Abstract
Cognitive aging processes are underpinned by multiple processes including genetic factors. The brain-derived neurotrophic factor (BDNF) has been suggested to be involved in age-related cognitive decline in otherwise healthy individuals. The gender-specific role of the BDNF gene in cognitive aging remains unclear. The identification of genetic biomarkers might be a useful approach to identify individuals at risk of cognitive decline during healthy aging processes. The aim of this study was to investigate the associations between three single-nucleotide polymorphisms (SNPs) in the BDNF gene and domains of cognitive functioning in normal cognitive aging. The sample, comprising 369 participants (M = 72.7 years, SD = 4.45 years), completed an extensive neuropsychological test battery measuring memory, motor function, and perceptual speed. The relationships between the SNPs rs6265, rs7103411, and rs7124442 and cognitive domains were examined. While significant main effects of BDNF SNPs on cognitive function were found for the association between rs7103411 and memory performance, gender-specific analyses revealed for females significant main effects of rs7103411 for memory and of rs6265 for perceptual speed independent of the APOE*E4 status and education. The finding for the association between rs6265 and perceptual speed in females remained significant after Bonferroni correction for multiple comparisons. None of the analyses showed significant results for males. This study is the first to implicate that the SNPs rs6265 and rs7103411 affect cognitive function in the elderly in a gender-specific way.
Keywords: BDNF, BDNF gene, Memory, Motor function, Perceptual speed, Elderly, Gender
Introduction
Many healthy aging individuals exhibit a decline in cognitive function, including memory, motor function, and perceptual speed. Research has implicated one of the neurotrophic factors, brain-derived neurotrophic factor (BDNF), to be involved in age-related cognitive decline. A hypothesis has emerged that aging is associated with a decreased BDNF signalling capacity in the CNS (Mattson et al. 2004). Age-related impaired cognitive function might therefore reflect decreases in BDNF in subregions of the brain that are primarily affected by age-related diseases. Studies of higher-cognitive-functioning animals including macaque monkeys (Hayashi et al. 1997, 2001) support this hypothesis. In humans, BDNF levels have been shown to decrease in brain regions involved in age-related neurodegenerative diseases: in Alzheimer’s disease in the frontal cortex (Ferrer et al. (1999), the hippocampus (Phillips et al. 1991), the parietal cortex (Hock et al. 2000), and the entorhinal cortex (Narisawa-Saito et al. 1996). However, there are fewer data on the effects of BDNF levels in elderly humans during normal cognitive aging. Some authors found that elderly females (but not elderly males) with poorer cognitive performance had lower BDNF levels than better performers (Komulainen et al. 2008).
Long-term potentiation (LTP) is evident in the hippocampus (Lynch 2004) and has been proposed as a cellular mechanism for memory (Morris 1989; Tang et al. 2001). LTP has been observed as being directly related to BDNF levels in BDNF manipulated mice (Korte et al. 1995, 1996, 1998) with progressive decrease with genotype: wild-type mice, heterozygous mice, and homozygous mice (Korte et al. 1995). Linnarsson et al. (1997) found a similar relationship in Morris water maze learning capacity of wild-type mice versus BDNF mutant mice.
The BDNF gene encodes a precursor peptide that is cleaved to form the mature BDNF protein (Seidah et al. 1996, as cited in Egan et al. 2003). The BDNF gene, located on chromosome 11 (Jones and Reichardt 1990), comprises many single-nucleotide polymorphisms (SNPs). Research focusing on BDNF SNPs suggests an association between BDNF genotype and BDNF protein expression. Egan et al. (2003) found that hippocampal neurons of individuals exhibiting the minor allele in the BDNF SNP rs6265 (val66met) had a reduced number of granules, suggesting that the minor allele in this SNP may lead to impaired regulated secretion of the BDNF protein. Human studies have also focused on rs6265 and possible associations with hippocampal function and memory. Hariri et al. (2003) found that, during encoding and retrieval, major allele participants had increased memory-related hippocampal activity, compared with met-BDNF carriers. Moreover, in healthy adults, the met allele was associated with poorer processing speed and memory performance (Miyajima et al. 2008; Raz et al. 2009). Research also implicates the met-BDNF allele in working (short-term) memory impairments in bipolar patients (Rybakowski et al. 2003, 2006). Recently, it was suggested that the presence of the met-BDNF allele, particularly in association with apolipoprotein E (APOE*E4), may predict a worse cognitive outcome in patients with mild cognitive impairment (MCI; Forlenza et al. 2010) and may negatively impact on cognitive function in Parkinson’s disease (Guerini et al. 2009). In contrast, however, studies on patients with other diseases suggest that the met allele in the rs6265 SNP may play a protective role against the decline of processing speed function. Met-BDNF carriers with systemic lupus erythematosus had significantly better processing speed, executive function, and motor function performance (Oroszi et al. 2006), and those with multiple sclerosis had better processing speed and divided attention performance (Zivadinov et al. 2007).
In addition to memory, the BDNF protein and BDNF gene have been implicated in other age-sensitive cognitive functions, including motor function. Parkinson’s disease (PD) patients had a weaker expression of BDNF mRNA as compared with their normal counterparts (Howells et al. 2000) and protein (Parain et al. 1999) in substantia nigra neurons. Additionally, rs6265 met-BDNF carriers with PD versus major allele patients exhibited better cognitive performance in the Tower of London test (Foltynie et al. 2005) and showed a significantly higher risk of developing levodopa-induced dyskinesia (Foltynie et al. 2009), a disorder involving motor dysfunction.
The research on memory, motor function, and processing speed demonstrates (a) a reduced expression of the BDNF protein and mRNA in humans with impaired functioning in these age-sensitive cognitive domains and (b) effects of the BDNF genotype on cognitive performance. This supports the hypothesis that decreased expression of BDNF may reflect age-sensitive impaired cognitive function. Better cognitive performance in major allele participants may reflect a healthy secretion of BDNF protein, versus impaired secretion in minor allele carriers.
This study aimed to replicate and strengthen previous studies, by supporting the hypothesis that cognitive performance (specifically memory, motor function, and perceptual speed) associates with BDNF SNPs. We focused on the rs6265 SNP, as well as BDNF SNPs rs7103411 and rs7124442, selected for their involvement in previously shown associations between BDNF and age-sensitive cognitive functioning (Miyajima et al. 2008; Xiromerisiou et al. 2007; Huang et al. 2007).
The overall aim was to better understand the role of the BDNF gene in domain-specific cognitive functioning and to apply this knowledge to a model of cognitive functioning in the elderly general population.
It was hypothesised that three BDNF SNPs, rs6265, rs7103411, and rs7124442, would associate with performance in the three cognitive domains (a) memory, (b) motor function, and (c) perceptual speed, and that participants exhibiting the major allele genotype would have better cognitive performance, followed by the heterozygous and homozygous genotypes, respectively.
Materials and method
Sample
The sample comprised participants who were originally part of the 1989/1990 WHO MONICA Survey Augsburg, Germany (“Monitoring trends and determinants in cardiovascular disease”; Keil et al. 1998). Participants of the MONICA Survey aged 65 years and older were re-contacted as part of a follow-up study, the “Memory and Morbidity in Augsburg Elderly” (MEMO-Study; Schmidt et al. 2004). The principal aim of the MEMO-Study was to investigate the risk factors, notably cognitive and cardiovascular function, of neurodegenerative diseases in an elderly cohort. Of those re-contacted, 60.6% responded for the MEMO-Study, resulting in a total of 385 participants (M = 72.7 years, SD = 4.45 years). The data presented in this analysis are based on N = 369 participants with complete data and genetic analyses. Approval for the study was given by the ethics committee at the University of Muenster, Germany. Approval for the genetic analysis of the MEMO-Study DNA was given by James Cook University, Townsville, Australia. All participants gave written informed consent. All tests were conducted in the study centre under controlled conditions.
Measures
Cognitive test battery
The cognitive test battery comprised several individual tests that measured global cognitive function, processing speed, attention, motor function, and short- and long-term memory (Nilsson et al. 2005).
Processing speed and attention were assessed by two traditional tasks. The first was the Stroop test (Stroop 1935) which consists of three subtests that assess word reading, colour naming, and interference performance. The Stroop test measures how easily a person can adjust their perceptual set in accordance with changing demands. More time required to complete the Stroop test is associated with poorer cognitive performance. An interference score was determined by subtracting the word reading (Stroop Subtest I) response time from the colour-word series (Stroop Subtest III) response time. Previous research has found that age does not usually affect performance in word reading and may only yield a slight decline in performance in colour naming. However, a dramatic age decline is seen in interference performance (Houx et al.1993, as cited in Nilsson et al. 2005).
The second task used to assess cognitive speed and attention was the Letter Digit Substitution Test (Salthouse 1978). This test is a revised version of the digit symbol test used in the Wechsler Adult Intelligence Sales (WAIS) battery (Wechsler 1981, as cited in Nilsson et al. 2005). A lower score on this test represents poorer cognitive performance.
The Purdue Pegboard test (Tiffin 1948; Costa et al. 1963; Desrosiers et al. 1995), a motor function test, was included to assess gross and fine motor coordination, and processing function. Participants were required to place pins in holes as rapidly as possible during three 30-s trials. During the first trial, they used their right hand, in the second trial, their left hand, and in the last trial, both hands simultaneously. A score on this test is the sum of pins correctly inserted by each single hand and the number of pairs of pins correctly inserted using both hands. A lower score reflects poorer motor speed.
Short- and long-term memories were assessed using three word recall tests, originally used in the Betula Study (Nilsson et al. 2005). In each word list, 12 words were presented on a tape recorder. The words were carefully chosen (a) based on their frequencies of use in the spoken language, and (b) to be as similar as possible with respect to word length. In Word List 1 (WL1), the words were presented every 2 s with no delay. Participants were then instructed to freely recall as many of the presented words as possible. All three word lists assess memory through recall of studied events, but WL1 also served as a baseline to which performance in the other two lists can be compared. In Word List 2, a different list of words was presented at half the speed (every 4 s), thus manipulating the speed of encoding. In Word List 3, the words were presented every 2 s and were presented simultaneously with a distractor task, in which participants were required to sort a number of cards into two piles according to colour. Due to the divided attention aspect of this word list, it is also a measure of working memory and executive functioning. The number of recalled words was assessed in all three lists. A higher score represents better memory performance.
The production component of semantic memory was also assessed using a category Word Fluency test (Beckman and Nilsson 1996; Nyberg et al. 2003). In this task, participants were asked to name as many animals as possible in a 1-min period. A lower score in this test reflects poorer cognitive performance.
Principal component analysis
All individual cognitive and motor test scores were transformed to standardised Z-scores in a precursor study (Baune et al. 2006). A principal component analysis using a Varimax rotation was carried out on these Z-scores, revealing three main factors. All test variables with a factor loading of 0.4 or greater were considered significant contributors to one of these factors. The first factor was labelled ‘memory performance’ and included the number of correctly recalled words from Word Lists 1–3 (factor loadings, 0.52–0.70) plus the total number of animal names recalled in the word fluency test (factor loading, 0.50). The second factor was labelled ‘perceptual speed’ and included the difference in time needed to complete Stroop Subtest III versus Subtest I (factor loading 0.74), plus the number of correct cells in the Letter Digit Substitution Test (factor loading 0.47). The third factor was labelled ‘motor function’ and included the sum of pegs sorted in the Purdue Pegboard Test for the right, left, and both hands (factor loadings 0.75–0.76).
Genotyping
Three BDNF SNPs, including the functional val66met polymorphism (rs6265) (position: chr11:27,636,492) previously investigated in relation to cognitive function, rs7124442 (position: chr11:27,633,617) and which has shown to be functionally related to BDNF plasma levels in eating disorder subjects (Mercader et al. 2007), as well as an intronic SNP rs7103411 (position: chr11:27,656,701), whose potential function for cognition has not been described yet, were investigated.
Genotyping of the three SNPs was carried out following published protocols for the multiplex genotyping assay iPLEX™ for use with the MassARRAY platform (Oeth et al. 2007). The genotyping completion rate was 98% for rs6265 (N = 360/369), 97% for rs7103411 (N = 356/369), and 93% for rs7124442 (N = 344/369) due to genotyping errors. Genotypes were determined by investigators blinded to the purposes of this study. Genotype determination of the APOE*E4 gene was carried out using a multi-locus assay (Cheng et al. 1999) developed by Roche Molecular Systems Alameda, USA (see details in Baune et al. 2008).
Linkage disequilibrium was moderately high between SNPs rs6265 and rs7103411 (D′ = 0.968, r2 = 0.762) and low between SNPs rs6265 and rs7124442 (D′ = 0.936, r2 = 0.078) and SNPs rs7103411 and rs7124442 (D′ = 0.906, r2 = 0.088; haploview 4.2).
Statistical analyses
Hardy–Weinberg equilibrium was examined using the programme Finetti provided as an online source (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl; Wienker TF & Strom TM). Differences between men and women in categorical variables were tested by chi-square test or Fisher’s exact test if less than five participants were in any group. Linkage disequilibrium for the three BDNF SNPs (D′ and r2) was calculated using Haploview 4.2. Differences in performance in the three cognitive domains (motor function, memory, and perceptual speed) across genotypes were tested for each of the three SNPs using analysis of covariance (ANCOVA). Age, gender, and education were considered as covariates in these analyses.
The effects of the variable ‘gender’ in previous related research (Foltynie et al. 2005; Raz et al. 2009; Komulainen et al. 2008) provided a rationale for entering this variable as a covariate in the present study. Age and education (measured as number of years of education) were entered as covariates because they significantly correlated to the three cognitive domains (Z-scores). We also applied multivariable linear regression analyses to evaluate if differences, between genotypes of a specific SNP follow a linear trend. In these models, the respective cognitive domain score was the dependent variable and age, gender, education, and a categorical variable for genotype (coded 0 for major allele, 1 for heterozygous, and 2 for minor allele genotype) were the independent variables. The models were performed for each domain and each of the three SNPs. All ANCOVAs and multivariable linear regression analyses were also carried out in gender-stratified groups, in order to reveal possible gender differences. In these analyses, the variable gender was not included in the models.
Additional sensitivity analyses considered APOE*E4 status as covariate together with gender and educational years. Furthermore, since a strong correlation between cognitive ability and length of education has been described previously, education was withdrawn as a covariate from the linear regression models during sensitivity analyses. Statistical analyses were performed using SPSS software (version 17.0). Bonferroni correction for multiple comparisons was calculated for three SNPs and three cognitive tests leading to a corrected p value of p = 0.006.
Power calculations
Power calculations were carried out to determine the minimal effect size this study could detect for effects of BDNF SNP on cognitive function in the whole sample and in gender-stratified subgroups. In the whole sample, according to genotyping rates per SNP of N = 360 (rs6265), N = 356 (rs7103411), and N = 343 (rs7124442) and an assumed power = 0.8, α err prob = 0.05 and three genotype groups, it was possible to detect minimal effect sizes of f = 0.165 for rs6265, f = 0.166 for rs7103411, and f = 0.169 for rs7124442. Assuming the same power of 0.8, α err prob = 0.05 and three genotype groups, gender-stratified power calculations indicated SNP-specific minimal detectable effect sizes f for men (f = 0.23 for rs6265; f = 0.228 for rs7103411; f = 0.23 for rs7124442) and women (f = 0.24 for rs6265; f = 0.245 for rs7103411; f = 0.25 for rs7124442).
Partial η2 from ANOVA analyses were utilised to calculate empirical effect sizes as presented in Table 2. Comparisons between minimal detectable and empirically observed effect sizes were made for data interpretation. Power calculations with G*Power version 3.1 were carried using the statistical test ANOVA (fixed effects, one-way).
Table 2.
Results of one-way ANCOVAs and linear regression analyses corrected for age and education in gender-stratified groups
| ANCOVA | Linear regression model | |||||||
|---|---|---|---|---|---|---|---|---|
| F value | p Value | η2 value | B value | CI (95%) | p Value | |||
| Lower bound | Upper bound | |||||||
| rs6265; n = 360 (98%); males, n = 189; females, n = 171 | ||||||||
| Motor function | Males | 2.126 | 0.122 | 0.024 | 0.244 | −0.384 | 0.872 | 0.444 |
| Females | 1.213 | 0.243 | 0.003 | 0.185 | −0.418 | 0.789 | 0.545 | |
| Memory | Males | 0.981 | 0.377 | 0.011 | −0.195 | −0.926 | 0.537 | 0.600 |
| Females | 2.472 | 0.088 | 0.030 | −0.778 | −1.470 | −0.086 | 0.028 | |
| Perceptual speed | Males | 0.953 | 0.387 | 0.011 | 0.234 | −0.182 | 0.651 | 0.269 |
| Females | 5.226 | 0.006 | 0.061 | −0.622 | −1.056 | −0.188 | 0.005 | |
| rs7103411; n = 356 (97%); males, n = 190; females, n = 166 | ||||||||
| Motor function | Males | 0.211 | 0.810 | 0.002 | −0.007 | −0.599 | 0.585 | 0.982 |
| Females | 0.033 | 0.968 | <0.001 | −0.030 | −0.642 | 0.582 | 0.923 | |
| Memory | Males | 0.825 | 0.440 | 0.009 | −0.221 | −0.871 | −0.429 | 0.503 |
| Females | 5.103 | 0.007 | 0.063 | −0.981 | −1.600 | −0.217 | 0.010 | |
| Perceptual speed | Males | 0.473 | 0.624 | 0.005 | 0.183 | −0.197 | 0.563 | 0.344 |
| Females | 3.377 | 0.037 | 0.042 | −0.581 | −1.022 | −0.141 | 0.010 | |
| rs7124442; n = 343 (93%); males, n = 185; females, n = 158 | ||||||||
| Motor function | Males | 0.610 | 0.544 | 0.007 | −0.297 | −0.826 | 0.232 | 0.270 |
| Females | 0.572 | 0.565 | 0.008 | 0.268 | −0.320 | 0.856 | 0.368 | |
| Memory | Males | 0.061 | 0.941 | 0.001 | −0.049 | −0.607 | 0.509 | 0.862 |
| Females | 0.471 | 0.625 | 0.006 | 0.329 | −0.341 | 0.999 | 0.333 | |
| Perceptual speed | Males | 0.884 | 0.415 | 0.010 | −0.171 | −0.499 | 0.156 | 0.303 |
| Females | 1.785 | 0.172 | 0.024 | 0.256 | −0.170 | 0.681 | 0.237 | |
CI confidence interval
Results
Characteristics of the sample and genotype distribution
Table 1 presents the characteristics of the study participants stratified by gender. The sample consisted of 203 males and 182 females. Men had significantly more years of education and poorer scores in all three cognitive domains than women, with the difference in the motor function domain being statistically significant. The major allele distributions of the three examined SNPs are also presented in Table 1. The major allele genotype was carried by about two thirds of the participants for SNPs rs6265 (GG; 68.9%) and rs7103411 (TT; 63.8%), and by about half for rs7414442 (TT; 51.0%). No significant differences were found in the distributions of SNP genotypes between both genders. The distributions of rs6265, rs7103411, and rs7124442 genotypes did not significantly differ from the expected numbers calculated on the basis of observed allele frequencies according to the Hardy–Weinberg equilibrium.
Table 1.
Characteristics of the MEMO-Study participants (N = 369)
| Characteristic | Male (n = 197) | Female (n = 172) | Total (N = 369) | p Value |
|---|---|---|---|---|
| Age, years (mean, SD) | 73.01 (4.46) | 72.35 (4.41) | 72.70 (4.45) | 0.144* |
| Education, years (mean, SD) | 11.26 (2.52) | 10.01 (1.92) | 10.67 (2.34) | 0.000* |
| Motor function (Z-score)a | −0.76 (2.57) | 0.84 (2.47) | 0.00 (2.64) | 0.000* |
| Memory (Z-score)a | −0.21 (3.10) | 0.23 (3.03) | 0.00 (3.07) | 0.171* |
| Perceptual speed (Z-score)a | −0.14 (1.65) | 0.16 (1.81) | 0.00 (1.73) | 0.101* |
| rs6265, n = 360 (98%) | ||||
| GG | 124 (65.6) | 124 (72.5) | 248 (68.9) | |
| GA | 59 (31.2) | 38 (22.2) | 97 (26.9) | |
| AA | 6 (3.2) | 9 (5.3) | 15 (4.2) | |
| Total | 189 (100.0) | 171 (100.0) | 360 (100.0) | 0.119** |
| rs7103411, n = 356 (97%) | ||||
| TT | 119 (62.6) | 108 (65.1) | 227 (63.8) | |
| TC | 60 (31.6) | 50 (30.1) | 110 (30.9) | |
| CC | 11 (5.8) | 8 (4.8) | 19 (5.3) | |
| Total | 190 (100.0) | 166 (100.0) | 356 (100.0) | 0.861** |
| rs7124442, n = 343 (93%) | ||||
| TT | 97 (52.4) | 78 (49.4) | 175 (51.0) | |
| TC | 67 (36.2) | 68 (43.0) | 135 (39.4) | |
| CC | 21 (11.4) | 12 (7.6) | 33 (9.6) | |
| Total | 185 (100.0) | 158 (100.0) | 343 (100.0) | 0.299** |
Percent (%) genotype completion rate
aA Z-score of 0 represents the population mean; a score of 1.0 represents better performance than the mean by one standard deviation
*p Value of Student’s t test for continuous variables
**p Value of χ2-test for categorical variables
Association of genotypes and cognitive performance
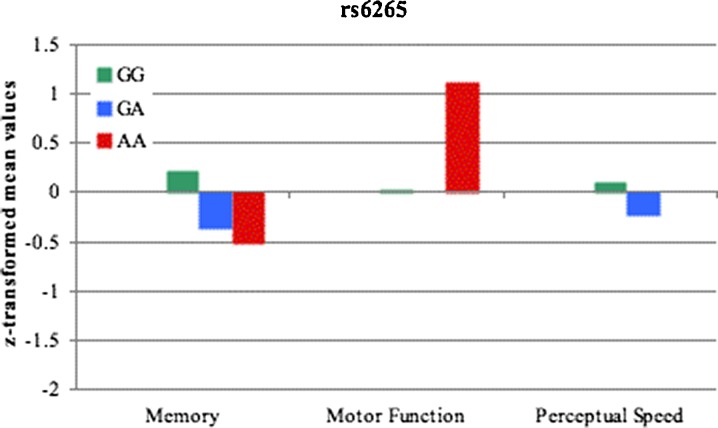
Figures 1 and 2 present the associations between the rs6265, rs7103411, and rs7124442 SNPs and cognitive performance after being adjusted for age, gender, and years of education. Figure 1 presents the mean Z-scores for the three cognitive domains (motor function, memory, and perceptual speed) according to rs6265 genotype. Results from one-way ANCOVAs showed no significant main effect of rs6265 genotype for (a) motor function (F(2,340) = 1.574, p = 0.209, η2 = 0.009 ), (b) memory (F(2,338) = 1.898, p = 0.151, η2 = 0.011), or (c) perceptual speed (F(2,340) = 1.416, p = 0.244, η2 = 0.008).
Fig. 1.
Cognitive performance by rs6265 genotype for memory, motor function, and perceptual speed domains. A Z-score of 0 represents the population mean; a score of −1.0 represents poorer performance than the mean by one standard deviation, and a score of +1.0 represents better performance than the mean by one standard deviation. Covariates included age, gender and years of education
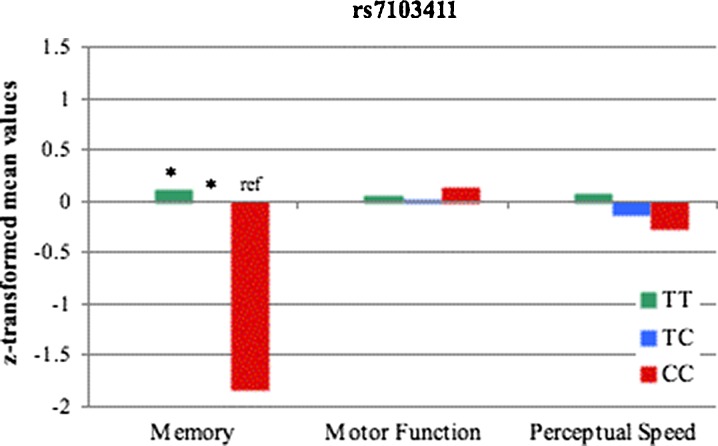
Fig. 2.
Cognitive performance by rs7103411 genotype for memory, motor function and perceptual speed domains. A Z-score of 0 represents the population mean; a score of −1.0 represents poorer performance than the mean by one standard deviation, and a score of +1.0 represents better performance than the mean by one standard deviation. Covariates included age, gender and years of education
Additional linear regression analysis revealed that the decrease in memory performance with each additional A allele showed a close to significant trend (p = 0.059). Trends for the other two cognitive domains were non-significant.
Figure 2 presents the mean Z-scores for the three cognitive domains (motor function, memory, and perceptual speed) according to rs7103411 genotype. Results from one-way ANCOVAs showed a significant main effect of genotype for memory (F(2,333) = 4.613, p = 0.011, η2 = 0.027; effect size f = 0.166), but no significant main effect for genotype for (a) motor function (F(2,335) = 0.026, p = 0.975, η2 = <0.001) or (b) perceptual speed (F(2,334) = 0.721, p = 0.487, 4η2 = 0.004).
Trend analyses provided a significant trend for a decrease in memory performance with every increase of the C allele (p = 0.03), whereas no significant trends were found for the other cognitive domains.
Results from one-way ANCOVAs for the relationship between mean Z-scores for the three cognitive domains (motor function, memory, and perceptual speed) and the rs7124442 genotypes showed no significant main effect of genotype for (a) motor function (F(2,321) = 0.081, p = 0.922, η2 = 0.001), (b) memory (F(2,319) = 0.171, p = 0.843, η2 = 0.001), or (c) perceptual speed (F(2,320) = 0.015, p = 0.985, η2 = <0.001). Additional trend analyses showed no significant results.
Gender differences
In female participants, one-way ANCOVA, corrected for age and education, showed a significant main effect of rs6265 genotype for perceptual speed (empirical effect size f = 0.255 vs minimal detectable effect size f = 0.24) and a close to significant main effect for memory (Table 2). Similarly, the SNP rs7103411 showed a significant main effect with memory (empirical effect size f = 0.259 vs minimal detectable effect size f = 0.245) and speed domains (empirical effect size f = 0.21 vs minimal detectable effect size f = 0.245). Linear regression analysis confirmed these results and showed a significant trend of decrease in memory performance and perceptual speed with the increasing number of mutant alleles in case of rs6265 and rs7103411 in females. However, neither of the aforementioned associations were present in males (Table 2). We found no significant effect of rs7124442 genotype on any of the cognitive domains either in males or in females (Table 2).
Sensitivity analyses
Investigating the effects of the genetic APOE*E4 status (not including BDNF SNPs in ANCOVA) on the three domains of cogntive performance showed no significant effect of the apolipoprotein E risk allele on poorer cognitive performance either in the whole or gender-stratified samples (with or without eudcation as covariate). When the BDNF SNPs, together with the APOE*E4 status were included as a covariates in the ANCOVA models for the association between BDNF SNPs and cognitive function, the previously reported female-specific significant associations between rs6265 and perceptual speed (F(2,161) = 3.27; p = 0.04) and between rs7103411 and memory (F(2,154) = 4.57; p = 0.01) remained signifcant independent of APOE*E4 status; however, the previously reported effects of rs7103411 on perceptual speed (F(2,155) = 1.83; p = 0.16) disappeared.
Finally, when education in years was removed as a covariate from these gender-specific analyses, the reported associations between rs6265 and perceptual speed (F(2,161) = 4.44; p = 0.01) remained the same, whereas the association between rs7103411 and memory (F(2,155) = 5.10; p = 0.007) was strengthened.
Discussion
This study investigated associations between three SNPs of the BDNF gene (rs6265, rs7103411, and rs7124442) and three domains of cognitive function for memory, motor function, and perceptual speed in a community-based elderly population. The age-adjusted results indicate a gender-specific association between rs6265 and perceptual speed and between rs7103411 and memory independent of APOE*E4 and education status, with major allele participants having better performance. In the female subgroup, the observed empirical effect sizes for the associations between rs6265 and perceptual speed as well as for the relationship between rs7103411 and memory (but not for rs7103411 and perceptual speed) were higher than the minimal detectable effect sizes. These findings indicate that the relatively small study had sufficient statistical power to detect smaller effects of the BDNF gene on the performance of these cognitive domains in gender-stratified analyses.
The first main finding is that, like previous research (Miyajima et al. 2008; Foltynie et al. 2005, 2009; Raz et al. 2009), the current study demonstrated a significant association between the rs6265 SNP and cognitive performance. The hypothesis that the rs6265 SNP would significantly associate with memory was supported; however, the hypothesis that the rs6265 SNP would significantly associate with motor function or perceptual speed was not supported. The results also indicated a trend favouring the major allele participants in memory and perceptual speed performance. This trend echoes previous studies that found better performance in major allele participants (Rybakowski et al. 2003, 2006). Additionally, the pattern of performance (better performance in major allele participants, followed by poorer performance in heterozygous and homozygous participants, respectively) in the memory and perceptual speed domains is in line with the notion that the BDNF gene modulates cognitive performance, by virtue of its effects on BDNF protein levels. Specifically, increased levels of the met allele in the rs6265 SNP may increasingly impair regulated secretion of the BDNF protein as suggested by Egan et al. (2003). This may explain the impaired memory and perceptual speed performance observed in the met-BDNF carriers of this aging cohort. Our findings are supported by a previous study showing that the presence of the met-BDNF allele may predict a worse cognitive outcome in patients with MCI (Forlenza et al. 2010) and cognitive impairment in Parkinson patients (Guerini et al. 2009).
The second main finding of this study is that, as was hypothesised, the rs7103411 SNP was significantly associated with memory performance. This result differs from the previous study by Miyajima et al. (2008) that found no significant associations between this SNP and cognitive performance, however, which might be related to the mainly haplotype approach employed by Miyajima et al. In our study, participants exhibiting the major allele versus heterozygous and minor allele genotypes had significantly better global memory performance. Sub-analyses of associations between the rs7103411 SNP and the individual memory tests indicated that this effect was strongest for the Word Fluency task. This supports previous research implicating the BDNF gene in long-term memory (Hariri et al. 2003). Sub-analyses not described in the results also revealed a genotype effect on performance for Word List 3, which supports previous results implicating the BDNF gene in short-term memory (Rybakowski et al. 2003, 2006). However, unlike these studies that found better short-term memory performance in major allele participants, the current study found better performance in heterozygous participants for Word List 3. This may reflect a difference in mental health as the participants in the previous studies suffered from bipolar disorder.
The third main finding of this study is that the hypothesis that the three BDNF SNPs would significantly associate with motor function and perceptual speed was not supported. As already mentioned, a non-significant trend favouring major allele participants for better perceptual speed performance was observed in the rs6265 SNP; this was also evident in the rs7103411 SNP. Another non-significant trend favouring homozygous participants for better motor function performance was observed in the rs6265 and rs7103411 SNPs. This corroborates the results from the prior study by Oroszi et al. (2006) who hypothesised that the BDNF minor allele plays a protective role against motor function decline.
The fourth main finding is that the rs712442 SNP was not significantly associated with performance in any of the cognitive domains. Furthermore, the patterns of performance according to rs712442 genotype greatly differed from the performance patterns observed in the rs6265 and rs7103411 SNPs. This suggests that the rs712442 SNP may not play an important role in cognitive functioning in a normal cognitive aging.
Finally, the gender-specific analyses of the aforementioned associations between the BDNF gene and cognitive function, derived primarily from the female subgroup, suggests, for the first time, that female aging participants with the minor allele of either rs6265 or rs7103411 had an increased risk of poorer cognitive performance (memory and perceptual speed) as compared with the major allele carrier. Although this observation was validated by sufficient statistical power in this study and the sample size in our study is comparable to or higher than several other studies in the field, it needs to be considered that other genetic association studies on cognitive function have reported smaller effect sizes. When applying an α err prob = 0.01 (1% instead of 5%) for three genotype groups per SNP to the gender-specific power calculations (assumed power of 80%), the observed effected sizes for all SNPs were smaller than the expected effect sizes (SNP-specific range between 0.28 and 0.3). Thus, a false-positive finding in our study cannot be ruled out. After applying Bonferroni correction for multiple comparisons yielding a corrected p value of p = 0.006 (3 SNPs × 3 cognitive domains), the female-specific finding for the association between rs6265 and perceptual speed remained significant, whereas the other findings did not withstand this Bonferroni-corrected p value. However, it is important to recognise that the Bonferroni correction can be overly conservative for non-independent tests (Perneger 1998). Overall, this study requires a replication study in independent samples which should aim for a comparable cognitive phenotype. Homogenous cognitive phenotypes across replication samples would help to reduce the probability of false-positive/negative findings. The use of individual cognitive tests rather than compound measures derived from various single cognitive tests as in our study might be best suited for a replication study for reasons of comparability of cognitive phenotypes.
The mechanisms for this gender-specific observation—although indirectly supported by a recent study in male failing to show an association between the Val66Met polymorphisms and cognitive function assessed by various cognitive domains (Tsai et al. 2008)—remain unclear and require replication and further assessment in independent and larger samples utilising comparable cognitive phenotypes. In contrast to our finding, a larger Scottish study on cognitive aging showed an association between the Met66 allele and improved reasoning skills; however, reasoning skills describe different cognitive abilities than our cognitive phenotypes, which make the studies not directly comparable. In addition, this study showed no gender by genotype interaction as compared with the Scottish study (Harris et al. 2006). Again, the lack of comparability on the gender finding between this and our study may be due to the use of different cognitive phenotypes.
The underlying notion tested in the current study was that the BDNF gene affects the BDNF protein, which in turn affects cognitive performance. The absence of the knowledge of the BDNF protein levels for each participant was a major limitation. Additionally, there was no imaging of the brain structures relevant to the cognitive domains being tested (e.g. hippocampus and substantia nigra). Therefore, the linking of the BDNF gene to cognitive functioning via its effects on BDNF protein and brain functioning can only be inferred. The reality of this system may include interactions of the BDNF gene with other genes or proteins, or some undetermined neurological pathway involving the BDNF gene. Therefore, future research should aim to include measures of (a) BDNF protein levels, (b) brain imaging, (c) BDNF genotyping, and (d) cognitive functioning. The synthesis of such data would reveal a clearer picture of the BDNF gene’s role in cognitive functioning, including its possible effects on BDNF protein levels, brain functioning, and cognitive functioning. In addition, linking BDNF with brain morphology and cognitive function is of special relevance as recently demonstrated in a twin study using diffusion tensor imaging that showed the BDNF gene (rs6265) may affect the intellectual performance by modulating the white matter development (Chiang et al. 2011).
In conclusion, the current study suggests a relationship between the BDNF gene and (a) memory and (b) perceptual speed functioning but not (c) motor function in humans during normal aging. Although trends have been observed, the current findings suggest that the BDNF gene plays a less important role in motor function and perceptual speed in normal cognitive aging humans than in diseased aging humans. The current findings support previous research implicating the BDNF gene in memory functioning and provide the first results implicating the rs7103411 SNP in affecting memory performance in this population. In addition, this study indicates that female participants may be at a higher risk of poorer memory and perceptual speed when being carrier of minor allele risk genotypes of SNPs of the BDNF gene independent of the APOE*E4 status. Future research should further investigate these particular SNPs, as well as investigate more robustly the genotypic effects of the BDNF SNPs on short- versus long-term memory. Finally, future research should aim to further explore the relationship between the BDNF gene and BDNF protein to clarify the relationship between the BDNF gene and age-sensitive cognitive functioning.
Acknowledgements
The MEMO-Study is supported by the German Research Society (Deutsch Forschungsgemeinschaft, Grant: BE1996/1-1). Data assessment was done within the framework of the Cooperative Health Research in the Augsburg Region (KORA). Bernhard Baune had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
References
- Baune BT, Suslow T, Engelien A, Arolt V, Berger K. The association between depressive mood and cognitive performance in an elderly general population - the MEMO Study. Dement Geriatr Cogn Disord. 2006;22(2):142–149. doi: 10.1159/000093745. [DOI] [PubMed] [Google Scholar]
- Baune B, Ponath G, Rothermundt M, Riess O, Funke H, Berger K. Association between genetic variants of IL-1 [beta], IL-6 and TNF-[alpha] cytokines and cognitive performance in the elderly general population of the MEMO-Study. Psychoneuroendocrinology. 2008;33:68–76. doi: 10.1016/j.psyneuen.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Beckman L, Nilsson L. Semantic memory functioning across the adult life span. Eur Psychol. 1996;1:27–33. doi: 10.1027/1016-9040.1.1.27. [DOI] [Google Scholar]
- Cheng S, Grow M, Pallaud C, Klitz W, Erlich H, Visvikis S, Chen J, Pullinger C, Malloy M, Siest G. A multilocus genotyping assay for candidate markers of cardiovascular disease risk. Genome Res. 1999;9:936. doi: 10.1101/gr.9.10.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Toga AW, Medland SE, Hansell NK, James MR, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. BDNF gene effects on brain circuitry replicated in 455 twins. NeuroImage. 2011;55:448–454. doi: 10.1016/j.neuroimage.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa L, Vaughan H, Jr, Levita E, Farber N. Purdue Pegboard as a predictor of the presence and laterality of cerebral lesions. J Consult Psychol. 1963;27:133–137. doi: 10.1037/h0040737. [DOI] [PubMed] [Google Scholar]
- Desrosiers J, Hebert R, Bravo G, Dutil E. The Purdue Pegboard Test: normative data for people aged 60 and over. Disabil Rehabil. 1995;17:217–224. doi: 10.3109/09638289509166638. [DOI] [PubMed] [Google Scholar]
- Egan M, Kojima M, Callicott J, Goldberg T, Kolachana B, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/S0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Ferrer I, Marin C, Rey M, Ribalta T, Goutan E, Blanco R, Tolosa E, Marti E. BDNF and full-length and truncated TrkB expression in Alzheimer disease. Implications in therapeutic strategies. J Neuropathol Exp Neurol. 1999;58:729. doi: 10.1097/00005072-199907000-00007. [DOI] [PubMed] [Google Scholar]
- Foltynie T, Lewis S, Goldberg T, Blackwell A, Kolachana B, Weinberger D, Robbins T, Barker R. The BDNF Val 66 Met polymorphism has a gender specific influence on planning ability in Parkinson’s disease. J Neurol. 2005;252:833–838. doi: 10.1007/s00415-005-0756-5. [DOI] [PubMed] [Google Scholar]
- Foltynie T, Cheeran B, Williams-Gray C, Edwards M, Schneider S, Weinberger D, Rothwell J, Barker R, Bhatia K. BDNF val66met influences time to onset of levodopa induced dyskinesia in Parkinson′s disease. J Neurol Neurosurg Psychiatry. 2009;80:141. doi: 10.1136/jnnp.2008.154294. [DOI] [PubMed] [Google Scholar]
- Forlenza OV, Diniz BS, Teixeira AL, Ojopi EB, Talib LL, Mendonca VA, Izzo G, Gattaz WF. Effect of brain-derived neurotrophic factor Val66Met polymorphism and serum levels on the progression of mild cognitive impairment. World J Biol Psychiatry. 2010;11:774–780. doi: 10.3109/15622971003797241. [DOI] [PubMed] [Google Scholar]
- Guerini FR, Beghi E, Riboldazzi G, Zangaglia R, Pianezzola C, Bono G, Casali C, Di Lorenzo C, Agliardi C, Nappi G, Clerici M, Martignoni E. BDNF Val66Met polymorphism is associated with cognitive impairment in Italian patients with Parkinson’s disease. Eur J Neurol. 2009;16:1240–1245. doi: 10.1111/j.1468-1331.2009.02706.x. [DOI] [PubMed] [Google Scholar]
- Hariri A, Goldberg T, Mattay V, Kolachana B, Callicott J, Egan M, Weinberger D. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J Neurosci. 2003;23:6690. doi: 10.1523/JNEUROSCI.23-17-06690.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S, Fox H, Wright A, Hayward C, Starr J, Whalley L, Deary I. The brain-derived neurotrophic factor Val66Met polymorphism is associated with age-related change in reasoning skills. Mol Psychiatry. 2006;11:505–513. doi: 10.1038/sj.mp.4001799. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Yamashita A, Shimizu K. Somatostatin and brain-derived neurotrophic factor mRNA expression in the primate brain: decreased levels of mRNAs during aging. Brain Res. 1997;749:283–289. doi: 10.1016/S0006-8993(96)01317-0. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Mistunaga F, Ohira K, Shimizu K. Changes in BDNF-immunoreactive structures in the hippocampal formation of the aged macaque monkey. Brain Res. 2001;918:191–196. doi: 10.1016/S0006-8993(01)03002-5. [DOI] [PubMed] [Google Scholar]
- Hock C, Heese K, Hulette C, Rosenberg C, Otten U. Region-specific neurotrophin imbalances in Alzheimer disease: decreased levels of brain-derived neurotrophic factor and increased levels of nerve growth factor in hippocampus and cortical areas. Arch Neurol. 2000;57:846. doi: 10.1001/archneur.57.6.846. [DOI] [PubMed] [Google Scholar]
- Howells D, Porritt M, Wong J, Batchelor P, Kalnins R, Hughes A, Donnan G. Reduced BDNF mRNA expression in the Parkinson’s disease substantia nigra. Exp Neurol. 2000;166:127–135. doi: 10.1006/exnr.2000.7483. [DOI] [PubMed] [Google Scholar]
- Huang R, Huang J, Cathcart H, Smith S, Poduslo SE. Genetic variants in brain-derived neurotrophic factor associated with Alzheimer′s disease. J Med Genet. 2007;44:e66. doi: 10.1136/jmg.2006.044883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones K, Reichardt L. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci. 1990;87:8060. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keil U, Liese A, Hense H, Filipiak B, Doring A, Stieber J, Lowel H. Classical risk factors and their impact on incident non-fatal and fatal myocardial infarction and all-cause mortality in southern Germany: results from the MONICA Augsburg Cohort Study 1984–1992. Eur Heart J. 1998;19:1197. doi: 10.1053/euhj.1998.1089. [DOI] [PubMed] [Google Scholar]
- Komulainen P, Pedersen M, Hanninen T, Bruunsgaard H, Lakka T, Kivipelto M, Hassinen M, Rauramaa T, Pedersen B, Rauramaa R. BDNF is a novel marker of cognitive function in ageing women: the DR’s EXTRA Study. Neurobiol Learn Mem. 2008;90:596–603. doi: 10.1016/j.nlm.2008.07.014. [DOI] [PubMed] [Google Scholar]
- Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci. 1995;92:8856. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte M, Griesbeck O, Gravel C, Carroll P, Staiger V, Thoenen H, Bonhoeffer T. Virus-mediated gene transfer into hippocampal CA1 region restores long-term potentiation in brain-derived neurotrophic factor mutant mice. Proc Natl Acad Sci USA. 1996;93:12547. doi: 10.1073/pnas.93.22.12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte M, Kang H, Bonhoeffer T, Schuman E. A role for BDNF in the late-phase of hippocampal long-term potentiation. Neuropharmacology. 1998;37:553–559. doi: 10.1016/S0028-3908(98)00035-5. [DOI] [PubMed] [Google Scholar]
- Linnarsson S, Bjorklund A, Ernfors P. Learning deficit in BDNF mutant mice. Eur J Neurosci. 1997;9:2581–2587. doi: 10.1111/j.1460-9568.1997.tb01687.x. [DOI] [PubMed] [Google Scholar]
- Lynch MA. Long-term potentiation and memory. Physiol Rev. 2004;84:87–136. doi: 10.1152/physrev.00014.2003. [DOI] [PubMed] [Google Scholar]
- Mattson M, Maudsley S, Martin B. A neural signaling triumvirate that influences ageing and age-related disease: insulin/IGF-1, BDNF and serotonin. Ageing Res Rev. 2004;3:445–464. doi: 10.1016/j.arr.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Mercader J, Ribasés M, Gratacòs M, González J, Bayés M, de Cid R, Badía A, Fernández-Aranda F, Estivill X. Altered brain-derived neurotrophic factor blood levels and gene variability are associated with anorexia and bulimia. Genes Brain Behav. 2007;6:706–716. doi: 10.1111/j.1601-183X.2007.00301.x. [DOI] [PubMed] [Google Scholar]
- Miyajima F, Ollier W, Mayes A, Jackson A, Thacker N, Rabbitt P, Pendleton N, Horan M, Payton A. Brain-derived neurotrophic factor polymorphism Val66Met influences cognitive abilities in the elderly. Genes Brain Behav. 2008;7:411–417. doi: 10.1111/j.1601-183X.2007.00363.x. [DOI] [PubMed] [Google Scholar]
- Morris R. Synaptic plasticity and learning: selective impairment of learning rats and blockade of long-term potentiation in vivo by the N-methyl-d-aspartate receptor antagonist AP5. J Neurosci. 1989;9:3040. doi: 10.1523/JNEUROSCI.09-09-03040.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narisawa-Saito M, Wakabayashi K, Tsuji S, Takahashi H, Nawa H. Regional specificity of alterations in NGF, BDNF and NT-3 levels in Alzheimer’s disease. Neuroreport. 1996;7:2925. doi: 10.1097/00001756-199611250-00024. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Söderlund H, Berger K, Breteler M, de Ridder M, Dufouil C, Fuhrer R, Giampaoli S, Hofman A, Pajak A. Cognitive test battery of CASCADE: tasks and data. Aging Neuropsychol Cogn. 2005;12:32–56. doi: 10.1080/13825580590925099. [DOI] [Google Scholar]
- Nyberg L, Maitland S, Ronnlund M, Backman L, Dixon R, Wahhn A, Nilsson L. Selective adult age differences in an age-invariant multifactor model of declarative memory. Psychol Aging. 2003;18:149–160. doi: 10.1037/0882-7974.18.1.149. [DOI] [PubMed] [Google Scholar]
- Oeth P, Beaulieu M, Park C, Kosman D, del Mistro G, van den Boom D, Jurinke C (2007) iPLEX™ assay: increased plexing efficiency and flexibility for MassARRAY system through single base primer extension with mass-modified terminators. Online access: http://www.agrf.org.au/docstore/snp/iPlex.pdf
- Oroszi G, Lapteva L, Davis E, Yarboro C, Weickert T, Roebuck-Spencer T, Bleiberg J, Rosenstein D, Pao M, Lipsky P. The Met66 allele of the functional Val66Met polymorphism in the brain-derived neurotrophic factor gene confers protection against neurocognitive dysfunction in systemic lupus erythematosus. Ann Rheum Dis. 2006;65:1330. doi: 10.1136/ard.2006.051623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parain K, Murer M, Yan Q, Faucheux B, Agid Y, Hirsch E, Raisman-Vozari R. Reduced expression of brain-derived neurotrophic factor protein in Parkinson’s disease substantia nigra. Neuroreport. 1999;10:557. doi: 10.1097/00001756-199902250-00021. [DOI] [PubMed] [Google Scholar]
- Perneger TV. What’s wrong with Bonferroni adjustments. BMJ. 1998;316:1236–1238. doi: 10.1136/bmj.316.7139.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H, Hains J, Armanini M, Laramee G, Johnson S, Winslow J. BDNF mRNA is decreased in the hippocampus of individuals with Alzheimer’s disease. Neuron. 1991;7:695–702. doi: 10.1016/0896-6273(91)90273-3. [DOI] [PubMed] [Google Scholar]
- Raz N, Rodrigue K, Kennedy K, Land S. Genetic and vascular modifiers of age-sensitive cognitive skills: effects of COMT, BDNF, ApoE and hypertension. Neuropsychology. 2009;23:105. doi: 10.1037/a0013487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybakowski JK, Borkowska A, Czerski PM, Skibiska M, Hauser J. Polymorphism of the brain-derived neurotrophic factor gene and performance on a cognitive prefrontal test in bipolar patients. Bipolar Disord. 2003;5:468–472. doi: 10.1046/j.1399-5618.2003.00071.x. [DOI] [PubMed] [Google Scholar]
- Rybakowski J, Borkowska A, Skibinska M, Szczepankiewicz A, Kapelski P, Leszczynska-Rodziewicz A, Czerski P, Hauser J. Prefrontal cognition in schizophrenia and bipolar illness in relation to Val66Met polymorphism of the brain-derived neurotrophic factor gene. Psychiatry Clin Neurosci. 2006;60:70–76. doi: 10.1111/j.1440-1819.2006.01462.x. [DOI] [PubMed] [Google Scholar]
- Salthouse T. The role of memory in the age decline in digit-symbol substitution performance. J Gerontol. 1978;33:232. doi: 10.1093/geronj/33.2.232. [DOI] [PubMed] [Google Scholar]
- Schmidt W, Roesler A, Kretzschmar K, Ladwig K, Junker R, Berger K. Functional and cognitive consequences of silent stroke discovered using brain magnetic resonance imaging in an elderly population. J Am Geriatr Soc. 2004;52:1045–1050. doi: 10.1111/j.1532-5415.2004.52300.x. [DOI] [PubMed] [Google Scholar]
- Stroop J. Studies of interference in serial verbal reactions. J Exp Psychol. 1935;18:643–662. doi: 10.1037/h0054651. [DOI] [Google Scholar]
- Tang YP, Wang H, Feng R, Kyin M, Tsien JZ. Differential effects of enrichment on learning and memory function in NR2B transgenic mice. Neuropharmacology. 2001;41:779–790. doi: 10.1016/S0028-3908(01)00122-8. [DOI] [PubMed] [Google Scholar]
- Tiffin J. Manual for the Purdue Pegboard. Chicago (IL): Science Research Associates; 1948. [Google Scholar]
- Tsai SJ, Gau YT, Liu ME, Hsieh CH, Liou YJ, Hong CJ. Association study of brain-derived neurotrophic factor and apolipoprotein E polymorphisms and cognitive function in aged males without dementia. Neurosci Lett. 2008;433:158–162. doi: 10.1016/j.neulet.2007.12.057. [DOI] [PubMed] [Google Scholar]
- Xiromerisiou G, Hadjigeorgiou G, Eerola J, Fernandez H, Tsimourtou V, Mandel R, Hellström O, Gwinn-Hardy K, Okun M, Tienari P. BDNF tagging polymorphisms and haplotype analysis in sporadic Parkinson’s disease in diverse ethnic groups. Neurosci Lett. 2007;415:59–63. doi: 10.1016/j.neulet.2006.12.038. [DOI] [PubMed] [Google Scholar]
- Zivadinov R, Weinstock-Guttman B, Benedict R, Tamaño-Blanco M, Hussein S, Abdelrahman N, Durfee J, Ramanathan M. Preservation of gray matter volume in multiple sclerosis patients with the Met allele of the rs6265 (Val66Met) SNP of brain-derived neurotrophic factor. Hum Mol Genet. 2007;16:2659. doi: 10.1093/hmg/ddm189. [DOI] [PubMed] [Google Scholar]