flegel WA. Clinical relevance of D category IV comprising phylogenetically heterogeneous alleles. Transfusion. 2009;49(Suppl. s3):119A. Ref Type: Abstract. [Google Scholar]
- 58.Fong C, Vege S, Weber KM, Denomme GA, Velliquette RW, Lomas-Francis C, Westhoff C. An RHD*455C allele encodes a partial D phenotype associated with production of allo anti-D. Transfusion. 2012;52(Suppl. S3):166A. Ref Type: Abstract. [Google Scholar]
- 59.Wagner FF, Eicher NI, Jorgensen JR, Lonicer CB, Flegel WA. DNB: a partial D with anti-D frequent in Central Europe. Blood. 2002 Sep 15;100(6):2253–6. doi: 10.1182/blood-2002-03-0742. [DOI] [PubMed] [Google Scholar]
- 60.Körmöczi GF, Legler TJ, Daniels GL, Green CA, Struckmann R, Jungbauer C, Moser S, Flexer M, Schönitzer D, Panzer S, et al. Molecular and serologic characterization of DWI, a novel “high-grade” partial D. Transfusion. 2004 Apr;44(4):575–80. doi: 10.1111/j.1537-2995.2003.03318.x. [DOI] [PubMed] [Google Scholar]
- 61.Salmon C, Gerbal A, Liberge G, Sy B, Tippett P, Sanger R. Le complexe génique DIV (C)- [The gene complex DIV (C)-]. Rev.Fr.Transfus. 1969 Jun;12(2):239–47. doi: 10.1016/s0035-2977(69)80066-0. [DOI] [PubMed] [Google Scholar]
- 62.Cartron JP, Rouillac C, Le Van Kim C, Mouro I, Colin Y. Tentative model for the mapping of D epitopes on the RhD polypeptide. Transfus.Clin.Biol. 1996;6:497–503. doi: 10.1016/s1246-7820(96)80070-x. [DOI] [PubMed] [Google Scholar]
- 63.Scott ML, Voak D, Jones JW, Avent ND, Liu W, Hughes-Jones N, Sonneborn H-H. A structural model for 30 Rh D epitopes based on serological and DNA sequence data from partial D phenotypes. Transfus.Clin.Biol. 1996;3:391–6. doi: 10.1016/s1246-7820(96)80051-6. [DOI] [PubMed] [Google Scholar]
- 64.Hipsky CH, Hue-Roye K, Lomas-Francis C, Huang CH, Reid ME. Molecular basis of the rare gene complex, DIVa(C)-, which encodes four low-prevalence antigens in the Rh blood group system. Vox Sang. 2012;102(2):167–70. doi: 10.1111/j.1423-0410.2011.01519.x. [DOI] [PubMed] [Google Scholar]
- 65.Bruce DG, Poole J, Tilley L, Wallis JP, Todd A. Immune alloanti-D in a patient with a novel RHD mutation. Transfusion Medicine. 2005;15(S1):52. Ref Type: Abstract. [Google Scholar]
- 66.Touinssi M, Chapel-Fernandes S, Granier T, Bokilo A, Bailly P, Chiaroni J. Molecular analysis of inactive and active RHD alleles in native Congolese cohorts. Transfusion. 2009;49(7):1353–60. doi: 10.1111/j.1537-2995.2009.02161.x. [DOI] [PubMed] [Google Scholar]
- 67.Westhoff CM, Vege S, Halter HC, Hue-Roye K, Copeland T, Velliquette RW, Horn T, Lomas-Francis C, Reid ME. RHCE*ceTI encodes partial c and partial e and is often in cis to RHD*DIVa. Transfusion. 2012 Jul 13;:10–2995. doi: 10.1111/j.1537-2995.2012.03800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu X, Wagner FF, Witter B, Flegel WA. Outliers in RhD membrane integration are explained by variant RH haplotypes. Transfusion. 2006;46(8):1343–51. doi: 10.1111/j.1537-2995.2006.00902.x. [DOI] [PubMed] [Google Scholar]
- 69.Pham BN, Peyrard T, Juszczak G, Dubeaux I, Gien D, Blancher A, Cartron JP, Rouger P, Le Pennec PY. Heterogeneous molecular background of the weak C, VS+, hr B-, Hr B- phenotype in black persons. Transfusion. 2009 Mar;49(3):495–504. doi: 10.1111/j.1537-2995.2008.02005.x. [DOI] [PubMed] [Google Scholar]
- 70.Westhoff CM. Rh complexities: serology and DNA genotyping. Transfusion. 2007;47(Suppl 1):17S–22S. doi: 10.1111/j.1537-2995.2007.01305.x. [DOI] [PubMed] [Google Scholar]
- 71.Endres W, Flegel WA, Helmbold W, Kasulke D, Montag-Lessing T, Poschmann A, Sonneborn H-H. Überlegungen zum Vorgehen bei der Bestimmung der D-Eigenschaft. Infusionsther.Transfusionsmed. 1996;23:172–5. [Google Scholar]
- 72.Hyland CA. The challenge and paradox in serology RhD typing for blood donors and patients. Blood Transfus. 2013:1–2. doi: 10.2450/2012.0107-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Flegel WA, von Zabern I, Doescher A, Wagner FF, Vytiskova J, Pisacka M. DCS-1, DCS-2 and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion. 2008 Jan;48(1):25–33. doi: 10.1111/j.1537-2995.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 74.Denomme GA, Wagner FF, Fernandes BJ, Li W, Flegel WA. Partial D, weak D types, and novel RHD alleles among 33,864 multiethnic patients: implications for anti-D alloimmunization and prevention. Transfusion. 2005;45(10):1554–60. doi: 10.1111/j.1537-2995.2005.00586.x. [DOI] [PubMed] [Google Scholar]
- Flegel WA. The Rhesus Immunization Surveillance. DRK-Blutspendedienst Baden-Württemberg - Hessen; Ulm: 1998 - 2009. http://www.uni-ulm.de/~wflegel/RH/RIR/rirres.html. [Google Scholar]
- Wagner FF. The RhesusBase. DRK-Blutspendedienst Baden-Württemberg - Hessen; Ulm: 1998 - 2012. http://www.uni-ulm.de/~fwagner/RH/RB2/ [Google Scholar]