Abstract
The Saccharomyces cerevisiae Slx5/8 complex is the founding member of a recently defined class of SUMO-targeted ubiquitin ligases (STUbLs). Slx5/8 has been implicated in genome stability and transcription, but the precise contribution is unclear. To characterise Slx5/8 function, we determined genome-wide changes in gene expression upon loss of either subunit. The majority of mRNA changes are part of a general stress response, also exhibited by mutants of other genome integrity pathways and therefore indicative of an indirect effect on transcription. Genome-wide binding analysis reveals a uniquely centromeric location for Slx5. Detailed phenotype analyses of slx5Δ and slx8Δ mutants show severe mitotic defects that include aneuploidy, spindle mispositioning, fish hooks and aberrant spindle kinetics. This is associated with accumulation of the PP2A regulatory subunit Rts1 at centromeres prior to entry into anaphase. Knockdown of the human STUbL orthologue RNF4 also results in chromosome segregation errors due to chromosome bridges. The study shows that STUbLs have a conserved role in maintenance of chromosome stability and links SUMO-dependent ubiquitination to a centromere-specific function during mitosis.
Introduction
Accurate inheritance of chromosomes during each cell division is crucial for cell survival. Genome instability is disadvantageous and directly associated with many diseases including cancer [1]. Cells have a wide variety of regulatory mechanisms that monitor the fidelity of DNA replication and mitosis. At a molecular level, control of genome stability is an intrinsic process that depends on proper posttranslational modification of many proteins. This includes ubiquitination and sumoylation. Ubiquitin and SUMO (small ubiquitin-like modifier) are small peptides that can be covalently attached to substrates through a three-step enzymatic cascade that activates (E1), conjugates (E2) and ligates (E3) the peptide to a substrate [2]–[4]. In S. cerevisiae, the genes SLX5 and SLX8 encode the heterodimeric protein complex Slx5/8, which is required for maintenance of genome integrity in yeast [5], [6]. Biochemical characterisation of the complex has revealed that Slx5/8 is a SUMO-dependent ubiquitin E3 ligase [7], [8]. Both Slx5 and Slx8 have a C-terminal zinc finger RING domain, commonly found in many ubiquitin E3 ligases [9]. Slx5/8 also has multiple SUMO-interaction motifs, which confers a unique ability to ubiquitinate and degrade sumoylated proteins [7], [8]. This indicates that SUMO-targeted ubiquitin ligases (STUbLs) control the turnover of sumoylated proteins via ubiquitin-dependent protein degradation to ensure appropriate cellular levels of sumoylated proteins [10].
The cellular role of STUbLs is less well characterised. SLX5 and SLX8 were originally identified in a screen for genes that are synthetic lethal with deletion of SGS1, a DNA helicase of the RecQ family, indicating a role for Slx5/8 in genome stability [5]. Inactivation of STUbLs leads to a broad spectrum of genome instability phenotypes in S. cerevisiae and S. pombe. These include a strong cell cycle delay, DNA damage checkpoint activation, sensitivity to genotoxic stress, gross chromosomal rearrangements and increased rates of DNA mutation and recombination [6], [11]–[15]. The Slx5/8 complex resides at sites of DNA damage and replication, and contributes to DNA repair by relocating double stranded DNA breaks to the nuclear pore [11], [12], [16]. How the role of Slx5/8 in genome stability ties in with its function as STUbL is unclear and other cellular roles have also been proposed. STUbL orthologues have now been identified from fission yeast (Rfp1, Rfp2, spSlx8) to human (RNF4), indicating an evolutionarily conserved and important function for ubiquitin-dependent degradation of sumoylated proteins [14], [15], [17], [18]. In humans, there is evidence that RNF4 regulates transcription with several transcription regulators identified as targets for SUMO-dependent ubiquitination [19]–[21]. In yeast, Slx5/8 has also been implicated in transcription regulation [8], [22]–[24], indicating that STUbL function may extend beyond genome stability.
To better characterise the function of STUbLs, the phenotypes of slx5Δ and slx8Δ deletion mutants in transcription and genome stability were investigated in detail. We show that changes in mRNA expression in slx5/8 mutants are largely associated with a general stress response that is likely due to genome instability rather than a direct transcriptional defect. Determination of the genomic location of the Slx5/8 complex reveals that Slx5 locates at centromeres. Loss of SLX5 or SLX8 is accompanied by accumulation of Rts1 at centromeres during metaphase. Moreover, the slx5Δ and slx8Δ mutants display a variety of mitotic defects, supporting a role for Slx5/8 in chromosome stability. This is distinct from previously reported roles of Slx5/8 and gives a better insight into how genome instability arises in slx5/8 mutants. Analysis of human RNF4 shows that the role of Slx5/8 in chromosome stability is evolutionarily conserved, further underscoring the importance of STUbL function in genome stability during mitosis.
Results
Slx5Δ and slx8Δ Display a General Stress Response that is Shared With Mutants of Various Genome Integrity Pathways
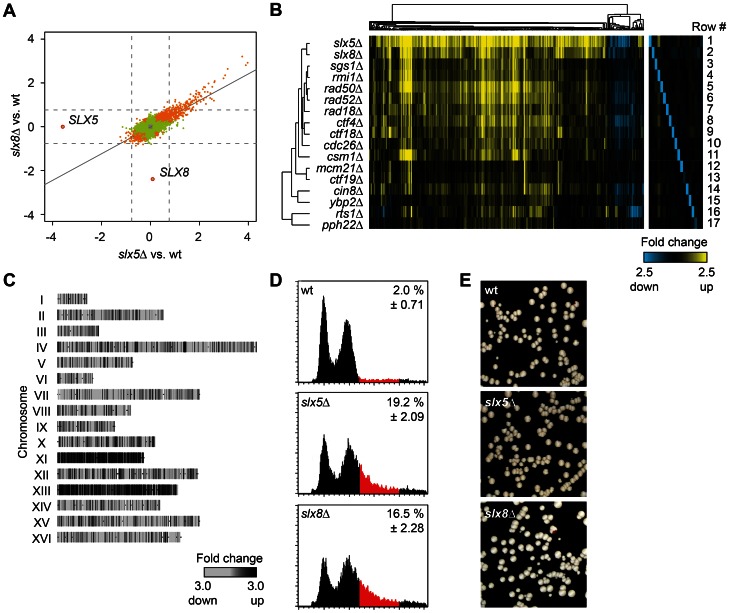
To investigate the role of Slx5/8 in transcription, genome-wide mRNA levels in SLX5 and SLX8 deletion strains were compared to wild type (wt), all grown to mid-log phase under standard growth conditions. A strong transcriptional response is observed upon SLX5 deletion, resulting in changed expression of 321 genes (fold change (FC) >1.7, p<0.05, Figure 1A). The response in slx8Δ is quantitatively weaker (132 genes, FC >1.7, p<0.05), but correlates highly with slx5Δ (r = 0.81, Figure 1A). The similarity is also readily observed upon visual inspection of individual genes and consists largely of upregulated expression (Figure 1B; rows 1–2). The differentially expressed genes are enriched for various Gene Ontology (GO) terms (Table S1), some of which have previously also been associated with the environmental stress response in yeast [25]. The slx5Δ and slx8Δ mutant profiles were therefore compared with previously published DNA microarray datasets of various stress responses, which all share a similar expression response [25]. This reveals a significant correlation between slx5Δ and slx8Δ expression profiles and stress responses such as heat shock (r = 0.48, p = 2.95E-200), indicating constitutive activation of a stress response in both slx5Δ and slx8Δ under normal growth conditions.
Figure 1. Slx5Δ and slx8Δ display a genome instability-induced stress response.
(A) Scatter plot comparing the changes in mRNA expression levels in slx5Δ and slx8Δ mutants. Fold change (FC) in expression is the average of four measurements for each mutant (two independent cultures each measured twice), plotted as log2 values of mutant over wt. Genes changing significantly (p<0.05) are indicated in orange (significant in both mutants) or green (significant in one mutant). Deleted genes are indicated. Solid line indicates the regression line. Dashed lines mark a 1.7 FC threshold. (B) Heatmap and cluster diagram of the gene expression profiles of deletion mutants, showing all significant genes (FC >1.7, p<0.05) that change at least once in any mutant. FC expression of mutant over wt is indicated by the colour scale, with yellow for upregulation, blue for downregulation and black for no change. Deleted genes are depicted in the right-hand panel. (C) Microarray expression profile of an aneuploid slx8Δ mutant with duplications of chromosome XI and XIII. Genes are mapped per chromosome. The grey scale indicates FC expression in the slx8Δ strain versus wt. (D) Flow cytometric profiles of asynchronous populations of wt, slx5Δ and slx8Δ. Cell population with a >2N DNA content, indicated in red, is quantified (± s.d., n = 3). (E) Chromosome loss assay of wt, slx5Δ and slx8Δ cells. Red-sectoring of colonies reflects loss of the reporter chromosome. Frequency of chromosome missegration events is 0.03% (n = 1 out of 3415 colonies) for wt, 0.06% (n = 2 out of 3254, p = 0.247) for slx5Δ, and 0.11% (n = 4 out of 3555, p = 0.022) for slx8Δ. Frequencies are quantified by colony half-sector analysis and significance of the difference of mutant compared to wt was determined using the binomial test.
Although Slx5/8 have previously been implicated in regulating transcription, one interpretation of the observed changes in gene expression is that these are indirectly caused by cellular stress as a consequence of the genome instability known to occur in slx5Δ and slx8Δ. To investigate this, DNA microarray expression profiles were generated for deletion mutants of various genome integrity pathways (Figure 1B). These mutants include components of the Sgs1-Rmi1-Top3 DNA helicase complex (sgs1Δ, rmi1Δ), DNA repair factors (rad18Δ, rad50Δ, rad52Δ), Cohesin components (ctf4Δ, ctf18Δ), a component of the anaphase promoting complex (cdc26Δ), a kinesin motor protein (cin8Δ), kinetochore-associated proteins (csm1Δ, ctf19Δ, mcm21Δ, ybp2Δ) and protein phosphatase 2A subunits (pph22Δ, rts1Δ). The transcription responses of all these mutants show a high degree of similarity to each other and to slx5Δ and slx8Δ, differing mainly in the degree of upregulation, rather than in the affected genes (Figure 1B). The differentially expressed genes are enriched in DNA binding sites for the stress response transcription factors Msn2 and Msn4 (Table S1) [26], [27]. Perturbation of several different genome integrity pathways therefore result in a similar gene expression response that is related to stress. A likely explanation for a large part of the gene expression response in slx5Δ and slx8Δ is therefore that this results indirectly from genome instability-induced stress rather than from a direct defect at the level of transcription of all these genes.
Slx5Δ and slx8Δ Mutants are Aneuploid
The DNA microarray analyses of slx5Δ and slx8Δ also reveals a second unanticipated phenotype, the occurrence of whole chromosome aneuploidy, an example of which is shown in Figure 1C. Microarray analyses were performed on liquid cultures derived from independent colonies for each mutant. In slx5Δ and slx8Δ, aneuploidy of various chromosomes (VII, XI, XII and XIII) was observed in the form of an apparent upregulation of all genes from one or more of these chromosomes. Detection of aneuploidy in this way has been described before [28]. Note that the expression profiles shown in Figure 1A and B are the average of two independent colonies per mutant where no aneuploidy was observed. Since the detection of aneuploidy in DNA microarray experiments depends on singular events in the starting colonies, flow cytometric profiles were generated to examine the DNA content of individual cells (Figure 1D). In asynchronous cell cultures, slx5Δ and slx8Δ mutants are characterised by a large fraction of the cell population having a DNA content higher than 2N (19.2%, 16.5% and 2% in slx5Δ, slx8Δ and wt respectively). The flow cytometry therefore agrees with the aneuploidy observed in individual slx5Δ and slx8Δ microarray experiments. A colony colour assay was performed to measure the chromosome stability in slx5Δ and slx8Δ [29]. Wt, slx5Δ and slx8Δ cells, bearing the ochre mutation ade2-101, were complemented with a reporter chromosome bearing the SUP11 gene that suppresses the red colour (Figure 1E). Chromosome missegregation events of the reporter chromosome were analyzed. Although slx8Δ shows a statistically significant increase of missegregation events (p = 0.022), this increase is only marginal and not statistically significant for slx5Δ (p = 0.247). The slx5Δ, slx8Δ aneuploidy is therefore not likely caused by chromosome loss or nondisjunction.
Slx5 Resides at the Centromere
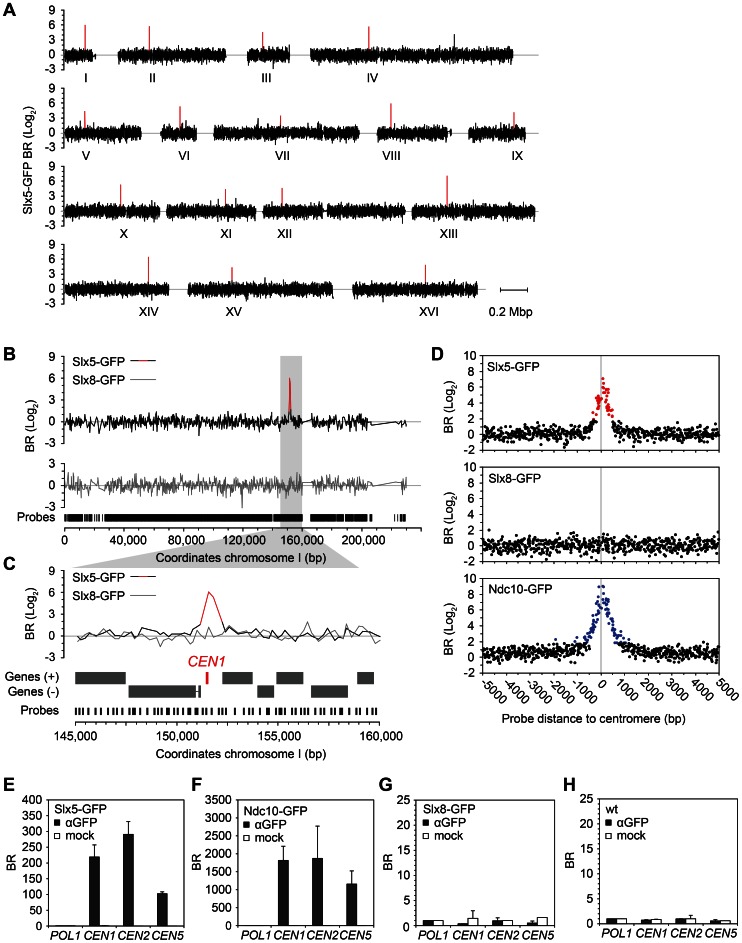
To further elucidate the function of Slx5/8, their location on DNA was investigated by genome-wide chromatin immunoprecipitation (ChIP-chip). This was motivated by the observation that previously reported roles of Slx5/8, such as transcription [8], [22]–[24] and DNA repair [11], [12], [16], may be associated with location on DNA. Slx5 and Slx8 were C-terminally fused to GFP by genomic integration, resulting in expression at endogenous levels. ChIP-chip reveals the presence of 17 distinct binding peaks for Slx5 (Figure 2A). Strikingly, each Slx5 peak maps to a different chromosome and coincides exactly with the location of the centromere. One exception is chromosome IV, where a second smaller Slx5 peak is detected (Figure 2A). As opposed to Slx5, Slx8 did not show enrichment at centromeres. For example, whereas chromosome I shows a single centromeric Slx5 peak, we did not detect any coinciding Slx8 signal (Figure 2B and C). Other genomic locations, such as ORFs, promoters, (sub-)telomeres, ARS, or rDNA, do not show notable enrichment for Slx5 or Slx8.
Figure 2. Slx5 resides at centromeres.
(A) ChIP-chip analysis of Slx5. Binding ratios (BR) of individual probes are mapped to the 16 chromosomes of the S. cerevisiae genome. BR is expressed as log2 value of αGFP ChIP/input with subtraction of the mock/input signal. The positions of the centromeres are marked I to XVI. (B) Binding profile of Slx5 and Slx8 at chromosome 1. The genomic region shown in grey is magnified in (C). (D) Average centromeric binding profiles of Slx5, Slx8 and Ndc10. Signals from microarray probes are mapped relative to their position to the centromere and all probes are included that map to within 5000 bp of all 16 centromeres. Probes with a BR (log2) >2 are shown in red (Slx5) or blue (Ndc10). (E–H) ChIP-qPCR of Slx5 (E), Ndc10 (F), Slx8 (G) and wt (H). BRs at centromere 1, 2, and 5 are normalised to the control gene POL1 (± s.d., n = 3).
Centromeres play a key role in chromosome segregation. They provide the binding site for the kinetochore, which physically connects centromeres to microtubules, allowing segregation of sister chromatids during mitosis and meiosis [30]. The centromeres of budding yeast are small (∼120 bp) and known as point centromeres. Each Slx5 peak is characterised by enriched signals on three to five consecutive microarray probes that span a centromere (e.g. Figure 2C). An average centromere binding profile was generated by mapping all (peri-)centromeric probes relative to their respective centromere (Figure 2D). Slx5 enrichment is centred on the core centromere, without global enrichment of the entire 10 kb pericentromeric region. We compared the binding profile of Slx5 to a known kinetochore component, Ndc10, which is the centromere DNA binding subunit of the kinetochore [31]. Strikingly, the binding pattern of Slx5 is equivalent to Ndc10, showing that Slx5 is preferentially located at the core centromere (Figure 2D). Centromeric location of Slx5 was further confirmed by ChIP quantitative real-time PCR. Slx5 and Ndc10 are highly enriched at all centromeres tested (Figure 2E and F), in agreement with the genome-wide experiments. Mock ChIPs, Slx8 and an untagged wt strain show no centromeric enrichment (Figure 2E–H), confirming the specific location of Slx5 to centromeres.
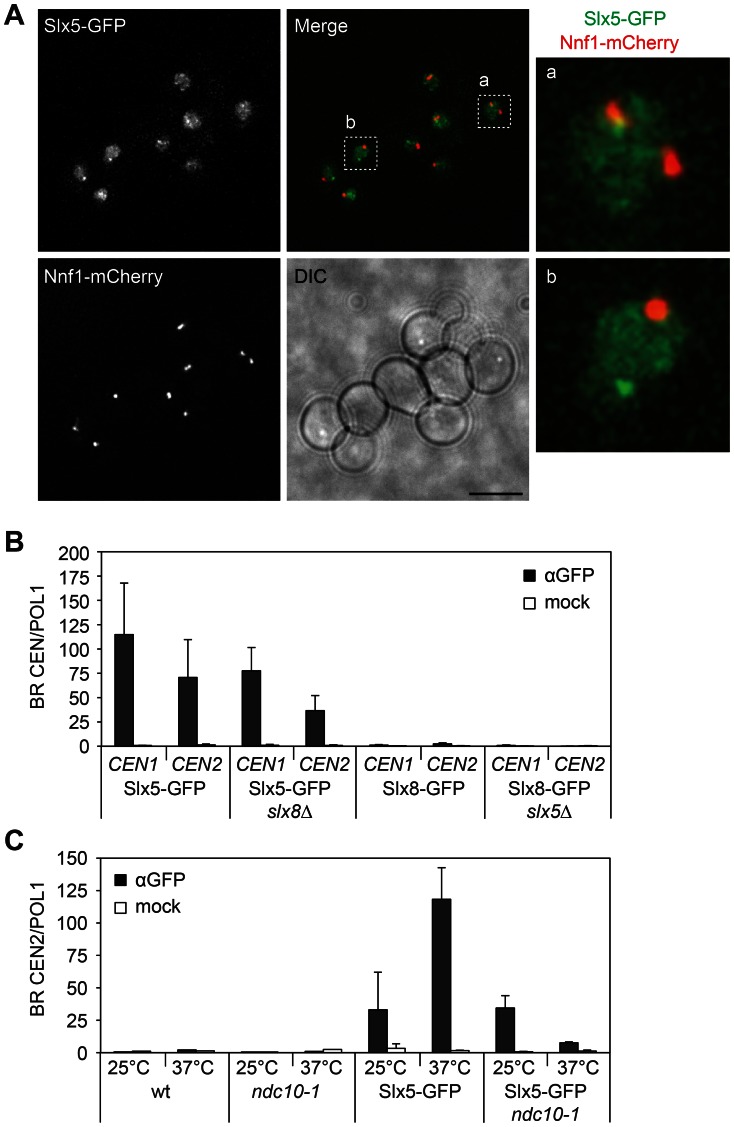
The subcellular localisation of Slx5 was investigated further by fluorescence microscopy (Figure 3A). Slx5 has a diffuse nuclear location with occasional subnuclear foci. This is similar to other studies, where Slx5 is shown to have a diffuse nuclear location with DNA foci at DNA repair centres [16]. We investigated whether Slx5 foci also colocalise with centromeric regions, as marked by kinetochore subunit Nnf1 (Figure 3A). Strict colocalisation of the Slx5 foci with Nnf1 was not observed. Although in rare cases Slx5 foci do overlap with the kinetochore (Figure 3A, cell a) it cannot be ruled out that they may represent cases where a DNA break is in close proximity to the centromere. This indicates that the centromeric pool of Slx5 cannot be distinguished visually by fluorescence microscopy and that it is likely part of the diffuse nuclear Slx5 pool (Figure 3A, cell b).
Figure 3. Slx5 binding to centromeres is largely kinetochore-dependent.
(A) Live cell microscopy of cells coexpressing Slx5-GFP and kinetochore protein Nnf1-mCherry. Right-hand panel shows a magnification of two nuclei, indicated in the merged image. Scale bars, 5 µm. (B) ChIP-qPCR of Slx5-GFP and Slx8-GFP in wt, slx5Δ or slx8Δ strains. Binding ratios (BR) at CEN1 and CEN2 are represented as enrichment over the control gene POL1 (± s.d., n = 4), based on two independent biological replicate experiments. (C–D) ChIP-qPCR of Slx5-GFP in wt and ndc10-1 strains at permissive (25°C) and nonpermissive (37°C) temperatures. Data is represented as enrichment at CEN2 over POL1 (Slx5-GFP and Slx5-GFP ndc10-1: ± s.d., n = 2) (wt and ndc10-1: n = 1, no enrichment).
We next tested the dependency of the Slx5 centromeric location on other proteins by ChIP. Slx5 binding to centromeres is hardly reduced in the absence of SLX8 (Figure 3B). This agrees with previous observations showing that Slx5 still forms nuclear foci and binds to DNA breaks without Slx8 [16]. Slx8 also remains absent from centromeres upon deletion of SLX5 (Figure 3B). To investigate whether centromere location of Slx5 is kinetochore-dependent, a ChIP was performed in the ndc10-1 mutant, which is defective in kinetochore assembly at a nonpermissive temperature [31]. Centromeric binding of Slx5 is nearly completely eliminated when shifting Slx5-GFP ndc10-1 cells from 25°C to 37°C (Figure 3C). There is some residual binding, indicating that although centromere location of Slx5 is largely kinetochore-dependent, additional kinetochore-independent interactions may also be involved.
Rts1 Accumulates at Kinetochores in slx5Δ and slx8Δ Metaphase Cells
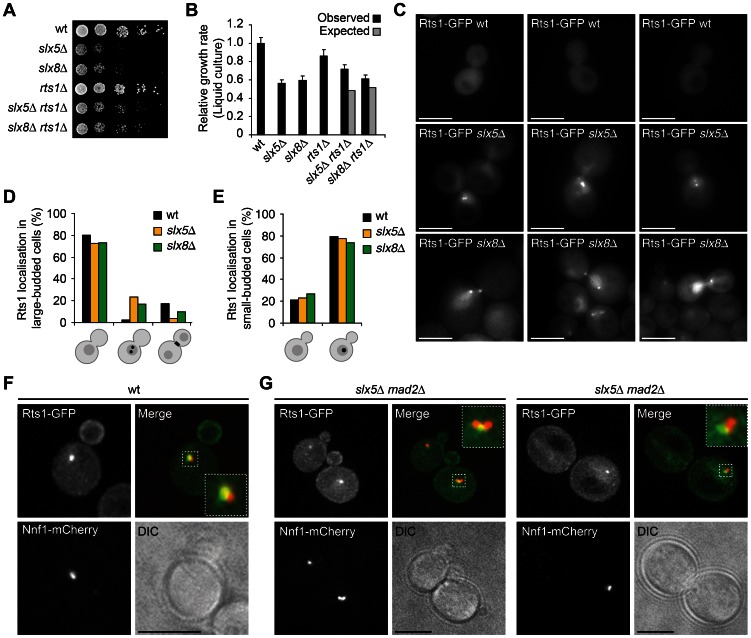
A role for Slx5/8 that is associated with a centromeric location is particularly interesting since this may better explain the genome stability defects of slx5Δ and slx8Δ mutants. Since many different regulatory pathways influence chromosome segregation, a candidate-based approach was adopted to determine factors that may be involved in the same pathway as Slx5/8, focusing in particular on potential targets. In yeast, kinetochore proteins Ndc10, Cep3 and Bir1 are sumoylated and Ndc10 interacts with Slx5 by yeast two-hybrid [32]. Changes in protein levels or subcellular location in slx5/8 mutants were not found for any of these candidates (Figure S1). Mutations in most centromere components display either no or only weak negative synthetic genetic interactions with slx5Δ and slx8Δ [33], [34]. Positive genetic interactions are more likely indicative of gene products functioning in the same protein complex or pathway. It was therefore of interest that RTS1, a regulatory subunit of the PP2A phosphatase [35], was found to have positive genetic interactions with both SLX5 and SLX8 in a high-throughput genetic interaction map [33]. Rts1Δ also has a similar mRNA expression phenotype as slx5Δ and slx8Δ (Figure 1B). To confirm the genetic interaction, single and double deletion strains were generated and growth was examined on solid medium (Figure 4A) and quantified in liquid cultures (Figure 4B). The double mutants slx5Δ rts1Δ and slx8Δ rts1Δ indeed grow better than is expected from the growth of single deletion mutants, confirming the positive synthetic genetic interactions of the pairs RTS1-SLX5 and RTS1-SLX8.
Figure 4. Slx5Δ and slx8Δ mutants accumulate Rts1 at kinetochores during metaphase.
(A) Growth rate of cells spotted in five-fold serial dilutions on YPD plates. Images are after two days of growth at 25°C. (B) Growth rate of yeast in liquid YPD media. Relative growth rate (mutant/wt) was quantified during mid-log phase (± s.d., n = 6). Expected relative growth rates (RGR) of the double deletion mutants are calculated by multiplying the observed RGRs of the single deletion mutants. (C) Live cell microscopy of large-budded wt, slx5Δ and slx8Δ cells expressing Rts1-GFP. Nuclear Rts1 foci are detected in slx5Δ and slx8Δ, which are absent in large-budded wt cells. Scale bars, 5 µm. (D–E) Quantification of subcellular Rts1 location in wt, slx5Δ and slx8Δ cells. An asynchronous cell population (n >200 cells) was morphologically divided in small-budded (G1-S phase) and large-budded (G2/M phase) cells. Diffuse Rts1 location in nucleus and cytoplasm is schematically indicated in grey. Presence of one or two Rts1 foci is schematically indicated as black nuclear dots. Enrichment of Rts1 at the bud neck is indicated as a black bar. Rts1 foci were not detected in nonbudded cells. (F–G) Live cell fluorescence microscopy of wt (F) and slx5Δ mad2Δ cells (G), expressing Rts1-GFP and kinetochore protein Nnf1-mCherry. The small-budded wt has a normal centromeric Rts1 focus. Note that Rts1 is also enriched at the bud membrane. The left panel in (G) shows two small-budded slx5Δ mad2Δ cells with normal Rts1 foci that colocalise with kinetochores. The right panel in (G) shows a large-budded slx5Δ mad2Δ cell with an aberrant, mislocalised centromeric Rts1 focus during metaphase. Scale bars, 5 µm.
Rts1 is a very dynamic centromeric protein that localises to specific subcellular sites in mitotic cells in a cell cycle-dependent manner [36], [37]. Rts1 localises to kinetochores in small-budded cells, then disappears and relocates to the bud neck in large-budded cells during cytokinesis [37]. We investigated the localisation of Rts1-GFP in asynchronous wt, slx5Δ and slx8Δ cells using live cell fluorescence microscopy. Strikingly, a subpopulation of large-budded slx5Δ and slx8Δ cells are characterised by the presence of aberrant Rts1 foci (Figure 4C and D). These Rts1 foci are rarely observed in large-budded wt cells. The cells presenting these foci appear arrested in metaphase and typically have one or two Rts1 foci. The Rts1 foci in small-budded slx5Δ and slx8 cells are indistinguishable from wt (Figure 4E). The foci colocalise with the kinetochore protein Nnf1 (Figure 4F and G), demonstrating that Rts1 accumulates at centromeres during metaphase in slx5Δ and slx8Δ cells. Deletion of the Spindle Assembly Checkpoint (SAC) component MAD2 does not affect the location of Rts1 (Figure 4G), indicating that the centromeric location of Rts1 during metaphase is independent of the SAC.
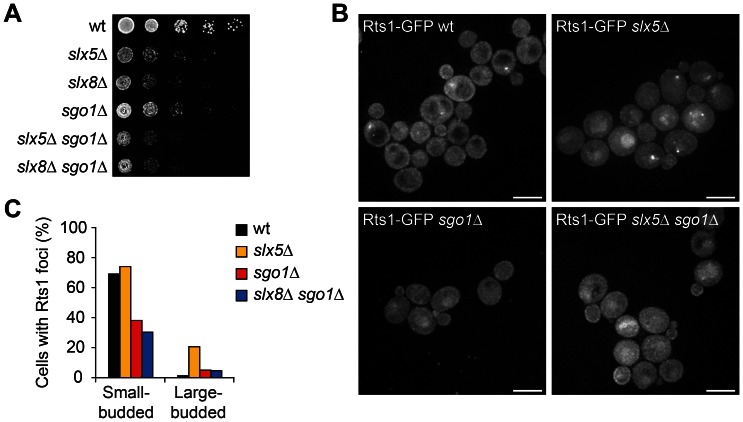
The centromeric accumulation of Rts1 in metaphase suggests the presence of a defect in slx5/8 mutants that prevents the removal of centromeric Rts1 after recruitment during S-phase. Since the recruitment of Rts1 in meiotic cells is dependent on the centromere cohesion regulator Shugoshin (Sgo1) [38], we also investigated whether this is the case in mitotic cells. Deletion of SGO1 results in a slow growth phenotype (Figure 5A). The Rts1 foci in sgo1Δ cells are less bright, indicating that Sgo1 promotes Rts1 recruitment to kinetochores in mitotic cells too (Figure 5B and C). Similarly, deletion of SGO1 in slx5Δ cells results in Rts1 foci in large-budded cells that are less bright and also reduced in number (Figure 5B and C), showing that the aberrant Rts1 foci in slx5Δ are also partially Shugoshin-dependent.
Figure 5. Rts1 foci are partially Shugoshin (Sgo1)-dependent.
(A) Growth rate assay of cells spotted in five-fold serial dilutions on YPD plates. Images are after two days growth at 25°C. (B) Live cell fluorescence microscopy of asynchronous wt, slx5Δ, sgo1Δ and slx5Δ sgo1Δ cells expressing Rts1-GFP. Scale bars, 5 µm. (C) Quantification of Rts1 foci in cells, shown in (B). Quantification is based on an asynchronous cell population (n = 89–203), that was morphologically divided in a small-budded and large-budded cell population.
Slx5Δ and slx8Δ have Aberrant Spindle Positioning, Morphology and Elongation Kinetics
In addition to the accumulation of Rts1 at the kinetochore during metaphase, a change in mitotic spindle morphology was observed in both slx5Δ and slx8Δ. The spindle morphology and dynamics were investigated in more detail using time-lapse video microscopy of asynchronous cell populations expressing GFP-Tub1 (Figure 6A). Cells were imaged at 2 minute intervals to capture the progression from metaphase into anaphase. Slx5Δ and slx8Δ arrest temporarily in mitosis as large-budded cells with short mitotic spindles and have an average mitotic delay of 80 minutes compared to wt (Figure S2A). During mitotic arrest, the cells are characterised by a spindle positioning defect in which the mitotic spindle fails to position itself stably at the bud neck (Figure 6A). Instead, the spindle oscillates heavily and frequently dislocates completely away from the bud neck, either shooting back into the mother cell or into the daughter bud. The mispositioned spindles are often accompanied by elongated astral microtubules (Figure 6A and B), which contribute to spindle positioning [39]. Around 30% of the slx5Δ and slx8Δ cells show spindle dislocation prior to entry into anaphase. This suggests that the mitotic delay in slx5Δ and slx8Δ is due to a failure in spindle positioning.
Figure 6. Mitotic spindle defects in slx5Δ and slx8Δ mutants.
(A) Time-lapse video microscopy of wt, slx5Δ and slx8Δ cells expressing GFP-Tub1. The upper panel shows a metaphase spindle in a wt cell at 2 minute intervals. Spindle elongation is initiated at t = 18′. The two panels below show examples of aberrant positioning of metaphase spindles in slx5Δ and slx8Δ cells during a temporary metaphase arrest. Arrows indicate spindle dislocation from the bud neck. Contours of cells are marked with a white line and are based on the DIC image. Scale bars, 5 µm. (B) Example of a spindle in slx8Δ, followed from metaphase to late anaphase. Colours are inverted to increase visibility of the astral microtubules at the outer tips of the spindle. The cell has a prolonged metaphase (t = 0′–52′) during which the spindle dislocates into the bud (t = 4′). Entry into anaphase is initiated at t = 52′, followed by spindle extension (t = 54′–80′). Formation of a fish hook spindle is apparent during late anaphase (t = 76′–80′). Scale bar, 5 µm. (C) Examples of fish hook spindles in slx5Δ and slx8Δ and a normal elongated spindle in wt during late anaphase. Scale bars, 5 µm. (D) Benomyl sensitivity assay. Growth rate of yeast cells is measured on YPD plates complemented with benomyl or DMSO (control). Images are after two days growth at 30°C. The benomyl-sensitive SAC mutant mad2Δ is included as control. (E) Quantification of spindle length, defined as the distance between two spindle pole bodies in wt, slx5Δ and slx8Δ expressing Spc42-GFP. Spindle length (n >100) was quantified from 20 minutes before anaphase onset to completion of anaphase. Grey lines depict the spindle length of individual cells. Black lines represent the average wt spindle length ± s.d., which is also shown as reference in the slx5Δ and slx8Δ plots. (F) Quantifications of spindle phenotypes in wt, slx5Δ and slx8Δ during metaphase and anaphase.
A second aberrant spindle phenotype was observed during anaphase. So-called ‘fish hook’ spindles form in 11% and 16% of the slx5Δ and slx8Δ anaphase cells respectively (Figure 6C). The formation of fish hook spindles can be a consequence of overstable microtubules and are observed in mutants of microtubule-associated proteins and kinetochore components [40]. Neither slx5Δ nor slx8Δ shows sensitivity or resistance to treatment with the microtubule-destabilising agent benomyl, suggesting that the stability of microtubules is actually normal (Figure 6D). Also the duration of anaphase in the mutants is not significantly different from wt (Figure S2B). Moreover, slx5Δ and slx8Δ do not interact genetically with SAC component MAD2 and deletion of MAD2 is not sufficient to overcome the mitotic arrest in slx5Δ and slx8Δ (Figure S2C and D). This indicates that microtubule-kinetochore interactions in slx5Δ and slx8Δ are normal and that Slx5/8 does not take part in the SAC signalling pathway.
The aberrant spindle morphology in slx5Δ and slx8Δ cells prompted us to investigate spindle dynamics. To analyse spindle elongation dynamics during chromosome segregation, strains expressing Spc42-GFP were imaged by time-lapse video microscopy. Spc42-GFP fluorescently labels the spindle pole bodies at the outer ends of the spindle. The spindle length was measured by determining the spindle pole body distance from metaphase until late anaphase (Figure 6E and F). The average wt spindle length in metaphase is 1.44 µm ±0.34. Upon entry into anaphase this quickly increases to a maximum length of 8.13 µm ±1.02 after ∼20 minutes. In contrast, the spindle length in slx5Δ and slx8Δ is very heterogeneous during metaphase and anaphase (Figure 6E). In metaphase, the average spindle length is 30% longer than wt, 1.94 µm ±0.81 and 1.79 µm ±0.55 for slx5Δ and slx8Δ respectively. During anaphase, the maximum spindle length measured in slx5Δ and slx8Δ is on average not significantly different from wt, but in mid-anaphase, the spindle length clearly deviates (Figure 6E and F). In S. cerevisiae, spindle elongation during anaphase B occurs in two phases, a quick elongation during early anaphase and a slower elongation during mid- and late anaphase [39]. Interestingly, slx5Δ and slx8Δ mutants show a continuous quick spindle elongation during mid-anaphase, whereas the wt reduces its spindle elongation speed (Figure 6E). The average spindle elongation speed was measured during mid-anaphase and is 0.28 µm/min ±0.13 and 0.28 µm/min ±0.10 for slx5Δ and slx8Δ respectively. This is 40% faster than wt, which has a spindle elongation rate of 0.20 µm/min ±0.11 (Figure 6F). The duration of anaphase in the mutants was not significantly different from wt (Figure S2B). The aberrant spindle morphology in slx5/8 mutants is therefore associated with changes in spindle kinetics during anaphase.
Slx5Δ and slx8Δ Phenotypes Persist in the Absence of the 2 µm Plasmid
Slx5Δ and slx8Δ mutants are known to have six-fold higher levels of extrachromosomal 2 µm plasmids compared to wt, resulting in clonal lethality [11]. We investigated the hypothesis that increased levels of native 2 µm plasmids may interfere with the spindle apparatus, resulting in defective chromosome segregation. Strains were cured of the 2 µm DNA (Cir0) and aneuploidy and growth rate phenotypes were examined (Figure S4A, B and C). Flow cytometry clearly indicates that slx5Δ Cir0 and slx8Δ Cir0 strains still suffer from aneuploidy (Figure S4B), which is accompanied with a reduction in growth rate compared to wt Cir0 strains (Figure S4C). This implies that the aneuploidy and growth phenotypes of slx5Δ Cir+ and slx8Δ Cir+ are not merely a result of overload of 2 µm DNA. The phenotypes are slightly reduced in severity compared to the Cir+ mutant strains, indicating that the presence of 2 µm DNA does aggravate the slx5Δ and slx8Δ phenotypes. As a final check, we also investigated the effect of 2 µm DNA on spindle dynamics in slx8Δ and wt strains (Figure S4D and E). As expected, slx8Δ Cir0 displays a similar defect in spindle dynamics as slx8Δ Cir+ strains. This again supports the conclusion that the presence of 2 µm DNA is not the main factor responsible for the mitotic defects of slx5Δ and slx8Δ.
Loss of hRNF4 Results in Chromosome Missegregation Due to Chromosome Bridges
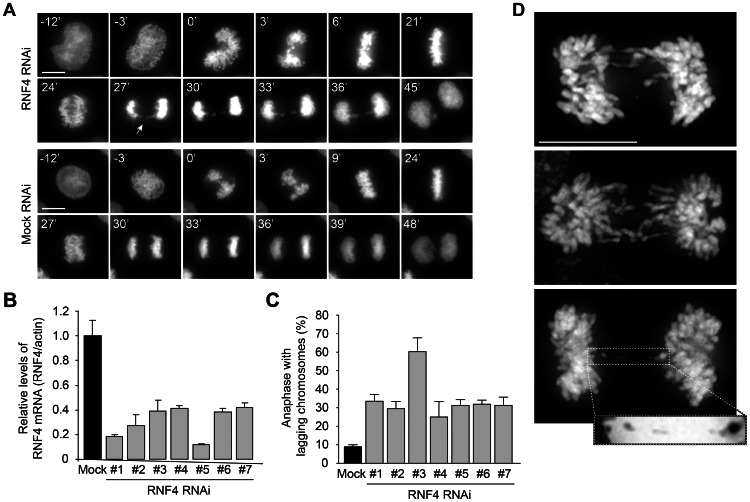
The changes in spindle morphology and elongation dynamics in slx5/8 mutants indicate a defect during anaphase. Since the microtubule stability itself is unaffected, it suggests that this phenotype is more likely a response to a defect during chromosome segregation. The formation of fish hook spindles, in combination with an increase in spindle elongation speed, suggests that slx5Δ and slx8Δ cells have an increased need for spindle pulling force in order to separate their sister chromatids during anaphase. The small size of S. cerevisiae does not readily allow high-resolution morphological examination of the sister chromatids during chromosome segregation to test this hypothesis. We therefore used human HeLa cells, also to investigate whether the STUbL orthologue hRNF4 has a related role in chromosome segregation. Biochemically, hRNF4 functions in a manner that is analogous to the Slx5/8 complex in S. cerevisiae [17], [18], but there is as yet little evidence for a role of hRNF4 in genome stability. RNF4 was depleted from H2B-EYFP expressing HeLa cells (Figure 7A and B). While there are no discernible defects in chromosome alignment, mitotic timing or mitotic checkpoint function, the frequency of lagging chromosomes in anaphase increased three- to six-fold with different siRNA oligos (Figure 7C). Though most segregation defects are minor, anaphase bridges persisting into telophase can be discerned (Figure 7A). More detailed examination in fixed cells shows lagging chromosomes in early anaphase, with persistent chromosome bridges in late anaphase (Figure 7D). The percentage of anaphases with lagging chromosomes in fixed cells resembles that observed in live cell imaging (Figure S3). The genomic instability during anaphase and telophase upon RNF4 knockdown agrees with the phenotypes of yeast slx5Δ and slx8Δ mutants. This demonstrates that Slx5/8 and hRNF4 have an evolutionarily conserved role in maintaining genome integrity during mitosis.
Figure 7. RNF4 depletion causes chromosome segregation errors.
(A) Time-lapse video microscopy of H2B-EYFP HeLa cells transfected with RNF4 siRNA or mock. Arrow indicates a chromosome bridge. Time (minutes) is given relative to the first time frame in prometaphase. Scale bars, 10 µm. (B) RNF4 knockdown efficiency determined by reverse transcription qPCR. cDNA was prepared from H2B-EYFP HeLa cells transfected with siRNAs targeting RNF4 or mock siRNA. mRNA levels of RNF4 and β-ACTIN were analysed by qPCR. Graph represents a single experiment, showing three technical replicates (± s.d.). (C) Quantification of chromosome segregation defects of H2B-EYFP HeLa cells, as shown in (A), transfected with different siRNAs as indicated. Graph represents the average of two independent experiments per siRNA (± s.d.) and at least 72 cells per siRNA. (D) Three examples of anaphase cells with lagging chromosomes in fixed HeLa cells transfected with RNF4 siRNA oligos. Inset (negative stain) shows a persistent chromatin bridge. Scale bars, 10 µm.
Discussion
SUMO-dependent ubiquitin ligases (STUbLs) such as Slx5/8 and RNF4 are a relatively new class of modifying enzymes, special in their ability to ubiquitinate proteins that have already been modified through sumoylation [7], [8]. Key questions regarding their cellular function and mechanism of action are unanswered. The results presented here address the cellular role of Slx5/8 in particular. Previous studies have postulated a role for Slx5/8 in regulation of transcription, through contributions to silencing [22] and turnover of transcription factors [8], [23], [24]. Although the DNA microarray analyses show that loss of either SLX5 or SLX8 results in upregulation of numerous genes, the majority of this transcriptional response is similar to the environmental stress response [25] and is in fact also common to inactivation of several different genome integrity pathways. Although this does not completely rule out a role for Slx5/8 in regulating gene expression, an important conclusion is that under the conditions of these experiments, the majority of gene expression changes observed upon deletion of SLX5 or SLX8 is likely an indirect effect of genome instability-induced stress.
An important finding presented here is the location of Slx5 at centromeres, since this may provide more focus for seeking relevant in vivo substrates. Interestingly, RNF4 has recently been shown to regulate the turnover of the human kinetochore protein CENP-I [41]. In yeast, the kinetochore protein Ndc10 has been reported to be sumoylated and to interact with Slx5 [32]. We have already undertaken several candidate-based approaches to find in vivo targets, focusing on candidates with roles at centromeres, with synthetic genetic interactions with SLX5 and SLX8, and that are known to be ubiquitinated and/or sumoylated. Changes in protein levels, modifications or subcellular location in slx5Δ and slx8Δ mutants were not found for any of the candidates tested, including the kinetochore proteins Ndc10, Cep3 or Bir1. The PP2A regulatory subunit RTS1 was found to genetically interact with SLX5/8 and aberrantly accumulates at kinetochores during metaphase in slx5Δ and slx8Δ. Rts1 is involved in different cellular pathways, including the Spindle Positioning Checkpoint (SPOC) [42]. The SPOC is important for inhibiting mitotic exit when the anaphase spindle is misaligned along the polarity axis of the yeast cell [43]. Both slx5Δ and slx8Δ display aberrant spindle elongation kinetics and fish hook spindles in anaphase, but the positioning of the anaphase spindle is normal. There is also no delay in mitotic exit. The spindle positioning defects are exclusively observed during metaphase, which makes it unlikely that SPOC activation by PP2ARts1 explains the anaphase defects of slx5Δ and slx8Δ. A more likely hypothesis is that the centromeric accumulation of PP2ARts1, which is partly dependent on Sgo1, involves the tension sensing pathway. Sgo1 senses whether cells have established correct kinetochore-microtubule interactions and regulates their progression into mitosis. Cells with unattached sister chromatids lack tension at the centromeres, leading to activation of the spindle assembly checkpoint and inhibition of entry into anaphase [44]. Rts1 is recruited to the centromere by Sgo1 to protect centromeric sister chromatid cohesion [38]. Possibly, slx5Δ and slx8Δ activate the tension sensing pathway to prevent cleavage of cohesion and thereby arrest in mitosis. Indeed, observation of the Rts1 foci in slx5Δ and slx8Δ metaphase cells reveal that there is a large variability in the distance between the centromeric Rts1 foci of the sister chromatids (Figure 4C), which suggests that there is loss of centromere tension. The underlying cause that potentially triggers the tension checkpoint in slx5Δ and slx8Δ is still unclear. Slx5/8 themselves are unlikely to take part as mitotic checkpoint components, since they do not show genetic interactions with MAD2 and are insensitive to benomyl treatment.
Several studies point to a role for Slx5/8 in the DNA damage response [5], [6], [11], [12], [16]. Loss of Slx5/8 function is thought to predominantly affect DNA replication, as this process greatly depends on accurate repair of DNA lesions that naturally occur during replication. It is therefore striking to see that the slx5Δ and slx8Δ mutant phenotypes revealed here, such as aberrant spindles and Rts1 retention, are exclusively observed during mitosis. High-resolution morphological examination of HeLa cells after RNF4 knockdown reveals the presence of lagging chromosomes and chromosome bridges, indicating that DNA damage arises during mitosis. It is unclear whether chromosome bridges also form in slx5Δ and slx8Δ. However, the altered kinetics of spindle elongation in slx5Δ and slx8Δ is suggestive of a defective separation of sister chromatids, reminiscent of chromosome bridges upon RNF4 knockdown. The source of DNA damage in slx5Δ and slx8Δ may therefore actually be the impaired segregation of chromosomes rather than defective repair of DNA replication-associated damage. We do not rule out that the origin of the defects may lie in S-phase. For instance, defective DNA decatenation during DNA replication may go unnoticed until the DNA is physically pulled apart during mitosis, ultimately leading to chromosome breaks [45]. Given the synthetic lethal genetic interactions of SLX5/8 with members of the RecQ family of DNA helicases [5], Slx5/8 may contribute to DNA decatenation, which may lead to chromosome segregation errors and DNA breaks. Moreover, the slx5Δ and slx8Δ mutants arrest in metaphase, which clearly indicates a cellular defect that precedes the separation of sister chromatids. Previously reported roles of Slx5/8 in DNA repair and replication may therefore be directly linked to the mitotic defects observed in this study.
It is also possible that defects in DNA repair or replication and the mitotic defects represent distinct functions of Slx5/8. Whereas ChIP analysis shows that Slx5 resides at centromeres, localisation studies using fluorescence microscopy show a predominantly diffuse nuclear localisation with occasional occurrence of subnuclear foci that do not strictly colocalise with kinetochores. The foci likely represent DNA replication and repair centres [16], which would agree with distinct functions for Slx5/8 in various cellular processes. Loss of Slx5/8 function also results in general accumulation of SUMO-conjugated protein species [7], [8], suggesting that Slx5/8 targets multiple substrates for proteasomal degradation, rather than controlling a single substrate or pathway. The lack of Slx5 and Slx8 colocalisation at centromeres is unexpected given that Slx5/8 is thought to function as a heterodimeric complex [7], [8]. It suggests that only Slx5 is stably associated with the centromere, where it may function independently of Slx8 or serve to recruit Slx8 in a transient manner. We favour the latter hypothesis, based on the complete overlap of mitotic phenotypes of slx5Δ and slx8Δ mutants. Regulatory control of proteins through sumoylation is well-established to be important for several (nuclear) processes, including transcription, DNA repair and chromosome organisation [3]. The results presented here will therefore also aid future studies aimed at identification of relevant in vivo substrates of Slx5/8 and RNF4, by focusing on the centromere-specific location and mitotic defects reported here.
Materials and Methods
Yeast Strains and Media
All strains are isogenic with S288c. Yeast strains and their genotypes are listed in Table S2. Deletion strains used for microarray expression profiling were from the Saccharomyces Genome Deletion library (Open Biosystems; Euroscarf) and are in the genetic background of the wt parental strain BY4742. SLX5 and SLX8 deletion mutants were generated by PCR-based gene disruption using pFA6a deletion cassettes [46]. Single and double deletion mutants used for assaying synthetic genetic interactions were created by PCR-based gene disruption of SLX5 or SLX8 in the heterozygote diploid deletion strains MAD2/mad2Δ, RTS1/rts1Δ and SGO1/sgo1Δ (BY4743; Open Biosystems), followed by tetrad dissection of sporulated diploids using standard genetic techniques. We noted that spores derived from SGO1/sgo1Δ had a strong reduction in viability and aberrant segregation of the mutant alleles. Accurate gene disruption and absence of wt alleles were confirmed by PCR. Slx5-GFP and Slx8-GFP strains were constructed by C-terminal genomic integration of a pFA6a-GFP-His3MX6 cassette [46]. All epitope- or fluorescent-tagged strains exhibited wt growth with the exception of Slx5-GFP. All attempts to fuse SLX5 to a variety of tags, either C- or N-terminally, resulted in strains with slow growth. All other epitope-tagged strains exhibited wt growth. Rts1-GFP was obtained from the GFP-tagged yeast collection [47]. Nnf1-mCherry was constructed by replacing the GFP tag from NNF1-GFP::His3MX6 [47] for mCherry::KanMX4. Ndc10-GFP, Cep3-GFP and Bir1-GFP strains were a kind gift of B. Montpetit [32]. Spc42-GFP (YYB3283) and GFP-Tub1 (YYB2327) are derived from previously described strains [48], [49]. The ndc10-1 mutation was described previously [31]. Coloured colony strains were generated by backcrossing slx5Δ and slx8Δ (BY4742) twice in the genetic background of strain YYB3085 [50]. Cir+ yeast strains were cured of the 2 µm plasmid by overexpression of FLP1 from the pBIS-Gal-KFLP-TRP1 and pBIS-Gal-KFLP-URA1 plasmids, and selection of Cir0 strains as described previously [51]. The loss of native 2 µm plasmids was monitored by PCR, using REP1 amplifying primers (Table S3). Experiments were performed in synthetic complete (SC) or yeast extract-peptone-dextrose (YPD) media (US Biologicals) containing 2% glucose.
Gene Expression Profiling
Microarray expression profiling was performed as described previously [52]. In brief, mutant and wt strains were grown at 30°C in SC media with 2% glucose and harvested in early mid-log phase. Dual-channel 70-mer oligonucleotide arrays were employed with a common reference wt RNA. All steps after RNA isolation were automated using robotic liquid handlers. After quality control, normalisation and dye-bias correction [53], statistical analysis was performed for each mutant versus a collection of 200 wt cultures. The reported FC is an average of four replicate mutant gene expression profiles versus the average of all wts. Fifty-eight genes that showed stochastic changes in wt profiles (wt variable genes) [54] were excluded from further downstream analyses. Clustering of the microarray expression profiles was performed using an unsupervised hierarchical cosine correlation, based on all significant genes (FC >1.7, p<0.05), excluding wt variable genes and deleted genes. The data is visualised using JavaTreeview [55] and GeneSpring (Agilent) software. Microarray data has been deposited in the public data repositories ArrayExpress and GEO under accession numbers E-TABM-1221 and GSE33929. The data are also available in flat-file from http://www.holstegelab.nl/publications/slx5_slx8.
Functional Enrichment Analyses
Enrichment analysis of GO-terms [56] and transcription factor binding sites [27] was performed on all significant genes in slx5Δ and slx8Δ (FC >1.7, p<0.05), excluding wt variable genes and deleted genes. The background gene population was set to 6,182 (the number of genes represented on the microarray) and p values are Bonferroni-corrected for multiple testing. Comparison with the general stress response is performed by a Pearson correlation analysis of the average slx5Δ and slx8Δ profile with a 30-minute heat shock condition [25].
ChIP-chip Analysis
ChIP-chip was performed essentially as described previously [57], with minor modifications. Cells were grown in 500 ml SC medium to mid-log phase at 30°C. For analysis of the temperature sensitive ndc10-1 mutant, cells were grown overnight at 25°C and subsequently for 6 hours at either 25°C or 37°C. Cells were crosslinked with 1% formaldehyde for 20 min at room temperature (RT). Glycine (300 mM) was added for 5 min at RT. Cells were harvested by centrifugation for 5 min at 4000 rpm at 4°C. The cell pellet was washed twice with cold TBS pH 7.5 (150 mM NaCl, 10 mM Tris), once with cold FA lysis buffer (50 mM HEPES-KOH pH 7.5, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS), and resuspended in 1.5 ml FA lysis buffer complemented with Complete Protease Inhibitor Cocktail (Roche). Cells were disrupted with the Disrupter Genie (Scientific Industries) using 0.5 ml zirconia beads (BioSpec Products Inc; ∅ 0.5 mm) at 4°C. The cell lysate was centrifuged 2 min at 4000 rpm at 4°C. The supernatant was centrifuged 15 min at 14,000 rpm at 4°C to collect the chromatin. The chromatin pellet was washed 30 min in 1.5 ml FA lysis buffer at 4°C, resuspended in 1.5 ml FA lysis buffer, and sonicated (Bioruptor, Diagenode: 10 cycles, 30 sec on/off, medium setting) to an average DNA fragment size of ∼400 bp. The lysate was centrifuged 20 min at 14,000 rpm at 4°C after which the supernatant (chromatin extract; CE) was collected for ChIP. ChIPs were performed by incubating 200 µl chromatin extract and 125 µg BSA to 20 µl Protein G-Agarose beads (Roche), coupled to rabbit polyclonal αGFP antibodies, for 2 h at RT. In parallel, mock ChIPs (no antibody) were performed on the same extracts. The beads were washed twice with 0.5 ml FA lysis buffer, twice in wash buffer 1 (FA lysis buffer, 500 mM NaCl), twice in wash buffer 2 (10 mM Tris pH, 0.25 mM LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA), and once in TE 10/1 (10 mM Tris pH 8, 1 mM EDTA). The beads were eluted twice in 50 µl TE 1% SDS (10 mM Tris pH 8, 1 mM EDTA, 1% SDS) for 10 min at 65°C. ChIP and input (20 µl CE) samples were incubated overnight in 100 µl TE 1% SDS and 10 µg ribonuclease A (Sigma) to reverse the formaldehyde cross-links. Samples were incubated with 400 µg proteinase K (Roche) for 2 hours at 37°C. For ChIP-chip, the proteinase K step was preceded by shrimp alkaline phosphatase (SAP) treatment by adding 1 ul of SAP (Roche) for 2 hours at 37°C. DNA was extracted with phenol-chloroform-isoamylalcohol (Sigma) and cleaned on PCR purification columns (Qiagen). Input and ChIP DNA was amplified using a robotically automated double-round T7 RNA polymerase-based amplification procedure [57]. Cy5-labelled ChIP samples were hybridised with cy3-labelled input DNA to a high-resolution 44K 4-pack yeast array (Agilent Technologies). The microarray data was quantified and normalised using a density lowess-normalisation algorithm [58]. ChIP/input and mock/input binding ratios were mapped to the ENSEMBL yeast genome EF 3 (February, 2011).
ChIP Quantitative Real-time PCR
Non-amplified input and ChIP DNA were analysed by qPCR using the iQ SYBR Green Supermix (Bio-Rad) in a CFX96 Real-Time PCR detection system (Bio-Rad). The PCR program was 95°C/10 min, 2 cycles of 95°C/15 sec, 50°C/30 sec, 72°C/30 sec and 45 cycles of 95°C/15 sec, 58°C/30 sec, 72°C/30 sec, followed by a melting curve to check for primer specificity. Primer sequences are listed in Table S3. Binding ratios at centromeres are based on ΔCt-values (Ct ChIP/Ct input) and are presented as fold occupancy over the control gene POL1.
Flow Cytometry
Cells were grown to early mid-log phase and either harvested directly as asynchronous cell population or synchronised in G1-phase by α-factor treatment (5 µg/ml; Zymo Research). Cells were released after 2.5 h in pre-warmed SC medium and harvested at 30 minute intervals. Cells (OD600 1.0) were washed twice in 1 ml FACS buffer (200 mM Tris, 20 mM EDTA), resuspended in 100 µl ribonuclease A (1 mg/ml in FACS buffer; Sigma) and incubated for 2 h at 37°C at 800 rpm. Cells were washed in phosphate-buffered saline (PBS) and stained in 100 µl propidium iodide (50 µg/ml in PBS; Molecular Probes) for 1 h at RT. Sample volume was increased to 1 ml with PBS and sonicated for 10 sec at 25% amplitude (Hielscher UP200S). DNA content was quantified by flow cytometry (FACSCalibur) and analysed using CellQuest 5.2.
Chromosome Loss Assay
Strains were grown in selective SC media lacking uracil for maintenance of the reporter chromosome and plated to single colonies on nonselective YPD plates. Colonies were allowed to grow for four days at 30°C. Red colour development was stimulated by incubating the plates one week at 4°C. The frequency of chromosome missegregation was quantified by colony half-sector analysis [59].
Yeast Live Cell Imaging
Cells were grown asynchronously in SC medium to early mid-log phase at 30°C. Cells were transferred to a pre-warmed 8-well chambered glass-bottom Lab-TEK slide (Nunc) and covered with pre-warmed solid SC medium (5% agar). Cells were imaged on a DeltaVision RT system (Applied Precision), equipped with a heated chamber at 30°C, using a 100×/1.42-numerical aperture (NA) PlanApoN objective (Olympus). Images were acquired using Softworx software for deconvolution and are maximum intensity projections of all Z planes stacked at 0.3 µm distance. Time-lapse video microscopy was performed by acquiring Z-stacks at 2 minute intervals for 2–3 hours. Images are processed in ImageJ and Adobe Photoshop CS2. Spindle length was quantified in ImageJ and defined as the distance between two spindle pole bodies (Spc42-GFP) from pixel to pixel with the highest intensity using Z-stack maximum intensity projections. Only cells for which a complete mitosis was captured were included in the analyses. Measurements were started 20 minutes before entry into anaphase and continued for maximally one hour. Metaphase cells are scored to have mispositioned spindles if the spindle showed one or more dislocation events from the bud neck into the bud or mother cell. Spindle oscillation at the bud neck without full dislocation was scored as normal. Spindle length in metaphase cells was measured 2–4 minutes before entry into anaphase. Cells displaying a bended spindle for at least two time frames were scored as having a fish hook spindles. Maximum spindle length in anaphase cells is measured in the last time frame before spindle shortening. Spindle elongation speed was measured in mid-anaphase from 4 to 12 minutes after start of anaphase.
Tissue Culture, Transfections and Treatments
HeLa cells and HeLa cells stably expressing H2B-EYFP were grown in DMEM supplemented with 9% FBS and pen/strep (50 µg/ml). Asynchronous cells were transfected twice with 40 nM siRNA (Table S4) using HiPerfect (Qiagen). Following the first transfection, cells were treated with 2 mM thymidine (Sigma) for 24 h. Subsequently, cells were transfected a second time and released into regular culture medium for 10 h. Cells were then treated with thymidine for 24 h and subsequently released into regular culture medium. For immunofluorescence microscopy, cells were fixed 12 h after the second release. RNF4 knockdown efficiency was measured by reverse transcription qPCR. Total rRNA was extracted using the RNeasy kit (Qiagen) including a DNase treatment step. Total RNA (250 ng) was used for cDNA synthesis (SuperScript II, Invitrogen). Expression of RNF4 and β-ACTIN was analysed by qPCR (Table S3) and normalised against a standard reference cDNA from untreated H2B-EYFP HeLa cells.
Immunofluorescence Microscopy and Live Cell Imaging of HeLa Cells
Cells, plated on 12-mm coverslips, were fixed in 3.7% Shandon Zinc Formal-Fixx (Thermo Scientific) for 10 min and permeabilised for 15 min with 0.5% Triton X-100 in PBS and washed with 0.1% Triton X-100 in PBS. Coverslips were washed and submerged in PBS containing DAPI, then washed again and mounted using ProLong antifade (Molecular Probes). Image acquisition was done using a DeltaVision RT system with a 60×/1.40NA UPlanSApo objective (Olympus) for acquiring images and SoftWorx software for deconvolution and projections. Images are maximum intensity projections of deconvolved stacks. For live cell imaging, cells were plated in eight-well chambered glass-bottomed slides (LabTek), transfected, and imaged in a heated chamber (37°C and 5% CO2) using a 60×/1.40NA UPlanSApo objective on an Olympus IX-81 microscope, controlled by Cell-M software (Olympus). Sixteen-bit yellow fluorescent images were acquired every 3 minutes using a Hamamatsu ORCA-ER camera. Images of H2B-EYFP were maximum intensity projections of all Z-planes and were processed using Cell-M software.
Supporting Information
Live cell fluorescence microscopy of wt, slx5Δ and slx8Δ. Slx5Δ and slx8Δ have longer anaphase spindles and normal localisation of kinetochore components Ndc10, Cep3 and Bir1 at the centromeres and along the mitotic spindle. Scale bars, 5 µm.
(TIF)
Slx5Δ and slx8Δ have a mitotic delay that cannot be relieved by deletion of MAD2. (A) Cumulative frequency graph of the duration of G1- to M-phase. Calculation is based on the time from spindle duplication in G1-phase to spindle pole body separation in anaphase, as measured by time-lapse video microscopy of cells (n = 100) expressing Spc42-GFP. (B) Cumulative frequency graph of the duration of anaphase. Calculation is based on the time from start of spindle elongation to spindle depolymerisation, as measured by time-lapse video microscopy of cells (n = 100) expressing Tub1-GFP. (C) Growth rate assay of cells spotted in five-fold serial dilutions on YPD plates. Images are after two days growth at 30°C. (D) Cell cycle progression of synchronized cells. DNA content was measured by flow cytometry at 30 minute intervals over a period of four hours after release from α-factor arrest in G1-phase. Arrows indicate cell populations with 1N (G1-phase) and 2N (G2/M-phase) DNA content.
(TIF)
Quantification of chromosome segregation defects in fixed HeLa cells. Graph represents the average of two independent experiments (± s.d.) and at least 110 cells per siRNA. Anaphases with chromosome segregation defects other than lagging chromosomes were infrequent in both mock and RNF4 knockdown situation and not considered for these analyses.
(TIF)
Comparison of slx5/8 phenotypes in the presence (Cir+) or absence (Cir0) of the 2 µm plasmid. (A) PCR verification of the loss of the 2 µm plasmid in three independent strains of wt, slx5Δ and slx8Δ. (B) Flow cytometric profiles of asynchronous populations of Cir+ and Cir0 wt, slx5Δ and slx8Δ strains. Cell population with a >2N DNA content, indicated in red, is quantified (± s.d., n = 3). (C) Growth rate of yeast cultures in liquid YPD media. Relative growth rate (mutant/wt) was quantified during mid-log phase (± s.d., n = 3). (D) Quantification of spindle length in wt Cir0 and slx8Δ Cir0 strains, expressing Spc42-GFP. Spindle length is quantified as described in Figure 6E. (E) Comparison of spindle phenotypes in Cir+ and Cir0 wt and slx8Δ strains during metaphase and anaphase.
(TIF)
GO and TFBS enrichment analyses. Gene Ontology (GO) and transcription factor binding site (TFBS) enrichment analyses of differentially expressed genes (mutant vs. wt, FC >1.7, p<0.05). The dataset is provided as separate Excel file.
(XLS)
Yeast strains.
(DOCX)
Primer sequences.
(DOCX)
SiRNA sequences.
(DOCX)
Acknowledgments
We thank M. Gartenberg for providing plasmids; L. Kleij and M. Groot Koerkamp for technical assistance; J. Benschop and T. Lenstra for contributing microarray data; P. Kemmeren, S. van Hooff and K. Sameith for bioinformatics.
Funding Statement
This work was funded by the Netherlands Bioinformatics Centre (NBIC) and the Netherlands Organization of Scientific Research (NWO), grants 016.108.607, 817.02.015, 050.71.057, 911.06.009. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hoeijmakers JH (2001) Genome maintenance mechanisms for preventing cancer. Nature 411: 366–374 doi:10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- 2. Kerscher O, Felberbaum R, Hochstrasser M (2006) Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu Rev Cell Dev Biol 22: 159–180 doi:10.1146/annurev.cellbio.22.010605.093503 [DOI] [PubMed] [Google Scholar]
- 3. Johnson ES (2004) Protein modification by SUMO. Annu Rev Biochem 73: 355–382 doi:10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- 4. Geiss-Friedlander R, Melchior F (2007) Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol 8: 947–956 doi:10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- 5. Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ (2001) Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics 157: 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang C, Roberts TM, Yang J, Desai R, Brown GW (2006) Suppression of genomic instability by SLX5 and SLX8 in Saccharomyces cerevisiae. DNA Repair (Amst) 5: 336–346 doi:10.1016/j.dnarep.2005.10.010 [DOI] [PubMed] [Google Scholar]
- 7. Uzunova K, Göttsche K, Miteva M, Weisshaar SR, Glanemann C, et al. (2007) Ubiquitin-dependent proteolytic control of SUMO conjugates. J Biol Chem 282: 34167–34175 doi:10.1074/jbc.M706505200 [DOI] [PubMed] [Google Scholar]
- 8. Xie Y, Kerscher O, Kroetz MB, McConchie HF, Sung P, et al. (2007) The yeast Hex3.Slx8 heterodimer is a ubiquitin ligase stimulated by substrate sumoylation. J Biol Chem 282: 34176–34184 doi:10.1074/jbc.M706025200 [DOI] [PubMed] [Google Scholar]
- 9. Ii T, Fung J, Mullen JR, Brill SJ (2007) The yeast Slx5-Slx8 DNA integrity complex displays ubiquitin ligase activity. Cell Cycle 6: 2800–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perry JJP, Tainer JA, Boddy MN (2008) A SIM-ultaneous role for SUMO and ubiquitin. Trends Biochem Sci 33: 201–208 doi:10.1016/j.tibs.2008.02.001 [DOI] [PubMed] [Google Scholar]
- 11. Burgess RC, Rahman S, Lisby M, Rothstein R, Zhao X (2007) The Slx5-Slx8 complex affects sumoylation of DNA repair proteins and negatively regulates recombination. Mol Cell Biol 27: 6153–6162 doi:10.1128/MCB.00787-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nagai S, Dubrana K, Tsai-Pflugfelder M, Davidson MB, Roberts TM, et al. (2008) Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science 322: 597–602 doi:10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kosoy A, Calonge TM, Outwin EA, O’Connell MJ (2007) Fission yeast Rnf4 homologs are required for DNA repair. J Biol Chem 282: 20388–20394 doi:10.1074/jbc.M702652200 [DOI] [PubMed] [Google Scholar]
- 14. Sun H, Leverson JD, Hunter T (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J 26: 4102–4112 doi:10.1038/sj.emboj.7601839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJP, et al. (2007) SUMO-targeted ubiquitin ligases in genome stability. EMBO J 26: 4089–4101 doi:10.1038/sj.emboj.7601838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cook CE, Hochstrasser M, Kerscher O (2009) The SUMO-targeted ubiquitin ligase subunit Slx5 resides in nuclear foci and at sites of DNA breaks. Cell Cycle 8: 1080–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tatham MH, Geoffroy M-C, Shen L, Plechanovova A, Hattersley N, et al. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat Cell Biol 10: 538–546 doi:10.1038/ncb1716 [DOI] [PubMed] [Google Scholar]
- 18. Lallemand-Breitenbach V, Jeanne M, Benhenda S, Nasr R, Lei M, et al. (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat Cell Biol 10: 547–555 doi:10.1038/ncb1717 [DOI] [PubMed] [Google Scholar]
- 19. Guo B, Sharrocks AD (2009) Extracellular signal-regulated kinase mitogen-activated protein kinase signaling initiates a dynamic interplay between sumoylation and ubiquitination to regulate the activity of the transcriptional activator PEA3. Mol Cell Biol 29: 3204–3218 doi:10.1128/MCB.01128-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Martin N, Schwamborn K, Schreiber V, Werner A, Guillier C, et al. (2009) PARP-1 transcriptional activity is regulated by sumoylation upon heat shock. EMBO J 28: 3534–3548 doi:10.1038/emboj.2009.279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Van Hagen M, Overmeer RM, Abolvardi SS, Vertegaal ACO (2010) RNF4 and VHL regulate the proteasomal degradation of SUMO-conjugated Hypoxia-Inducible Factor-2alpha. Nucleic Acids Res 38: 1922–1931 doi:10.1093/nar/gkp1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Darst RP, Garcia SN, Koch MR, Pillus L (2008) Slx5 promotes transcriptional silencing and is required for robust growth in the absence of Sir2. Mol Cell Biol 28: 1361–1372 doi:10.1128/MCB.01291-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Z, Prelich G (2009) Quality control of a transcriptional regulator by SUMO-targeted degradation. Mol Cell Biol 29: 1694–1706 doi:10.1128/MCB.01470-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nixon CE, Wilcox AJ, Laney JD (2010) Degradation of the Saccharomyces cerevisiae mating-type regulator alpha1: genetic dissection of cis-determinants and trans-acting pathways. Genetics 185: 497–511 doi:10.1534/genetics.110.115907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, et al. (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martínez-Pastor MT, Marchler G, Schüller C, Marchler-Bauer A, Ruis H, et al. (1996) The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE). EMBO J 15: 2227–2235. [PMC free article] [PubMed] [Google Scholar]
- 27. MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, et al. (2006) An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics 7: 113 doi:10.1186/1471-2105-7-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hughes TR, Roberts CJ, Dai H, Jones AR, Meyer MR, et al. (2000) Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 25: 333–337 doi:10.1038/77116 [DOI] [PubMed] [Google Scholar]
- 29. Hieter P, Mann C, Snyder M, Davis RW (1985) Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell 40: 381–392. [DOI] [PubMed] [Google Scholar]
- 30. McAinsh AD, Tytell JD, Sorger PK (2003) Structure, function, and regulation of budding yeast kinetochores. Annu Rev Cell Dev Biol 19: 519–539 doi:10.1146/annurev.cellbio.19.111301.155607 [DOI] [PubMed] [Google Scholar]
- 31. Goh PY, Kilmartin JV (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J Cell Biol 121: 503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montpetit B, Hazbun TR, Fields S, Hieter P (2006) Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol 174: 653–663 doi:10.1083/jcb.200605019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, et al. (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 doi:10.1038/nature05649 [DOI] [PubMed] [Google Scholar]
- 34. Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, et al. (2010) The genetic landscape of a cell. Science 327: 425–431 doi:10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhao Y, Boguslawski G, Zitomer RS, DePaoli-Roach AA (1997) Saccharomyces cerevisiae homologs of mammalian B and B’ subunits of protein phosphatase 2A direct the enzyme to distinct cellular functions. J Biol Chem 272: 8256–8262. [DOI] [PubMed] [Google Scholar]
- 36. Gentry MS, Hallberg RL (2002) Localization of Saccharomyces cerevisiae protein phosphatase 2A subunits throughout mitotic cell cycle. Mol Biol Cell 13: 3477–3492 doi:10.1091/mbc.02-05-0065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dobbelaere J, Gentry MS, Hallberg RL, Barral Y (2003) Phosphorylation-dependent regulation of septin dynamics during the cell cycle. Dev Cell 4: 345–357. [DOI] [PubMed] [Google Scholar]
- 38. Riedel CG, Katis VL, Katou Y, Mori S, Itoh T, et al. (2006) Protein phosphatase 2A protects centromeric sister chromatid cohesion during meiosis I. Nature. 441: 53–61 doi:10.1038/nature04664 [DOI] [PubMed] [Google Scholar]
- 39. Straight AF, Marshall WF, Sedat JW, Murray AW (1997) Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277: 574–578. [DOI] [PubMed] [Google Scholar]
- 40. Vizeacoumar FJ, van Dyk N, Vizeacoumar FS, Cheung V, Li J, et al. (2010) Integrating high-throughput genetic interaction mapping and high-content screening to explore yeast spindle morphogenesis. J Cell Biol 188: 69–81 doi:10.1083/jcb.200909013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mukhopadhyay D, Arnaoutov A, Dasso M (2010) The SUMO protease SENP6 is essential for inner kinetochore assembly. J Cell Biol 188: 681–692 doi:10.1083/jcb.200909008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chan LY, Amon A (2009) The protein phosphatase 2A functions in the spindle position checkpoint by regulating the checkpoint kinase Kin4. Genes Dev 23: 1639–1649 doi:10.1101/gad.1804609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Caydasi AK, Ibrahim B, Pereira G (2010) Monitoring spindle orientation: Spindle position checkpoint in charge. Cell Div 5: 28 doi:10.1186/1747-1028-5-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Indjeian VB, Stern BM, Murray AW (2005) The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science 307: 130–133 doi:10.1126/science.1101366 [DOI] [PubMed] [Google Scholar]
- 45. Wang JC (2002) Cellular roles of DNA topoisomerases: a molecular perspective. Nat Rev Mol Cell Biol 3: 430–440 doi:10.1038/nrm831 [DOI] [PubMed] [Google Scholar]
- 46. Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, et al. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 doi:;10.1002/(SICI)1097-0061(199807)14:10<953: AID-YEA293>3.0.CO;2-U [DOI] [PubMed] [Google Scholar]
- 47. Huh W-K, Falvo JV, Gerke LC, Carroll AS, Howson RW, et al. (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 doi:10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- 48. Grava S, Schaerer F, Faty M, Philippsen P, Barral Y (2006) Asymmetric recruitment of dynein to spindle poles and microtubules promotes proper spindle orientation in yeast. Dev Cell 10: 425–439 doi:10.1016/j.devcel.2006.02.018 [DOI] [PubMed] [Google Scholar]
- 49. Neurohr G, Naegeli A, Titos I, Theler D, Greber B, et al. (2011) A midzone-based ruler adjusts chromosome compaction to anaphase spindle length. Science 332: 465–468 doi:10.1126/science.1201578 [DOI] [PubMed] [Google Scholar]
- 50. Spencer F, Gerring SL, Connelly C, Hieter P (1990) Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics 124: 237–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tsalik EL, Gartenberg MR (1998) Curing Saccharomyces cerevisiae of the 2 micron plasmid by targeted DNA damage. Yeast 14: 847–852 doi:;–10.1002/(SICI)1097-0061(19980630)14:9<847: AID-YEA285>3.0.CO;2–9 [DOI] [PubMed] [Google Scholar]
- 52. Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NACH, et al. (2011) The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42: 536–549 doi:10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, et al. (2009) Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol Syst Biol 5: 266 doi:10.1038/msb.2009.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Van Wageningen S, Kemmeren P, Lijnzaad P, Margaritis T, Benschop JJ, et al. (2010) Functional overlap and regulatory links shape genetic interactions between signaling pathways. Cell 143: 991–1004 doi:10.1016/j.cell.2010.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Saldanha AJ (2004) Java Treeview-extensible visualization of microarray data. Bioinformatics 20: 3246–3248 doi:10.1093/bioinformatics/bth349 [DOI] [PubMed] [Google Scholar]
- 56. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 doi:10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Van Bakel H, van Werven FJ, Radonjic M, Brok MO, van Leenen D, et al. (2008) Improved genome-wide localization by ChIP-chip using double-round T7 RNA polymerase-based amplification. Nucleic Acids Res 36: e21 doi:10.1093/nar/gkm1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Van Werven FJ, van Bakel H, van Teeffelen HAAM, Altelaar AFM, Koerkamp MG, et al. (2008) Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes Dev 22: 2359–2369 doi:10.1101/gad.1682308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hyland KM, Kingsbury J, Koshland D, Hieter P (1999) Ctf19p: A novel kinetochore protein in Saccharomyces cerevisiae and a potential link between the kinetochore and mitotic spindle. J Cell Biol 145: 15–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Live cell fluorescence microscopy of wt, slx5Δ and slx8Δ. Slx5Δ and slx8Δ have longer anaphase spindles and normal localisation of kinetochore components Ndc10, Cep3 and Bir1 at the centromeres and along the mitotic spindle. Scale bars, 5 µm.
(TIF)
Slx5Δ and slx8Δ have a mitotic delay that cannot be relieved by deletion of MAD2. (A) Cumulative frequency graph of the duration of G1- to M-phase. Calculation is based on the time from spindle duplication in G1-phase to spindle pole body separation in anaphase, as measured by time-lapse video microscopy of cells (n = 100) expressing Spc42-GFP. (B) Cumulative frequency graph of the duration of anaphase. Calculation is based on the time from start of spindle elongation to spindle depolymerisation, as measured by time-lapse video microscopy of cells (n = 100) expressing Tub1-GFP. (C) Growth rate assay of cells spotted in five-fold serial dilutions on YPD plates. Images are after two days growth at 30°C. (D) Cell cycle progression of synchronized cells. DNA content was measured by flow cytometry at 30 minute intervals over a period of four hours after release from α-factor arrest in G1-phase. Arrows indicate cell populations with 1N (G1-phase) and 2N (G2/M-phase) DNA content.
(TIF)
Quantification of chromosome segregation defects in fixed HeLa cells. Graph represents the average of two independent experiments (± s.d.) and at least 110 cells per siRNA. Anaphases with chromosome segregation defects other than lagging chromosomes were infrequent in both mock and RNF4 knockdown situation and not considered for these analyses.
(TIF)
Comparison of slx5/8 phenotypes in the presence (Cir+) or absence (Cir0) of the 2 µm plasmid. (A) PCR verification of the loss of the 2 µm plasmid in three independent strains of wt, slx5Δ and slx8Δ. (B) Flow cytometric profiles of asynchronous populations of Cir+ and Cir0 wt, slx5Δ and slx8Δ strains. Cell population with a >2N DNA content, indicated in red, is quantified (± s.d., n = 3). (C) Growth rate of yeast cultures in liquid YPD media. Relative growth rate (mutant/wt) was quantified during mid-log phase (± s.d., n = 3). (D) Quantification of spindle length in wt Cir0 and slx8Δ Cir0 strains, expressing Spc42-GFP. Spindle length is quantified as described in Figure 6E. (E) Comparison of spindle phenotypes in Cir+ and Cir0 wt and slx8Δ strains during metaphase and anaphase.
(TIF)
GO and TFBS enrichment analyses. Gene Ontology (GO) and transcription factor binding site (TFBS) enrichment analyses of differentially expressed genes (mutant vs. wt, FC >1.7, p<0.05). The dataset is provided as separate Excel file.
(XLS)
Yeast strains.
(DOCX)
Primer sequences.
(DOCX)
SiRNA sequences.
(DOCX)