Abstract
Verrucomicrobium spinosum is a Gram-negative bacterium that is related to bacteria from the genus Chlamydia. The bacterium is pathogenic towards Drosophila melanogaster and Caenorhabditis elegans, using a type III secretion system to facilitate pathogenicity. V. spinosum employs the recently discovered l,l-diaminopimelate aminotransferase biosynthetic pathway to generate the bacterial cell wall and protein precursors diaminopimelate and lysine. A survey of the V. spinosum genome provides evidence that the bacterium should be able to synthesize peptidoglycan de novo, since all of the necessary genes are present. The enzyme UDP-N-acetylmuramoyl-l-alanyl-d-glutamate: meso-2,6-diaminopimelate ligase (MurE) (E.C. 6.3.2.15) catalyzes a reaction in the cytoplasmic step of peptidoglycan biosynthesis by adding the third amino acid residue to the peptide stem. The murE ortholog from V. spinosum (murE Vs) was cloned and was shown to possess UDP-MurNAc-l-Ala-d-Glu:meso-2,6-diaminopimelate ligase activity in vivo using functional complementation. In vitro analysis using the purified recombinant enzyme demonstrated that MurEVs has a pH optimum of 9.6 and a magnesium optimum of 30 mM. meso-Diaminopimelate was the preferred substrate with a K m of 17 µM, when compared to other substrates that are structurally related. Sequence alignment and structural analysis using homology modeling suggest that key residues that make up the active site of the enzyme are conserved in MurEVs. Our kinetic analysis and structural model of MurEVs is consistent with other MurE enzymes from Gram-negative bacteria that have been characterized. To verify that V. spinosum incorporates diaminopimelate into its cell wall, we purified peptidoglycan from a V. spinosum culture; analysis revealed the presence of diaminopimelate, consistent with that of a bona fide peptidoglycan from Gram-negative bacteria.
Introduction
The bacterial cell wall plays an integral role in withstanding stress from external and internal forces in addition to maintaining the shape of bacteria. As such, the cell wall is essential for cell viability due to its overarching function in providing physical support for the cytoplasmic membrane. The cell wall of bacteria is mainly composed of a cross-linked polymer known as peptidoglycan (PG). PG contains glycan chains and peptide stems, and its monomer unit consists of a disaccharide tetrapeptide (Fig. 1) [1]. Its synthesis is divided into three main steps. In the first step, the nucleotide sugar-linked precursors UDP-N-acetylmuramyl-pentapeptide (UDP-MurNAc-pentapeptide) and UDP-N-acetylglucosamine (UDP-GlcNAc) are synthesized in the cytoplasm. In the second step, precursor lipid intermediates (lipids I and II) are synthesized at the cytoplasmic membrane. The polymerization of newly synthesized disaccharide-peptide units and incorporation into the growing PG by penicillin-binding proteins (PBPs) is the third and final step of the pathway [2].
Figure 1. The monomer unit of the peptidoglycan structure.

The disaccharide moiety is composed of the amino sugars N-acetylglucosamine (GlcNAc) and N-acetylmuramic (MurNAc) linked via a β-1,4 glycosidic bond. The amino acid at position 3 of the stem peptide is meso-diaminopimelic acid (R = COOH) in most Gram-negative bacteria and l-lysine (R = H) in most Gram-positive bacteria.
Verrucomicrobium spinosum is a Gram-negative heterotrophic bacterium that is generally found in fresh water and soil. The morphology of V. spinosum is very interesting in that it possesses protruding wart-like and tube-like appendages known as prosthecae that are an extension of the cell membrane (Fig. 2). The bacterium has garnered a lot of interest from the scientific community due to its close evolutionarily relationship with bacteria from the genus Chlamydia [3]. Annotation of the genome suggests that the bacterium employs a protein secretion system known as Type III that is involved in pathogenicity [4]. A recent study shows that V. spinosum is pathogenic to Drosophila melanogaster and Caenorhabditis elegans [5].
Figure 2. Scanning electron microscopy of V. spinosum DSM 4136T.
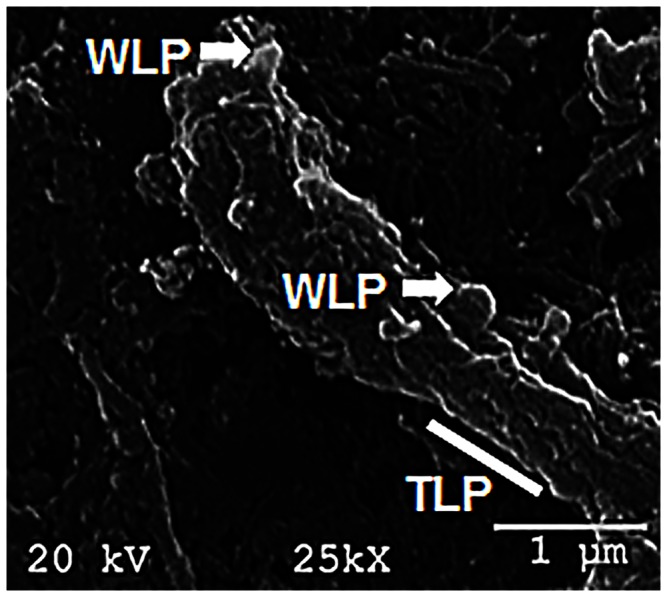
The white arrows show the wart-like prosthecae (WLP) and the white bar depicts a tube-like prosthecae (TLP). The picture was taken at 25 K magnification. The scale bar is 1 µm.
V. spinosum was found to employ the recently discovered l,l-diaminopimelate aminotransferase (DapL) pathway [6], [7], [8], [9] as the sole route for the synthesis of diaminopimelate (A2pm) and l-lysine (l-Lys), based on biochemical and bioinformatical evidence [10]. In the anabolism of PG, the penultimate intermediate in the l-lysine biosynthesis pathway, meso-diaminopimelate (meso-A2pm), serves as one of the cross-linking amino acids in Gram-negative bacteria, and l-Lys serves the same purpose in many Gram-positive bacteria [11].
The enzyme UDP-N-acetylmuramoyl-l-alanyl-d-glutamate:meso-2,6-diaminopimelate ligase (MurE) (E.C. 6.3.2.15) catalyzes the addition of the third amino acid residue to the peptide stem of PG in the cytoplasmic step of PG synthesis. In most bacteria, this third residue is either meso-A2pm or l-Lys (Fig. 1). In particular species, other amino acids can be found, such as l-ornithine, meso-lanthionine, l,l-A2pm, l-diaminobutyric acid or l-homoserine [1], [12], [13]. Since the third residue in the bacterial cell wall is involved in PG cross-linking, the lack of or incorrect substrate incorporation into the PG macromolecule can lead to improperly constructed PG and ultimately to cell death via lysis due to inability of the bacterium to maintain osmotic pressure [14], [15].
Here we report the first characterization of a Mur ligase from the genus Verrucomicrobium, namely MurE from V. spinosum (MurEVs). In vivo analysis demonstrates that the enzyme is able to functionally complement an Escherichia coli strain that harbors a mutation in the murE gene. Using in vitro analyses, we show that MurEVs is a meso-A2pm-adding enzyme. Furthermore, we present a structural analysis of the enzyme using protein sequence alignment and homology modeling, which shows that key amino acids for substrate binding and/or catalysis are conserved in MurEVs. Together, these experiments contribute to the further understanding of the kinetic, physical and structural properties of the Mur ligase involved in the synthesis of PG from the organism V. spinosum. Finally, V. spinosum PG was purified and analyzed; its composition in which A2pm is one of the main constituents is similar to that of most Gram-negative bacteria.
Materials and Methods
V. spinosum growth conditions
V. spinosum DSM 4136T was cultured in R2A medium at 26°C [10].
PCR amplification and cloning of the V. spinosum murE open reading frame (ORF) for protein expression and purification
The open reading frame annotated by the locus tag (VspiD_010100019130) UDP-N-acetylmuramoyl-l-alanyl-d-glutamate:meso-2,6-diaminopimelate ligase was amplified by PCR. The following forward and reverse primers were used: murE Vs-forward 5′-CACCATGACCATTTTGCGCGATCTTATCGAGGGT-3′ and murEVs-reverse 5′-GTCGAC TCACTGACGGTCATCCCTCCTTTGGCGTGC-3′ (the underlined sequence represents the restriction enzyme site used to facilitate sub-cloning of the ORF while the bold and italicized sequences represent initiation and termination codons). The PCR reaction contained 12 pmol of forward and reverse primers, 1 mM MgSO4, 0.5 mM of each of the four deoxynucleotide triphosphates, 0.5 ng of genomic DNA and 1 unit of Platinum Pfx DNA polymerase (Invitrogen Corporation, Carlsbad, CA, USA). PCR conditions were: 1 cycle at 94°C for 2 min, followed by 30 cycles of 94°C for 15 s, 60°C for 30 s and 72°C for 2 min. The murE PCR fragment was ligated into the plasmid pET100D/topo (Invitrogen Corporation, Carlsbad, CA, USA) to produce the plasmid pET100D::murE Vs. The recombinant protein encoded by this plasmid carries a MRGSHHHHHHGMASMTGGQQMGRDLYDDDDKDHPFT sequence containing a hexa-histidine tag derived from the pET100D plasmids at the amino terminus. To confirm the fidelity of the PCR reaction, the murE ORF was sequenced from pET100D using the T7 promoter primer, 5′-TAATACGACTCACTATAGGG-3′ and the T7 reverse primer, 5′-TAGTTATTGCTCAGCGGTGG-3′. The cloned murE ORF was 100% identical to the sequences deposited in the Integrated Microbial Genomes public database (http://img.jgi.doe.gov/cgi-bin/w/main.cgi).
Protein expression and purification of the recombinant MurEvs
The E. coli BL21-CodonPlus® (DE3)-RIPL (Agilent Technologies, USA) strain was transformed with the plasmid pET100D::murEVs and grown in LB broth containing 50 µg⋅mL−1 ampicillin and 34 µg⋅mL−1 chloramphenicol at 37°C to an OD600 of 0.5. Protein expression was induced in 1 L of culture using isopropyl β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM for 8 h at 20°C. The cell pellet was lysed by sonication in a buffer consisting of 50 mM sodium phosphate, pH 8.0, and 300 mM NaCl. The soluble extract was incubated with 1 mL bed volume of TALON Metal Affinity Resin (Clontech, Mountain View CA, USA) for 30 min at 4°C. The resin was washed 5 times with 30 mL of sonication buffer containing 10 mM imidazole for 15 min each. The enzyme was eluted with 10 mL of sonication buffer containing 250 mM imidazole. The hexa-histidine tag was not removed after protein purification. The pure protein was concentrated in an Amicon Ultra 10,000 molecular weight cutoff filter unit replacing the elution buffer with 20 mM potassium phosphate, pH 7.2, 1 mM dithiothreitol (DTT), 1 mM EDTA and 10% (v/v) glycerol. The protein concentration was determined by quantitative amino acid analysis as described below.
Preparation of substrates and MurE activity assay
UDP-MurNAc-l-Ala-d-Glu, UDP-MurNAc-l-Ala-d-[14C]Glu and the three isomers of A2pm were prepared according to published procedures [16], [17], [18].
The standard MurE activity assay measured the formation of UDP-MurNAc-l-Ala-γ-d-[14C]Glu-meso-A2pm in mixtures (final volume, 40 µL) containing 100 mM Tris-HCl, pH 9.6, 30 mM MgCl2, 0.25 mg⋅mL−1 bovine serum albumin, 5 mM ATP, 150 µM UDP-MurNAc-l-Ala-d-[14C]Glu (350 Bq), 150 µM meso-A2pm, and enzyme (20 µL of an appropriate dilution in 20 mM phosphate buffer, pH 7.2, containing 1 mM DTT). After 30 min at 37°C, the reaction was stopped by the addition of glacial acetic acid (8 µL), followed by lyophilization. The radioactive substrate and product were separated on a Nucleosil 100 C18 5 U column (150×4.6 mm; W. R. Grace S. A.) using 50 mM ammonium formate, pH 4.4, as the mobile phase at a flow rate of 0.6 mL⋅min−1. Radioactivity was detected with a flow detector (model LB506-C1, Berthold) using the Quicksafe Flow 2 scintillator (Zinsser Analytic) at 0.6 mL⋅min−1. Quantification was performed with the Radiostar software (Berthold).
Identical assay conditions were used when the l,l and d,d isomers of A2pm were tested as substrates. With lysine or ornithine, l-[14C]-Lys or l-[14C]-Orn, respectively, was used as the labeled substrate; in that case, radioactive substrate and product were separated by thin-layer chromatography on silica gel plates LK6D (Whatman) using 1-propanol/ammonium hydroxide/water (6∶3∶1; v/v) as the mobile phase, and the radioactive spots were located and quantified with a radioactivity scanner (Rita Star, Raytest Isotopenmeβgeräte GmbH).
Determination of the kinetic constants
For the determination of the kinetic constants, the same assay was used with various concentrations of one substrate and fixed concentrations of the others. In all cases, the enzyme concentration was chosen so that substrate consumption was <20%, the linearity being ensured within this interval even at the lowest substrate concentration. Data were fitted to the equation v = V max S/(K m+S) by the Levenberg-Marquardt method [19], where v is the initial velocity and S is the substrate concentration, and values ± standard deviation at 95% of confidence were calculated. The MDFitt software developed by M. Desmadril (UMR 8619, CNRS, Orsay, France) was used for this purpose.
Sequence alignment and homology modeling
A multiple amino acid sequence alignment between the Mur ligase enzymes of V. spinosum (ZP_02928794.1), Mycobacterium tuberculosis (CCE37632.1), E. coli (NP_414627.1) Chlamydia trachomatis (NP_219774.1) and Pectobacterium carotovorum (ZP_03831119.1) was generated using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) with the Gonnet scoring matrix.
The homology model of the MurEVs protein was generated using the SWISS-MODEL Protein Modeling Server [20], [21], [22] (http://swissmodel.expasy.org/) using the E. coli MurE structure as a template PDB id: 1E8C [23], which was identified using a PSI-BLAST search of the MurEVs protein sequence against proteins in the Protein Data Bank using the web server: (http://blast.ncbi.nlm.nih.gov/). The model was examined by hand for clashes and appropriate geometry using the visualization software PyMOL (The PyMOL Molecular Graphics System, Schrödinger, LLC).
Purification and analysis of V. spinosum PG
PG was prepared and analyzed essentially according to Mengin-Lecreulx et al. [24]. Cells from 1 L of culture were harvested at 4°C and resuspended in 4% (w/v) sodium dodecyl sulfate (SDS) (10 mL⋅g−1 of cell wet weight) under constant and vigorous stirring at 100°C for 30 min. The suspension was incubated overnight at 25°C followed by centrifugation for 1 h at 17,000 rpm. The pellet containing crude PG was washed 5 times with 10 mL of sterile water and stored in water for further analysis.
Half of the preparation was used to obtain purified PG. Briefly, the following treatments at 37°C were performed: (i) overnight incubation with 0.05% (w/v) pancreatin in 0.1 M potassium phosphate buffer (pH 7.4); (ii) overnight incubation with 0.02% (w/v) pronase in 0.01 M Tris-HCl buffer (pH 7.4); (iii) overnight incubation with 0.02% (w/v) trypsin in 0.02 M potassium phosphate buffer (pH 7.4). Finally, after centrifugation and several washes with 8 M lithium chloride containing 0.1 M EDTA, and water, the pellet was stored in water. Aliquots of crude and purified PGs were hydrolyzed as described below.
Amino acid and hexosamine analysis
Samples were hydrolyzed in 6 M HCl containing 0.05% (v/v) 2-mercaptoethanol at 105°C for 24 h (proteins), or in 6 M HCl at 95°C for 16 h (PG). After evaporation of the acid, the pellet was dissolved with 67 mM trisodium citrate-HCl buffer (pH 2.2) and injected into a Hitachi L-8800 amino acid analyzer equipped with a 2620MSC-PS column (ScienceTec). Amino acids and hexosamines were detected after post-column reaction with ninhydrin.
Results
The genome of V. spinosum contains the full complement of genes necessary for the de novo synthesis of peptidoglycan
The V. spinosum genome was searched from the Integrated Microbial Genomes (IMG) database (http://www.jgi.doe.gov/) using the annotated PG synthesis pathway from Kyoto Encyclopedia of Genes and Genomes (KEGG). The search resulted in the identification of 20 genes that are known to be involved in PG metabolism. Importantly, the search identified orthologs of all the genes necessary for the de novo synthesis of PG in V. spinosum (Table 1).
Table 1. List of genes involved in PG metabolism of V. spinosum DSM 4136T.
| Locus Tag | Protein Symbol | Annotated Gene Product Name | EC Number |
| VspiD_010100024635 | PBP | d-Alanyl-d-alanine carboxypeptidase-class C | 3.4.16.4 |
| VspiD_010100022270 | PBP | Multimodular transpeptidase-transglycosylase-class A | 2.4.1.129 |
| VspiD_010100006740 | PBP | Penicillin-binding protein 1C- class A | 2.4.1.129 |
| VspiD_010100020475 | PBP | Penicillin-binding protein 2-class A | 2.4.1.129 |
| VspiD_010100007940 | PBP | Peptidoglycan transpeptidase-class B | 2.4.1.129 |
| VspiD_010100017450 | PBP | Peptidoglycan transpeptidase-class B | 2.4.1.129 |
| VspiD_010100019135 | PBP | Peptidoglycan synthetase FtsI-class B | 2.4.1.129 |
| VspiD_010100018680 | PBP | Cell elongation specific d,d-transpeptidase- class B | 2.4.1.129 |
| VspiD_010100019120 | MraY | Phospho-N-acetylmuramoyl-pentapeptide-transferase | 2.7.8.13 |
| VspiD_010100011745 | MurA | UDP-N-acetylglucosamine 1-carboxyvinyltransferase | 2.5.1.7 |
| VspiD_010100019100 | MurG | UDP-N-acetylglucosamine-N-acetylmuramoyl-(pentapeptide) pyrophosphoryl-undecaprenol N-acetylglucosamine transferase | 2.4.1.227 |
| VspiD_010100019125 | MurF | UDP-N-acetylmuramoyl-tripeptide:d-alanyl-d-alanine ligase | 6.3.2.10 |
| VspiD_010100018175 | Ddl | d-Alanine:d-alanine ligase | 6.3.2.4 |
| VspiD_010100019115 | MurD | UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase | 6.3.2.9 |
| VspiD_010100019130 | MurE | UDP-N-acetylmuramoyl-l-alanyl-d-glutamate:meso-2,6-diaminopimelate ligase | 6.3.2.13 |
| VspiD_010100026230 | UppP | Undecaprenyl pyrophosphate phosphatase | 3.6.1.27 |
| VspiD_010100018130 | MurB | UDP-N-acetylenolpyruvoylglucosamine reductase | 1.1.1.158 |
| VspiD_010100018130 | MurC | UDP-N-acetylmuramate:l-alanine ligase | 6.3.2.8 |
| VspiD_010100000100 | AlaR | Alanine racemase | 5.1.1.1 |
| VspiD_010100008415 | MurI | Glutamate racemase | 5.1.1.3 |
The annotated gene product names are from NCBI (www.ncbi.nlm.nih.gov/protein/) queried of February 28, 2013. The pencillin-binding proteins (PBP) class designations are denoted by activity based on protein family (pfam) domains. Class A and class B PBPs are high-molecular mass PBPs while class C PBPs are low-molecular mass PBPs. Class A PBPs are predicted to have both transglycosylase and transpeptidase activities; class B PBPs are predicted to have only transpeptidase activity; class C PBPs are predicted to have d,d-carboxypeptidase activity.
Identification of the MurE ortholog from V. spinosum
The orthologous MurE protein from V. spinosum was initially identified using the MurE protein sequence from C. trachomatis (NP_219774) as a query. The BlastP algorithm from the Integrated Microbial Genomes (IMG) database was employed. The search resulted in the identification of a putative MurE from V. spinosum annotated by the locus tag VspiD_010100019130, which is 37% identical to the C. trachomatis MurE [10].
Overproduction and purification of murE Ligase from V. spinosum
The murE Vs gene was cloned into the pET100D/topo plasmid, allowing expression of the protein with an N-terminal short peptide extension comprising a hexa-histidine tag (see Materials and Methods). E. coli BL21 DE3-CodonPlus-RIPL cells, transformed with the resulting vector pET100D::murE Vs, were grown and subjected to IPTG induction. Extraction and purification afforded 2.5 mg of MurEVs per liter of culture. The protein was homogenous by SDS-PAGE: a band at ∼59 kDa could be observed, in keeping with a calculated molecular mass of 59,578 Da. (Fig. 3). Its identity was further confirmed by MALDI-TOF mass spectrometry: peaks at m/z 59,568 and 29,774 Da, corresponding to the [MH]+ and [M+2H]2+ ions, respectively, were observed (Fig. S1).
Figure 3. Expression and purification of recombinant MurEVs using His-tag affinity chromatography.
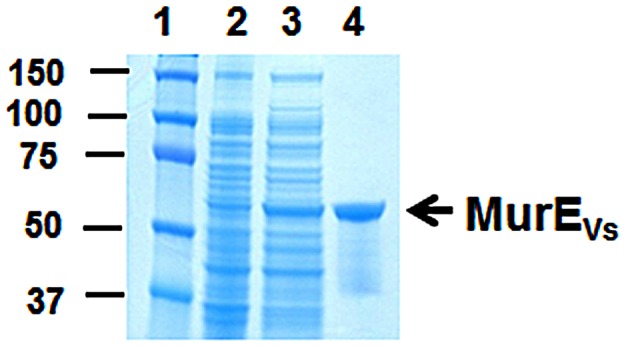
Lane (1) protein makers (kDa); Lane (2) 10 µg of soluble protein from uninduced cells; Lane (3) 10 µg of soluble protein from induced cells; Lane (4) 1 µg of purified recombinant MurEVs. The proteins were resolved on 10% (w/v) acrylamide gel and were stained using Coomassie blue.
Kinetic properties of the MurE ligase from V. spinosum
The optimal pH, temperature and magnesium concentration for MurEVs were found to be 9.6, 44–46°C, and 30 mM, respectively. In vitro assays were thus performed at pH 9.6 and with 30 mM MgCl2, the usual temperature of 37°C being used. With meso-A2pm and UDP-MurNAc-l-Ala-d-Glu as substrates, the maximum velocity was 36±2 µmol⋅min−1⋅mg−1. The K m values for the substrates were: ATP, 290±70 µM; UDP-MurNAc-l-Ala-d-Glu, 24±6 µM; and meso-A2pm, 17±3 µM. The enzyme proved to be stereospecific for meso-A2pm, since the rate of incorporation of the l,l or d,d isomer was <1% that of the meso isomer. No incorporation of l-lysine or l-ornithine could be detected, even with a significant amount of purified recombinant enzyme (Table 2).
Table 2. Specificity of MurEVs for the amino acid substrate.
| Substrate | Enzymatic activity (μmol.min−1.mg−1)a |
| meso-A2pm | 36 |
| l,l-A2pm | 0.18 |
| d,d-A2pm | 0.043 |
| l-Lysine | NDb |
| l-Ornithine | ND |
Determined as described in Materials and Methods with fixed concentrations of ATP (5 mM), UDP-MurNAc-l-Ala-d-Glu (0.15 mM) and amino acid (0.15 mM).
ND, no activity detected after 30 minutes with 11 µg of enzyme.
Sequence alignment and homology modeling
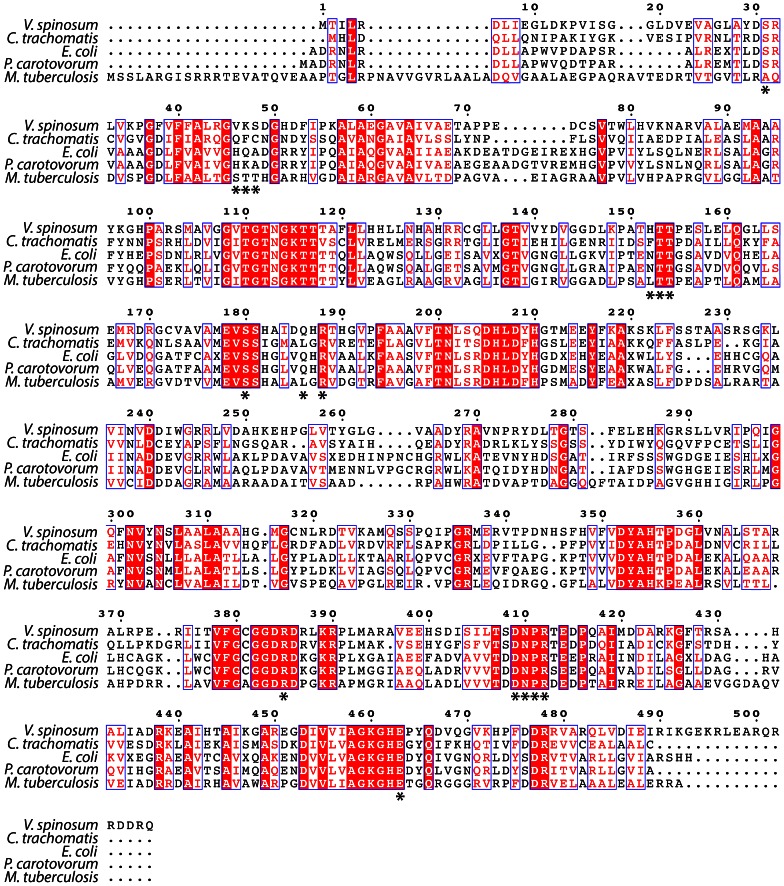
To identify conserved regions of the enzyme and motifs employed during catalysis, a multiple amino acid sequence alignment was performed between MurE enzymes from V. spinosum, M. tuberculosis, E. coli, C. trachomatis and P. carotovorum using ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Ten of the 16 putative active site residues thought to be involved in substrate binding were conserved among all five sequences. The key DNPR motif [23], [25], [26] which comprises residues 409–412 in the V. spinosum sequence, was identical across all five sequences.
To examine both the sequence and consider the consequences of differences within the MurEVs active site, we developed a MurEVs homology model. Using the MurEVs amino acid sequence as a template, we performed a PSI-BLAST search against proteins with known structure in the Protein Data Bank. The top hits were the MurE enzymes from M. tuberculosis and E. coli, with an identity of 38% and 37%, respectively. We chose the E. coli MurE structure (PDB id: 1E8C) as a template since this ortholog was well characterized [23], and generated a homology model for MurEVs using SWISS-MODEL (http://swissmodel.expasy.org/). The QMEAN score of the homology model was 0.55 (range is between 0 to 1) and the QMEAN Z score is −3.51 (Fig. S2) [27].
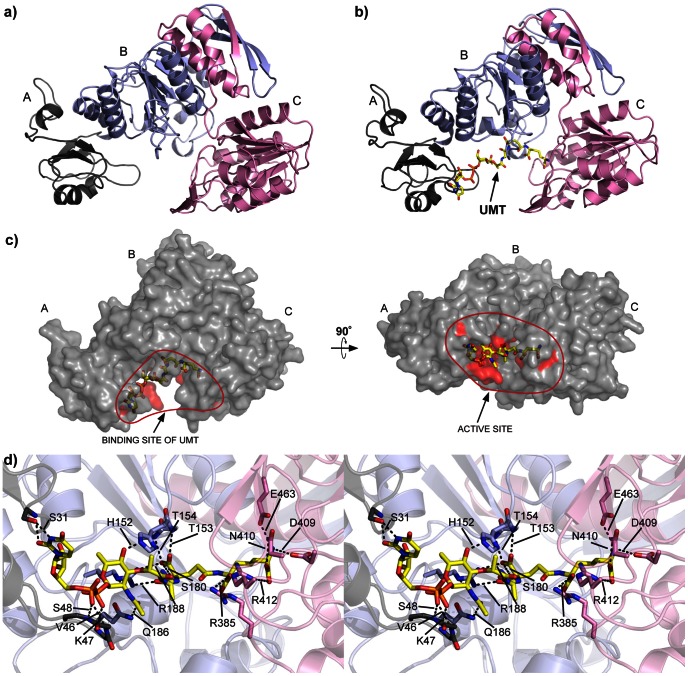
As annotated in the MurEEc structure, the MurEVs homology model is predicted to have three domains: A, B and C (Fig. 4a). Domain A comprises residues 1–84 and consists of a four-stranded parallel β-sheet, compared to the five-stranded β-sheet in the template MurEEc, and the β-sheet is flanked by two helices, as in template structure. Domain B comprises residues 85–289 and consists of a central six-stranded parallel β-sheet surrounded by six α-helices. The MurEEc template has seven α-helices and in the homology model the seventh α-helix is broken into two small helices. Additionally, there are two antiparallel strands that interact with domain C, which comprises residues 290–507 and consists of a six-stranded β-sheet with five parallel strands and one anti-parallel strand flanked by six α-helices.
Figure 4. Homolgy model of MurEVs.
(a) The homology model of MurEVs highlighting domains A (grey), B (violet) and C (pink). (b) Shows the structure model of MurEVs bound to UDP-MurNAc-tripeptide (UMT) product (yellow). (c) Active site residue hypothesized to bind to UMT product is shown in red. The structure has been rotated 90° on the right panel for the better viewing of the binding pocket. (d) Cross eye stereo view showing the interaction between amino acid residues of the binding site and UMT product.
An overlay of our MurEVs homology model with the ligand-bound MurEEc template structure highlights how the UDP-MurNAc-tripeptide (UMT) product (albeit in the conformation that fits within the MurEEc active site) is proposed to interact with residues in the putative active site of MurEVs (Fig. 4b and c). A comparison of the active site binding residues suggests that all three domains of MurEVs are involved in the interaction with the product (Fig. 4d). Most interactions already found between MurEEc and UMT [23] are conserved with MurEVs. In particular, hydrogen bonds between the ε-carboxyl group of meso-A2pm and N410 and R412 are predicted. These two H-bonds have been proposed to be responsible for the meso-A2pm/l-Lys discrimination [23]. A direct sequence comparison between the MurEVs and MurEEc active sites suggests that only 12 out of 16 active site residues are conserved in MurEVs (Fig. 5). However, three of the five non-conserved residues (K47, S48, H152, V. spinosum numbering) employ their main-chain atoms for binding and may therefore be more tolerant to mutation with minimal effects on substrate/product binding.
Figure 5. Multiple amino acid sequence alignment of five representative sequences of MurE.
The residues that are predicted to be involved in binding in the active site are marked with a star below the sequence. The sequence identity score against MurE from V. spinosum was: C. trachomatis, 37%; E. coli, 35%; P. carotovorum, 36%; and M. tuberculosis. The multiple amino acid sequence alignment figure was generated using the ESPript 2.2 server (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi).
Isolation and analysis of V. spinosum PG
Peptidoglycan has been indirectly detected in V. spinosum in a recent study using in situ probing via florescent D-amino acids [28]. To directly ascertain that V. spinosum does in fact possess authentic PG; cells were submitted to boiling SDS, a treatment used to isolate PG from other bacteria [24], [29]. Analysis of the SDS-insoluble material (Table 3) revealed that it contained Mur and A2pm; Mur is a specific component of all PGs, and A2pm is found in PG from Gram-negative bacteria. However, this crude PG was contaminated with proteogenic amino acids. Most of these amino acids could be removed by protease treatment. Molar ratios of the main PG constituents in the purified polymer were: Glu, 0.9; Ala, 1.5; A2pm, 1.1; Mur, 0.8; GlcN, 1.0; other amino acids had molar ratios ≤0.06 (Table 3). Therefore, this experiment shows that V. spinosum possesses a PG that is similar to the one of E. coli [24] and indeed other Gram-negative bacteria [1]. From the quantitative determination of A2pm in the crude PG preparation, an A2pm content of the PG of V. spinosum of 1.5×10−11 µmol/cell was estimated. This is of the same order of magnitude as the one found for E. coli PG (8.2×10−12 µmol/cell [24].
Table 3. Analysis of crude and purified PG from V. spinosum DSM 4136T.
| Constituent | Molar ratio (Calculated with GlcN = 1.0) | |
| Crude PGa | Purified PGa | |
| Asp | 1.22 | 0.04 |
| Thr | 0.62 | 0.03 |
| Ser | 0.63 | 0.03 |
| Mur | 0.79 | 0.80 |
| Glu | 2.33 | 0.91 |
| Pro | 0.47 | 0.02 |
| Gly | 0.95 | 0.06 |
| Ala | 2.47 | 1.51 |
| Val | 0.54 | 0.04 |
| A2pm | 1.02 | 1.10 |
| Met | 0 | 0 |
| Ile | 0.42 | 0.03 |
| Leu | 0.86 | 0.05 |
| Tyr | 0.34 | 0.03 |
| Phe | 0.39 | 0.03 |
| GlcN | 1.0 | 1.0 |
| Lys | 0.76 | 0.04 |
| His | 0.22 | 0.02 |
| Arg | 0.43 | 0.02 |
Crude and purified PG designate the macromolecule before and after, respectively, treatment with pancreatin, pronase and trypsin (see Materials and Methods).
Discussion
The heterotrophic Gram-negative bacterium V. spinosum has recently garnered significant interest from the scientific community, since the genome has been sequenced, annotated and is publically available. In addition, the bacterium was found to be pathogenic towards D. melanogaster and C. elegans, two model invertebrate organisms [5].
A recent study from our laboratories confirmed the presence of the plant-like biosynthetic pathway for diaminopimelate and l-lysine in V. spinosum through the partial characterization of the enzyme l,l-diaminopimelate aminotransferase (DapL) [10]. In the same study, we identified the MurE ortholog and showed that the enzyme was able to functionally complement an E. coli mutant that harbors a mutation in the murE gene [10].
The genus Verrucomicrobium is evolutionarily related to the genus Chlamydia [3]. Interestingly, we were able to identify all the genes that are involved in the de novo anabolism of PG from the annotated genome of V. spinosum (Table 1). The MurE ortholog from Chlamydia trachomatis was identified and was shown to be an authentic MurE enzyme, even though PG cannot be detected from the bacterium using methods developed thus far [26]. Unlike C. trachomatis, we were able to isolate and detect PG from V. spinosum in addition to quantifying all the major components of the macromolecule. V. spinosum is an attractive candidate model organism to address questions relating to: i) the chlamydial PG paradox; and ii) the feasibility and plausibility of whether the newly discovered DapL enzyme is a potential target for antibiotic development given the fact the enzyme is involved in the synthesis of both PG and lysine.
MurEVs shares 37% and 35% amino acid identity to the MurE orthologs from C. trachomatis and E. coli, respectively. With regards to the substrate specificity of the enzyme, MurEVs resembles that of the C. trachomatis and E. coli orthologs by showing preference for meso-A2pm. The enzyme incorporated very weakly the two other stereoisomers of A2pm; it was unable to incorporate l-lysine and l-ornithine, two structurally related diamine compounds. Therefore, MurEVs is highly specific for meso-A2pm.
The enzyme's optimum catalytic profile with respect to pH, temperature and [Mg2+] was examined to define optimum assay conditions and also gauge its similarity with other known MurE enzymes. MurEVs displays maximum activity at pH 9.6, which is slightly higher than those found in E. coli (pH 8.0–9.2) and C. trachomatis (pH 8.0–8.6) Mur ligases [15]. The optimal temperature for MurEVs (44–46°C) seems somewhat high but difficult to compare with other orthologs and paralogs since this parameter is almost never mentioned. These unusual values for MurEVs might be attributed to environmental factors such as the natural habitat(s) of the organism. As for the optimal [Mg2+] concentration, it falls within the range (5–100 mM) found for E. coli and C. trachomatis Mur ligases [15], [26], [30].
The maximum velocity of 36 µmol⋅min−1⋅mg−1 for the MurEVs using saturating levels of all substrates is approximately 110, 26 and 14 times more than those of MurECt, MurEEc and MurE from Pseudomonas aeruginosa, respectively [15], [26], [31]. Whereas the higher specific activity of MurEVs with respect to MurECt can easily be explained by the fact that Chlamydiae are slow-growing, primarily intracellular organisms [26], we have no explanation for the difference between MurEVs and the orthologs from E. coli and P. aeruginosa.
Primary sequence analysis showed that MurEVs contains ten out of the sixteen amino acids that make up the active site of the enzyme including the DNPR motif. The DNPR motif is conserved among MurEs that have been experimentally authenticated. A homology model, based on the well characterized MurEEc enzyme (PDB id: 1E8C), was developed to examine the sequence further and consider the consequences of differences within the MurEVs active site. The MurEVs enzyme is likely to comprise three domains, A, B and C, each of which contribute amino acid residues to the active site. Nearly all of the active site moieties (10 of 16) known to interact with the substrates and products are conserved in the MurEVs active site. Overall, the homology model is entirely consistent with our validated function of MurEVs and suggests that the enzyme binds the substrates in a similar way to other known MurE enzymes.
Even though the diaminopimelate/l-lysine pathway have been the subject and focus of numerous studies regarding the development of antibiotics, no novel antibiotics have been developed or identified thus far that target any enzyme belonging to the four variants of the anabolic pathways [6], [32]. To this end, we are interested in the essentiality of the DapL enzyme in eubacteria that defines one of the four anabolic variants identified thus far. The enzyme converts tetrahydrodipicolinate to l,l-diaminopimelate in one step circumventing three enzymatic steps in the E. coli acyl pathways [6]. l,l-Diaminopimelate is subsequently converted to the meso isomer by an epimerase; this facilitates the synthesis of lysine via a decarboxylation reaction for protein synthesis in addition to cell wall biosynthesis via MurE in many Gram-positive bacteria. The inhibition of DapL or other enzymes in the diaminopimelate/l-lysine pathway would affect bacterial growth in two different ways. First the bacteria will be unable to grow because of the lack of protein synthesis due to the absence of l-lysine. Second, PG biosynthesis will be impaired due to the lack of meso-A2pm. Presumably, this will result in a bacteriostatic effect, as already observed for other enzymes of the diaminopimelate/l-lysine pathway [33], [34].
The genomes of animals and particularly humans do not possess the genetic machinery to facilitate the biosynthesis of diaminopimelate/l-lysine de novo. Therefore, animals must acquire l-lysine through dietary means. Thus there is a unique opportunity to assess the essentiality of enzymes that are important for cell wall and protein synthesis from eubacteria. V. spinosum is an attractive model bacterial system based on the fact that the organism is closely related to Chlamydia, which was found to use the DapL pathway to diaminopimelate/l-lysine. Bioinformatic analysis shows that the sequenced and annotated genomes of bacteria belonging to the genus Chlamydia contain putative dapL orthologs (data not shown). V. spinosum is aerobic and facile to culture using commercially available media because it is not an obligate intracellular bacterium as is the case with Chlamydia. Importantly, the bacterium is not pathogenic to mammals based on what we currently know. Since the genome of the organism can be genetically modified using transposon mutagenesis, analysis of genes that are essential for V. spinosum that are involved in the diaminopimelate/l-lysine biosynthesis can be the focus of future studies [10], [35].
Here we present the identification and characterization of the first Mur ligase namely, MurE from the bacterium V. spinosum. Bioinformatic and biochemical analyses provide evidence that the bacterium is able to synthesize PG de novo. In vivo analysis shows that MurEVs is an authentic meso-A2pm adding enzyme. This was further validated by in vitro analyses that show that the kinetic and physical properties are consistent with MurE orthologs that have been experimentally confirmed. Finally, primary amino acid sequence and structural analysis based on protein modeling show that key amino acids that are involved in substrate binding and or catalysis are conserved in MurEVs.
Supporting Information
MALDI-TOF mass spectrometry analysis of purified MurEVs. Matrix: sinapinic acid. Peaks with m/z ratios consistent with the His6-tagged protein (calculated mass, 59,578 Da) are shown.
(TIF)
Homology model quality statistics. The cartoon structure shows the quality of model by coloring the residues according to the error. The coloring is from blue (reliable region) to red (potentially unreliable region). The residue error plot depicts the local model reliability with estimated pre-residue inaccuracies along the sequence.
(TIF)
Acknowledgments
The authors thank Dr. Richard Hailstone from the Chester F. Carlson Center for Imaging Science at RIT for acquisition of the scanning electron microscopy image. AOH thank Dr. Naomi Ward from the University of Wyoming for graciously providing the genomic DNA of Verrucomicrobium spinosum DSM 4136T to facilitate this work. The authors acknowledge Matt Walters for help with image presentation. HS acknowledges the Royal Society of New Zealand and the Japan Society for the Promotion of Science Researcher Exchange Program.
Funding Statement
This research was supported by a National Science Foundation (NSF) (MCB-1120541) and a Rochester Institute of Technology (RIT) College of Science 2012 Dean's Research Initiation Grant to AOH. MAS acknowledges a RIT College of Science Faculty Development (FEAD-2012) grant. SEM and DTP were supported by the NSF MCB-1120541 awarded to AOH as undergraduate students in the Bioinformatics program in the Thomas H. Gosnell School of Life Sciences and the Biochemistry program in the School of Chemical and Material Sciences at RIT. RCJD acknowledges: 1) the C.R. Roper Bequest for Fellowship support; 2) the New Zealand Royal Society Marsden Fund for funding support, in part (contract UOC1013); and 3) the U.S. Army Research Laboratory and U.S. Army Research Office under contract/grant number W911NF-11-1-0481 for supported, in part. DP and DB were supported by grants from the Centre National de la Recherche Scientifique (UMR 8619). HS acknowledges the Royal Society of New Zealand and the Japan Society for the Promotion of Science for salary support by the FY 2012 Researcher Exchange Program. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Vollmer W, Blanot D, de Pedro MA (2008) Peptidoglycan structure and architecture. FEMS Microbiol Rev 32: 149–167. [DOI] [PubMed] [Google Scholar]
- 2. Scheffers DJ, Pinho MG (2005) Bacterial cell wall synthesis: new insights from localization studies. Microbiol Mol Biol Rev 69: 585–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wagner M, Horn M (2006) The Planctomycetes, Verrucomicrobia, Chlamydiae and sister phyla comprise a superphylum with biotechnological and medical relevance. Curr. Opin. Biotechnol 17: 241–249. [DOI] [PubMed] [Google Scholar]
- 4. Hueck CJ (1998) Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev 62: 379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sait M, Kamneva OK, Fay DS, Kirienko NV, Polek J, Shiasus-Hiza MM, et al. (2011) Genomic and experimental evidence suggests that Verrucomicrobium spinosum interacts with eukaryotes. Front. Microbiol 2: 211 doi:10.3389/fmicb.2011.00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hudson AO, Singh BK, Leustek T, Gilvarg C (2006) An L,L-diaminopimelate aminotransferase defines a novel variant of the lysine biosynthesis pathway in plants. Plant Physiol 140: 292–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCoy AJ, Adams NE, Hudson AO, Gilvarg C, Leustek T, et al. (2006) Diaminopimelate aminotransferase a trans-kingdom enzyme shared by Chlamydia and plants for synthesis of diaminopimelate/lysine. Proc. Natl. Acad. Sci. USA 103: 17909–17914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hudson AO, Gilvarg C, Leustek T (2008) Biochemical and phylogenetic characterization of a novel diaminopimelate biosynthesis pathway in prokaryotes identifies a diverge from of L,L-diaminopimelate aminotransferase. J. Bacteriol 190: 3256–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, White RH, Whitman WB (2010) Methanococci use the diaminopimelate aminotransferase (DapL) pathway for lysine biosynthesis. J. Bacteriol 192: 3304–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nachar VR, Savka FC, McGroty SE, Donovan KA, North RA, et al. (2012) Genomic and biochemical analysis of the diaminopimelate and lysine biosynthesis pathway in Verrucomicrobium spinosum: identification and partial characterization of L,L-diaminopimelate aminotransferase and UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6-meso-diaminopimelate ligase. Front. Microbiol 3: 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hutton CA, Perugini MA, Gerrard JA (2007) Inhibition of lysine biosynthesis: an evolving antibiotic strategy. Mol. Biosyst 3: 458–465. [DOI] [PubMed] [Google Scholar]
- 12. Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell wall and their taxonomic implications. Bacterol Rev 36: 407–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barreteau H, Kovač A, Boniface A, Sova M, Gobec S, et al. (2008) Cytoplasmic steps of peptidoglycan biosynthesis. FEMS Microbiol Rev 32: 168–207. [DOI] [PubMed] [Google Scholar]
- 14. Mengin-Lecreulx D, Falla T, Blanot D, van Heijenoort J, Adams DJ, et al. (1999) Expression of the Staphylococcus aureus UDP-N-acetylmuramoyl- L-alanyl-D-glutamate:L-lysine ligase in Escherichia coli and effects on peptidoglycan biosynthesis and cell growth. J. Bacteriol 181: 5909–5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Patin D, Boniface A, Kovač A, Hervé M, Dementin S, et al. (2010) Purification and biochemical characterization of Mur ligases from Staphylococcus aureus . Biochimie 92: 1793–1800. [DOI] [PubMed] [Google Scholar]
- 16.Babič A, Patin D, Boniface A, Hervé M, Mengin-Lecreulx D, et al.. (2007) Chemoenzymatic synthesis of the nucleotide substrates of the Mur ligases, 1–4. In D. Kikelj, 5th Joint Meeting on Medicinal Chemistry, 17 to 21 June, Portoro?, Slovenia. Medimond Srl, Bologna, Italy.
- 17. Michaud C, Mengin-Lecreulx D, van Heijenoort J, Blanot D (1990) Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramoyl-l-alanyl-d-glutamate:meso-2,6-diaminopimelate ligase from Escherichia coli . Eur. J. Biochem 194: 853–861. [DOI] [PubMed] [Google Scholar]
- 18. van Heijenoort J, Bricas E (1968) Contribution à l′étude des isomères de l′acide α,α′-diaminopimélique. Bull. Soc. Chim. Fr 7: 2828–2831. [Google Scholar]
- 19.Press WH, Flannery BP, Teukolsky SA, Vetterling WT (1986) Numerical recipes:.The art of scientific computing, Cambridge University Press, Cambridge, UK.
- 20. Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 21. Kiefer F, Arnold K, Künzli M, Bordoli L, Schwede T (2009) The SWISS-MODEL repository and associated resources. Nucleic Acids Research 37: 387–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Peitsch MC (1995) Protein Modeling by E-mail. Nat Biotech 13: 658–660. [Google Scholar]
- 23. Gordon E, Flouret B, Chantalat L, van Heijenoort J, Mengin-Lecreulx D, et al. (2001) Crystal structure of UDP-N-acetylmuramoyl-L-alanyl-D-glutamate: meso-diaminopimelate ligase from Escherichia coli . J. Biol Chem 276: 10999–11006. [DOI] [PubMed] [Google Scholar]
- 24. Mengin-Lecreulx D, Flouret B, van Heijenoort J (1982) Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli . J. Bacteriol 151: 1109–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Boniface A, Bouhss A, Mengin-Lecreulx D, Blanot D (2006) The MurE synthetase from Thermotoga maritima is endowed with an unusual D-lysine adding activity. J Biol Chem 281: 15680–15686. [DOI] [PubMed] [Google Scholar]
- 26. Patin D, Bostock J, Blanot D, Mengin-Lecreulx D, Chopra I (2009) Functional and biochemical analysis of the Chlamydia trachomatis ligase MurE. J. Bacteriol 191: 7430–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27: 343–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuru E, Hughes HV, Brown PJ, Hall E, Tekkam S (2012) In situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent d-amino acids. Angew. Chem Int. Ed. 51 : doi: 10.1002/anie.201206749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Girardin SE, Boneca IV, Viala J, Chamaillard M, Labigne A (2003) Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem 278: 8869–8872. [DOI] [PubMed] [Google Scholar]
- 30. Patin D, Bostock J, Chopra I, Mengin-Lecreulx D, Blanot D (2012) Biochemical characterization of the chlamydial MurF ligase, and possible sequence of the chlamydial peptidoglycan pentapeptide stem. Arch. Microbiol 194: 505–512. [DOI] [PubMed] [Google Scholar]
- 31. Paradis-Bleau C, Lloyd A, Sanschagrin F, Maaroufi H, Clarke T (2009) Pseudomonas aeruginosa MurE amide ligase: enzyme kinetics and peptide inhibitor. Biochem. J. 421: 263–272. [DOI] [PubMed] [Google Scholar]
- 32. Cox RJ (1996) The DAP pathway to lysine as a target for antimicrobial agents. Nat. Prod. Rep 13: 29–43. [DOI] [PubMed] [Google Scholar]
- 33. Berges DA, DeWolf DE, Dunn GL, Grappel SF, Newman DJ (1986) Peptides of 2-aminopimelic acid: antibacterial agents that inhibit diaminopimelic acid biosynthesis. J. Med. Chem 29: 89–95. [DOI] [PubMed] [Google Scholar]
- 34. Le Roux P, Blanot D, Mengin-Lecreulx D, van Heijenoort J (1991) Peptides containing 2-aminopimelic acid. Synthesis and study of in vitro effect on bacterial cells. Int. J. Peptide Protein Res 37: 103–111. [PubMed] [Google Scholar]
- 35. Domman DB, Steven BT, Ward NL (2011) Random transposon mutagenesis of Verrucomicrobium spinosum DSM 4136(T). Arch. Microbiol 193: 307–312. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MALDI-TOF mass spectrometry analysis of purified MurEVs. Matrix: sinapinic acid. Peaks with m/z ratios consistent with the His6-tagged protein (calculated mass, 59,578 Da) are shown.
(TIF)
Homology model quality statistics. The cartoon structure shows the quality of model by coloring the residues according to the error. The coloring is from blue (reliable region) to red (potentially unreliable region). The residue error plot depicts the local model reliability with estimated pre-residue inaccuracies along the sequence.
(TIF)