Abstract
To assess the consequences of inactivation of heat shock factor 1 (HSF1) during aging, we analyzed the effect of HSF1 K80Q, a mutant unable to bind DNA, and of dnHSF1, a mutant lacking the activation domain, on the transcriptome of cells 6 and 24 h after heat shock. The primary response to heat shock (6 h recovery), of which 30 % was HSF1-dependent, had decayed 24 h after heat shock in control cells but was extended in HSF1 K80Q and dnHSF1 cells. Comparison with literature data showed that even the HSF1 dependent primary stress response is largely cell specific. HSF1 K80Q, but not HSF1 siRNA-treated, cells showed a delayed stress response: an increase in transcript levels of HSF1 target genes 24 h after heat stress. Knockdown of NRF2, but not of ATF4, c-Fos or FosB, inhibited this delayed stress response. EEF1D_L siRNA inhibited both the delayed and the extended primary stress responses, but had off target effects. In control cells an antioxidant response (ARE binding, HMOX1 mRNA levels) was detected 6 h after heat shock; in HSF1 K80Q cells this response was delayed to 24 h and the ARE complex had a different mobility. Inactivation of HSF1 thus affects the timing and nature of the antioxidant response and NRF2 can activate at least some HSF1 target genes in the absence of HSF1 activity.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-012-0400-0) contains supplementary material, which is available to authorized users.
Keywords: Heat shock factor 1, NRF2, Stress response, HSPA1A, HSPA6
Introduction
Cells respond to cytoplasmic proteotoxic stress by producing additional chaperones, the heat shock proteins (HSP). This heat shock response plays an important role in maintaining proteostasis (reviewed by Fujimoto and Nakai 2010; Hartl et al. 2011; Morimoto 2008). The heat shock response is mainly regulated at the level of transcription by heat shock factor 1 (HSF1). Under normal circumstances HSF1 is monomeric and complexed with chaperones. Upon stress, when unfolded proteins accumulate and chaperones become scarce, HSF1 trimerizes, binds to the heat shock element (HSE) and activates transcription (reviewed in Fujimoto and Nakai 2010; Anckar and Sistonen 2011). HSF1 is required for longevity (Hsu et al. 2003; Steinkraus et al. 2008) and its inactivation, for example, during the DNA damage response (Kim et al. 2012) or the amino acid starvation response (Hensen et al. 2012), is linked to senescence. During aging, the activity of HSF1 declines (Jurivich et al. 2005; Ambra et al. 2004; Lee et al. 2009), although the protein is still present. This aging-related failure of HSF1 interferes with an organism’s ability to combat proteotoxic stress, which results in increased susceptibility to protein folding diseases (Balch et al. 2008; Broadley and Hartl 2009; Satyal et al. 2000; Bailey et al. 2002; Fujimoto et al. 2005; Calderwood et al. 2009). Furthermore, accumulating evidence indicates that HSF1 also regulates gene expression under non-stress conditions. For example, the circadian clock gene Per2 is an HSF1 target (Kornmann et al. 2007; Tamaru et al. 2011). In addition, HSF1 regulates a transcriptional circuit distinct from the proteotoxic stress induced pathway, which has been recruited by malignant cells (Mendillo et al. 2012). Thus, a decline in HSF1 activity may cause phenotypic defects in the absence of exogenous stress (Slavotinek and Biesecker 2003). Previously we found that the expression of an HSF1 mutant retaining the DNA binding domain but lacking the activation domain (dnHSF1) reduced the expression level of ten genes in non-stressed HEK293 cells, amongst which the genes for the chaperones Hsp90, HSPA6, DNAJB1 (Hsp40) and HSPB1; expression of dnHSF1 did not result in increased transcript levels (Heldens et al. 2010). HeLa cells treated with siRNA directed against HSF1 showed changed expression levels of 378 genes in the absence of stress (Page et al. 2006), where 80 % of the affected genes showed increased transcript levels. A comparison of the transcriptome of HSF1−/− mouse embryonic fibroblasts (MEFs) with that of wild type MEF cells resulted in 49 genes (19 related to immune response) that were expressed at reduced levels in MEF HSF1−/− cells (Inouye et al. 2004). The aging cell differs from the HSF1−/− cells in that the cell still contains HSF1, although not active, and differs from the dnHSF1 cells in that HSF1 is no longer bound to its target promoters. In this study, we have investigated the effect of heat stress on the transcriptome changes in two stable cell lines, one with a tet-inducible dnHSF1 mutant and one with tet-inducible expression of an HSF1 mutant in which lysine 80 in the DNA binding region is replaced by glutamine (HSF1 K80Q), thus impairing DNA binding (Westerheide et al. 2009). Unexpectedly, we detected a delayed stress response, i.e., an increase in transcript levels of HSF1 dependent genes in HSF1 K80Q cells 24 h after heat stress, suggesting that there are alternative routes to activation of transcription of these genes when the HSF1-directed transcription fails. We noted that the antioxidant response is delayed in heat-stressed HSF1 K80Q cells and found NRF2, a transcription factor directing the antioxidant response, to be responsible for the increase in HSPA1A and HSPA6 mRNA levels in HSF1 K80Q cells 24 h after heat stress.
Materials and methods
Cell culture
Flp-In T-REx-293 cells (Invitrogen) were manipulated according to the manufacturer’s instructions using the T-REx system (Invitrogen) to generate the stable cell lines HEK-dnHSF1, HEK-HSF1K80Q, HEK-wtHSF1 and HEK-pcDNA5 that carry a single copy of the tetracycline-inducible plasmids pcDNA5-dnHSF1, pcDNA5-HSF1K80Q, pcDNA5-wtHSF1 and pcDNA5-FRT/TO, respectively. The cells were cultured at 37 °C/5 % CO2 in high glucose DMEM medium supplemented with 10 % fetal calf serum,100 U/ml penicillin and 100 μg/ml streptomycin. Blasticidin (1.65 μg/ml; Invitrogen) and 100 μg/ml hygromycin were also added to the culture medium during maintenance of the cell lines, but were omitted during experiments.
Plasmid construction, transfections and reporter gene assays
The expression vectors pcDNA5-dnHSF1, pcDNA5-wtHSF1 and pcDNA5-HSF1 K80Q have been described earlier (Heldens et al. 2010; Hensen et al. 2012). Transient transfections were performed using FuGENE-6 (Roche) according to the manufacturer’s instructions. Cells were seeded on 24-well plates and on the next day transfected with 0.2 μg SV40-luc per well and treated with doxycycline to express HSF1 K80Q. 24 h after transfection cells were pre-heat shocked for 30 min at 45 °C. 14 h later, cells were harvested or heat shocked again for 30 min at 45 °C in the presence of 20 μg/ml CHX to inhibit translation and harvested immediately or after 1 h of recovery. Cells were lysed in 200 μl reporter lysis mix (25 mM Bicine, 0.05 % Tween 20, 0.05 % Tween 80) for 10 min. For the luciferase assay, 20 μl cell lysate was mixed with 50 μl luciferin solution (Promega) and luminescence was measured with the Lumat LB 9507 tube luminometer (Berthold). All reporter gene assays were performed in triplicate.
Electrophoretic mobility shift assay
HEK-HSF1K80Q or HEK-wtHSF1 cells were cultured for 48 h in the presence or absence of doxycycline and subsequently heat shocked for 30 min at 45 °C. Cells were harvested at the indicated times after heat shock and nuclear extracts were prepared using NE-per nuclear and cytoplasmic reagents (Pierce). Extracts were aliquoted and stored at −80 °C. Oligonucleotide probes were end-labeled with 32P. The sequences of the oligonucleotides used in EMSA are listed in Table S1. The EMSA protocol was adapted from (Klok et al. 1998; Thiaville et al. 2008). A mixture containing 5 μg nuclear extract and 3 μg poly dIdC in binding buffer (20 mM HEPES pH 7.9, 100 mM KCl, 1 mM EDTA, 1 mM DTT, 4 % (v/v) Ficoll, 1× PhosSTOP; Roche) was incubated for 20 min on ice. Then, 0.01 pmol radiolabeled oligonucleotide was added, and the samples were incubated for 20 min at room temperature. DNA–protein complexes were separated on a pre-run 4 % polyacrylamide gel in 0.25× TBE with recirculation of the buffer. The gel was dried and signals were visualized using a PhosphorImager.
Chromatin immunoprecipitation
HEK-HSF1 K80Q cells were cultured in the absence or presence of doxycycline and heat stressed for 30 min at 45 °C. After 2 or 18 h of recovery cells were subjected to chromatin immunoprecipitation (ChIP), performed as described in (Denissov et al. 2007) except that cells were cross-linked for 15 min with 1 % formaldehyde. After quenching with 125 mM glycine, cells were washed twice with ice cold PBS and resuspended in ice cold lysis buffer (50 mM HEPES–KOH pH 7.6, 140 mM NaCl, 1 mM EDTA pH 8.0, 1 % (v/v) Triton X-100, 0.1 % NaDOC and 1× protease inhibitor complete). Sonicated chromatin was centrifuged for 5 min at 4 °C and then incubated overnight in incubation buffer (final concentration; 12 mM HEPES–KOH pH 7.6, 90 mM NaCl, 0.6 mM EDTA, pH 8.0, 0.09 % SDS, 0.6 % Triton X-100, 0.1 % BSA) together with purified anti-HSF1 antibody (SPA-901; Stressgen) and protein A/G beads (Santa Cruz Biotechnology). Negative control without adding antibody was included. Beads were washed six times with different buffers at 4 °C: twice with 0.1 % SDS, 0.1 % NaDOC, 1 % Triton X-100, 150 mM NaCl, HEG (1 mM EDTA, 0.5 mM EGTA and 20 mM HEPES–KOH pH 7.6), once with the same buffer but with 500 mM NaCl, once with 0.25 M LiCl, 0.5 % NaDOC, 0.5 % NP-40, HEG and twice with HEG. Precipitated chromatin was eluted with 400 μl of elution buffer (1 % SDS, 0.1 M NaHCO3), incubated at 65 °C for 4 h in the presence of 200 mM NaCl, phenol extracted and precipitated with 20 μg of glycogen at −20 °C overnight. ChIP experiments were analyzed by quantitative PCR (QPCR). Efficiency of ChIP was calculated as percentage of input. The primers used are listed in Table S1.
RNA interference
ATF4 (CREB-2) (sc-35112), HSF1 (sc-35611), c-Fos (sc-29221) and FosB (sc-35403) siRNAs were purchased from Santa Cruz Biotechnology. The control siRNA against luciferase (5′-CGUACGCGGAAUACUUCGAdTdT-3′), NRF2 siRNA (5′-CAGCAUGCUACGUGAUGAAdTdT-3′), NRF2 siRNA#2 (5′-CCAGUGGAUCUGCCAACUAdTdT-3′) and EEF1D siRNA (5′-CUGGCUCAGCAAGCCUGCCUAdTdT-3′) were purchased from Eurogentec. HEK293 cells were cultured in 6-well plates and transfected with 50 nM siRNA using oligofectamine transfection reagent (Invitrogen) according to the manufacturer’s instructions. Forty-eight hours after transfection, cells were re-transfected as described above. Cells were left at 37 °C or heat-shocked for 30 min at 45 °C and allowed to recover for 6 or 24 h. All cells were harvested simultaneously 48 h after re-transfection.
Western blot analysis
Cells were harvested in lysis buffer (25 mM Tris–HCl pH 7.5, 100 mM KCl, 1 mM DTE, 2 mM EDTA, 0.5 mM PMSF, 0.05 % NP-40, 1X PhosSTOP [Roche], 1× protease inhibitor cocktail [Complete Mini, Roche]) and protein concentration was determined using a Bradford protein assay (Bio-Rad). Then 4x sample buffer (200 mM Tris–HCl 6.8, 20 % β-mercaptoethanol, 8 % SDS, 40 % glycerol and 0.4 % Bromophenol blue) was added, and the lysates were incubated at 95°C for 5 min. Protein samples were separated in 10 % polyacrylamide gels and transferred to nitrocellulose transfer membrane (Protran). For Western blot analysis, the following antibodies were used: mouse monoclonal β-actin antibody (AC-15; Sigma; 1:5,000), rabbit polyclonal HSF1 antibody (SPA-901; Stressgen; 1:1,000), mouse monoclonal Hsp70 antibody 4G4 (ab5444; Abcam; 1:5,000), rabbit polyclonal ATF4 antibody (sc-200X; Santa Cruz Biotechnology) and polyclonal HSPB1 antibody (obtained from Dr. A. Zantema; 1:400). Next, blots were incubated with fluorescent secondary antibodies IRDye® 800CW conjugate goat anti-rabbit IgG and IRDye® 680 conjugated goat anti-mouse IgG (926-32211 and 926-32220, respectively; LI-COR Biosciences) according to the manufacturer’s instructions and scanned using a LI-COR Odyssey infrared scanner.
QPCR analysis
RNA was isolated with TRIzol (Invitrogen) according to manufacturer’s recommendation. First, 1 μg of RNA was treated with DNaseI (amplification grade; RNase-free; Invitrogen). Subsequently, 5 mM MgCl2, RT buffer, 1 mM dNTPs, 18.75 units AMV reverse transcriptase, 20 units RNase inhibitors and 1.25 μM oligo(dT) were added to a total volume of 20 μl. Reverse transcription was performed for 10 min at 25 °C, 60 min at 42 °C and 5 min at 95 °C. For QPCR analysis, cDNA was 10-fold diluted. Quantitative real-time PCR was performed using the StepOnePlus™ Real-Time PCR System with Power SYBR® Green PCR Master mix (Applied Biosystems) using the following amplification protocol: 10 min at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. Per reaction 3 μl of diluted cDNA was used and the DNA was amplified using primers for the sequences of interest, listed in Table S1.
Microarray analysis
HEK-pcDNA, HEK-HSF1 K80Q, HEK-wtHSF1 and HEK-dnHSF1 cells were treated with doxycycline for a total of 48 h. Cells were left at 37 °C or heat shocked for 30 min at 45 °C and harvested either 6 or 24 h after heat shock. The transcriptomes of HEK-pcDNA cells and HEK-HSF1 K80Q cells, HEK-HSF1 K80Q 6 h after heat shock versus unstressed HEK-HSF1 K80Q cells or HEK-pcDNA5 6 h after heat shock versus unstressed HEK-pcDNA5 cells were compared. HEK-HSF1 K80Q cells or HEK-dnHSF1 cells 24 h after heat stress were compared with control cells under non-stress conditions. Total RNA was isolated using Trizol according to the manufacturer’s instructions (Invitrogen) and copied into Cy3-labeled or Cy5-labeled cRNA using the Agilent Low RNA Input Linear Amp Kit PLUS, and reverse labeled for the repeat array. Labeled cRNA samples were hybridized to an Agilent Whole Human Genome Microarray Kit (4 × 44K). The arrays were scanned using an Agilent Microarray Scanner. Image analysis and feature extraction were done with Feature Extraction (version 9.5.1, Agilent). We used a cut-off level of 2-fold changed expression and an arbitrarily chosen signal cut-off of >50.
Results
Characterization of the HEK-HSF1 K80Q cell line
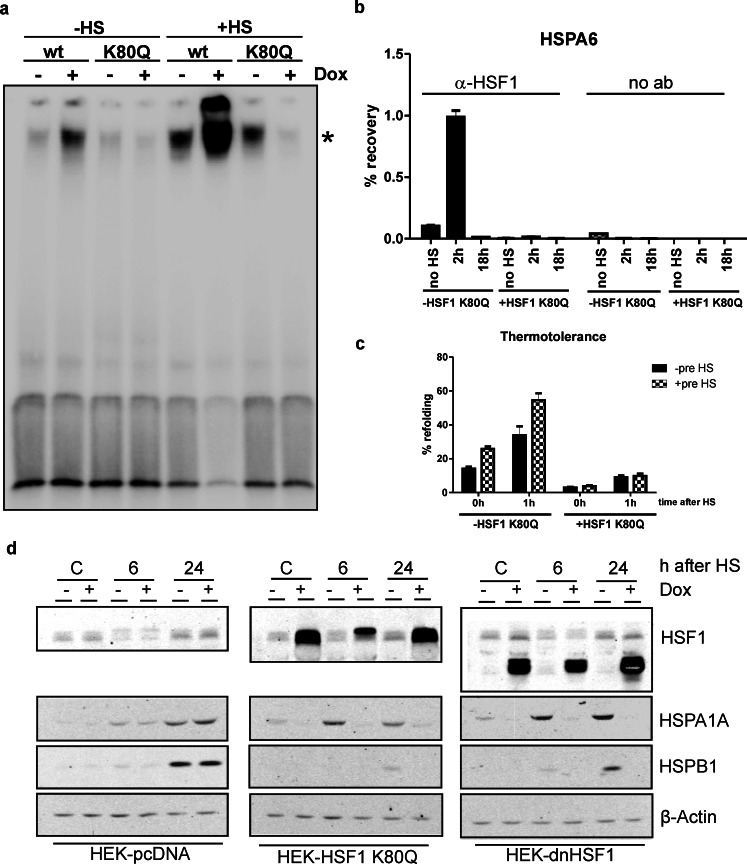
In the HSF1 K80Q mutant lysine 80 in the DNA binding region is replaced by glutamine and because of this mutation HSF1 loses its DNA-binding activity (Westerheide et al. 2009). HSF1 K80Q is expected to trimerize with endogenous HSF1, resulting in a strong reduction of binding competent HSF1 trimers. Using nuclear extracts of HSF1 K80Q expressing cells indeed only a weak signal of HSF1 binding to the HSE was detected (Fig. 1a). With nuclear extracts from cells overexpressing wild-type HSF1 (wtHSF1) increased binding of HSF1 to the HSE was observed, as expected, even when cells were unstressed (Fig. 1a). The bandshifts shown in Fig. 1a could be supershifted by an antibody to HSF1, indicating that it is indeed HSF1 that was bound (data not shown). Extracts from heat shocked cells showed a more intense bandshift signal and thus an increase in binding competent HSF1. Expression of HSF1 K80Q blocked this increase (Fig. 1a). The loss of binding of HSF1 to the HSE in the presence of HSF1 K80Q was confirmed by ChIP (Fig. 1b). In control cells, i.e., HEK-HSF1 K80Q cells cultured in the absence of doxycycline, HSF1 was bound to the HSPA6 promoter region 2 h after heat shock. The binding of HSF1 is transient and as expected we did not observe HSF1 binding 18 h after heat stress. When HSF1 K80Q expression was induced, no bound HSF1 could be detected either 2 or 18 h after heat shock (Fig. 1b).
Fig. 1.
Characterization of HEK-HSF1 K80Q cells. a Nuclear extracts were made of HEK-wtHSF1 cells and HEK-HSF1 K80Q cells either non-stressed (−HS) or exposed to heat shock (30 min, 45 °C, +HS). An electrophoretic mobility shift assay was performed with a double-stranded oligo with the HSE sequence. Where indicated (+Dox), doxycyclin was added to induce expression of either wtHSF1 or HSF1 K80Q. b Chromatin immunoprecipitation using nuclear extracts from control and HSF1 K80Q expressing cells was performed with an HSF1 antibody or no antibody added. Bound chromatin was analyzed by QPCR using a primer set surrounding the HSE of the HSPA6 promoter. Cells were either non-stressed or harvested 2 or 18 h after heat shock, as indicated. c HEK-HSF1 K80Q cells were transfected with SV40-luc and 24 h after transfection cells were pre-heat shocked for 30 min at 45 °C. Fourteen hours later, cells were harvested or heat-shocked again for 30 min at 45 °C in the presence of 20 μg/ml CHX to inhibit translation and harvested immediately or after 1 h of recovery and a luciferase assay was performed. d HEK-pcDNA, HEKdnHSF1 or HEK-HSF1 K80Q cells were cultured in the presence or absence of doxycycline and exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Cell lysates were subjected to SDS-PAGE and levels of HSF1, HSPA1A, and HSPB1 were determined by Western blotting. β-Actin was used as a loading control. Bars indicate SD
When cells are exposed to heat stress they increase their chaperone levels and become more resistant to a subsequent heat stress, a process known as thermotolerance. HSF1 knockout or knockdown or overexpression of a dominant negative HSF1 mutant has been shown to inhibit the acquisition of thermotolerance (Yin et al. 2005; Heldens et al. 2012; Zhang et al. 2002). To determine whether expression of HSF1 K80Q also inhibits the development of thermotolerance we heat stressed HEK-HSF1 K80Q cells and analyzed the refolding of luciferase after a second heat shock. As shown in Fig. 1c, pre-heat shocked control cells showed increased refolding activity compared to naïve control cells. In cells overexpressing HSF1 K80Q, no difference in refolding activity was found between pre-heat shocked and naïve cells, indicating that these cells do not develop thermotolerance. Expression of HSF1 K80Q also inhibited luciferase refolding after a single heat shock (Fig. 1c), suggesting that HSF1 K80Q expression lowers the chaperoning capacity of the non-stressed cells as well.
We also analyzed the heat-induced expression levels of HSPA1A and HSPB1 and found that their increase was inhibited by expression of HSF1 K80Q, similar to the effect of an HSF1 mutant lacking the activation domains, dnHSF1 (Fig. 1d). Together, these data show that the HSF1 K80Q mutant blocks the HSF1 directed transcriptional heat shock response.
Transcriptome changes in the presence of HSF1 K80Q in non-stressed cells
If HSF1 plays a role in the absence of stress, then expression of a non-DNA binding mutant should change the transcriptome. We used microarrays to analyze the effect of HSF1 K80Q on the transcriptome in the absence of stress. As overexpression of the HSF1 protein may have secondary effects, for example, by sequestering chaperones, we also analyzed the effect of overexpressing wild type HSF1 on the transcriptome of non-stressed cells. Table 1 shows the list of the 26 genes of which the transcript level changed at least 2-fold upon expression of HSF1 K80Q in non-stressed cells (relative to the level in both control cells and in cells overexpressing wild type HSF1). For 18 genes, we noted a decrease in transcript level; for eight genes, an increase was noted, particularly in that of the SEPP1 gene. We have not attempted to verify the increase in the SEPP1 transcript level as the signal of this transcript in the microarray was just above background. Note that none of these genes is a canonical heat shock gene (see also Table S2) and that there is no overlap between the transcriptome changes in HSF1 K80Q expressing cells and in dnHSF1 expressing cells (Heldens et al. 2010). We also compared our microarray data with previously reported results obtained by HSF1 knockdown in HeLa cells (Page et al. 2006). For 16 of the 26 genes, transcript levels were not significantly affected in the siRNA-treated HeLa cells; one (CRLF1, cytokine receptor-like factor 1) had lower transcript levels, like in the HEK-HSF1 K80Q cells, but this could not be confirmed by QPCR (Fig. S2a); for the remaining nine genes, data were not available. Together these data show that HSF1 does control the level of a limited set of transcripts in the non-stressed cells. If depletion of HSF1 by siRNA and blocking HSF1 activity by expression of HSF1 K80Q can be equated (see also below), then this set of genes is largely cell type specific.
Table 1.
Transcriptome changes in non-stressed HEK293 cells upon expression of HSF1 K80Q
| Accession no. | Gene | K80Q/Cntrl 37 °C | Description |
|---|---|---|---|
| NM_004695 | SLC16A5 | 0.1 | Mnocarboxylic acid transporter 6 |
| NM_001163335 | SYTL5 | 0.2 | Synaptotagmin-like 5 |
| NM_000735 | CGA | 0.2 | Glycoprotein hormones, alpha polypeptide |
| NM_017527 | LY6K | 0.2 | Lymphocyte antigen 6 complex, locus K |
| NM_017671 | FERMT1 | 0.2 | Fermitin family homolog 1 |
| NM_199441 | ZNF334 | 0.3 | Zinc finger protein 334 |
| NM_016378 | VCX2 | 0.3 | Variable charge, X-linked 2 |
| NM_001007125 | C20orf201 | 0.3 | Chromosome 20 open reading frame 201 |
| NM_002523 | NPTX2 | 0.3 | Neuronal pentraxin II |
| NM_001195 | BFSP1 | 0.4 | Beaded filament structural protein 1, filensin |
| NM_021785 | RAI2 | 0.4 | Retinoic acid induced 2 |
| NM_001030059 | PPAPDC1A | 0.4 | Phosphatidic acid phosphatase type 2 domain containing 1A |
| NM_152349 | KRT222 | 0.5 | Keratin 222 |
| NM_001077489 | GNAS | 0.5 | GNAS complex locus |
| NM_004750 | CRLF1 | 0.5 | Cytokine receptor-like factor 1 |
| NM_181803 | UBE2C | 0.5 | Ubiquitin-conjugating enzyme E2C |
| NM_015432 | PLEKHG4 | 0.5 | Pleckstrin homology domain containing, family G |
| XM_002345507 | LOC100292909 | 0.5 | Hypothetical protein |
| NM_032876 | JUB | 2.1 | Jub, ajuba homolog |
| NM_001673 | ASNS | 2.2 | Asparagine synthetase |
| NM_004563 | PCK2 | 2.3 | Phosphoenolpyruvate carboxykinase 2 (mitochondrial) |
| NM_014331 | SLC7A11 | 2.3 | Ccationic amino acid transporter, y + system |
| NM_001902 | CTH | 2.4 | Cystathionase (cystathionine gamma-lyase) |
| NM_003714 | STC2 | 2.4 | Stanniocalcin 2 |
| NM_022445 | TPK1 | 2.9 | Thiamin pyrophosphokinase 1 |
| NM_005410 | SEPP1 | 37.0 | Selenoprotein P, plasma, 1 |
Overexpression of wild type HSF1 leads to elevated levels of activated HSF1 and our microarray data thus identify potential HSF1 targets. For ten genes we found a significantly lower transcript level (relative to that in control cells and in HSF1 K80Q expressing cells) and for 32 an increase in transcript level (Table S2). Eleven of these genes were also heat shock inducible, whereas for one of these genes the transcript level decreased 6 h after heat shock. Only three (CRYAB, DHRS2 and SEPW1) of these 42 potential HSF1 targets in non-stressed HEK293 cells were also identified as HSF1 target genes by exogenous expression of a constitutively active HSF1 mutant in HeLa cells (Hayashida et al. 2010). Transcriptional activation by HSF1 in the absence of stress is thus constrained by cell specific factors.
The effect of HSF1 K80Q and dnHSF1 expression on the transcriptome of heat shocked cells — 6 h after heat shock
The classical role of HSF1 is transcription activation in heat-stressed cells. In HEK293 cells allowed to recover from a heat shock for 6 h, we found an increase of at least 2-fold in the transcript levels of 180 genes. Expression of HSF1 K80Q and/or dnHSF1 inhibited the increase in transcript level of 53 of these genes by at least 2-fold (Fig. 2a, Table S3). Among these genes are the canonical HSF1 target genes, such as HSPA1A/B, HSPA6, DNAJB1, HSPH1, HSP90AA1, BAG3 and ATF3. For the canonical HSF1-dependent genes, the expression of HSF1 K80Q or dnHSF1 had a similar effect: it inhibited the heat shock induced increase in transcript levels, where expression of dnHSF1 was usually more effective than that of HSF1 K80Q. Note that for the highly expressed heat shock genes, such as HSPA6, the transcript levels still increased significantly in both HSF1 K80Q and dnHSF1 expressing cells, although the increase was far less than in control cells (Table S3). For other genes the effect of HSF1 K80Q or dnHSF1 expression was different. In a number of cases, such as ACTA1 or AOC3, expression of HSF1 K80Q had no effect or even enhanced expression levels, while that of dnHSF1 inhibited (Table S3). Possibly binding of dnHSF1 blocks access of other transcription factors to the promoter; expression of HSF1 K80Q would leave an HSF1 binding site empty.
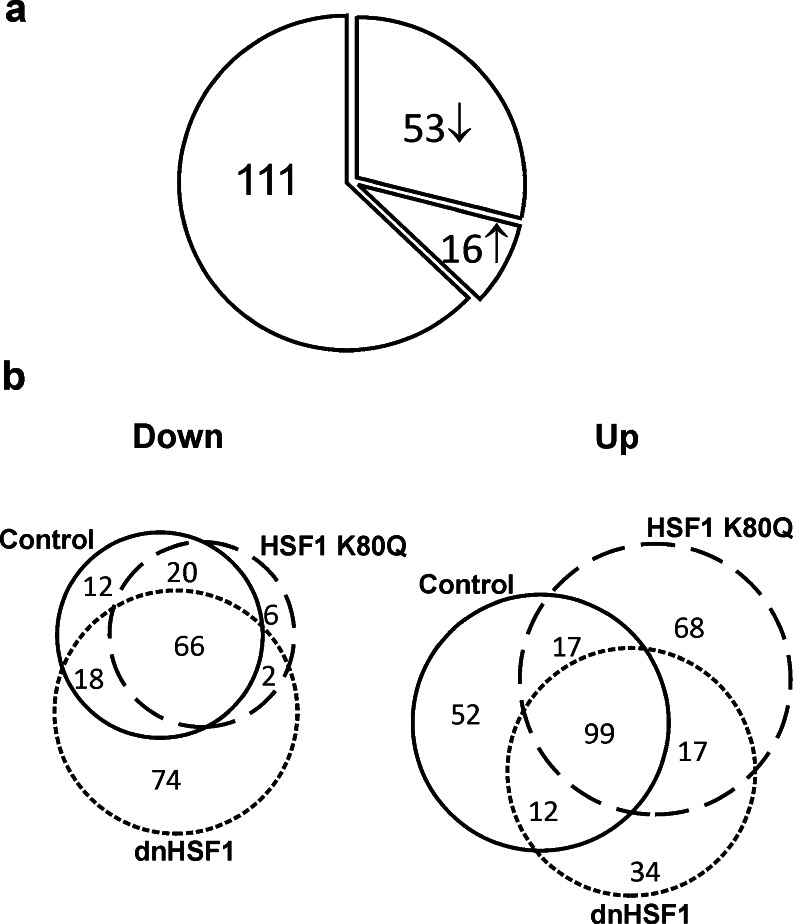
Fig. 2.
a Pie diagram illustrating the effect of expression of HSF1 K80Q and dnHSF1 on the increase in transcript levels in HEK293 cells 6 h after heat shock. An ↓ indicates a ≥2-fold decrease in level; an ↑ a ≥2-fold increase in level. For details, see Fig. S1 and Table S3. b Venn diagram illustrating the overlap in transcriptome changes in control, HSF1 K80Q and dnHSF1 cells 6 h after heat shock. “Down” indicates that the transcript levels were ≥2-fold lower than in non-stressed cells; “up” indicates that the transcript levels were ≥2-fold higher than in non-stressed cells. The solid line circle represents control cells, the dotted line HSF1 K80Q cells and the dashed line dnHSF1 cells. For details, see Fig. S3
Paradoxically, for 16 heat shock induced genes, expression levels increased significantly in either HSF1 K80Q or dnHSF1 cells (Table S4). The explanation for this is not clear, but it does suggest that HSF1 partially represses stress induced transcription mediated by other transcription factors. This suggestion is supported by the finding that about a third of the genes of which the transcript levels are increased in HSF1 K80Q cells and/or dnHSF1 cells did not respond significantly to heat stress in control cells (Fig. 2b). Alternatively, the impaired chaperoning capacity of the HSF1 K80Q (Fig. 1c) and dnHSF1 cells (Heldens et al. 2012) could result in a stronger stress response.
A heat stress is known to shift the transcription pattern to heat shock genes leading to a decrease in activity of non-heat shock promoters, and it is thus expected that the transcript levels of some genes decrease in heat shocked cells. Indeed, in control cells that had recovered for 6 h from the heat shock, the level of the transcripts of 116 genes was at least 2-fold lower than in non-stressed cells. A response of similar magnitude was seen in HSF1 K80Q cells; in dnHSF1 cells the effect was larger (Fig. 2b and Fig. S3).
When we compared our data for HEK293 cells with the published data for heat-shocked HeLa cells (Page et al. 2006), we found only 20 genes that were heat shock-responsive in both cell lines (note that the heat shock conditions differ). The transcript level of one gene (ZNF264) decreased in both cell lines (Fig. S1 and Table 2) but we could not confirm this decrease by QPCR in HEK293 cells (Fig. S2a). For three genes, the transcript levels decreased in HeLa cells, while they increased in HEK293 cells. Of the 16 genes of which the transcript levels increased in both heat-shocked HeLa and HEK293 cells, four appeared to be not regulated by HSF1 (Table 2). The common set of heat shock responsive genes consists mostly of the well known canonical heat shock genes, such as HSPA1A and DNAJB1, and general stress responsive genes such as GADD45B and PPP1R15A (GADD34).
Table 2.
Comparison between the effect of HSF1 K80Q expression in HEK293 cells and that of HSF1 siRNA treatment in HeLa cells
| Accession no. | Gene | Cntrl 6 h hs | K80Q 6 h hs | dnHSF 6 h hs | HeLa 4 h hsa | siRNA 4 h hsa | Description |
|---|---|---|---|---|---|---|---|
| NM_002155 | HSPA6 | 61.9 b | 27.4c | 18.1 | 11.3 | 5.1 | Heat shock 70 kDa protein 6 |
| NM_006145 | DNAJB1 | 32.6 | 3.4 | 0.8 | 2.9 | 1.5 | DnaJ (Hsp40) homolog, subfamily B, member 1 |
| NM_015675 | GADD45B | 32.0 | 29.9 | 13.4 | 2.0 | 1.5 | Gowth arrest and DNA damage-inducible, beta |
| NM_001885 | CRYAB | 24.3 | 2.4 | 3.6 | 2.2 | 0.6 | Crystallin, alpha B |
| NM_005345 | HSPA1A | 13.7 | 1.2 | 1.0 | 2.0 | 1.6 | Heat shock 70 kDa protein 1A |
| NM_006644 | HSPH1 | 9.8 | 1.9 | 1.0 | 2.8 | 1.3 | Heat shock 105 kDa/110 kDa protein 1 |
| NM_004281 | BAG3 | 6.7 | 2.2 | 1.3 | 4.3 | 1.6 | BCL2-associated athanogene 3 |
| NM_001124 | ADM | 6.2 | 6.6 | 3.6 | 2.3 | 1.1 | Adrenomedullin |
| NM_006472 | TXNIP | 4.4 | 2.8 | 4.6 | 2.6 | 0.9 | Thioredoxin interacting protein |
| NM_004419 | DUSP5 | 4.3 | 2.7 | 1.8 | 2.6 | 1.6 | Dual specificity phosphatase 5 |
| NM_002228 | JUN | 4.0 | 1.8 | 1.4 | 2.0 | 1.8 | Jun oncogene |
| NM_001040619 | ATF3 | 3.7 | 1.6 | 2.5 | 2.4 | 2.2 | Activating transcription factor 3 |
| NM_001554 | CYR61 | 7.8 | 4.0 | 5.0 | 2.0 | 1.9 | Cysteine-rich, angiogenic inducer, 61 |
| NM_002923 | RGS2 | 19.8 | 12.4 | 13.7 | 2.0 | 1.1 | Regulator of G-protein signaling 2, 24 kDa |
| NM_014330 | PPP1R15A | 8.0 | 6.9 | 6.1 | 2.6 | 1.6 | GADD34; protein phosphatase 1, regulatory (inhibitor) subunit 15A |
| NM_020127 | TUFT1 | 2.2 | 2.8 | 2.0 | 2.9 | 2.8 | Tuftelin 1 |
The effect of HSF1 K80Q and dnHSF1 expression on the transcriptome of heat-shocked cells 24 h after heat shock: an extended primary stress response and a secondary stress response
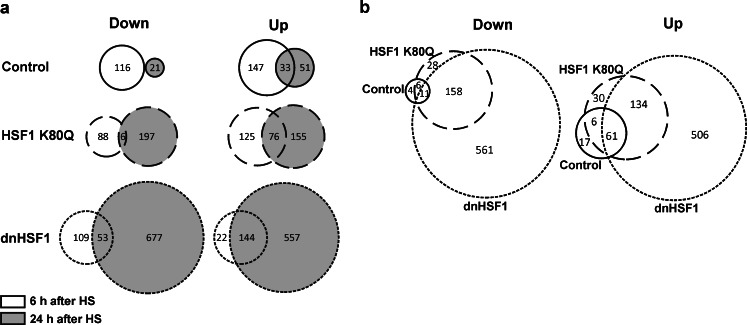
The synthesis of HSF1-dependent chaperones serves as a feedback mechanism to dampen the heat shock response and to restore homeostasis (Zou et al. 1998; Guo et al. 2001; Shi et al. 1998; Voellmy and Boellmann 2007; Balch et al. 2008; Morimoto 2008). As in the HSF1 K80Q and the dnHSF1 cells, the expression of these chaperones is inhibited, we analyzed the effect of these HSF1 mutants on the transcriptome of cells that had been allowed to recover for 24 h from the heat shock. In control cells the primary transcriptional response to the heat shock had largely decayed: the levels of most transcripts were back to the level in non-stressed cells or at least decreased relative to the level in cells 6 h after heat shock (Fig. 3a and Fig. S4, Tables S3 and S4). In HSF1 K80Q and dnHSF1 cells, the primary response decayed as well, although to a lesser extent than that in control cells (Fig. 3a and Fig. S4, Tables S3 and S4). These cells thus show an extended primary stress response. We also saw a secondary response, i.e., a change in transcript level of genes which did not respond significantly to a heat shock initially (Fig. 3a), with the transcript level of 21 genes decreased and that of 51 genes increased (Table S5). This relatively small transcriptome change in control cells partially overlapped with the much stronger secondary response in HSF1 K80Q and dnHSF1 cells (Fig. 3b and Fig. S5) with the transcript levels of 197 genes down and 155 up in HSF1 K80Q cells and those of 677 genes down and of 551 up in dnHSF1 cells (Fig. 3a). The much larger secondary response in cells lacking active HSF1 shows that HSF1 directed macromolecular synthesis plays a major role in restoring homeostasis. The genes of which the transcript levels are increased in HSF1 K80Q or dnHSF1 cells 24 h after heat stress are enriched for the GO category transcription and transcription regulation (http://david.abcc.ncifcrf.gov/), suggesting that these cells mount an additional transcriptional response in an attempt to restore homeostasis.
Fig. 3.
Venn diagrams illustrating the changes in transcriptome changes in control, HSF1 K80Q and dnHSF1 cells between 6 and 24 h after heat shock (a) and the overlap in transcriptome changes in control, HSF1 K80Q and dnHSF1 cells 24 h after heat shock (b). “Down” indicates that the transcript levels were ≥2-fold lower than in non-stressed cells; “up” indicates that the transcript levels were ≥2-fold higher than in non-stressed cells. The solid line circle represents control cells, the dotted line HSF1 K80Q cells and the dashed line dnHSF1 cells. The shading indicates 24 h after heat shock. For details, see Figs. S4 and S5 and Tables S3 and S4
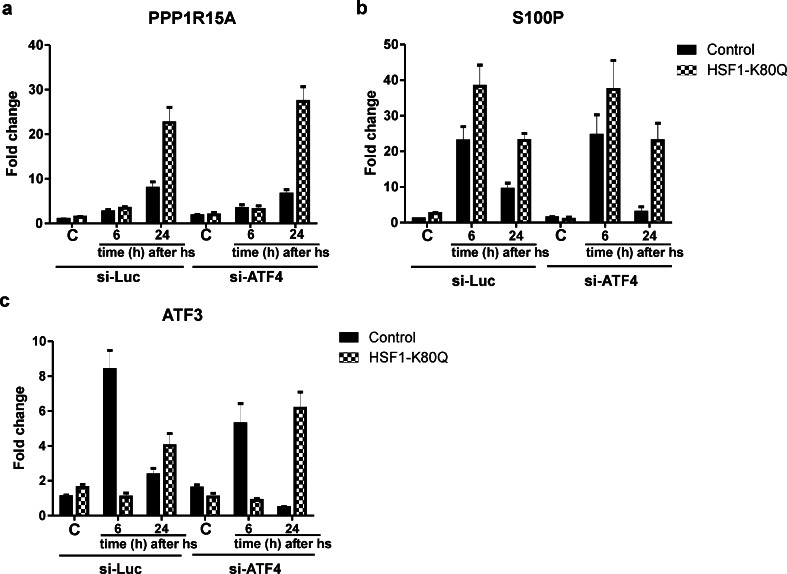
Among the genes of which the transcript levels remained high in HSF1 K80Q cells were a number of ATF4 target genes, e.g., PPP1R15A (GADD34) (Ron and Walter 2007), S100P (Namba et al. 2009) and ATF3 (Pan et al. 2007). We therefore tested whether ATF4 activity was responsible for the high transcript levels of these genes. The level of ATF4 did increase markedly in heat shocked cells, but equally so in control or HSF1 K80Q cells. 24 h after heat stress the level of ATF4 was lower than 6 h after heat stress but still higher than in non-stressed cells (Fig. S6b). To examine the role of ATF4 further, we knocked down ATF4 mRNA with siRNA (Fig. S6c–d) and determined the effect on the mRNA levels of the ATF4 target genes PPP1R15A (GADD34), S100P and ATF3 (Fig. 4) by QPCR. ATF4 knockdown did not block the expression of these genes, although the exact effect on the transcript levels varied. These data show that ATF4 does not play a major role in heat shocked cells and suggest that the extended primary stress response of ATF4 target genes in HSF1 K80Q cells is not ATF4 dependent.
Fig. 4.
ATF4 is not involved in the extended primary stress response of its target genes in HSF1 K80Q cells. a–c HEK-HSF1 K80Q cells were cultured in the presence (HSF1 K80Q) or absence (Control) of doxycycline and transfected for 96 h with siRNA against ATF4 or luciferase as a control with a re-transfection at 48 h. Cells were exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for 6 or 24 h as indicated before harvesting. Total RNA was isolated and transcript levels of the genes indicated relative to GAPDH mRNA levels were measured by QPCR. The fold change of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
The effect of HSF1 K80Q and dnHSF1 expression on the transcriptome of heat shocked cells 24 h after heat shock: a delayed stress response
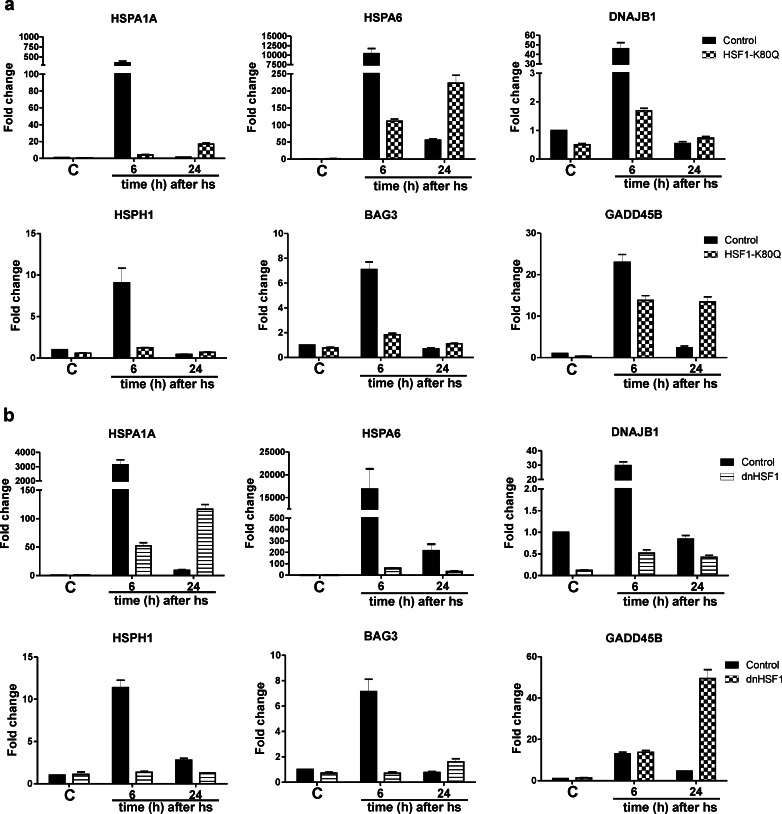
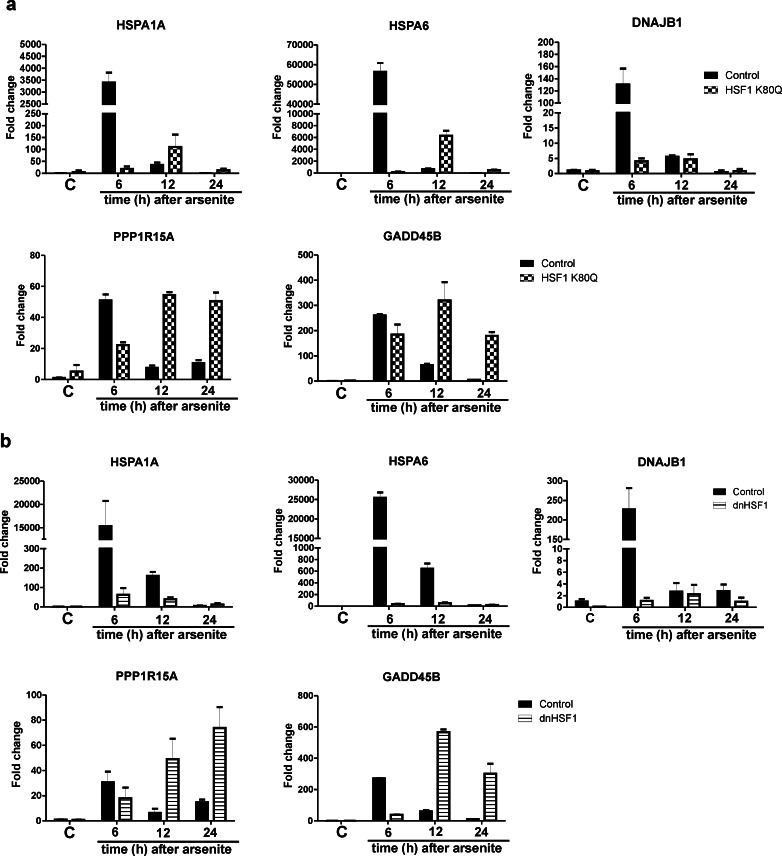
Surprisingly, the transcript levels of a large fraction of the HSF1 dependent genes stayed the same or were even higher in HSF1 K80Q and dnHSF1 cells 24 h after heat shock, while they decreased in control cells (Table S3). To confirm that at least some apparently HSF1 dependent genes do show a higher transcript level in HSF1 K80Q or dnHSF1 cells 24 h after heat stress, we examined the changes in the transcript level of a set of HSF1 dependent genes by QPCR (Fig. 5). Noteworthy is the increase in transcript level of the HSPA1A and HSPA6 genes in HSF1 K80Q cells. The transcript level of the HSPA1A gene but not that of the HSPA6 gene also went up in dnHSF1 cells. This delayed stress response is not limited to heat shocked cells expressing an inactive HSF1 mutant: arsenite stressed HSF1 K80Q or dnHSF1 cells also showed the delayed stress response (Fig. 6).
Fig. 5.
A delayed stress response in HSF1 K80Q cells. HEK-HSF1 K80Q (a) or dnHSF1 (b) cells were cultured in the presence (HSF1 K80Q or dnHSF1) or absence of doxycycline (Control) and exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Total RNA was isolated and transcript levels relative to GAPDH mRNA levels were measured by QPCR. The fold induction of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
Fig. 6.
Relative changes in transcript levels of various genes in arsenite stressed HSF1 K80Q or dnHSF1 cells. HEK-HSF1 K80Q (a) or HEK-dnHSF1 (b) cells were cultured in the presence (HSF1 K80Q/dnHSF1) or absence of doxycycline (Control) and treated with 0.5 mM of arsenite for 1.5 h. Medium was washed off and cells were allowed to recover for the indicated time before harvesting. Total RNA was isolated and transcript levels relative to GAPDH mRNA levels were measured by QPCR. The fold induction of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
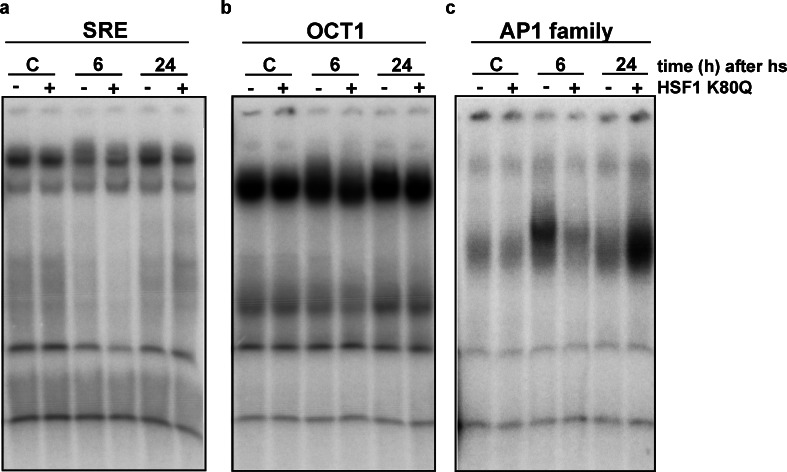
These data suggest that HSF1 can be bypassed for at least some HSF1-dependent genes. To investigate which transcription factors could be involved, we analyzed the transcriptome changes by Gene Set Enrichment Analysis (GSEA) to assess whether certain transcription factor binding sites were over-represented in the promoter regions of the genes of which the transcript level was elevated 24 h after heat stress. The transcription factors that scored the highest were serum response factor (SRF), octamer transcription factor OCT1 and members of the AP1 family. To determine whether these transcription factors were indeed activated 24 h after heat stress in HSF1 K80Q cells, we analyzed whether they showed increased DNA binding activity. We used probes with the DNA binding sequences for these transcription factors and performed an electrophoretic mobility shift assay with nuclear extracts of control or HSF1 K80Q expressing cells before, 6 or 24 h after heat stress. No differences in signals were found between control and HSF1 K80Q cells either 6 or 24 h after heat shock for the SRE (serum response element) and OCT1 probes (Fig. 7a–b). When we used a generic AP1 family probe, clear differences were observed (Fig. 7c). A strong band shift signal was found using extracts of control cells 6 h after heat stress but not using similar extracts of HSF1 K80Q cells. When we used extracts isolated from cells 24 h after heat stress, a much stronger band shift signal was detected using extracts of HSF1 K80Q expressing cells compared to extracts of control cells. This complex consistently migrated slightly faster than the complex detected using extracts from control cells 6 h after heat stress and thus might represent a different DNA–protein complex.
Fig. 7.
Binding to the SRF, OCT1 or AP1 consensus sequence. HEK-HSF1 K80Q cells were cultured in the presence (+) or absence (−) of doxycycline and exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Nuclear extracts were used in an electrophoretic mobility shift assay with a double-stranded oligonucleotide with the SRE (a), OCT1 (b) or AP1 family (c) sequence
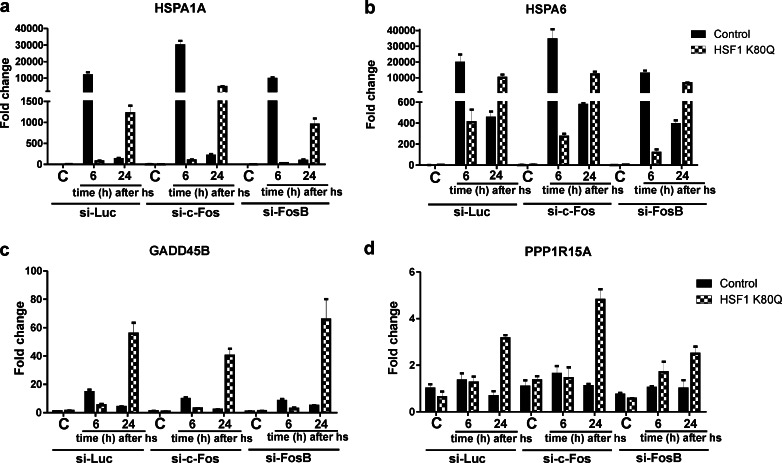
Our microarray data showed that the transcript levels of the AP1 family members c-Fos and FosB were elevated 24 h after heat shock in the presence of HSF1 K80Q (Table S3, Fig. S7a–c). c-Fos and FosB are both early response genes and have been implicated in the regulation of proliferation and differentiation and their transcript levels increase upon several stimuli, such as growth factors and stress, including oxidative stress (Piechaczyk and Blanchard 1994; Milde-Langosch 2005). To test whether c-Fos or FosB plays a role in the delayed stress response, we knocked down c-Fos and FosB mRNA (Fig. S7b–c) and examined the effect on HSPA1A and HSPA6 mRNA levels, as well as on the GADD45B and PPP1R15A mRNA levels. Knockdown of c-Fos resulted in an increase in HSPA1A mRNA levels in control cells 6 h after heat shock and in HSF1 K80Q cells 24 h after heat shock (Fig. 8a). c-Fos thus appears to inhibit rather than enhance expression of the HSPA1A gene. Knockdown of c-Fos did not alter the expression pattern of the HSPA6, GADD45B or PPP1R15A genes significantly either in control or in HSF1 K80Q cells, except for a slight increase in PPP1R15A mRNA in HSF1 K80Q cells 24 h after heat stress (Fig. 8b–d). Knockdown of FosB had little effect on HSPA1A, GADD45B or PPP1R15A mRNA levels. Only the increase in HSPA6 mRNA levels 6 h after heat stress, but not that 24 h after heat stress, was slightly inhibited (Fig. 8b). These data show that neither c-Fos nor FosB is responsible for the delayed and/or extended primary stress response in HSF1 K80Q cells.
Fig. 8.
c-Fos and FosB are not involved in activating transcription of the HSPA1A and HSPA6 genes 24 h after heat shock in HSF1 K80Q cells. a–d HEK-HSF1 K80Q cells were cultured in the presence (HSF1 K80Q) or absence (Control) of doxycycline and transfected for 96 h with siRNA against c-Fos, FosB or luciferase as a control with a re-transfection at 48 h. Cells were exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Total RNA was isolated and transcript levels of the genes indicated relative to GAPDH mRNA levels were measured by QPCR. The fold change of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
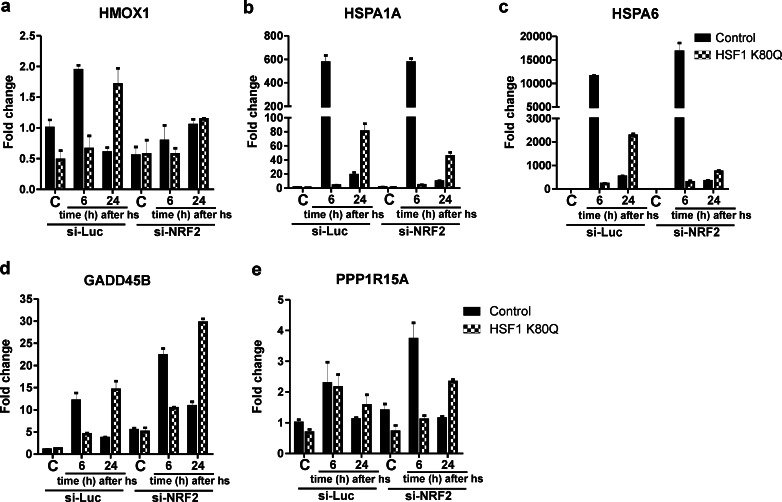
The generic AP1 probe used in the EMSA shown in Fig. 7 can also be bound by NRF2, a transcription factor involved in the antioxidant response. To determine a possible involvement of NRF2 in the delayed stress response in HSF1 K80Q cells after heat shock, we analyzed the binding to a probe with the antioxidant response element (ARE) sequence, the consensus binding sequence for NRF2, in extracts of heat-stressed control or HSF1 K80Q cells and found the same pattern as that seen when the generic AP1 probe was used (Fig. 7 and Fig. S8a). The sequence of the CRE (cAMP responsive element; CREB protein consensus binding sequence) or TRE (12-O-tetradecanoyl phorbol13-acetate (TPA)-responsive element; AP1 consensus binding sequence) overlaps with the ARE. However, competition with a 10-fold molar excess of unlabeled probes for either the CRE or the TRE did not compete away the signal of the ARE complex, whereas the signal almost completely disappeared when unlabeled ARE probe was the competitor (Fig. S8b). These data show that the extracts of control and HSF1 K80Q cells 6 and 24 h after heat stress differ in an ARE binding activity and suggest a role for NRF2 in the stress response of HSF1 K80Q cells. To confirm the differential pattern of NRF2 activity, we analyzed the transcript levels of the NRF2 target gene heme oxygenase 1 (HMOX1). HMOX1 transcript levels increased in control cells 6 h after heat stress, but not in HSF1 K80Q cells (Fig. 9a). In control cells 24 h after heat stress the HMOX1 mRNA level had decayed, while in HSF1 K80Q cells the HMOX1 mRNA level had increased. The changes in HMOX1 mRNA levels thus correspond to the changes in the ARE binding pattern. Treatment with NRF2 siRNA blocked the heat shock induced changes in HMOX1 mRNA levels in both control and HSF1 K80Q cells (Fig. 9a and Fig. S9), showing that NRF2 indeed regulates HMOX1 expression in response to heat shock.
Fig. 9.
Transcript levels in NRF2 siRNA-treated cells. a–e HEK-HSF1 K80Q cells were cultured in the presence (HSF1 K80Q) or absence (Control) of doxycycline and transfected for 96 h with siRNA against NRF2 or luciferase as a control with a re-transfection at 48 h. Cells were exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Total RNA was isolated and transcript levels of the genes indicated relative to GAPDH mRNA levels were measured by QPCR. The fold change of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
To look further into the role of NRF2 in the delayed and/or extended primary stress response in HSF1 K80Q cells, we investigated the effect of NRF2 siRNA treatment on HSPA1A and HSPA6 mRNA levels, as well as on the GADD45B and PPP1R15A mRNA levels. Knockdown of NRF2 mRNA did not have any effect on HSPA1A and HSPA6 mRNA levels 6 h after heat shock (Fig. 9b–c). In HSF1 K80Q cells 24 h after heat stress the level of HSPA6 mRNAs was significantly lower in NRF2 siRNA-treated cells compared to cells treated with control siRNA. The HSPA1A mRNA level was also decreased, but not as markedly as that of HSPA6 mRNA. These results were confirmed using a second NRF2 siRNA (Fig. S10). These data indicate that NRF2 is involved in the delayed stress response of HSF1 target genes in HSF1 K80Q cells 24 h after heat stress. GADD45B mRNA levels in control cells appeared to be increased when cells were treated with NRF2 siRNA (Fig. 9d); this could, however, not be confirmed with the second siRNA (Fig. S10). The increase in GADD45B transcript 24 h after heat shock in the presence of HSF1 K80Q was not affected by NRF2 knockdown. For the PPP1R15A mRNA, results were also somewhat variable: the first, but not the second, NRF2 siRNA enhanced PPP1R15A transcript levels in control cells 6 h after heat stress (Fig. 9e); the second, but not the first, NRF2 siRNA decreased PPP1R15A mRNA levels in HSF1 K80Q cells both 6 and 24 h after heat stress (Fig. S10). However, irrespective of which NRF2 siRNA was used, the PPP1R15A mRNA level was always higher in HSF1 K80Q cells than in control cells 24 h after heat shock. Together, these data indicate that NRF2 is not involved in setting the GADD45B and PPP1R15A mRNA levels in HSF1 K80Q cells 24 h after heat shock.
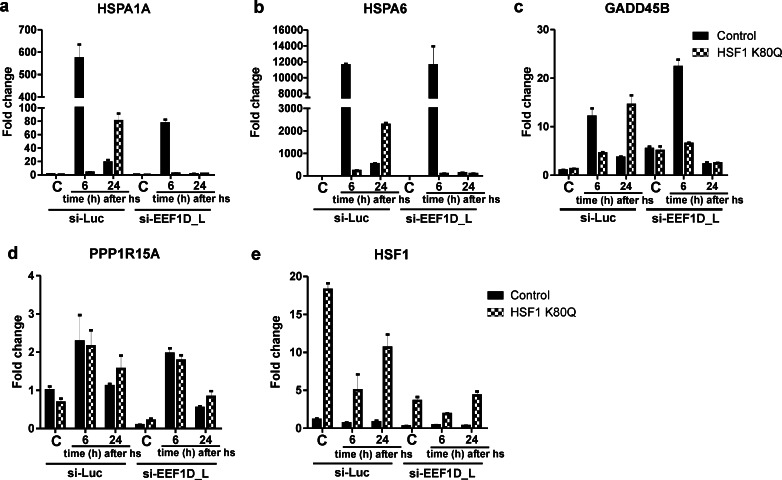
Recently, a shared co-factor for HSF1 and NRF2 has been described. Kaitsuka et al. (2011) reported that the long spliced variant of the eukaryotic elongation factor 1δ (EEF1D_L) is a heat shock transcription factor. This variant was shown to bind to the HSE in the promoter of the HSPA6 gene and activate transcription. In addition, they showed that the N-terminal domain of EEF1D_L interacts with NRF2 and that EEF1D_L is an essential protein for NRF2-dependent (HMOX1) gene induction. We therefore investigated whether EEF1D_L is involved in the delayed and/or extended primary stress response in HSF1 K80Q cells. We treated cells with the EEF1D_L siRNA described (Kaitsuka et al. 2011; Fig. S11), and examined HSPA1A and HSPA6 mRNA levels. Remarkably, the transcript levels of the HSPA1A and HSPA6 genes responded differently to EEF1D_L siRNA. In contrast to the findings of Kaitsuka et al. (2011), we did not see an effect of EEF1D_L siRNA on the increase in HSPA6 mRNA levels in control cells 6 h after heat shock (Fig. 10b), but the increase of HSPA1A mRNA levels was diminished (Fig. 10a). In the HSF1 K80Q cells, however, the increase in both the HSPA1A and HSPA6 mRNA levels 24 h after heat stress was strongly inhibited by EEF1D_L siRNA. EEF1D_L siRNA treatment also abolished the increase in GADD45B and PPP1R15A mRNA levels in HSF1 K80Q cells 24 h after heat stress (Fig. 10c–d), but the data reported above (Figs. 5 and 9) show that HSF1 and NRF2, the suggested partners of EEF1D_L, are not involved. The interpretation of these data is complicated by our finding that EEF1D_L siRNA treatment also decreased HSF1 mRNA levels (Fig. 10e) and EEF1D_L siRNA thus has off-target effects. Unfortunately, we were unable to knockdown EEF1D_L mRNA using other siRNAs, including the second siRNA described by Kaitsuka et al. (2011).
Fig. 10.
Transcript levels in EEF1D_L siRNA-treated cells. a–e HEK-HSF1 K80Q cells were cultured in the presence (HSF1 K80Q) or absence (Control) of doxycycline and transfected for 96 h with siRNA against EEF1D_L or luciferase as a control with a re-transfection at 48 h. Cells were exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Total RNA was isolated and transcript levels of the genes indicated relative to GAPDH mRNA levels were measured by QPCR. The fold change of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
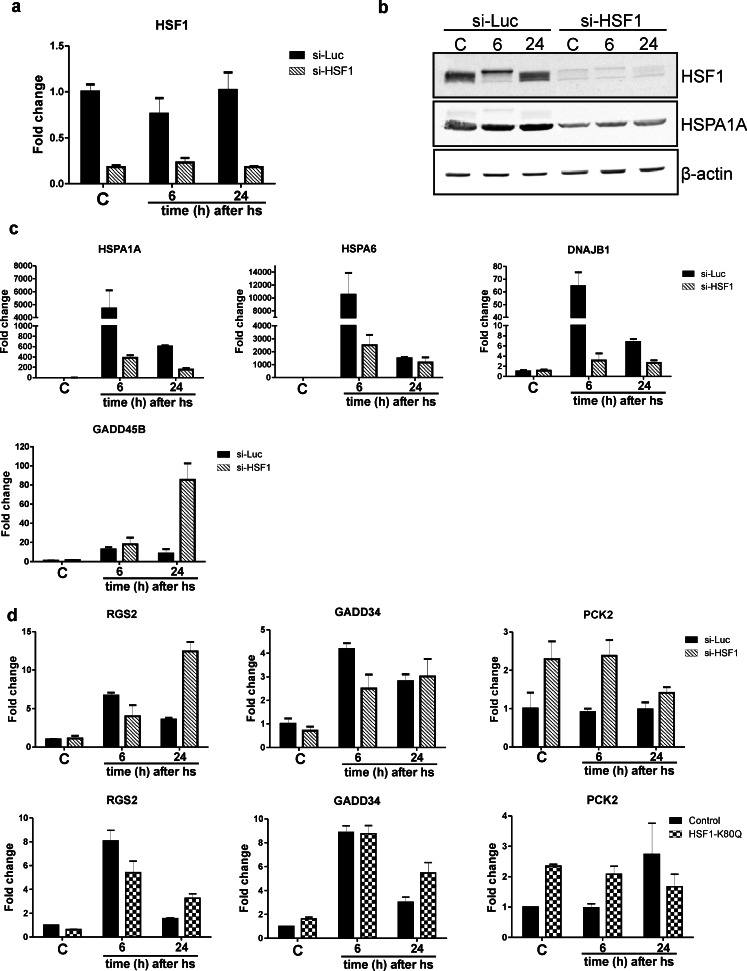
Comparison of transcriptome changes in the presence of HSF1 K80Q or siRNA HSF1
We demonstrated above (Fig. 5) that the transcript levels of the HSPA6 gene increase in HSF1 K80Q cells but not in dnHSF1 cells 24 h after heat stress. This suggests that the HSE must be free of HSF1 to allow access of other transcription factors to this promoter. To determine if this is indeed the case, we treated HEK293 cells with HSF1 siRNA, which resulted in efficient knockdown of HSF1 (Fig. 11a–b). The lack of HSF1 activity in siRNA-treated cells was also evident 6 h after heat shock: the increase in the HSPA1A, HSPA6 and DNAJB1 transcript levels was strongly inhibited (Fig. 11c). Surprisingly, we saw no increase in the levels of these transcripts 24 h after heat shock. In fact, these were even lower 24 h after heat shock than 6 h after heat shock. HSF1 siRNA-treated cells thus did not show the delayed response of the HSPA1A and HSPA6 genes seen in HSF1 K80Q cells. These data suggest that the delayed response of these HSF1 target genes somehow requires the HSF1 protein.
Fig. 11.
Relative changes in transcript levels of various genes in stressed and non-stressed HSF1 siRNA-treated cells. a HEK293 cells were transfected with siRNA against luciferase or HSF1 for 96 h to decrease HSF1 levels. Cells were exposed to a heat shock for 30 min at 45 °C or left at 37 °C (C). When heat shocked, cells were allowed to recover for the indicated time before harvesting. Total RNA was isolated and HSF1 transcript levels relative to GAPDH mRNA levels were measured by QPCR. The fold induction of mRNA levels is plotted relative to the level in non-stressed control cells. b HSF1 siRNA-treated cells were harvested and lysates were subjected to SDS-PAGE and Western blot analysis using antibodies against the indicated proteins. c–e mRNA levels were determined by QPCR analysis and are shown relative to GAPDH mRNA levels. The fold induction of mRNA levels is plotted relative to the level in non-stressed control cells. Bars indicate SD
Next, we investigated whether the HSF1-independent stress response (see Table 2 and Table S3) differed between HSF1 siRNA and HSF1 K80Q expressing cells. For the RGS2 (regulator of G-protein signaling 2) and PPP1R15A (GADD34) genes, the response was similar, although quantitatively somewhat larger for the PPP1R15A (GADD34) gene in HSF1 K80Q cells (Fig. 11d; for the response in dnHSF1 cells, see Fig. S2b). Finally, we determined whether treatment with HSF1 siRNA also caused an increase in the level of the PCK2 (mitochondrial phosphoenolpyruvate carboxykinase 2) in unstressed HEK293 cells, just as expression of HSF1 K80Q did. The results in Fig. 11d demonstrate that PCK2 transcript levels indeed increased in HSF1 siRNA-treated cells. In conclusion, assaying an, admittedly limited, number of transcripts showed no difference between lack of HSF1 and expression of an non-DNA binding HSF1 mutant with the important exception of the delayed stress response seen 24 h after heat shock.
Discussion
HSF1 was originally discovered as the transcription factor required for the synthesis of additional cytoplasmic chaperones, the heat shock proteins, during the proteotoxic stress response. Later HSF1 was also shown to have a physiological role in non-stressed cells, for example the circadian clock gene Per2 is an HSF1 target (Kornmann et al. 2007; Tamaru et al. 2011). Tumor cells rely on active HSF1 (Mendillo et al. 2012) and blocking HSF1 activity in HeLa cells causes cell death (Dai et al. 2007). HEK293 cells are less sensitive to a lack of HSF1 activity (Dai et al. 2007), possibly because the HSPA1A promoter is activated by E1A (Wu et al. 1986), the transforming protein in HEK293 cells. Accordingly, blocking HSF1 activity in non-stressed HEK293 cells by overexpression of the non-DNA binding HSF1 K80Q mutant led to only a limited change in the transcriptome — a change that is apparently mostly cell-specific as judged from a comparison with the published data for the effect of depletion of HSF1 by siRNA in HeLa cells (Page et al. 2006). None of the canonical HSF1 target genes appear to rely on HSF1 activity for expression in non-stressed HEK293 cells as for none of these genes the transcript level decreased more than 2-fold (see also Table S2). Yet expression of HSF1 K80Q does affect the chaperoning capacity of the cells adversely as judged from the luciferase refolding assays (Fig. 1c). It is possible that overexpression of HSF1 does sequester cytoplasmic chaperones. As expected, HSF1 plays a major role in the response of HEK293 cells to proteotoxic stress and regulates about 30 % of genes of which the transcript level increases. The effect of a lack of HSF1 activity is most apparent in cells that had been allowed to recover from a heat stress for 24 h. In control cells the transcriptome has largely returned to normal, while in HSF1 K80Q and particularly in dnHSF1 cells the transcriptome has diverged even further from the non-stressed transcriptome than in cells 6 h after heat shock (Fig. 3). HSF1-directed chaperone synthesis is thus required for heat-shocked cells to regain homeostasis. The lack of chaperone synthesis in a cell expressing the HSF1 K80Q mutant is reflected in an extended primary stress response, the continued high level of stress induced transcripts. The simplest model is that the activity of some heat stress activated transcription factor(s) remains high in HSF1 K80Q cells but decays in control cells. Our results do not support such a model, at least as judged from the changes in the transcript levels of the GADD45B and PPP1R15A genes. We could block the increase in level of these transcripts in HSF1 K80Q cells 24 h after heat shock but not the increase in level 6 h after heat shock by treatment with an EEF1D_L siRNA. Different factor(s) must thus be involved in maintaining these transcript levels in cells 6 and 24 h after heat stress. Which factors regulate transcription of the GADD45B or the PPP1R15A genes in cells either 6 or 24 h after heat stress is not clear. The results reported above show that it is not ATF4, c-Fos, FosB or one of the suggested partners of EEF1D_L, HSF1 or NRF2. In cells 24 h after heat stress it must be either an as yet unidentified partner of EEF1D_L or a factor encoded by a transcript that is also targeted by the EEF1D_L siRNA. Unfortunately, in our hands other EEF1D_L siRNAs were not effective, so we cannot distinguish between these two possibilities. The transcription factor(s) involved in driving HSF1 independent transcription 6 h after heat stress also remain to be identified. In the case of the PPP1R15A gene, we had assumed this to be ATF4, but our results show that that is not the case. The GADD45B promoter has been shown to be targeted by NFY, Sp1, and Egr1 (Zumbrun et al. 2009). We have not examined whether any of these factors play a role in the extended primary stress response of HSF1 K80Q cells.
Next to the extended primary stress response, HSF1 K80Q cells mount what we have called a delayed stress response, i.e., an increase in transcript level of HSF1 target genes 24 h after heat shock. Curiously, this response does require HSF1 protein as we could not detect it in HSF1 siRNA-treated cells. Our data further show that NRF2 is involved in this delayed stress response. Previously, it has been reported that murine heat shock protein mRNA levels (i.e., Hspb1, Dnajb1 and Hsp90) are increased in an NRF2-dependent manner upon NRF2-activating compounds (Hu et al. 2006a, b). In addition, an NRF2 binding element has been found in the promoter of the Hsp70 gene in zebrafish and this element was conserved between mouse and zebrafish (Almeida et al. 2010). Finally, mammalian heat shock genes have been reported to be enriched in AP1/NRF2/Fos binding sites (Mendillo et al. 2012). The activity of NRF2 is regulated post-translationally. Under non-stress conditions the cysteine rich protein KEAP1 (Kelch-like ECH associated protein 1) retains NRF2 in the cytoplasm (Itoh et al. 1999) and maintains it at a low level through KEAP1-dependent ubiquitination and proteasomal degradation (Cullinan et al. 2004; Itoh et al. 2003; McMahon et al. 2003; Zhang and Hannink 2003; Zhang et al. 2004). Upon oxidative stress, cysteines in the KEAP1 protein are oxidatively modified, resulting in a conformational change and release of NRF2 (Itoh et al. 2003). NRF2 then binds to the ARE usually as a heterodimer with one of the small Maf proteins (for reviews, see Kannan et al. 2012; Sykiotis and Bohmann 2010; Eychene et al. 2008). The data reported above show that the formation of an ARE binding complex is delayed in HSF1 K80Q cells. Apparently, the formation of active NRF2 in cells recovering from heat stress for 6 h requires HSF1 or an HSF1 regulated function. The difference in mobility of the ARE binding complex formed in extracts of control cells 6 h after heat stress and that in extracts of HSF1 K80Q cells 24 h after heat stress suggests that it is the heteromeric partner of NFR2 that differs, for example because a different small Maf protein is involved. In our microarray analysis, we found no significant changes in the levels of the mRNAs encoding the small Maf proteins upon heat stress or upon expression of HSF1 K80Q. However, small Mafs can be (in)activated by post-translational modification (Kannan et al. 2012), and such a modification could be affected by heat stress and HSF1 activity. Our data thus suggest that the delayed stress response in HSF1 K80Q cells is a delayed antioxidant response and point to an interaction between HSF1 or an HSF1 regulated function and a partner of NRF2 in the timing of the antioxidant response. Aging cells are not only impaired in protein homeostasis but also in redox homeostasis. Our results imply that loss of HSF1 activity may be the common cause. Our results also support NRF2 as a promising target in treating age-related disease: it would not only redress the redox balance, but by enhancing the transcript level of at least some HSF1 target genes, also boost resistance to proteotoxic stress.
Electronic supplementary material
(DOCX 853 kb)
Acknowledgments
We thank Dr. A. Zantema for the HSPB1 antibody. We thank the microarray facility at the VU UMC (Amsterdam, The Netherlands) for performing the microarray experiments. This work was financially supported by AgentschapNL (project numbers IGE03018 and IGE07004, www.agentschapnl.nl).
Abbreviations
- HSP
Heat shock protein
- HSF1
Heat shock factor 1
- HSE
Heat shock element
- MEF
Mouse embryonic fibroblast
- NRF2
Nuclear factor (erythroid-derived 2)-related factor 2
- ARE
Antioxidant response element
- ATF
Activating transcription factor
- KEAP1
Kelch-like ECH associated protein 1
References
- Almeida DV, Nornberg BF, Geracitano LA, Barros DM, Monserrat JM, Marins LF. Induction of phase II enzymes and hsp70 genes by copper sulfate through the electrophile-responsive element (EpRE): insights obtained from a transgenic zebrafish model carrying an orthologous EpRE sequence of mammalian origin. Fish Physiol Biochem. 2010;36(3):347–353. doi: 10.1007/s10695-008-9299-x. [DOI] [PubMed] [Google Scholar]
- Ambra R, Mocchegiani E, Giacconi R, Canali R, Rinna A, Malavolta M, Virgili F. Characterization of the hsp70 response in lymphoblasts from aged and centenarian subjects and differential effects of in vitro zinc supplementation. Exp Gerontol. 2004;39(10):1475–1484. doi: 10.1016/j.exger.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Anckar J, Sistonen L. Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu Rev Biochem. 2011;80:1089–1115. doi: 10.1146/annurev-biochem-060809-095203. [DOI] [PubMed] [Google Scholar]
- Bailey CK, Andriola IF, Kampinga HH, Merry DE. Molecular chaperones enhance the degradation of expanded polyglutamine repeat androgen receptor in a cellular model of spinal and bulbar muscular atrophy. Hum Mol Genet. 2002;11(5):515–523. doi: 10.1093/hmg/11.5.515. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319(5865):916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583(16):2647–2653. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Calderwood SK, Murshid A, Prince T. The shock of aging: molecular chaperones and the heat shock response in longevity and aging—a mini-review. Gerontology. 2009;55(5):550–558. doi: 10.1159/000225957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24(19):8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J. 2007;26(4):944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eychene A, Rocques N, Pouponnot C. A new MAFia in cancer. Nat Rev Cancer. 2008;8(9):683–693. doi: 10.1038/nrc2460. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Nakai A. The heat shock factor family and adaptation to proteotoxic stress. FEBS J. 2010;277(20):4112–4125. doi: 10.1111/j.1742-4658.2010.07827.x. [DOI] [PubMed] [Google Scholar]
- Fujimoto M, Takaki E, Hayashi T, Kitaura Y, Tanaka Y, Inouye S, Nakai A. Active HSF1 significantly suppresses polyglutamine aggregate formation in cellular and mouse models. J Biol Chem. 2005;280(41):34908–34916. doi: 10.1074/jbc.M506288200. [DOI] [PubMed] [Google Scholar]
- Guo Y, Guettouche T, Fenna M, Boellmann F, Pratt WB, Toft DO, Smith DF, Voellmy R. Evidence for a mechanism of repression of heat shock factor 1 transcriptional activity by a multichaperone complex. J Biol Chem. 2001;276(49):45791–45799. doi: 10.1074/jbc.M105931200. [DOI] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475(7356):324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Hayashida N, Fujimoto M, Tan K, Prakasam R, Shinkawa T, Li L, Ichikawa H, Takii R, Nakai A. Heat shock factor 1 ameliorates proteotoxicity in cooperation with the transcription factor NFAT. EMBO J. 2010;29(20):3459–3469. doi: 10.1038/emboj.2010.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldens L, Dirks RP, Hensen SM, Onnekink C, van Genesen ST, Rustenburg F, Lubsen NH. Co-chaperones are limiting in a depleted chaperone network. Cell Mol Life Sci. 2010;67(23):4035–4048. doi: 10.1007/s00018-010-0430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldens L, van Genesen ST, Hanssen LL, Hageman J, Kampinga HH, Lubsen NH. Protein refolding in peroxisomes is dependent upon an HSF1-regulated function. Cell Stress Chaperones. 2012;17(5):603–613. doi: 10.1007/s12192-012-0335-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensen SM, Heldens L, van Enckevort CM, van Genesen ST, Pruijn GJ, Lubsen NH. Heat shock factor 1 is inactivated by amino acid deprivation. Cell Stress Chaperones. 2012;17(6):743–755. doi: 10.1007/s12192-012-0347-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300(5622):1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Gene expression profiles induced by cancer chemopreventive isothiocyanate sulforaphane in the liver of C57BL/6J mice and C57BL/6J/Nrf2 (−/−) mice. Cancer Lett. 2006;243(2):170–192. doi: 10.1016/j.canlet.2005.11.050. [DOI] [PubMed] [Google Scholar]
- Hu R, Xu C, Shen G, Jain MR, Khor TO, Gopalkrishnan A, Lin W, Reddy B, Chan JY, Kong AN. Identification of Nrf2-regulated genes induced by chemopreventive isothiocyanate PEITC by oligonucleotide microarray. Life Sci. 2006;79(20):1944–1955. doi: 10.1016/j.lfs.2006.06.019. [DOI] [PubMed] [Google Scholar]
- Inouye S, Izu H, Takaki E, Suzuki H, Shirai M, Yokota Y, Ichikawa H, Fujimoto M, Nakai A. Impaired IgG production in mice deficient for heat shock transcription factor 1. J Biol Chem. 2004;279(37):38701–38709. doi: 10.1074/jbc.M405986200. [DOI] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh K, Wakabayashi N, Katoh Y, Ishii T, O'Connor T, Yamamoto M. Keap1 regulates both cytoplasmic-nuclear shuttling and degradation of Nrf2 in response to electrophiles. Genes Cells. 2003;8(4):379–391. doi: 10.1046/j.1365-2443.2003.00640.x. [DOI] [PubMed] [Google Scholar]
- Jurivich DA, Choo M, Welk J, Qiu L, Han K, Zhou X. Human aging alters the first phase of the molecular response to stress in T-cells. Exp Gerontol. 2005;40(12):948–958. doi: 10.1016/j.exger.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Kaitsuka T, Tomizawa K, Matsushita M. Transformation of eEF1Bdelta into heat-shock response transcription factor by alternative splicing. EMBO Rep. 2011;12(7):673–681. doi: 10.1038/embor.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan MB, Solovieva V, Blank V. The small MAF transcription factors MAFF, MAFG and MAFK: current knowledge and perspectives. Biochim Biophys Acta. 2012;1823(10):1841–1846. doi: 10.1016/j.bbamcr.2012.06.012. [DOI] [PubMed] [Google Scholar]
- Kim G, Meriin AB, Gabai VL, Christians E, Benjamin I, Wilson A, Wolozin B, Sherman MY. The heat shock transcription factor Hsf1 is downregulated in DNA damage-associated senescence, contributing to the maintenance of senescence phenotype. Aging Cell. 2012;11(4):617–627. doi: 10.1111/j.1474-9726.2012.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klok EJ, van Genesen ST, Civil A, Schoenmakers JG, Lubsen NH. Regulation of expression within a gene family. The case of the rat gammaB- and gammaD-crystallin promoters. J Biol Chem. 1998;273(27):17206–17215. doi: 10.1074/jbc.273.27.17206. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5(2):e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Liu DJ, Lu J, Chen KY, Liu AY. Aberrant regulation and modification of heat shock factor 1 in senescent human diploid fibroblasts. J Cell Biochem. 2009;106(2):267–278. doi: 10.1002/jcb.21997. [DOI] [PubMed] [Google Scholar]
- McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- Mendillo ML, Santagata S, Koeva M, Bell GW, Hu R, Tamimi RM, Fraenkel E, Ince TA, Whitesell L, Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell. 2012;150(3):549–562. doi: 10.1016/j.cell.2012.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer. 2005;41(16):2449–2461. doi: 10.1016/j.ejca.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Morimoto RI. Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 2008;22(11):1427–1438. doi: 10.1101/gad.1657108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namba T, Homan T, Nishimura T, Mima S, Hoshino T, Mizushima T. Up-regulation of S100P expression by non-steroidal anti-inflammatory drugs and its role in anti-tumorigenic effects. J Biol Chem. 2009;284(7):4158–4167. doi: 10.1074/jbc.M806051200. [DOI] [PubMed] [Google Scholar]
- Page TJ, Sikder D, Yang L, Pluta L, Wolfinger RD, Kodadek T, Thomas RS. Genome-wide analysis of human HSF1 signaling reveals a transcriptional program linked to cellular adaptation and survival. Mol Biosyst. 2006;2(12):627–639. doi: 10.1039/b606129j. [DOI] [PubMed] [Google Scholar]
- Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem J. 2007;401(1):299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piechaczyk M, Blanchard JM. c-fos proto-oncogene regulation and function. Crit Rev Oncol Hematol. 1994;17(2):93–131. doi: 10.1016/1040-8428(94)90021-3. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8(7):519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97(11):5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12(5):654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slavotinek A, Biesecker LG. Genetic modifiers in human development and malformation syndromes, including chaperone proteins. Hum Mol Genet. 2003;12(Spec No 1):R45–R50. doi: 10.1093/hmg/ddg099. [DOI] [PubMed] [Google Scholar]
- Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, Kennedy BK, Kaeberlein M. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell. 2008;7(3):394–404. doi: 10.1111/j.1474-9726.2008.00385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sykiotis GP, Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci Signal. 2010;3(112):re3. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru T, Hattori M, Honda K, Benjamin I, Ozawa T, Takamatsu K. Synchronization of circadian Per2 rhythms and HSF1-BMAL1:CLOCK interaction in mouse fibroblasts after short-term heat shock pulse. PLoS One. 2011;6(9):e24521. doi: 10.1371/journal.pone.0024521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiaville MM, Dudenhausen EE, Zhong C, Pan YX, Kilberg MS. Deprivation of protein or amino acid induces C/EBPbeta synthesis and binding to amino acid response elements, but its action is not an absolute requirement for enhanced transcription. Biochem J. 2008;410(3):473–484. doi: 10.1042/BJ20071252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323(5917):1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu BJ, Kingston RE, Morimoto RI. Human HSP70 promoter contains at least two distinct regulatory domains. Proc Natl Acad Sci U S A. 1986;83(3):629–633. doi: 10.1073/pnas.83.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin C, Xi L, Wang X, Eapen M, Kukreja RC. Silencing heat shock factor 1 by small interfering RNA abrogates heat shock-induced cardioprotection against ischemia-reperfusion injury in mice. J Mol Cell Cardiol. 2005;39(4):681–689. doi: 10.1016/j.yjmcc.2005.06.005. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23(22):8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Huang L, Zhang J, Moskophidis D, Mivechi NF. Targeted disruption of hsf1 leads to lack of thermotolerance and defines tissue-specific regulation for stress-inducible Hsp molecular chaperones. J Cell Biochem. 2002;86(2):376–393. doi: 10.1002/jcb.10232. [DOI] [PubMed] [Google Scholar]
- Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24(24):10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R. Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell. 1998;94(4):471–480. doi: 10.1016/S0092-8674(00)81588-3. [DOI] [PubMed] [Google Scholar]
- Zumbrun SD, Hoffman B, Liebermann DA. Distinct mechanisms are utilized to induce stress sensor gadd45b by different stress stimuli. J Cell Biochem. 2009;108(5):1220–1231. doi: 10.1002/jcb.22354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 853 kb)