Abstract
Megsin is a Serine protease inhibitor (Serpin) that has known expression in kidney mesangial cells. Here, we report the generation and characterization of a bacterial artificial chromosome (BAC) transgene expressing Cre under the control of Megsin regulatory elements. When crossed to the ROSA26R-lacZ reporter mice, the Megsin-Cre transgene mediates loxP recombination primarily in the skin, forestomach and esophagus, but surprisingly not in the mesangial cells. Within the skin, cells in all epidermal layers and the hair follicle cells expressed Cre. This transgene also has uniform expression in the epithelium of the forestomach and esophagus. Conditional deletion of Adam10, a gene known to have important functions in skin development, by using this Megsin-Cre transgene led to severe skin defects. In addition, these mutants appear to have reduced folds and surface area in the forestomach. These results show that the Megsin-Cre transgene can mediate loxP-recombination in all epidermal layers of the skin, the hair follicle cells, as well as in the epithelium of the forestomach and esophagus, all of which have known expression of various keratins. This Megsin-Cre transgene can serve as a new tool for conditional genetic manipulation to study development and diseases in the skin and the upper digestive tract.
Keywords: Megsin, Adam10, epidermis, forestomach, esophagus
Introduction
Mesangial cells play critical roles in maintaining the structure and function of the glomerulus in the kidney (Herrera, 2006). Injury of mesangial cells can lead to various glomerular diseases with marked upregulation of Megsin (Miyata et al., 1998). Megsin, a member of the Serpin (serine protease inhibitor) superfamily, is highly conserved among different species (Inagi et al., 2003). It is expressed in mesangial cells and reportedly plays a role in Mesangial cell function and homeostasis (Miyata et al., 1998). Recent studies identified upregulation of Megsin in many human kidney diseases and the polymorphisms of Megsin gene are associated with the susceptibility and/or progression of kidney disease (Li et al., 2004; Miyata et al., 2007; Xia et al., 2006a; Xia et al., 2006b). There are several Cre lines showed reporter activity in mesangial cells, but the Cre expression directed by these transgenes is widespread, limiting their use as a cell type-specific driver in studies targeting mesangial cells (Cuttler et al., 2011; Humphreys et al., 2010). Considering the important roles of Mesangial cells in kidney development and chronic diseases, the need for generating a mesangial cell-specific Cre transgenic line is urgent. An earlier attempt using a segment of the Megsin promoter to generate a Cre line was unsuccessful due to the absence of Cre expression (Gawlik and Quaggin, 2004; Kohan, 2008; Miyata et al., 1998). We thought to change the approach by using BAC transgenes in our efforts to increase the probability of having strong and faithful expression of Cre in the mesangial cells.
The ADAMs (A Disintegrin and Metalloproteinase) are a family of peptidase proteins. They are transmembrane proteins belong to the zinc protease superfamily and contain a disintegrin and metalloprotease domain. The ADAMs have been implicated in a wide range of biological processes from the control of membrane fusion, cell fate determination, to pathologic processes such as cancer and inflammation (Seals and Courtneidge, 2003). Adam10 is capable of proteolytic cleavage of a wide range of substrates, including type IV collagen, EGF, Ephrin, chemokines, prion precursor protein, and many others (Klein and Bischoff, 2011). Most importantly, Adam10 mediates the S2 cleavage of the Notch ligands, effectively serving as an upstream activator of the Notch signaling pathway (Hartmann et al., 2002; Jorissen et al., 2010; Weber et al., 2011). Recent studies in transgenic models have shown that Adam10 is involved in the regulation of skin morphogenesis and homeostasis. Epidermal deletion of Adam10 during skin morphogenesis led to a precocious epidermal differentiation and hyperproliferation in adult epidermis (Weber et al., 2011). Conversely, overexpression or increased activation of Adam10 in human also led to chronic skin disease (Dumortier et al., 2010; Maretzky et al., 2008).
Conditional gene targeting using the Cre/loxP-mediated recombination system offers a powerful approach to manipulate gene function under the control of a tissue/cell specific promoter in a spatially and temporally specific manner. Here we generated a bacterial artificial chromosome (BAC) transgene expressing Cre under the control of Megsin regulatory elements. The Megsin-Cre transgene mediates loxP recombination occurred primarily in the skin, forestomach, and esophagus, but surprisingly not in the mesangial cells. Conditional deletion of Adam10, led to severe skin defects and forestomach epithelium anomalies. The Megsin-Cre transgene mainly mediate loxP-recombination in tissues that have known expression of various keratins. It can serve as a new tool for conditional genetic manipulation to study development and diseases in these tissues.
Results and Discussion
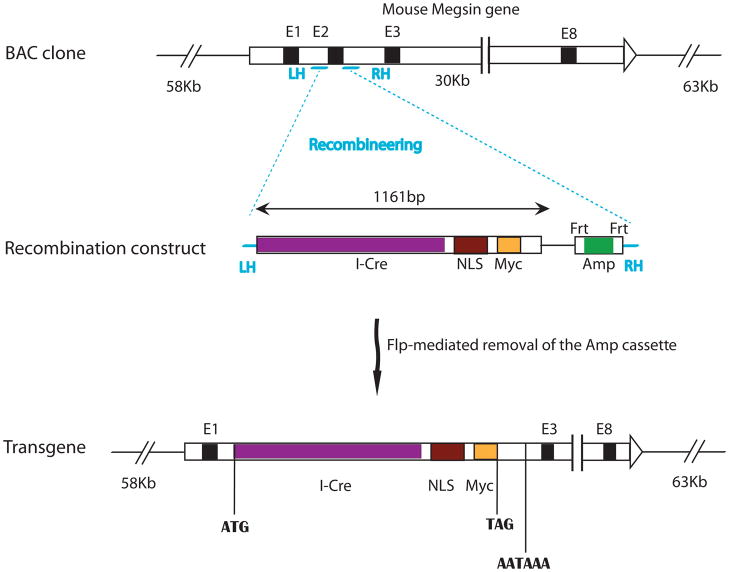
To provide a new genetic tool to investigate the development and pathology of the glomerular and kidney diseases, we set out to make a transgene that will direct Cre expression specifically under the promoter of the gene Megsin. Since BAC (Bacterial artificial chromosome) transgenes have higher capacity to carry most, if not all, of the regulatory sequences necessary to recapitulate the expression pattern of the endogenous gene, we set out to construct the Cre transgene in a BAC clone covering the Megsin gene but not any other full length known genes. We used recombineering (recombination-mediated genetic engineering) to replace the first coding exon of Megsin with the coding sequence of an I-Cre (codon Improved Cre) fused to a nuclear localization signal (NLS) (Fig. 1). Thus, Cre expression is effectively under the control of the regulatory elements of the Megsin gene. This cassette destroys the coding sequence of Megsin and terminates the transcription (by the transcription termination in the Cre cassette) upstream of exon 3 of the Megsin gene.
Figure 1. Generation of a Megsin-Cre transgene using recombineering in a BAC clone.
The entire Megsin gene resides in the middle portion of the BAC clone we used to build the Cre transgene. No other complete genes are present in this BAC clone. The recombination construct has two homology arms (LH: left homology; RH: right homology) that are homologous to the sequences immediately 5′ and immediately 3′ to exon 2 of Megsin, respectively. The recombination construct also has the coding sequence for a codon-improved Cre (I-Cre), a nuclear localization signal (NLS), and a Myc tag. In addition, it has an Frt-flanked ampicillin resistance gene cassette for selection. Exon 2 (E2) of Megsin in the BAC clone was replaced by the recombination construct between the two homologyarms after recombineering. An additional round of transient Flp expression eliminates the Amp cassette to avoid bringing unnecessary prokaryotic sequences into the mammalian genome.
After pronuclei injection, one healthy and fertile founder was produced in a C57Bl/6xCBA hybrid background. To test the efficiency and the specificity of the transgene expression, we crossed the founder and its offspring to the ROSA26R-lacZ reporter mice (Soriano, 1999). To our surprise, Megsin-Cre-mediated loxP recombination was not detected in the mesangial cells, revealed by histochemical detection of β-galactosidase activity (Data not shown). We are not certain about the reason for the lack of mesangial expression. Mutations in cis-acting element important for mesangial expression or the suppression of such elements by the local chromosomal environment are among the possible explanations.
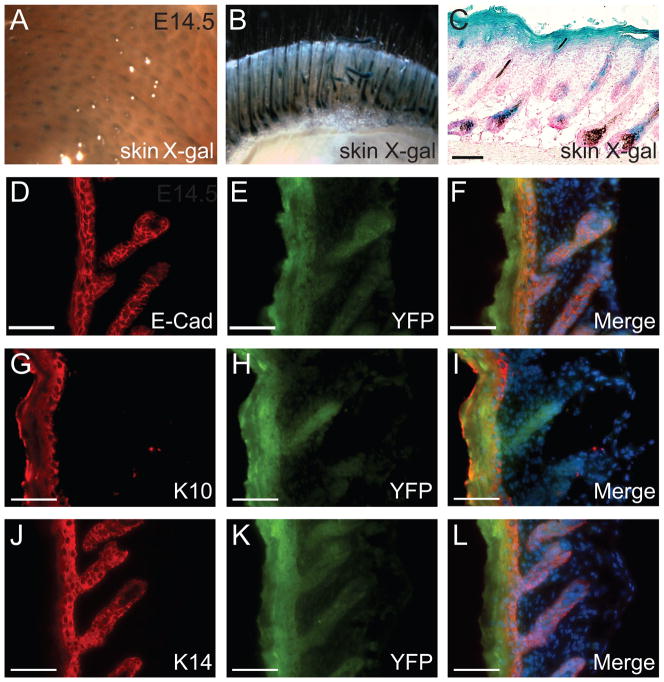
Although no expression in the mesangial cells, we found strong β-galactosidase activities in epidermis and hair follicles of both ventral and dorsal skin at E14.5 (Fig. 2A) and postnatally (Fig. 2B and C). The skin consists of several layers, the stratum corneum, the epithelial layer (epidermis), the connective tissue layer (dermis) and the adipose layer (hypodermis) underneath (Takahashi et al., 1998). The epidermis is a stratified squamous epithelium that contains several keratinocyte layers, including the granular layer, spinous layer and basal layer. Keratinocyte maturation is a process that relatively undifferentiated keratinocyte progenitors in the basal layer differentiate, in sequential order, into intermediate spinous layer, granular layer, and terminally differentiated corneocytes in the cornified layer. These layers can be identified by individual keratin markers. For example, Keratin10 (K10) is expressed in the suprabasal layer of the epidermis and Keratin 14 (K14) is expressed in the basal layer of epidermis (Koch and Roop, 2004; Margadant et al.). To study the expression of the Megsin-Cre transgene in the skin at the cellular level, we used K10, K14 and E-cadherin (E-Cad) to analyze skin sections from Megsin-Cre/+; ROSA26RlacZ mice and Megsin-Cre/+; ROSA26RYFP mice, respectively. The Megsin-Cre transgene was expressed in all layers of the epidermis, as indicated by colocalization of the YFP with the epithelial marker E-cadherin (Fig. 2D–F), with the suprabasal layer marker K10 (Fig. 2G–I), and with the basal layer marker K14 (Fig. 2J–L). Expression in the hair follicle epithelium was also noted (Fig. 2B–L).
Figure 2. The Megsin-Cre transgene has expression specifically in the skin.
Whole-mount β-galactosidase assay (A–B) and X-gal analysis on skin cryostat section (C) revealed the occurrence of Cre-mediated loxP recombination in the hair follicles and epidermis. A: E14.5 embryos. B: skin of P3 transgenic mouse. C: skin section from P3 transgenic mouse. Immunohistochemical analysis indicated Megsin was expressed in all epidermal layers (D-L). D-F: YFP colocalized with E-cadherin staining in the skin epithelium, G-I: YFP colocalized with K10 staining in the suprabasal layers of the epidermis and other cornified stratified epithelia, J-L: YFP colocalized with K14 staining in the basal layer of the epidermis. Scale bar: 100 μm.
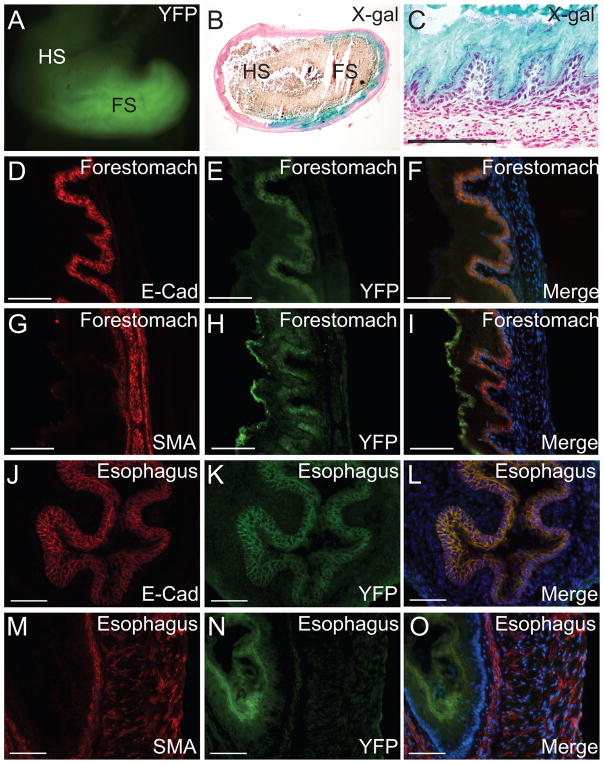
Besides skin expression, strong expression of the transgene was also detected in the mouse forestomach (Fig. 3A–I) and esophagus (Fig. 3J–O). Mouse forestomach is composed of a stratified squamous epithelium with high levels of similarity to the esophageal epithelium (Takahashi et al., 1998). The development of adult mouse stomach epithelium begins with the initiation of endoderm cytodifferentiation in late fetal life. E12.5 mouse gastric mucosa is lined with a simple undifferentiated epithelial monolayer that shows a pseudostratified appearance by E14.5. The keratinized stratified squamous forestomach epithelium starts to differentiate by E16.5 and organizes into primordial buds by E18, (Karam et al., 1997; Spencer-Dene et al., 2006). Megsin-Cre-mediated loxP recombination occurred in the epithelium of forestomach, but not in the glandular hindstomach, as shown by YFP signal and X-gal staining in Fig. 3A–C. To further determine the specific cell types where Megsin-Cre was expressed in the forestomach, we examined the transgene expression together with epithelial and smooth muscle markers, E-cadherin and SMA, respectively. We found that the Megsin-Cre-mediated loxP recombination only occurred in the epithelium, but not the muscle layers of the forestomach as revealed by the colocalization of YFP signal and E-cadherin staining (Fig. 3D–F), as well as the mutual exclusion of the YFP and SMA signals (Fig. 3G–I). Similar expression pattern was also found in esophagus epithelium, indicated by the colocalization of YFP signal and E-cadherin (Fig. 3J–L) and the mutual exclusion of YFP and SMA signals (Fig. 3M–O). The expression of Cre in the skin and forestomach may be driven by the Megsin transcriptional regulatory elements or influenced by regulatory sequences that are unrelated to Megsin but near the transgene insertion site. Reports of Megsin transcripts in the skin and the forestomach appear to suggest that expression in these tissues is related to Megsin promoter activity (Toulza et al., 2007).
Figure 3. Megsin-Cre expression is in the epithelium of the forestomach and esophagus.
Transgenic expression of Megsin-Cre in whole mouse stomach was detected in the forestomach indicated by YFP signals in whole-mount preparation from a Megsin-Cre/+, ROSA26RYFP/+ mouse (A) and β-galactosidase activity assays on sections from a Megsin-Cre/+, ROSA26RlacZ/+ mouse (B–C). It was undetectable in the glandular hindstomach. Expression of Megsin-Cre in the forestomach of transgenic mice (D–I). As indicated by the epithelial marker E-cadherin and the smooth muscle marker αSMA, Megsin-Cre expression was restricted to the epithelium of the forestomach (D–F) and no expression was seen in the smooth muscle layers (G–I). Colocalization of YFP and E-cadherin and mutual exclusion of YFP and α-SMA indicated the specific expression of Megsin-Cre in the esophagus epithelium of transgenic mice (Fig. 3J–O). FS: forestomach, HS: glandular hindstomach. Scale bar: 100 μm.
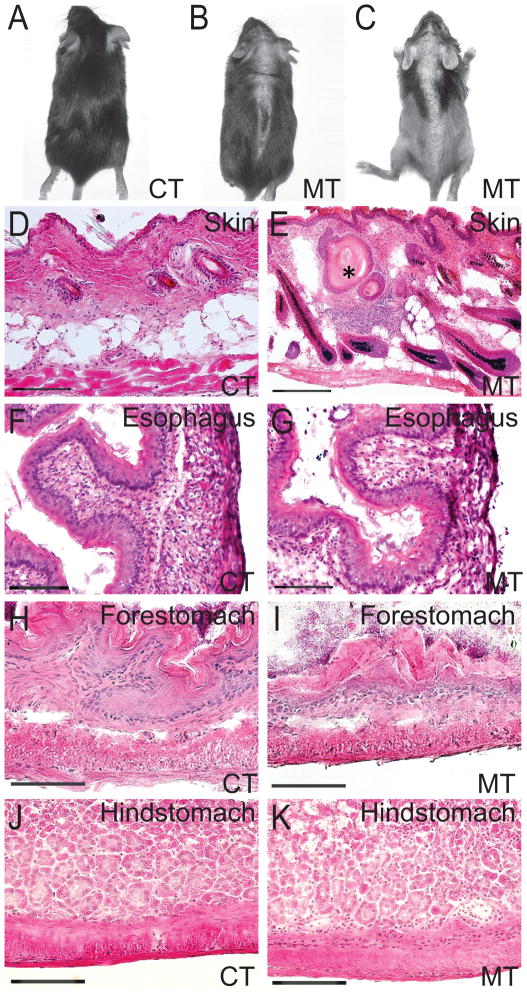
In order to show the Megsin-Cre transgenic can efficiently and specifically mediate loxP recombination for gene function studies, we have selectively crossed the Megsin-Cre mice with the mice carrying a floxed allele of Adam10. Previous study of Adam10 deletion using a similar transgenic Cre line, Keratin 14-Cre (K14-Cre), has demonstrated Adam10 as a central regulator of skin development and maintenance (Weber et al., 2011). The K14 promoter drives Cre expression specifically in the basal layer of the epidermis and its appendages, such as hair follicles and sebaceous glands (Hafner et al., 2004). The K14-Cre driven epidermal Adam10 deletion led to perinatal lethality due to a perturbed water barrier function. The mutants also had a strong reduction in epidermal thickness and absence of sebaceous glands (Weber et al., 2011). The Megsin-Cre mouse has hair follicle expression early (E14.5). In postnatal mice, Cre activity is detected in all epidermal layers of the skin and in the hair follicles in the dermis. Unlike the phenotypes shown in K14-Cre; Adam10 mutant mice, the Megsin-Cre; Adam10 mutant mice were able to survive into adulthood, with progressive hair loss. Hair loss started out in patchy areas at the midline of both dorsal and ventral skin, but progressed to more severe and broader hair loss throughout the body (Fig. 4A–C). The dynamic pattern of hair loss in these mutants may be caused by the combined effects of Adam10 deletion and the gradient anagen spreading waves of hair follicles (Plikus and Chuong, 2008). By H&E staining we did not detect any obvious reduction in the thickness of the epidermis or the absence of sebaceous glands, but we found the presence of keratin cysts in the dermis which may contribute to the destruction of hair follicles and the eventually hair loss (Fig. 4E). Keratin cyst formation is often interpreted as an adoption of an epidermal differentiation program. Adam10 serves as a key regulator of Notch signaling pathway that plays important roles in epidermal differentiation. The disruption of Notch signaling by Adam10 deletion in the developing hair follicle may lead to perturbation of terminal differentiation program, causing the diversion of Notch-deficient hair follicles to epidermal cysts (Demehri and Kopan, 2009; Vauclair et al., 2005; Weber et al., 2011). The differences in the phenotypes of the K14-Cre; Adam10 and the Megsin-Cre; Adam10 mutant mice appear to reflect the temporal-spatial differences of Cre activities from these two transgenes.
Figure 4. Megsin-Cre mediated deletion of Adam10 led to skin and stomach defects.
Megsin-Cre/+, Adam10/+ mouse exhibits a progressive hair loss starting from the midline in both ventral and dorsal skins (A–C). A: Macroscopic appearance of control mouse, B–C: Megsin-Cre/+, Adam10loxP/loxP mouse at Week 3(B) and Week 8(C). Morphological changes in the conditional mutant skin were found (D–E). Adult Megsin-Cre/+, Adam10loxP/loxP mutant mice display keratin cysts (star in E). Sagittal sections of adult control and mutant mice showing hypoplasia of squamous epithelium in the forestomach of Megsin-Cre/+, Adam10loxP/loxP mouse (I), when compared to the control (H). No morphological changes were detected in the esophagus (F–G) or glandular hindstomach (J–K). Scale bar: 100 μm.
Because of the finding of Megsin-Cre expression in the stomach and esophagus, we also examined the Megsin-Cre-Adam10 control and mutant mice for potential defects in these structures. None of the Megsin-Cre-Adam10 mutant mice showed overt digestive problems. All mutants grew at similar rates when compared to their littermates, as determined by body weight measured at regular intervals. Histological analyses showed no significant differences in esophagus and glandular hindstomach between the controls and the mutants (Fig. 4F-G, J-K). However, Megsin-Cre induced Adam10 deletion resulted in hypoplasia in the forestomach (Fig. 4H-I). The forestomach of these mutant mice showed decreased depth of gastric pits in the forestomach (mutants 62.5 ± 22.8 μm; controls 173.4 ± 54.2 μm; p<0.001) (Fig. 4I). These structural changes, however, were not sufficient to cause detectable digestive phenotypes in the mutants under normal diet and housing conditions. It remains to be investigated if these mutants are more susceptible to develop digestive problems when fed different diet regimens (for example, high-fiber or high-fat diets).
There are a number of transgenic mouse lines that can drive Cre expression in the skin (Schneider, 2012). Among them, the Krt1-15-CrePR1 transgene is very different from the Megsin-Cre since it used the Keratin 15 promoter to target the hair follicle bulge stem cells in an inducible manner (Morris et al., 2004). Two other Cre lines, K5-Cre and K14-Cre drive Cre expression in a similar group of cells that Megsin-Cre targets (Hafner et al., 2004; Ramirez et al., 2004), However, minor differences in the dynamics of Cre expression may lead to functional consequences, as evidenced by the phenotypic difference seen between the K14-Cre; Adam10 mutants and the Megsin-Cre; Adam10 mutants. In addition, these three lines seem to have very different expression pattern outside of skin. In particular, both K5-Cre and K14-Cre have expression in the oocytes, leading to ubiquitous Cre-mediated loxP recombination when the mother carries the Cre transgene. When the father carries the Cre transgene, Cre activities can be found in tongue, thymic epithelium, and other places (Hafner et al., 2004; Huang et al., 2009; Ramirez et al., 2004). These differences make each of these Cre lines with skin expression unique tools for genetic studies.
Taken together, we generated a new transgenic mouse line, Megsin-Cre, which exhibits Cre activity specifically in epithelial cells of the skin epidermis, hair follicles, forestomach, and esophagus. Conceivably, in addition to the gene manipulations similar to the Adam10 study presented here, this transgene can also be used for lineage tracing studies in normal development and in diseases states.
Methods
Generation of the BAC transgene construct by Recombineering
A BAC clone carrying the murine Megsin gene was purchased from Invitrogene (Clone # RP23-174I22). This BAC clone (MegsinBAC), carrying a chloramphenicol (Cm) resistant cassette, was transformed into DH10B host cells. We then introduced another plasmid pRedET, carrying a tetracycline (Tet) resistant cassette, into these DH10B cells that already have MegsinBAC. The presence of the pRedET plasmid restores the ability for selected types of recombination in the DH10B cells. At the same time, we used PCR to introduce 2 homology arms to the plasmid phCre2.myc.nuc.FRTN1.amp.FRT that expresses a codon-improved Cre with a small Myc tag and nuclear localization sequence (NLS) (a gift from Dr. Günther Schütz) (Casanova et al., 2001). The Frt.Amp.Frt cassette provides ampicillinresistance (Amp) for the intermediate cloning steps and was later removed before the construct was finalized. The homology arms were designed for replacing a stretch of the Megsin gene with the Cre cassette by recombineering. The left homology arm consists of the 50 nucleotides directly 5′ of the ATG start codon of Megsin. The right homology arm consists of a 50 nucleotides long sequence in intron 2 of Megsin. In this design, after recombineering, the coding sequence (CDS) in the first coding exon (exon 2) of Megsin would be replaced by the Cre-expressing cassette. Therefore, the expression of Cre would be subjected to the control of the Megsin promoter. The multiple transcriptional stop signals would ensure that the partial Megsin gene left on the BAC clone is not transcribed. The purified PCR product (Megsin.left.arm_phCre2.myc.nuc.FRTN1.amp.FRT_Megsin.right.arm) was electroporated into the competent DH10B cells that already had both MegsinBac and pRedET. The electroporated cells were selected on plates with Cm, Tet, Amp, and Arabinose (for inducing the required recombinases from the pRedET plasmid). The Megsin.left.arm_phCre.myc.nuc.FRT.AMP.FRT_Megsin.right.arm was a PCR product and could not be reproduced by the bacteria without recombination that incorporates it into the BAC. Thus, the Amp resistant clones were screened for the ones where recombination occurred between the homology arms of the PCR product and the corresponding Megsin sequence on the BAC. The Frt-Amp-Frt cassette was subsequently removed by the introduction of a p706FLPE plasmid (a gift from Dr. Stewart) (Zhang et al., 1998). Every cloning step was confirmed by restriction digestion, PCR, and/or sequencing. The final transgene construct (Megsin-Cre/BAC) was subjected to sequencing to ensure that everything proceeded as planned and no unwanted mutation was introduced.
Generation of the transgenic mice
All animal studies have been approved by IACUC (Institutional Animal Care and Use Committee) at Washington University School of Medicine. The transgene construct was purified by using the Qiagen large DNA Construct kit and was dialyzed by using the transgene injection buffer (10mM TRIS, pH7.4, 0.1mM EDTA). The construct was injected into the pronuclei of fertilized oocytes from C57Bl/6xCBA hybrids. The presence of the transgene in the founders and their offspring was detected by PCR using primers MCF 5′ CCATCCAACAGCACCTGGGCCAGCTCAACA 3′ and MCR 5′ CCACCATCGGTGCGGGAGATGTCCTTCACT 3′. The ROSA26R-lacZ reporter mice were described previously (Soriano, 1999) and were genotyped by using primers WS268 5′ GTTATCAGTAAGGGAGCTGCAGTGG 3″ WS270 5″ AAGACCGCGAAGAGTTTGTCCTC 3′ and WS271 5″ GGCGGATCACAAGCAATAATAACC 3′ to amplify a wild-type band of 500 bp and a band of 250 bp corresponding to the ROSA26R-lacZ allele. PCR conditions were: 95°C, 2′, 35 × (94°C 30″; 59.5°C, 30″ 72°C, 30″), 72°C, 5′. ROSA26R-LacZ mice were purchased from the Jackson Laboratories (Bar Harbor, ME). YFP mice were kindly provided by Dr. Frank Costantini (Srinivas et al., 2001).
Histology and Immunohistochemistry
10 μm cryostat sections of embryos or tissues were collected. 5-Bromo-4-chloro-3-indolyl-D-galactoside (Xgal) staining on cryostat sections was performed as described (Chang et al., 2004). Immunostaining on cryostat sections was performed as previously described (McDill et al., 2006). Antibodies used were: anti-α-SMA antibody (Sigma, St. Louis, MO, 1:500), anti-Keratin10 antibody (Covence, Princeton, NJ, 1: 1000), anti-Keratin 14 antibody (Covence, Princeton, NJ, 1: 1000), anti-E-cadherin antibody (Abcam, Cambridge, MA, 1:200). Appropriate fluorescent conjugated secondary antibodies (Molecular Probes, Invitrogen, Carlsbad, CA, 1:1000) were used to detect the corresponding primary antibodies.
Acknowledgments
We thank the Washington University Mouse Genetics Core and the Renal Disease Model Core within the George M. O’Brien Washington University Center for Kidney Disease Research (NIHP30DK079333) for assistance in the generation of the Megsin-Cre transgenic mice. We thank Drs. Radek Skoda, Ralph Tiedt, Günther Schütz, Francis Stewart, Gerald Crabtree and Ann Kuo for providing plasmids for the recombineering of the Megsin-Cre transgene construct; Dr. Frank Costantini for the kind gift of the ROSA26R-YFP reporter mice; and Dr. Meihua Lin for advice on the skin analyses. F.C. was supported in part by NIH grants (NIHR01DK081592 and NIHR01DK087960).
References
- Casanova E, Fehsenfeld S, Mantamadiotis T, Lemberger T, Greiner E, Stewart AF, Schutz G. A CamKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis. 2001;31:37–42. doi: 10.1002/gene.1078. [DOI] [PubMed] [Google Scholar]
- Chang CP, McDill BW, Neilson JR, Joist HE, Epstein JA, Crabtree GR, Chen F. Calcineurin is required in urinary tract mesenchyme for the development of the pyeloureteral peristaltic machinery. J Clin Invest. 2004;113:1051–1058. doi: 10.1172/JCI20049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuttler AS, LeClair RJ, Stohn JP, Wang Q, Sorenson CM, Liaw L, Lindner V. Characterization of Pdgfrb-Cre transgenic mice reveals reduction of ROSA26 reporter activity in remodeling arteries. Genesis. 2011;49:673–680. doi: 10.1002/dvg.20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demehri S, Kopan R. Notch signaling in bulge stem cells is not required for selection of hair follicle fate. Development. 2009;136:891–896. doi: 10.1242/dev.030700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A, Durham AD, Di Piazza M, Vauclair S, Koch U, Ferrand G, Ferrero I, Demehri S, Song LL, Farr AG, Leonard WJ, Kopan R, Miele L, Hohl D, Finke D, Radtke F. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PLoS One. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawlik A, Quaggin SE. Deciphering the renal code: advances in conditional gene targeting. Physiology (Bethesda) 2004;19:245–252. doi: 10.1152/physiol.00009.2004. [DOI] [PubMed] [Google Scholar]
- Hafner M, Wenk J, Nenci A, Pasparakis M, Scharffetter-Kochanek K, Smyth N, Peters T, Kess D, Holtkotter O, Shephard P, Kudlow JE, Smola H, Haase I, Schippers A, Krieg T, Muller W. Keratin 14 Cre transgenic mice authenticate keratin 14 as an oocyte-expressed protein. Genesis. 2004;38:176–181. doi: 10.1002/gene.20016. [DOI] [PubMed] [Google Scholar]
- Hartmann D, de Strooper B, Serneels L, Craessaerts K, Herreman A, Annaert W, Umans L, Lubke T, Lena Illert A, von Figura K, Saftig P. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet. 2002;11:2615–2624. doi: 10.1093/hmg/11.21.2615. [DOI] [PubMed] [Google Scholar]
- Herrera GA. Plasticity of mesangial cells: a basis for understanding pathological alterations. Ultrastruct Pathol. 2006;30:471–479. doi: 10.1080/01913120600932594. [DOI] [PubMed] [Google Scholar]
- Huang X, Bringas P, Jr, Slavkin HC, Chai Y. Fate of HERS during tooth root development. Dev Biol. 2009;334:22–30. doi: 10.1016/j.ydbio.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys BD, Lin S-L, Kobayashi A, Hudson TE, Nowlin BT, Bonventre JV, Valerius MT, McMahon AP, Duffield JS. Fate Tracing Reveals the Pericyte and Not Epithelial Origin of Myofibroblasts in Kidney Fibrosis. The American Journal of Pathology. 2010;176:85–97. doi: 10.2353/ajpath.2010.090517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagi R, Nangaku M, Miyata T, Kurokawa K. Mesangial cell-predominant functional gene, megsin. Clin Exp Nephrol. 2003;7:87–92. doi: 10.1007/s10157-003-0228-0. [DOI] [PubMed] [Google Scholar]
- Jorissen E, Prox J, Bernreuther C, Weber S, Schwanbeck R, Serneels L, Snellinx A, Craessaerts K, Thathiah A, Tesseur I, Bartsch U, Weskamp G, Blobel CP, Glatzel M, De Strooper B, Saftig P. The disintegrin/metalloproteinase ADAM10 is essential for the establishment of the brain cortex. J Neurosci. 2010;30:4833–4844. doi: 10.1523/JNEUROSCI.5221-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karam SM, Li Q, Gordon JI. Gastric epithelial morphogenesis in normal and transgenic mice. Am J Physiol. 1997;272:G1209–1220. doi: 10.1152/ajpgi.1997.272.5.G1209. [DOI] [PubMed] [Google Scholar]
- Klein T, Bischoff R. Active metalloproteases of the A Disintegrin and Metalloprotease (ADAM) family: biological function and structure. J Proteome Res. 2011;10:17–33. doi: 10.1021/pr100556z. [DOI] [PubMed] [Google Scholar]
- Koch PJ, Roop DR. The role of keratins in epidermal development and homeostasis--going beyond the obvious. J Invest Dermatol. 2004;123:x–xi. doi: 10.1111/j.0022-202X.2004.23495.x. [DOI] [PubMed] [Google Scholar]
- Kohan DE. Progress in gene targeting: using mutant mice to study renal function and disease. Kidney Int. 2008;74:427–437. doi: 10.1038/ki.2008.146. [DOI] [PubMed] [Google Scholar]
- Li Y-J, Du Y, Li C-X, Guo H, Leung JCK, Lam MF, Yang N, Huang F, Chen Y, Fang J-Q, Maxwell PH, Lai KN, Wang Y. Family-Based Association Study Showing that Immunoglobulin A Nephropathy Is Associated with the Polymorphisms 2093C and 2180T in the 3′ Untranslated Region of the Megsin Gene. Journal of the American Society of Nephrology. 2004;15:1739–1743. doi: 10.1097/01.asn.0000130858.00688.46. [DOI] [PubMed] [Google Scholar]
- Maretzky T, Scholz F, Koten B, Proksch E, Saftig P, Reiss K. ADAM10-mediated E-cadherin release is regulated by proinflammatory cytokines and modulates keratinocyte cohesion in eczematous dermatitis. J Invest Dermatol. 2008;128:1737–1746. doi: 10.1038/sj.jid.5701242. [DOI] [PubMed] [Google Scholar]
- Margadant C, Charafeddine RA, Sonnenberg A. Unique and redundant functions of integrins in the epidermis. The FASEB Journal. 2010;24:4133–4152. doi: 10.1096/fj.09-151449. [DOI] [PubMed] [Google Scholar]
- McDill BW, Li SZ, Kovach PA, Ding L, Chen F. Congenital progressive hydronephrosis (cph) is caused by an S256L mutation in aquaporin-2 that affects its phosphorylation and apical membrane accumulation. Proc Natl Acad Sci U S A. 2006;103:6952–6957. doi: 10.1073/pnas.0602087103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Li M, Yu X, Hirayama N. Megsin gene: its genomic analysis, pathobiological functions, and therapeutic perspectives. Curr Genomics. 2007;8:203–208. doi: 10.2174/138920207780833856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Nangaku M, Suzuki D, Inagi R, Uragami K, Sakai H, Okubo K, Kurokawa K. A mesangium-predominant gene, megsin, is a new serpin upregulated in IgA nephropathy. J Clin Invest. 1998;102:828–836. doi: 10.1172/JCI2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RJ, Liu Y, Marles L, Yang Z, Trempus C, Li S, Lin JS, Sawicki JA, Cotsarelis G. Capturing and profiling adult hair follicle stem cells. Nat Biotechnol. 2004;22:411–417. doi: 10.1038/nbt950. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Chuong CM. Complex hair cycle domain patterns and regenerative hair waves in living rodents. J Invest Dermatol. 2008;128:1071–1080. doi: 10.1038/sj.jid.5701180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez A, Page A, Gandarillas A, Zanet J, Pibre S, Vidal M, Tusell L, Genesca A, Whitaker DA, Melton DW, Jorcano JL. A keratin K5Cre transgenic line appropriate for tissue-specific or generalized Cre-mediated recombination. Genesis. 2004;39:52–57. doi: 10.1002/gene.20025. [DOI] [PubMed] [Google Scholar]
- Schneider MR. Genetic mouse models for skin research: Strategies and resources. Genesis. 2012 doi: 10.1002/dvg.22029. [DOI] [PubMed] [Google Scholar]
- Seals DF, Courtneidge SA. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 2003;17:7–30. doi: 10.1101/gad.1039703. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Spencer-Dene B, Sala FG, Bellusci S, Gschmeissner S, Stamp G, Dickson C. Stomach development is dependent on fibroblast growth factor 10/fibroblast growth factor receptor 2b-mediated signaling. Gastroenterology. 2006;130:1233–1244. doi: 10.1053/j.gastro.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Srinivas S, Watanabe T, Lin C-S, William C, Tanabe Y, Jessell T, Costantini F. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Imanaka T, Takano T. Spatial pattern of smooth muscle differentiation is specified by the epithelium in the stomach of mouse embryo. Dev Dyn. 1998;212:448–460. doi: 10.1002/(SICI)1097-0177(199807)212:3<448::AID-AJA12>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Toulza E, Mattiuzzo NR, Galliano MF, Jonca N, Dossat C, Jacob D, de Daruvar A, Wincker P, Serre G, Guerrin M. Large-scale identification of human genes implicated in epidermal barrier function. Genome Biol. 2007;8:R107. doi: 10.1186/gb-2007-8-6-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vauclair S, Nicolas M, Barrandon Y, Radtke F. Notch1 is essential for postnatal hair follicle development and homeostasis. Developmental Biology. 2005;284:184–193. doi: 10.1016/j.ydbio.2005.05.018. [DOI] [PubMed] [Google Scholar]
- Weber S, Niessen MT, Prox J, Lullmann-Rauch R, Schmitz A, Schwanbeck R, Blobel CP, Jorissen E, de Strooper B, Niessen CM, Saftig P. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development. 2011;138:495–505. doi: 10.1242/dev.055210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y, Li Y, Du Y, Yang N, Li C, Leung JC, Lam MF, Huang W, Chen S, Maxwell PH, Lai KN, Wang Y. Association of MEGSIN 2093C-2180T haplotype at the 3′ untranslated region with disease severity and progression of IgA nephropathy. Nephrol Dial Transplant. 2006a;21:1570–1574. doi: 10.1093/ndt/gfk096. [DOI] [PubMed] [Google Scholar]
- Xia YF, Huang S, Li X, Yang N, Huang J, Xue C, Zhang M, Leung JC, Lam MF, Li J. A family-based association study of megsin A23167G polymorphism with susceptibility and progression of IgA nephropathy in a Chinese population. Clin Nephrol. 2006b;65:153–159. doi: 10.5414/cnp65153. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]