Abstract
The development of tools to construct and investigate probabilistic maps of the adult human brain from MRI have led to advances in both basic neuroscience and clinical diagnosis. These tools are increasingly being applied to brain development in adolescence, childhood and even neonatal and premature neonatal imaging. Looking even earlier in development, parallel developments in clinical fetal Magnetic Resonance Imaging (MRI) have led to its growing use as a tool in challenging medical conditions. This has motivated new engineering developments that combine optimal fast MRI scans with techniques derived from computer vision that allow full 3D imaging of the moving fetal brain in utero without sedation. These promise to provide a new and unprecedented window into early human brain growth. This article will review the developments that have led us to this point, and examine the current state of the art in the fields of fast fetal imaging, motion correction and the tools to analyze dynamically changing fetal brain structure. New methods to deal with developmental tissue segmentation and the construction of spatio-temporal atlases will be examined, together with techniques to map fetal brain growth patterns.
Keywords: Motion Correction, Image Mosaic, Deformation Morphometry, Spatio-Temporal Atlas, Tissue Segmentation, Cortical Folding
1 Introduction
The combination of Magnetic Resonance Imaging (MRI) physics with mathematical and computational techniques that underpin modern brain image analysis has created a range of new tools for the neuroscientist and clinician. These allow the creation of maps quantifying both brain structure and function across human populations, and the examination of how clinical factors can influence the patterns of measurements. These developments have helped form the basis for the emergence of brain mapping (1–3) as a key research topic in both basic neuroscience and clinically focused brain research. In particular, computational morphometric methods have provided new insights into the way brain structure changes over time and how this process is influenced by disease in populations ranging from adults with dementia (4–6), psychiatric conditions (7), brain injury in substance abuse (8), to changes in the brain due to infectious disease (9). More recently, focus has shifted to the study of the developing brain (10,11,11–13) and large scale projects are underway to understand and map the process of brain growth from birth to adult (14). These studies have focused on postnatal mapping of growth. However, recent developments in clinical MRI, as illustrated in figure 1 have raised the possibility of pushing these studies back further, to image the process of human brain growth in utero. For the neuroscientist, the ability to image earlier, simpler structural configurations provides insights into the fully developed anatomy. By mapping the brain in its early form, prior to the process of anatomical individualization, and capturing its transformation into the individualized adult configuration, we hope to understand better how the adult brain itself is organized and functions. This article reviews the combination of engineering and clinical developments that are beginning to provide high quality and reproducible 3D images of early human brain growth, and examines some of the computational and algorithmic challenges in analyzing and modeling dynamically changing fetal neuroanatomy.
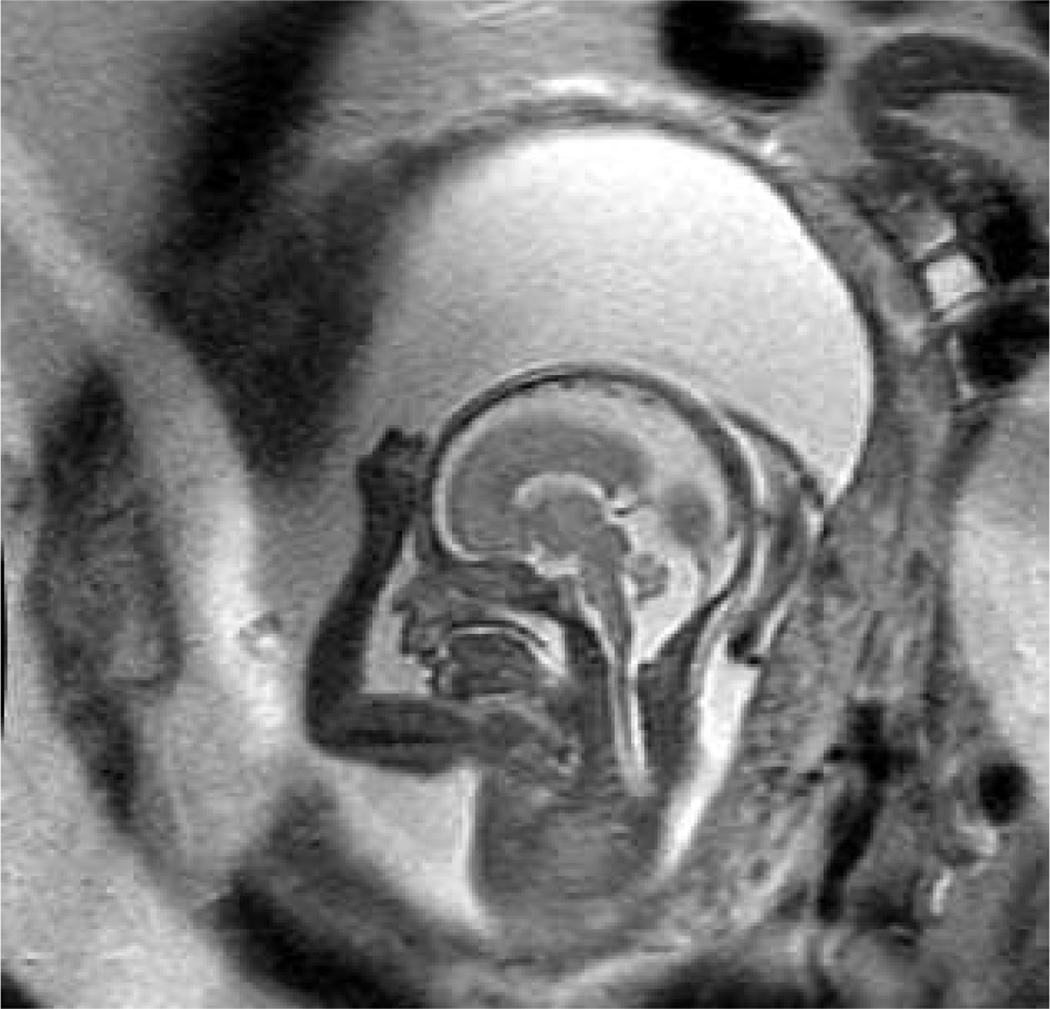
Figure 1.
Example slice from a clinical fast (SSFSE) T2W MRI image of a human fetus showing a mid-line sagittal view of the fetal brain.
2 Background: The Emergence of Clinical Fetal MRI
2.1 Fetal MRI Safety
A key factor in the development of brain mapping has been the availability of reproducible high quality Magnetic Resonance Imaging (MRI), enabling 3D and 4D mapping of quantitative structural and functional measurement within the living brain. Unlike X Ray computed tomography or nuclear medicine imaging used in adult brain studies, and also experimentally explored for human fetal imaging (15), MRI does not involve ionizing radiation. However it remained out of the toolbox of the clinical radiologist because of safety concerns for the developing fetus. A significant literature exists examining the possibility of MRI induced damage from radio frequency field (RF) and electromagnetic field (EMF) exposure (16), in terms of both static and changing fields (17). This early work was refined (18–20) using more accurate anatomical models (21, 22) for adult imaging. When clinical MRI during pregnancy was being considered, this work was extended with fetal specific RF-EMF models (16, 23–26). Previous studies examined concerns regarding thermal effects arising from the lack of surface cooling available to the fetus (27) and the effects of noise levels (28) resulting from the use of faster (and noisier) MRI sequences. The potential for MRI induced genetic mutations (29), and the consequences of maternal stress related to the MRI exam have also been examined (30). This work has been complemented by experimental investigations using animal models (31) to look at longer term effects of exposure to fields (32) and examine the thermal effects (33) on the fetus. Follow up studies of children studied with fetal MRI have also been carried out (34–36), and no significant longer term adverse effects have been discovered. As a result, fetal MRI has begun to be accepted more widely as a tool for clinical diagnosis.
2.2 Early Fetal Magnetic Resonance Imaging
The use of MRI with pregnant patients has been driven by the clinical radiologist and perinatologist, looking for better ways to analyze fetal health. However, from the beginning, motion was the key challenge. The first clinically driven studies included early T1 weighted (T1W) lower magnetic field imaging (37, 38) and T2 weighted (T2W) imaging (39–44). These early attempts used slower techniques that were highly susceptible to motion of the fetus, and were therefore largely dependent on the luck of the radiographic technician in catching the fetus while not in motion, and often were performed with shallow maternal breathing. In France, fetal motion was reduced by injection of a sedative into the umbilical vein (45) or via maternal sedation that crossed the placenta (46). Such fetal sedation was not practical for more general screening, although remains feasible for life threatening disorders. As a result of the general lack of reproducibility of methods, these promising early approaches were of limited use for general clinical practice.
The development of faster imaging techniques, combined with improved imaging hardware was the key to making MRI more generally applicable in clinical fetal studies (47–49). These include echo planar imaging (EPI) developed in the 1970’s (50), which was also used in early fetal studies (51, 52), providing a so called snap-shot image of the anatomy. Critically, this early work showed that a single slice could be acquired quickly enough to exclude most fetal motion and still produce an acceptable level of signal to noise to delineate anatomical detail. It also however illustrated that averaging of slices to increase signal without consideration for motion was still not possible due to between slice fetal movements.
2.3 Approaches to Faster and 3D MR Imaging
An MRI data acquisition samples the spatial frequency spectrum of the underlying anatomical measurements (k-space) and a spatial image is reconstructed using a Fourier transform (53). A single MR signal recording typically samples one line of this k-space over a few milliseconds, and can be acquired in such a way to collect 2D slices or full 3D volumes by encoding space using different temporally varying patterns of RF and magnetic field gradients (54). Full 3D volume imaging is typically employed in studies of adults and cooperative children and adolescents, as imaging time is still of the order of minutes. Motion during this time can induce ghosting or blurring as signal phase or frequency is corrupted. As a result, multislice 2D imaging is generally acquired in studies where motion is expected to be a problem. These allow pseudo 3D studies of the brain for fetal diagnosis (55) for fetuses exhibiting limited motion. However, motion can still induce artifacts when it occurs between data acquisition and the following RF excitation, or by inducing spin phase errors as the object moves through the magnetic field between excitation and the following data sampling acquisition. So called motion robust sequences have also been developed such as projection-reconstruction (56) and spiral (57,58) MRI sequences. However, they have so far been unable to provide clinically useful images in a short enough time interval for 3D fetal imaging.
2.4 Motion Correction Techniques in MRI
Even with the many advances in MRI, full 3D acquisitions that are fast enough to reduce the chance and impact of motion artifacts to an acceptable level are still not feasible. In adult imaging, a range of motion correction schemes have been developed to allow imaging in the presence of motion. Prospective motion correction techniques make use of independent estimates of anatomical positioning to update the acquisition process as it proceeds, by modifying the phase or frequency encoding being used to define the scan geometry (59), or to exclude corrupted components of the signal acquired at known times of motion. A range of such techniques have been explored using optical (60, 61) or MRI marker (62) based tracking of patient anatomy. However, the requirement for some form of attached marker or visible surface precludes these approaches from use in fetal imaging.
Motion estimates can also be derived from additional MRI measurements using so called navigator echos (63) which are rapid, simplified signal acquisitions interspersed within MR sequences. These have been used for both retrospective (63) and prospective correction (64) of motion, and can employ a variety of k-space trajectory shapes to measure motion in different axes, including linear (65), circular (66), spherical (67) for full 3D motion tracking (68, 69). To correct larger head motion in children recent methods (70) have combined spiral navigator echos with Kalmen filtering based tracking. In fetal MRI, navigator echos have been used to detect and trigger fast snapshot slice imaging (71) to times when the fetus is stationary along a selected axis. However these can be difficult to localize for estimation of full 3D motion in fetal MRI. An alternative approach, derived from radar imaging, is to retrospectively ’refocus’ the data in the k-space (Fourier) domain without separate motion estimates (72). However, because these algorithms operate on the k-space (Fourier) domain, it is difficult to adapt them to correct locally rigid motion, as required in 3D fetal imaging.
2.5 Current Multi-Slice Imaging and its Limitations
In place of earlier EPI methods, accelerated sequences such as fast spin echo (FSE) (73), the half Fourier single shot turbo spin echo (HASTE) (74) and single shot fast spin echo (SSFSE) (75) have been developed to provide improved tissue contrast. These require a second or less for one slice, and provide the clinician with a reliable route to getting selected slice views through unsedated fetal anatomy, albeit without accurately known 3D spatial relationships between consecutive slices. Studies typically involve the acquisition of multiple stacks of slices planned with different orthogonal orientations to provide complementary views of the anatomy. These can make use of real time planning to manually adjust to fetal motion (75) or gating to remove the influence of maternal breathing.
Clinical applications of faster multi-slice imaging rapidly expanded, particularly for neuroimaging (76), where ultrasound studies are limited by reverberations resulting from the bony structure of the fetal skull. Here MRI can provide valuable contrast between different tissue layers in the developing fetal brain which are invisible to ultrasound, such as the germinal matrix and sub-plate, as well as providing high resolution information about the folding of the cortical plate. MRI has found a key role in providing valuable complementary information in specific clinical conditions, particularly involving brain development (77). It has been shown to be extremely useful in evaluating causes of ventriculomegaly (78, 79), identifying cerebral malformations (80, 81), identifying infections (82), and identifying other fetal brain injuries (83, 84).
In addition to radiographic diagnosis, fetal Brain MRI has begun to be used in quantitative studies of brain growth in different clinical populations (79, 85–88).
For more quantitative studies of brain anatomy these types of imaging acquisition suffer from a number of important limitations. As well as the problem of the unknown anatomical relationships between successive slices, they also employ relatively thick slices to provide adequate in-plane resolution and signal to noise for radiological inspection, but hindering true 3D analysis. In addition, gating to the maternal breathing, although reducing the chance of within-slice motion, can also significantly extend the scan time, with little improvement in cases of significant fetal motion.
3 A Hybrid Solution to 3D Fetal MRI: Computer Vision Meets MR Physics
The current direction of research toward full 3D MR imaging of moving fetal anatomy has taken a hybrid approach of combining MR physics with computer vision techniques. This was motivated by the observation that clinical studies of the fetal brain commonly make use of multiple multi-slice acquisitions in different axes which are also often repeated because of fetal motion during a stack.
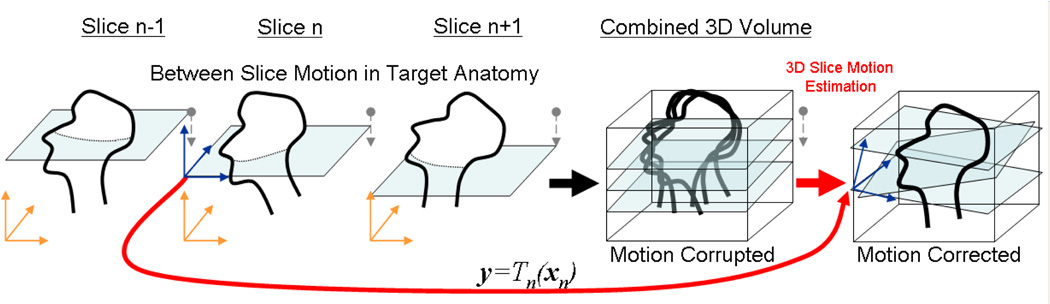
In computer vision, the field of image mosaicing (89) seeks to merge sub-images into a single image. This can provide either an enlarged field of view (90) by increasing the spatial extent of the data, or an improved resolution (91) by increasing the spatial frequency extent of the data. Mosaicing can be divided into the steps of transformation estimation and image fusion. It has been used in MRI using a context similar to that of many computer vision applications (92) to extend the field of view of an image, and in confocal microscopy, an elegant Lie group framework has been proposed for collective transformation estimation (93). In the context of fetal MRI, we need to combine regions of images containing a moving object (94) surrounded by confounding moving structure. As illustrated in figure 2, given a set of N 2D slice images In(xn), n ∈ {1 … N }, in order to make use of the partial data provided by each slice we need to estimate the full rigid 3D transformation y = Tn(xn) that maps from points xn in the brain in each 2D slice, to a consistent 3D anatomical coordinate frame y, in which a true 3D image V (y) is to be formed. Conventional 2D to 2D photographic mosaicing approaches can explicitly consider shared structure in overlapping images to refine alignment. However, here we have a 2D to 3D matching problem involving the collective alignment of slices within a volume space. Unlike previous work on slice to volume matching in medical imaging, where individual slices from an interventional MRI study (95), postmortem study (96), anatomical atlas (97), or functional MRI study (98) are matched to a full 3D volume acquired separately, here we do not have a reference 3D volume.
Figure 2.
The motion scattered multislice to volume matching problem: Combining fast multislice MRI acquisitions by retrospective fusion to a 3D volume: Each individual slice n making up the volume must be accurately located and orientated within the final 3D anatomical space by a full 3D rigid transformation Tn(xn) consisting of 3 rotations and 3 translations. Once these are estimated a final volume image can be reconstructed on a regular voxel lattice using scattered data interpolation techniques.
3.1 Reconstruction Based Slice Motion Estimation
The first practical approach to 3D fetal brain imaging (99, 100), proposed a two step process, by combining ideas from slice to volume matching and image mosaicing. The iterative approach involves the formation of a putative 3D volume image from the scattered slice data using a working estimate of individual slice transformations. Slice alignment to this volume is then refined and used to form an improved 3D volume for the following iteration. A 3D image can be formed from slice images and a given set of slice transformations, using adaptations of conventional scattered data interpolation schemes (101–103) to re-sample values from sets of arbitrarily orientated slices onto a regular voxel lattice. Refinement of alignment is achieved by defining an image similarity measure 𝒮(In(), Vi()) between the slice image intensities In(x) and the reconstructed volume image intensities Vi(y) at iteration i. A gradient ascent scheme is used to maximize 𝒮(In(), Vi()) with respect to Tn(x), for each slice.
The problem of estimating full rigid transformations (three rotations and three translations) for each individual slice is highly under determined and requires constraints to provide a tractable route to a solution. Image matching schemes often make use of an hierarchical multi resolution approach to avoid local optima, by first estimating coarse misalignment from lower resolution images. In a similar way, methods to estimate slice alignment can estimate bulk motion of all slices first, and then refine the estimate to smaller and smaller groups of slices (100) to recover fine scale structure in the motion trajectory. However, the slice acquisitions within a multislice fetal MRI study are commonly interleaved to avoid slice to slice crosstalk by acquiring all odd and then all evenly spatially indexed slices. Thus, hierarchical estimation of fetal trajectory from the slice stack must also account for temporal ordering of the slice acquisitions.
This two step reconstruction and motion estimation scheme essentially acts to sharpen the next reconstructed 3D volume, as slice alignment with the current volume is refined to remove inconsistent contributions from slices that can be explained by their spatial transformation parameters. This process can be related to retrospective k-space focusing methods of (72), but acting entirely within the spatial domain to allow localization of the motion correction to the anatomy of interest (here the fetal head).
This iterative ”reconstruction based” approach, making use of an intermediate volume, was emulated by later methods (104, 105), using different approaches to volume reconstruction. In (104), by implicitly assuming that through plane motions are unlikely to occur, the data acquisition requirement was simplified to a single plane orientation repeated over time.
3.2 Intersection Based Slice Motion Estimation
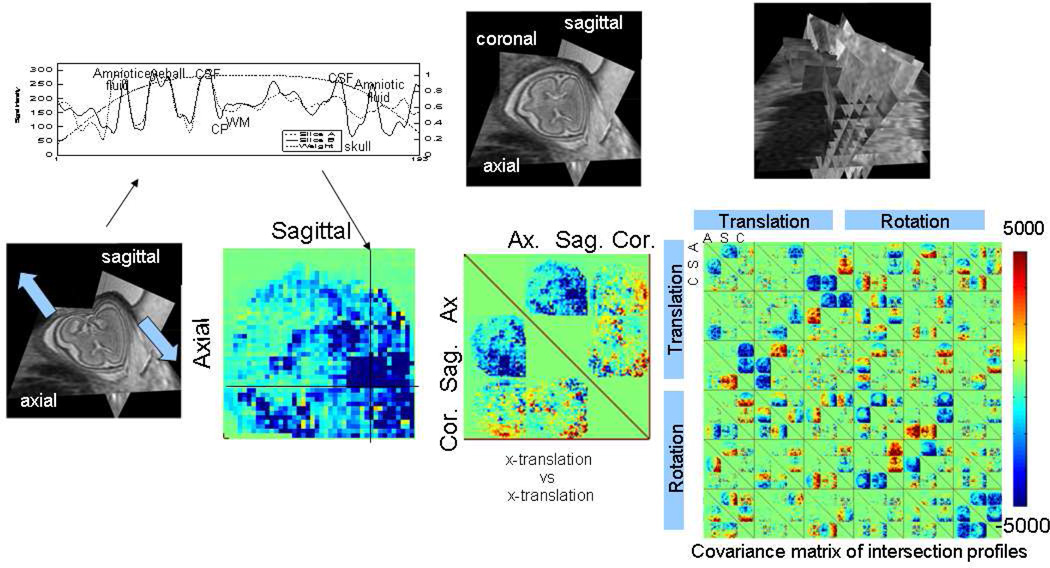
The reconstruction based motion estimation approaches make use of an iterative scheme alternating between reconstruction of a 3D image and matching slices to that image. Although this has been experimentally found to be effective, it suffers from a lack of proven convergence properties. Problems can occur where slice sample density falls during slice motion estimation, inducing local blurring in the putative volume reconstruction which then impacts the resulting slice alignment step. A more recent approach avoids these issues by removing the need for a volume reconstruction step (106, 107) for motion estimation. This approach, in effect, extends 2D image mosaicing to the extreme case of considering overlap at the intersection of any pairs of slices that have been acquired in approximately orthogonal orientations. As illustrated in Figure 3, for a single pair of intersecting slices, the image structure along this intersection must match when they are consistently mapped into the 3D space. This match provides partial geometric constraints on the relative location of the two slices (for example limiting the sliding along an intersection, but still allowing rotation as a hinge around the intersection). By considering all intersecting slice pairs, as illustrated in Figure 3, we can seek slice transformations that collectively resolves all relative mismatches in the acquired data. Using a sum of squared difference in image intensity as a matching criteria for the MR slices, it is possible to formulate the collective alignment in a least squares framework that is accessible to efficient optimization. This has the computational advantage of not requiring a volume 3D reconstruction at each step and potentially provides improved accuracy due to avoidance of the blurring which can occur in the 3D reconstruction process. It is interesting to relate the slice intersection approach to that of using an additional linear navigator echo to acquire motion information. Each intersection of a slice pair provides an equivalent one dimensional source of information about the object motion.
Figure 3.
An illustration of intersection based slice motion estimation: Any intersecting pairs of slices through the moving fetal head must match along their intersection when mapped correctly together into the 3D anatomical space. The square difference in intensity along an intersection is related to corresponding changes in the spatial transformation applied to each slice. By considering all slice pairs acquired in a multislice study consisting of many stacks of slices, a numerical optimization scheme can be formulated to minimize discrepancies, using the covariance matrix of all matches with respect to the slice transformation parameters. This collectively adjusts the 3 translations and 3 rotations of each slice to bring them into mutual alignment.
3.3 Anatomical Localization of Motion Estimates
In addition to the rigidly moving fetal head, fetal MRI slices generally include a large fraction of maternal anatomy and fluid, deforming or flowing around the fetal head which must be excluded from the motion estimation process. This can be achieved by approximate cropping of the data (100) around the fetal head, combined by the use of a robust image matching criteria. Later work (104) used a careful manual delineation of the fetal brain in each acquired slice to achieve local matching. More recently methods (107) have incorporated an explicit spatial windowing in the motion estimation process, which is aligned with the data during motion estimation.
3.4 Improving Volume Reconstruction
In the first approaches (100) to combining motion scattered multi-slice fetal brain slices, a Gaussian distance weighted interpolation was used to re-grid the scattered slice data onto a regular voxel lattice. This was motivated by the known slice profile and was robust to missing regions of data. Later work (104) used an alternative B-Spline scattered interpolation to provide a sharper rendition of structures from the combined slice data when many slice stacks were available. In (107), the general approach of kernel based interpolation was further refined using an edge weighted interpolation scheme. This acts to enhance the contributions of structure acquired in different slice orientations. In new work (105, 108, 109), the scattered data interpolation was extended further to incorporate concepts from super resolution image reconstruction to enhance the fidelity of the reconstructed image where the anatomy is significantly oversampled by many repeated acquisitions.
A second aspect of the volume reconstruction problem is that of changing MRI intensity inhomogeneity or ’bias’ across the field of view (110). This is particularly a problem in abdominal imaging where the anatomy of interest can be far from the phased array surface coils used in image formation. In fetal imaging, when the fetus moves with respect to the coils, the spatial relationship changes and therefore the in-slice distortions of the same anatomical region can be changed. When these images are combined to form a single volume, these differences induce artefactual structures that can obscure subtle underlying tissue boundaries present in the fetal anatomy such as the sub-plate (111), making them difficult to quantify. Rather than directly applying correction methods (112) to the final volume, we must consider the individual relative slice distortions contributing to volume reconstruction errors. To address this a relative slice bias correction scheme was incorporated into the final reconstruction (100). This assumed that one 3D stack contained limited fetal motion and could therefore be used as a reference for relative intensity correction of all remaining slices. This has then been developed further using a slice intersection framework (107) to provide a direct relative bias estimation for each slice. Here all individual slice bias fields are brought into agreement based on discrepancies along all slice pair intersections (113), removing the need to assume a minimal motion reference stack is present in the study.
3.5 Advantages of Retrospectively Combining Clinical Images
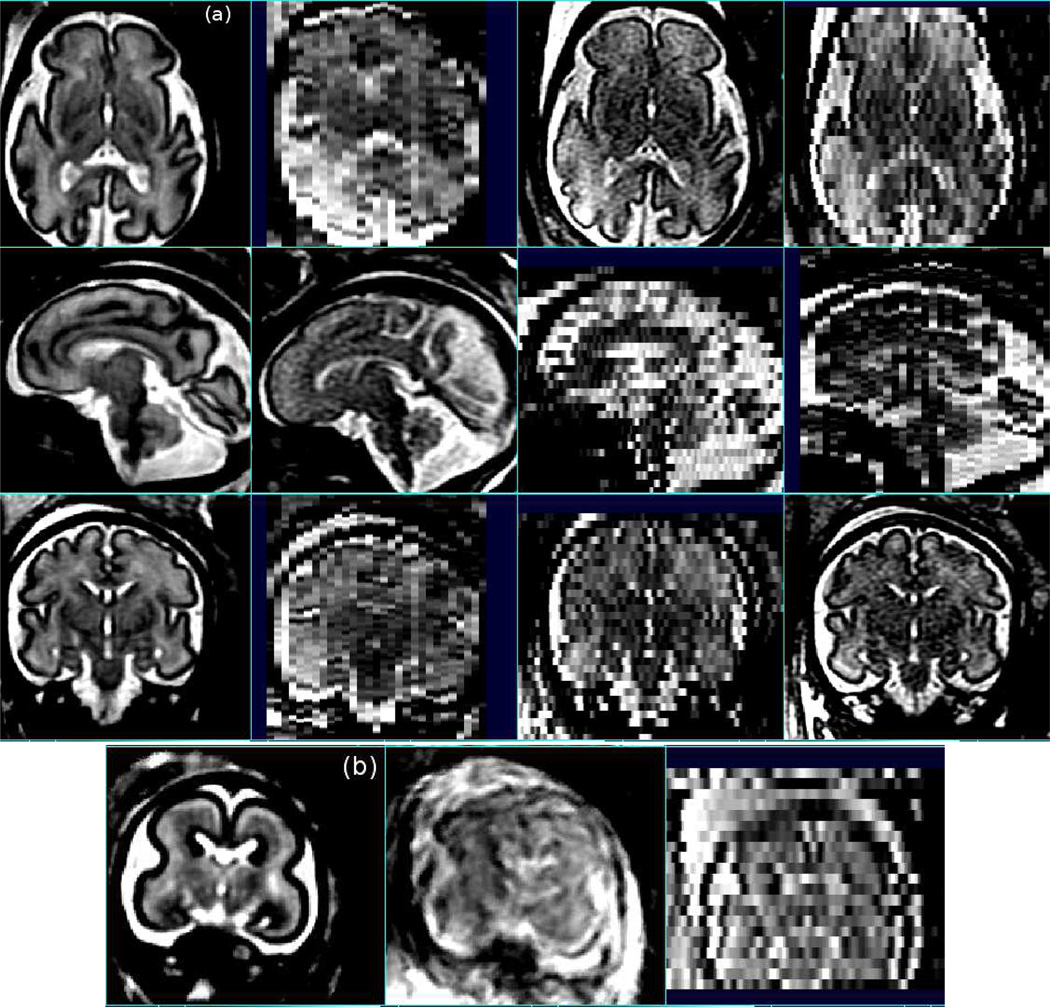
An example of the ability of retrospective multi-slice motion correction techniques to recover extreme motion in a study of a younger fetal brain is shown in Figure 4 (b). Critically, this step allows the clinician and neuroscientist to study normal fetal anatomy without biasing studies to those without motion artifact or who undergo sedation. The process of combining multiple clinical studies into a single coordinate system provides two key advantages in addition to 3D image formation, as illustrated by Figure 4 (a). Firstly, it allows signal averaging across multiple data acquisitions through the same anatomy, to improve contrast to noise and help delineation of subtle tissue boundaries such as those of the sub-plate (111). In addition, the original clinical slice data suffers from highly anisotropic resolution (i.e. fine in-plane resolution, but considerably greater slice thickness). This induces partial volume artifacts and poses considerable challenges when attempting to visualize and quantify cortical folding. By combining complementary structure delineated in slices with high in-plane resolution, but different orientation, improved isotropic resolution is possible (107). The need to re-plan a scan or manually ’chase’ a fetal head during a typical clinical study to ensure slices are in a specific orientation for radiological inspection is reduced by the post-hoc creation of a true 3D volume image, and this can significantly reduce study time for the mother and fetus. Finally, by making use of retrospective correction of scans, the clinical 2D slice are still available for radiological inspection where needed.
Figure 4.
(a) Example motion estimation (107), relative intensity bias correction (113) and reconstruction into a single 3D volume (orthogonal slices in left column) of individual sagittal, axial and coronal slice stacks (right three columns) acquired from a clinical fetal brain study using fast multi-slice imaging (note the original slices are shown in acquired coordinates before motion estimation and therefore are not precisely aligned). These illustrate the valuable increase (left column) in the isotropic resolution and contrast to noise resulting from the accurate fusion of multiple fast multi-slice imaging studies of the moving fetus. (b) Motion correction and fusion of MRI scans of a younger fetal brain with extreme motion shown: reconstructed with individual slice motion estimation (left) and without (center), together one of the original slice stacks (right).
4 Quantifying and Mapping Brain Growth in utero
4.1 Automated Fetal MRI Brain Tissue Segmentation
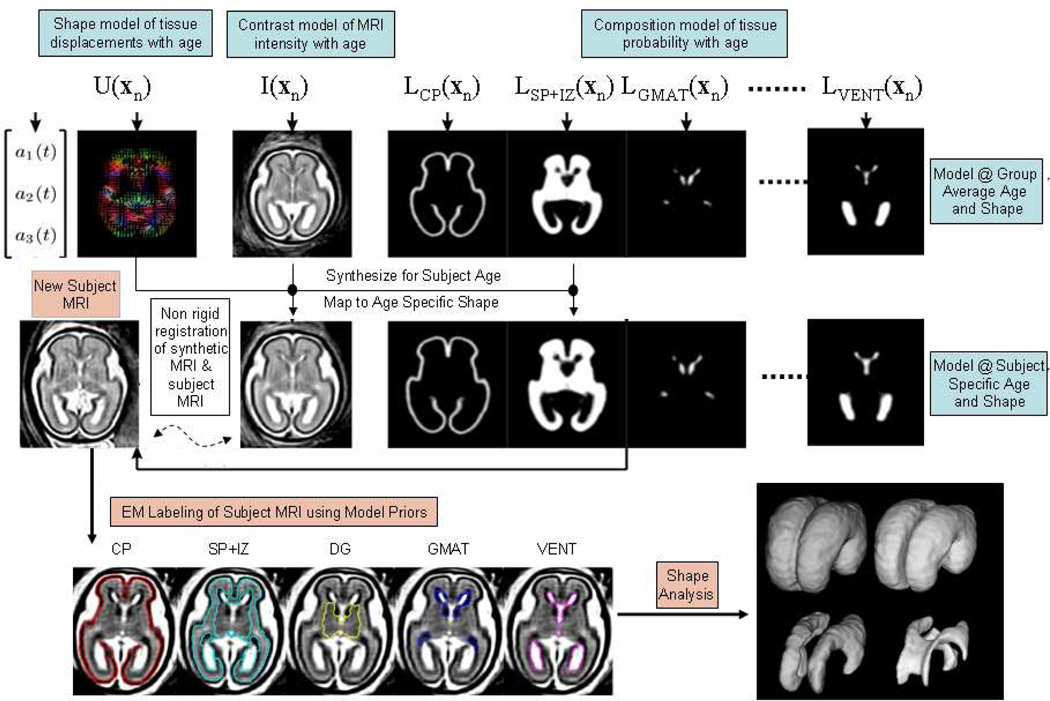
The first step in quantifying human brain anatomy from MRI scans is often to take the MRI scan and assign the voxels (or pixels) with labels indicating the most likely type of tissue at that location. Such MRI segmentation techniques, developed initially to automatically partition the adult human brain into tissue classes, have been extensively developed over the past 20 years (114). However, the challenges posed by the study of developing fetal brain anatomy have necessitated new directions in brain image segmentation methods. Many of the most accurate and robust approaches to adult brain tissue segmentation make use of parametric statistical models of the image contrast and noise (115, 116). These employ Gaussian mixture models (117), which capture the piecewise uniform nature of the tissue regions in the brain. The most widely used approaches also use atlas based priors on the probability of tissues over the brain to initialize and constrain a libeling of new MRI scans (118, 119) and provide spatial context to the labeling. These prior maps are estimated in a common canonical coordinate frame (120) and deformed into the space of a new unlabeled MRI scan using a non-rigid deformable image registration. Labeling is then adapted to the new brain MRI by combining the prior with a likelihood for the data conditioned on the labels and applying Maximum a posteriori (MAP) or maximum likelihood (ML) methods (121, 122).
In the most commonly used T1 weighted MRI of the adult and child, visible tissue classes commonly consist of gray matter and white matter and cerebrospinal fluid. However, in fetal brain imaging, T2 weighted (T2W) is typically used. At the time of initial clinical MR imaging (around 20–25 weeks), the fetal brain in utero consists of a layered structure of up to 7 different tissue zones (123), some of which are visible in clinical imaging at different stages of development. These zones, illustrated in Figure 5, include the germinal matrix, cortical plate (which will become cortical Grey matter), the sub-plate and the intermediate zone (which will become the White Matter). In early work on automated fetal tissue segmentation, approaches were first explored that extended atlases to contain priors on these developing tissues at a given developmental stage (124). These were built from carefully manually delineated MRI scans to provide an accurate statistical reference for a given age of development.
Figure 5.
An illustration of automated atlas based tissue segmentation of a 3D T2W fetal brain MRI using an age specific prior map of tissue probability, MRI contrast and brain shape and size. Labels assigned to each voxel dividing the brain into Cortical Plate, Intermediate Zone and Sub-Plate, Deep Grey Matter, Germinal Matrix and Ventricular CSF, are adapted from the atlas prior to fit the subject MRI scan using an iterative Expectation Maximization algorithm (132).
The use of subject or age specific templates, in both children (125,126), neonates (127–129), and even in adults (130), has been shown to significantly improve the tissue Labeling process. However, unlike clinical studies of adults or children, fetuses cannot easily be studied at a precise developmental stage. This is because the speed of changes in the developing brain occur on the order of weeks or days, combined with the uncertainty of the age of a fetus (131). Thus, the focus of research has been to develop continuous or computational atlases of the fetus, capable of modeling any given age. This parametrically models changes in shape, size, MRI contrast and tissue probability at every point in the fetal brain (132), as illustrated in figure 5. This enables the synthesis, for any given gestational age, of a specialized MRI template with representative tissue contrast and tissue probabilities. Given the difficulty and ethical considerations of carrying out repeat imaging on the same expectant mother, this atlas cannot be directly constructed from repeated longitudinal imaging as in adult analysis (1), but is naturally constructed from many different fetuses, each scanned at different ages. This allows the atlas to form a mean growth trajectory which is representative of a population and also encodes the natural variation of development together with uncertainties of estimation of fetal age (131). Further methodological research has also explored the development of fetal specific geometric priors in segmentation, which are aimed at improving tissue Labeling using the layered structure of the zones of tissues in the fetal brain. By making use of the fact that inner and outer boundaries of the brain may be delineated most easily from clinical scans, the more subtle tissue boundaries between them can be assumed to occur in a defined order from inside (germinal matrix) to outside (cortical plate). A statistical model of the occurrence of the tissues zones at different relative depths in the brain can then be used as an additional spatial prior to further improve the segmentation by helping resolve ambiguities in the labeling process (133). Such developmentally specific geometric models may provide routes to further improving many steps in fetal brain image analysis.
4.2 Mapping Regional and Local Patterns of Tissue Volume Increase
There have been a number of important studies of whole fetal brain tissue volume growth derived both from postmortem studies and in utero imaging. Manual segmentations of cerebral mantle tissue zones on MRI describe the different growth trajectories for the overall cerebral brain, germinal matrix, and ventricular volumes (134–136). Additionally, manual 2D measurements of laminar thickness suggest regionally varying thickness in cortex and sub-plate (137). The recent use of motion correction techniques to build 3D images has provided true volumetric estimates of whole brain (138) and subplate (111) in utero. Most recently, the development of automated tissue segmentation has begun to allow larger scale volumetric studies of tissue zones from motion corrected in utero imaging (124). While adult studies can parcellate the brain into regions (eg lobes) based on cortical landmarks, to study local volume changes, these landmarks are often absent or inconsistent in the smooth fetal brain, and therefore alternative and often more simple anatomical divisions must be used.
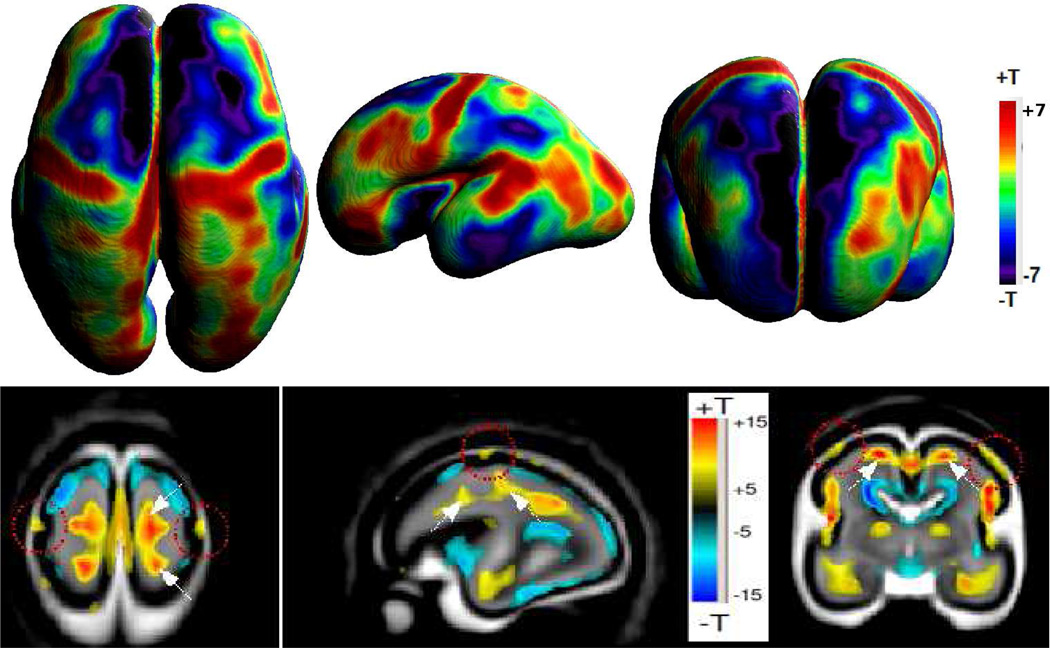
At a very local level, the growth of a sulcated neonatal brain from a smooth fetal brain requires a complex series of local tissue volume changes to form the individuals cortical folding pattern. Tensor based morphometry (139) (TBM) uses accurate spatial normalization of brain anatomy into a common coordinate frame to study patterns of size differences in local anatomy. Statistical analysis of properties of the Jacobian map of the non-rigid deformations required to bring each anatomy into the common coordinate system allows local tissue volume to be related to age and clinical variables. Such an approach requires modification to be used in the developing brain because of the dramatic changes in tissue contrast occurring over short time scales. Direct deformable registration of fetal MRI scans can induce artefactual deformations as image warping attempts to account for inconsistent tissue contrasts when mapping from one fetal age to another. By first employing automated tissue segmentation to extract age consistent tissue boundaries, recent work has shown it is possible to apply TBM successfully to map anatomical changes (140). Figure 6 illustrates the use of TBM to detect the locally varying pattern of expansion of tissue (regions that grow faster or slower than the overall brain), indicating where additional cortical complexity is being added.
Figure 6.
Example Tensor Based Morphometry of the pattern of tissue growth between 20 and 28 gestational weeks: Statistical significance maps show points where the rate of volume increase is greater or lesser than the rate of growth of the brain as a whole. The lower row shows slices within the brain volume, while the top row shows differences across the cortical plate only. These local variations in tissue growth rate create structural complexity as the primary sulci form.
4.3 Quantifying and Mapping Cortical Folding
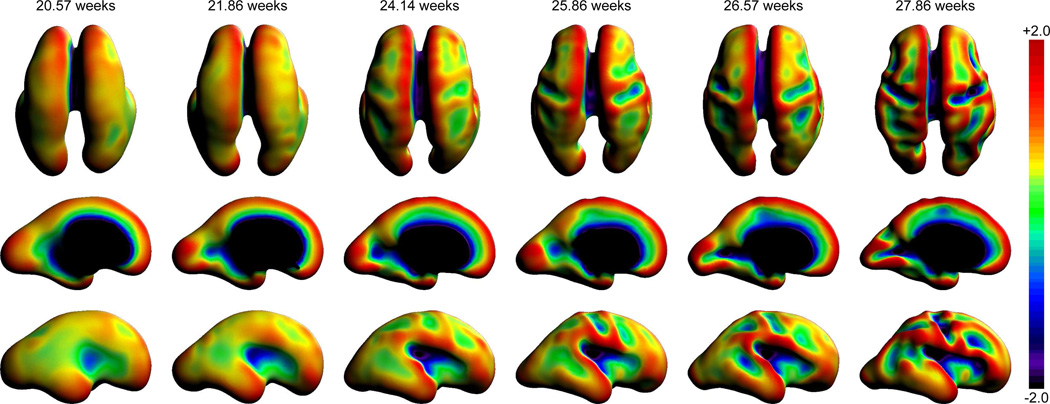
Postmortem studies have shown that the primary and secondary sulci form in a consistent spatial and temporal order during normal gestation, allowing the originally smooth fetal brain to dramatically increase cortical surface area. The timing of this has been considered an accurate marker of brain development (141) in clinical radiological inspection of MRI slices, with perturbations to this pattern providing potential stable biomarkers for abnormal functional development. Quantitative analysis of fetal brain folding from postmortem studies has shown the promise of full 3D surface curvature analysis (142). In particular, the two invariants of maximum and minimum curvature at each point on a surface give rise to a range of shape measures such as mean and Gaussian curvature (143) and shape index (144). These provide valuable quantitative ways of tracking folding that have been used in preterm brain studies (145). By creating surface based representations of the human fetal cortex from motion corrected in utero MRI, these methods have begun to be applied to the earliest stages of cortical folding. Using surface tessellation and quadric modeling of surface shape, the curvature of the fetal surface can be accurately quantified and mapped, forming a population based statistical model of the normal progression of brain surface curvature. Results show (146) that it is possible to use this approach to detect the subtle early stages of folding at the start of sulcal formation as illustrated in Figure 7 for normally developing fetuses at different developmental ages.
Figure 7.
Example maps of surface mean curvature of the boundary between the cortical plate and the sup-plate (equivalent to the gray matter/white matter interface in adults), evaluated on surface mesh representations automatically constructed from in utero MRI scans of fetuses with different gestational ages. These illustrate surface folding progression in terms of mean curvature increases (convexity) and decreases (concavity) as the early primary sulci common to all adults form. These include the sylvian fissure, the pre and post central sulci and the calcarine sulcus.
5 Future Directions
5.1 Imaging Developing Tissue Microstructure: Diffusion MRI
Imaging of tissue microstructural properties and connectivity of white matter tracts has emerged as a powerful structural modality both in adult brain studies and in childhood and adolescence. Diffusion weighted imaging (DWI), together with Diffusion tensor imaging (DTI) are MRI techniques (147, 148) that provide measures of microstructural tissue integrity (149, 150). These use fast MRI acquisitions to provide directionally sensitive measures of water diffusion within tissue. Research from clinical imaging has shown that DWI is feasible in some fetal brain studies (151–153) where there is limited motion, and that it can provide significant additional information in cases of suspected abnormalities (154, 155). However, studies have often been limited to a small number of slices and have limited anatomical coverage. DTI studies often involve the acquisition of larger numbers of directional measurement images to provide a more complete profile of water diffusion at each point in the brain, but are therefore even more sensitive to motion. In postmortem fetal imaging (156) it has shown great promise in studying the development of white matter tracts. Fetal DTI studies in utero, though requiring a longer imaging time, has also been successful (157) in providing insights into the formation of connections within the brain. However, much of this in utero research has been limited to studies of older fetuses or cases where the fetal head is engaged in the maternal pelvis, where fetal motion was constrained, and is still therefore not yet feasible for general clinical imaging or larger scale neuroscience studies.
Most recently, slice motion correction techniques for diffusion tensor imaging have been proposed (158) and adapted (159, 160). Since each directional diffusion measurement is with respect to a specific coordinate frame, motion of the fetal head between the acquisition of the images not only modifies the intended anatomical location but also the anatomical orientation of the diffusion measurement. By repeatedly acquiring multiple sets of each directionally weighted diffusion image, missing directions and locations in the scattered data can be filled in (158), at the expense of additional imaging time. Within slice motions cannot be easily be recovered since they corrupt the actual diffusion measurement, however the number of these are limited by the shorter slice acquisition time of EPI data, and they can be excluded as outliers in the fitting process. For the remaining data, different slice motion estimation methods have been proposed making use of slice to volume matching. One of the key challenges in this problem is the low signal to noise of the diffusion weighted imaging, which can impact the ability to accurately recover slice positioning. Approaches simplify the process by either first constructing a reference non-diffusion weighted image and estimating remaining slice motion relative to that (158), or by constructing a reference tissue map from conventional MRI and estimating motion using a maximum likelihood framework (159) which additionally allows the estimation of geometric slice distortion present in the DTI slices. The final reconstruction involves a more complex process which must combine spatially and directionally scattered diffusion measurements made during fetal motion into a single diffusion tensor field on a regular voxel lattice. This scattered data fitting process can be formulated in a least squares framework (158) with constraints to preserve the positive definite properties in the final diffusion tensors. However, given the scattered nature of the fetal diffusion measurements, it is natural to consider more general higher order diffusion models which can take into account the larger spread of diffusion measurements induced by fetal motion, and provide a route to resolving ambiguities in areas of crossing fibers. This approach has been shown to improve on FA measurements for fetal data (160). Once a regularly sampled set of diffusion profiles has been estimated, it can be analyzed using techniques such as DTI tractography to reconstruct the developing white matter tracts within the fetal brain.
5.2 Imaging Developing Brain Function: Functional MRI
A second area of key interest in brain development is the measurement of brain function. Functional MRI (fMRI) is used to provide a window into short term blood oxygen level changes in the brain (161) in adults and children that are related to underlying tissue function. These have been used to map both responses to external stimuli (eg visual or auditory) or to map so called resting state brain activations (162). Such capabilities are of significant interest in fetal imaging to provide an understanding of developing brain function. Work on resting state activations (without stimuli) have shown great promise in mapping the development of the so-called ’default mode’ in infants (163) and children (164,165), and extending this work to show earlier stages of this process is of significant interest. Early work on fetuses has shown that it is possible to apply stimulus driven methods, in cases of limited fetal motion, to study response to auditory stimuli (166), and visual stimuli (167, 168). All fMRI methods however rely on the detection of subtle temporal variations in signal at each location in the brain across many repeated scans, and as such are highly sensitive to motion. In adults even small motions need to be corrected for using techniques available in standard software tools to ensure no stimulus correlated motions remain. For the techniques to be generally applicable for use in fetuses, either clinically or for realistically sized and unbiased neuroscience studies, it is essential that large scale fetal head motion correction of the data be possible. Techniques developed for structural image motion correction should, in theory, contribute in this area, although adaptation of the scanning protocol (to include, for example, more repeated acquisitions to ensure full spatial-temporal coverage) may be required. An important additional factor is also that of possible changes in geometric distortions that can occur in echo planar imaging data. Finally, more work is also needed in the post-hoc functional data analysis, to address the possibility of missing or scattered spatio-temporal data.
6 Conclusion
This paper has reviewed the background to the development of MRI as a tool to image the human fetal brain in utero in 3D without sedation. These developments are beginning to open a new window into early human brain growth that has so far been inaccessible to the neuroscientist and clinician. The availability of true, high resolution 3D imaging of growing brain tissues will allow us to examine the key changes in the brain that form the basis for adult brain structure and function. These measurements include the precise spatial and temporal pattern of cortical folding and the process by which a unique folding pattern emerges in individuals. Such data may contribute to further refining and validating theories of brain folding (169–171). Measurements made in utero complement histological studies that provide a cellular level understanding of tissues which are unable to examine change and growth over time or to study 3D structural variability across large populations. An important new direction of future work will be to build stronger bridges between in utero measurements and histology. In terms of imaging methodology, two key directions for new work are on the development of fetal brain functional imaging and the imaging of the formation of white matter tracts from diffusion imaging. Together, these imaging methods will be particularly important in understanding the process of initiating normal brain function: i.e. understanding the processing of ’software booting’ of brain function that must occur when the underlying hardware has developed. Understanding the time-line of functional development and how it relates to structural changes in humans will be an interesting new area of research which may bring new understanding of adult brain function and its variation. Another interesting direction is that of linking genetic information to accurate growth maps in order to understand in detail how genes modulate the pattern and order of local growth on a macroscopic scale. Better knowledge of the spatial and temporal variability of these events in healthy fetal brain growth may allow the construction of biomarkers for abnormal cortical development, which may have important applications in poorly understood conditions such as ventriculomegaly (78, 79). In terms of direct clinical intervention, the increased interest in fetal surgery (172) driven by newer minimally invasive techniques (173) will depend on the availability of high quality imaging both for diagnosis, planning and guidance (174). In conclusion, the combination of MRI physics with computer vision and biomedical image analysis techniques promises to dramatically expand our understanding of the human brain and provide a range of powerful new diagnostic tools.
Acknowledgments
The author would like to thank the NIH for research support (NS 061957,NS 055064) during the preparation of this manuscript and the many contributions from the postdoctoral scholars who have worked in his lab including: Dr K. Kim, Dr P. Habas, Dr V. Rajagopalan, Dr J. Scott, Dr F. Rousseau, Dr C. Rodriguez and Dr C. Drapaca, and his collaborators Dr A.J. Barkovich Dr O.A. Glenn and Dr V. Cardenas for important contributions to this work.
Literature Cited
- 1.Toga Arthur W, Thompson Paul M. Temporal dynamics of brain anatomy. Annual Review of Biomedical Engineering. 2003;5:119–145. doi: 10.1146/annurev.bioeng.5.040202.121611. [DOI] [PubMed] [Google Scholar]
- 2.Mazziotta JC, Toga AW, Evans AC, Fox P, Lancaster J. A probabilistic atlas of the human brain: theory and rationale for its development. NeuroImage. 1995;2:89–101. doi: 10.1006/nimg.1995.1012. [DOI] [PubMed] [Google Scholar]
- 3.Miller Michael I, Trouve Alain, Younes Laurent. On the metrics and euler-lagrange equations of computational anatomy. Annual Review of Biomedical Engineering. 2002;4:375–405. doi: 10.1146/annurev.bioeng.4.092101.125733. [DOI] [PubMed] [Google Scholar]
- 4.Thompson Paul M, Mega1 Michael S, Woods Roger P, Zoumalan Chris I, Lindshield Chris J, Blanton Rebecca E, Moussai Jacob, Holmes Colin J, Cummings Jeffrey L, Toga Arthur W. Cortical change in alzheimer’s disease detected with a disease-specific population-based brain atlas. Cereb. Cortex. 2001;11(1):1–16. doi: 10.1093/cercor/11.1.1. [DOI] [PubMed] [Google Scholar]
- 5.Lerch JP, Pruessner JC, Zijdenbos A, Hampel H, Teipel SJ, Evans AC. Focal decline of cortical thickness in alzheimers disease identified by computational neuroanatomy. Cereb Cortex. 2005;15:995–1001. doi: 10.1093/cercor/bhh200. [DOI] [PubMed] [Google Scholar]
- 6.Cardenas Valerie A, Boxer Adam L, Chao Linda L, Gorno-Tempini Maria L, Miller Bruce L, Weiner Michael W, Studholme Colin. Deformation-based morphometry reveals brain atrophy in frontotemporal dementia. Arch. Neurol. 2007;64(6):873–877. doi: 10.1001/archneur.64.6.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narr KL, Bilder RM, Toga AW, Woods RP, Rex DE, Szeszko PR, et al. Mapping cortical thickness and gray matter concentration in first episode schizophrenia. Cereb Cortex. 2005;15:708–719. doi: 10.1093/cercor/bhh172. [DOI] [PubMed] [Google Scholar]
- 8.Cardenas VA, Studholme C, Gazdzinski S, Durazzo TC, Meyerhoff DJ. Deformation-based morphometry of brain changes in alcohol dependence and abstinence. Neuroimage. 2007;34(3):879–887. doi: 10.1016/j.neuroimage.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cardenas VA, Meyerhoff DJ, Studholme C, Kornak J, Rothlind J, Lampiris H, Neuhaus J, Grant RM, Chao LL, Truran D, Weiner MW. Evidence for ongoing brain injury in human immunodeficiency virus-positive patients treated with antiretroviral therapy. J. Neurovirol. 2009;15(4):324–333. doi: 10.1080/13550280902973960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lenroot Rhoshel K, Gogtay Nitin, Greenstein Deanna K, Molloy Wells Elizabeth, Wallace Gregory L, Clasen Liv S, Blumenthal Jonathan D, Lerch Jason, Zijdenbos Alex P, Evans Alan C, Thompson Paul M, Giedd Jay N. Sexual dimorphism of brain developmental trajectories during childhood and adolescence. NeuroImage. 2007;36:1065–1073. doi: 10.1016/j.neuroimage.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aljabar P, Bhatia KK, Murgasova M, Hajnal JV, Boardman JP, Srinivasan L, Rutherford MA, Dyet LE, Edwards AD, Rueckert D. Assessment of brain growth in early childhood using deformation-based morphometry. NeuroImage. 2008;39:348–358. doi: 10.1016/j.neuroimage.2007.07.067. [DOI] [PubMed] [Google Scholar]
- 12.Brun CC, Nicolson R, Lepore N, Chou YY, Vidal CN, DeVito TJ, Drost DJ, Williamson PC, Rajakumar N, Toga AW, Thompson PM. Mapping brain abnormalities in boys with autism. Human Brain Mapping. 2009;30:3887–3900. doi: 10.1002/hbm.20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gogtay Nitin, Giedd Jay N, Lusk Leslie, Hayashi Kiralee M, Greenstein Deanna, Vaituzis A. Catherine, Nugent Tom F, Herman David H, Clasen Liv S, Toga Arthur W, Rapoport Judith L, Thompson Paul M. Dynamic mapping of human cortical development during childhood through early adulthood. PNAS. 2004;101(21):8174–8179. doi: 10.1073/pnas.0402680101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans A. The NIH MRI study of normal brain development. Neuroimage. 2006;30:184–202. doi: 10.1016/j.neuroimage.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 15.Patterson JA, Gold WR, Sanz LE, McCullough DC. Antenatal evaluation of fetal hydrocephalus with computed tomography. American journal of obstetrics and gynecology. 1981;140(3):344–345. doi: 10.1016/0002-9378(81)90288-x. [DOI] [PubMed] [Google Scholar]
- 16.Pediaditis M, Leitgeb N, Cech R. RF-EMF exposure of fetus and mother during magnetic resonance imaging. Phys. Med. Biol. 2008;53:7187–7195. doi: 10.1088/0031-9155/53/24/012. [DOI] [PubMed] [Google Scholar]
- 17.Budinger TF. Nuclear magnetic resonance (NMR) in vivo studies: known thresholds for health effects. J Comput Assist Tomogr. 1981 Dec;5(6):800–811. doi: 10.1097/00004728-198112000-00003. [DOI] [PubMed] [Google Scholar]
- 18.Schaefer DJ. Safety aspects of radio-frequency power deposition in magnetic resonance. Magn. Reson. Imaging Clin. N. Amer. 1998;6:775–789. [PubMed] [Google Scholar]
- 19.Shellock FG. Radiofrequency-induced heating during MR procedures: A review. J. Magn. Reson. Imaging. 2000;12:30–36. doi: 10.1002/1522-2586(200007)12:1<30::aid-jmri4>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Gandhi OP, Chen XB. Specific absorption rates and induced current densities for an anatomy-based model of the human for exposure to time varying magnetic fields of MRI. Magn. Res. Med. 1999;41(5):816–823. doi: 10.1002/(sici)1522-2594(199904)41:4<816::aid-mrm22>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 21.Collins CM, Liu W, Wang J, Gruetter R, Vaughan JT, Ugurbill K, Smith MB. Temperature and specific absorption rate calculations for a human head within volume and surface coils at 64 and 300 MHz. J. Magn. Res. 2004;19:650–656. doi: 10.1002/jmri.20041. [DOI] [PubMed] [Google Scholar]
- 22.Nguyen UD, Brown S, Chang IA, Krycia J, Mirotznik MS. Numerical evaluation of heating of the human head due to magnetic resonance imaging. IEEE Trans. Biomed. Eng. 2004 Aug;51(8):1301–1309. doi: 10.1109/TBME.2004.827559. [DOI] [PubMed] [Google Scholar]
- 23.Wu Dagang, Shamsi Saad, Chen Ji, Kainz Wolfgang. Evaluations of specific absorption rate and temperature increase within pregnant femal models in magnetic resonance imaging birdcage coils. IEEE Transaction on Microwave Theory and Techniques. 2006 Dec;54(12) [Google Scholar]
- 24.Dimbylow Peter. SAR in the mother and foetus for RF plane wave irradiation. Phys. Med. Biol. 2007;52:3791–3802. doi: 10.1088/0031-9155/52/13/009. [DOI] [PubMed] [Google Scholar]
- 25.Hand JW, Li Y, Thomas EL, Rutherford MA, Hajnal JV. Pre diction of specific absorption rate in mother and fetus associated with MR examinations during pregnancy. Magnetic Resonance in Medicine. 2006;55:883–893. doi: 10.1002/mrm.20824. [DOI] [PubMed] [Google Scholar]
- 26.De Wilde JP, Rivers AW, Price DL. A review of the current us of magnetic resonance imaging in pregnancy and safety implications for the fetus. Prog Biophys Mol Bio. 2005;87:335–353. doi: 10.1016/j.pbiomolbio.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Gowland PA, De Wilde J. Temperature increase in the fetus due t radio frequency exposure during magnetic resonance scanning. Phys. Med Biol. 2008;53:L15–L18. doi: 10.1088/0031-9155/53/21/L01. [DOI] [PubMed] [Google Scholar]
- 28.Arulkumaran S, Talbert D, Hsu TS, et al. In-utero sound levels whe acoustic stimulation is applied to the maternal abdomen: an assessment of the possibility of cochlea damage in the fetus. Br J Obstet Gynaecol. 1992;99:43–45. doi: 10.1111/j.1471-0528.1992.tb14390.x. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber Wolfgang G, Teichmann Elke M, Schiffer Ilka, Hast Wahida Akbari Jochem, Georgi Hildegart, Graf Robert, Hehn Michael, Spi@articleManfred Thelen Han W, Oesch Franz, Hengstler Jan G. Lac of mutagenic and co-mutagenic effects of magnetic fields during magneti resonance imaging. Journal of Magnetic Resonance Imaging. 2001;14(6):779–788. doi: 10.1002/jmri.10010. [DOI] [PubMed] [Google Scholar]
- 30.Leithner Katharina, Pornbacher Susanne, Assem-Hilger Eva, Krampl Elisabeth, Ponocny-Seliger Elisabeth, Prayer Daniela. Psychological reactions in women undergoing fetal magnetic resonance imaging. Obstetrics & Gynecology. 2008 Feb;111(2) doi: 10.1097/AOG.0b013e3181610281. [DOI] [PubMed] [Google Scholar]
- 31.Foster MA, Knight CH, Rimmington JE, Mallard JR. Fetal imaging by nuclear magnetic resonance: a study in goats. Radiology. 1983;149:193–195. doi: 10.1148/radiology.149.1.6310681. [DOI] [PubMed] [Google Scholar]
- 32.Magin RL, Lee JK, Klintsova A, Carnes KI, Dunn F. Biological effects of long-duration, high-field (4T) MRI on growth and development in the mouse. J Magn Reson Imaging. 2000;12:140–149. doi: 10.1002/1522-2586(200007)12:1<140::aid-jmri15>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 33.Levine D, Zuo C, Faro CB, Chen Q. Potential heating effect in the gravid uterus during MR haste imaging. J. Magn. Reson. Imaging. 2001;13(6):856–861. doi: 10.1002/jmri.1122. [DOI] [PubMed] [Google Scholar]
- 34.Baker PN, Johnson IR, Harvey PR. A three-year follow up of children imaged in utero with echo-planar magnetic resonance. Am J Obstet Gynecol. 1994;170:32–33. doi: 10.1016/s0002-9378(94)70379-5. [DOI] [PubMed] [Google Scholar]
- 35.Clements H, Duncan KR, Fielding K, et al. Infants exposed to MRI in utero have a normal paediatric assessment at 9 months of age. Br J Radiol. 2000;73:190–194. doi: 10.1259/bjr.73.866.10884733. [DOI] [PubMed] [Google Scholar]
- 36.Kok RD, de Vries MM, Heerschap A, et al. Absence of harmful effects of magnetic resonance exposure in the third trimester of pregnancy: a follow up study. Magn Reson Imaging. 2004;22:851–854. doi: 10.1016/j.mri.2004.01.047. [DOI] [PubMed] [Google Scholar]
- 37.Thickman D, Mintz M, Mennuit M, Kressel HY. MR imaging of cerebral abnormalities in utero. J. Compt. Assisted Tomography. 1984 Dec;8(6):1058–1061. doi: 10.1097/00004728-198412000-00003. [DOI] [PubMed] [Google Scholar]
- 38.McCarthy SM, Filly RA, Stark DD, Hricak H, Brant-Zawadzki MN, Callen PW, Higgins CB. Obstetrical magnetic resonance imaging: fetal anatomy. Radiology. 1985 Feb;154(2):427–432. doi: 10.1148/radiology.154.2.3966129. [DOI] [PubMed] [Google Scholar]
- 39.Smith FW, Adam AH, Phillips WD. Nmr imaging in pregnancy. Lancet. 1983;1:61–62. doi: 10.1016/s0140-6736(83)91588-x. [DOI] [PubMed] [Google Scholar]
- 40.Levine Deborah, Barnes PatrickD, Edelman RobertR. Obstetric MR imaging. Radiology. 1999 Jun;211:609–617. doi: 10.1148/radiology.211.3.r99jn20609. [DOI] [PubMed] [Google Scholar]
- 41.Williamson Roger, Weiner Carl, Yuh William, Abu-yousef Monzer. Magnetic resonance imaging of anomalous fetuses. Obstetrics and Gynecology. 1989 Jun;73(6):952–956. doi: 10.1097/00006250-198906000-00009. [DOI] [PubMed] [Google Scholar]
- 42.Weinreb JC, Lowe TW, Santos-Ramos R, et al. Magnetic resonance imaging in obstetric diagnosis. Radiology. 1985;154:157–161. doi: 10.1148/radiology.154.1.3880601. [DOI] [PubMed] [Google Scholar]
- 43.Antuaco TL, Shah HR, Mattison DR, et al. MR imaging in high-risk obstetric patients: A valuable complement to US. Radiographics. 1992;12:91–109. doi: 10.1148/radiographics.12.1.1734485. [DOI] [PubMed] [Google Scholar]
- 44.Powell MC, Worthington BS, Buckley JM, et al. Magnetic resonance imaging (mri) in obstetrics. ii. fetal anatomy. Br J Obstet Gynaecol. 1988;95:38–46. doi: 10.1111/j.1471-0528.1988.tb06478.x. [DOI] [PubMed] [Google Scholar]
- 45.Daffos F, Forestier F, Mac Aleese J, Aufrant C, Mandelbrot L, Cabanis EA, Iba-Zizen MT, Alfonso JM, Tamraz Fetal curarization for prenatal magnetic resonance imaging. Prenatal Diagnosis. 1988;8:311–314. doi: 10.1002/pd.1970080412. [DOI] [PubMed] [Google Scholar]
- 46.Revel MP, Pons JC, Lelaidier C, Fournet P, Vial M, Musset D, Labrune M, Frydman R. Magnetic resonance imaging of the fetus: A study of 20 cases performed without curarization. Prenatal Diagnosis. 1993;13:775–799. doi: 10.1002/pd.1970130902. [DOI] [PubMed] [Google Scholar]
- 47.Haase A, Frahm J, Mattaei D, et al. Flash imaging: Rapid nmr imaging using low flip-angle pulses. J Magn Reson. 1986;67:256–266. doi: 10.1016/j.jmr.2011.09.021. [DOI] [PubMed] [Google Scholar]
- 48.Listerud J, Einstein S, Outwater E, et al. First principles of fast spin-echo. Magn Reson Q. 1992;8:199–244. [PubMed] [Google Scholar]
- 49.Hennig J, Friedburg H. Clinical applications and methodological developments of the RARE technique. Magn. Reson. Imaging. 1988;6:391–395. doi: 10.1016/0730-725x(88)90475-4. [DOI] [PubMed] [Google Scholar]
- 50.Manfield P. Multi-planar image formation using NMR spin echoes. J. Phys. C: Solid State Phys. 1977:10. [Google Scholar]
- 51.Echo planar imaging of the human fetus in utero at 0.5 T. The british journal of radiology. British Journal of Radiology. 1990 Nov;63(755) doi: 10.1259/0007-1285-63-755-833. [DOI] [PubMed] [Google Scholar]
- 52.Johnson R, Stehling MK, Coxon AMRJ, Howseman AM, Chapman B, Ordidge RJ, Mansfield P, Symonds EM, Worthington BS. Study of internal structure of the human fetus in-utero by echo-planar magnetic resonance imaging. Am J Obstet Gynecol. 1990;163:601–607. doi: 10.1016/0002-9378(90)91209-u. [DOI] [PubMed] [Google Scholar]
- 53.Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. New York: Wiley-Liss; 1999. [Google Scholar]
- 54.Bernstein Matt A, King Kevin F, Joe Zhou Xiaohong. Handbook of MRI Pulse Sequences. Elsevier; 2004. [Google Scholar]
- 55.Levine D. Three-dimensional fetal MR imaging: Will it fullfill its promise? Radiology. 2001;219:313–315. doi: 10.1148/radiology.219.2.r01ma46313. [DOI] [PubMed] [Google Scholar]
- 56.Glover GH, Pauly JM. Projection reconstruction techniques for reduction of motion effects in MRI. Magn Reson Med. 1992;28:275–289. doi: 10.1002/mrm.1910280209. [DOI] [PubMed] [Google Scholar]
- 57.Ahn CB, Kim JH, Cho ZH. High-speed spiral-scan echo planar nmr imaging-i. Medical Imaging, IEEE Transactions on. 1986 Mar;5(1):2–7. doi: 10.1109/TMI.1986.4307732. [DOI] [PubMed] [Google Scholar]
- 58.Meyer CH, Hu BS, Nishimura DG, Macovski A. Fast spiral coronary artery imaging. Magn Reson Med. 1992;28:202–213. doi: 10.1002/mrm.1910280204. [DOI] [PubMed] [Google Scholar]
- 59.Dold C, Zaitsev M, Speck O, Firle EA, Hennig J, Sakas G. Prospective head motion compensation by updating the MR-gradients and radio frequency during data acquisition. Medical Image Computing and Computer-Assisted Intervention. 2005;volume 8:482–489. doi: 10.1007/11566465_60. [DOI] [PubMed] [Google Scholar]
- 60.Zaitsev M, Dold C, Sakas G, Hennig J, Speck O. Magnetic resonance imaging of freely moving objects: prospective real-time motion correction using an external optical motion tracking system. Neuroimage. 2006;31(3):1038–1050. doi: 10.1016/j.neuroimage.2006.01.039. [DOI] [PubMed] [Google Scholar]
- 61.Tremblay M, Tam F, Graham SJ. Retrospective coregistration of functional magnetic resonance imaging data using external monitoring. Magn Reson Med. 2005;53(1):141–149. doi: 10.1002/mrm.20319. [DOI] [PubMed] [Google Scholar]
- 62.Peshkovsky A, Knuth KH, Helpern JA. Motion correction in MRI using an apparatus for dynamic angular position tracking (adapt) Magn Reson Med. 2003;49(1):138–143. doi: 10.1002/mrm.10329. [DOI] [PubMed] [Google Scholar]
- 63.Ehman R, Felmlee J. Adaptive technique for high-definition MR imaging of moving structures. Radiology. 1989;173(1):255–263. doi: 10.1148/radiology.173.1.2781017. [DOI] [PubMed] [Google Scholar]
- 64.Wang Y, Rossman PJ, Grimm RC, Riederer SJ, Ehman RL. Navigatorecho-based real-time respiratory gating and triggering for reduction of respiration effects in three-dimensional coronary MR angiography. Radiology. 1996;198:55–60. doi: 10.1148/radiology.198.1.8539406. [DOI] [PubMed] [Google Scholar]
- 65.Norris DG, Driesel W. Online motion correction for diffusion-weighted imaging using navigator echoes: application to RARE imaging without sensitivity loss. Magn. Reson. Med. 2001;45:729–733. doi: 10.1002/mrm.1099. [DOI] [PubMed] [Google Scholar]
- 66.Fu ZW, Wang Y, Grimm RC, Rossman PJ, Felmlee JP, Riederer SJ, Ehman RL. Orbital navigator echoes for motion measurements in magnetic resonance imaging. Magn. Reson. Med. 1995;34:746–753. doi: 10.1002/mrm.1910340514. [DOI] [PubMed] [Google Scholar]
- 67.Wong ST, Roos MS. A strategy for sampling on a sphere applied to 3D selective RF pulse design. Magn. Reson. Med. 1994;32:778–784. doi: 10.1002/mrm.1910320614. [DOI] [PubMed] [Google Scholar]
- 68.Brian Welch Edward, Manduca A, Grimm Roger C, Ward Heidi A, Jack Clifford R. Spherical navigator echoes for full 3D rigid body motion measurement in MRI. Magnetic Resonance in Medicine. 2002;47:32–41. doi: 10.1002/mrm.10012. [DOI] [PubMed] [Google Scholar]
- 69.van der Kouwe AJW, Benner T, Dale AM. Real-time rigid body motion correction and shimming using cloverleaf navigators. Magn Reson Med. 2006;56(5):1019–1032. doi: 10.1002/mrm.21038. [DOI] [PubMed] [Google Scholar]
- 70.Brown Timothy T, Kuperman Joshua M, Erhart Matthew, White Nathan S, Cooper Roddey J, Shankaranarayanan Ajit, Han Eric T, Rettmann Dan, Dale Anders M. Prospective motion correction of high-resolution magnetic resonance imaging data in children. NeuroImage. 2010;53:139–145. doi: 10.1016/j.neuroimage.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bonel H, Frei K, Raio L, Meyer-Wittkopf M, Remonda L, Wiest R. Prospective navigator-echo-based real-time triggering of fetal head movement for the reduction of artifacts. European Radiology. 2008;18:822–829. doi: 10.1007/s00330-007-0812-x. [DOI] [PubMed] [Google Scholar]
- 72.Atkinson D, Hill DL, Stoyle PN, Summers PE, Keevil SF. Automatic correction of motion artifacts in magnetic resonance images using an entropy focus criterion. IEEE Trans Med Imaging. 1997 Dec;16(6):903–910. doi: 10.1109/42.650886. [DOI] [PubMed] [Google Scholar]
- 73.Horton KM, Tempany CMC. MRI in pregnancy. In: Tempany CMC, editor. MR and imaging of the female pelvis. Mosby: 1995. pp. 235–260. Mosby. [Google Scholar]
- 74.Yamashita Y, Namimoto T, Abe Y, Takahashi M, Iwamasa J, Miyazaki K, Okamura H. MR imaging of the fetus by a HASTE sequence. Am J Roentgenol. 1997 Feb;168(2):513–519. doi: 10.2214/ajr.168.2.9016238. [DOI] [PubMed] [Google Scholar]
- 75.Busse RF, Riederer SJ, Fletcher JG, Bharucha AE, Brandt KR. Interactive fast spin-echo imaging. Magnetic Resonance in Medicine. 2000;44(3):339–348. doi: 10.1002/1522-2594(200009)44:3<339::aid-mrm1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 76.Glenn OA, Barkovich AJ. Magnetic resonance imaging of the fetal brain and spine: an increasingly important tool in prenatal diagnosis part 1. Am J Neuroradiol. 2006;27:1604–1611. [PMC free article] [PubMed] [Google Scholar]
- 77.Poutamo J, Vanninen R, Partanen K, et al. Magnetic resonance imaging supplements ultrasonographic imaging of the posterior fossa, pharynx and neck in malformed fetuses. Ultrasound Obstet Gynaecol. 1999;13:327–334. doi: 10.1046/j.1469-0705.1999.13050327.x. [DOI] [PubMed] [Google Scholar]
- 78.Bena Cerraf BR, Shipp TD, Bromley B, et al. What does magnetic resonance imaging add to the prenatal sonographic diagnosis of ventriculomegaly. J Ultrasound Med. 2007;26:1513–1522. doi: 10.7863/jum.2007.26.11.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kazan-Tannus JF, Dialani V, Kataoka ML, Chiang G, Feldman HA, Brown SJ, Levine D. MR volumetry of brain and CSF in fetuses referred for ventriculomegaly. American Journal of Radiology. 2007;189:145–151. doi: 10.2214/AJR.07.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ercole CD, Girard N, Cravello L, et al. Prenatal diagnosis of fetal corpus callosum agenesis by ultrasonography and magnetic resonance imaging. Prenat Diagn. 1998;18:247–253. [PubMed] [Google Scholar]
- 81.Rolo Liliam, Araujo Edward, Nardozza Luciano, de Oliveira Patrcia, Ajzen Srgio, Moron Antonio. Development of fetal brain sulci and gyri: assessment through two and three-dimensional ultrasound and magnetic resonance imaging. Archives of Gynecology and Obstetrics. 2010:1–10. doi: 10.1007/s00404-010-1691-y. [DOI] [PubMed] [Google Scholar]
- 82.James Barkovich A, Girard Nadine. Fetal brain infections. Child’s Nervous System. 2003;19(7–8):501–507. doi: 10.1007/s00381-003-0763-8. [DOI] [PubMed] [Google Scholar]
- 83.Girard Nadine, Gire Catherine, Sigaudy Sabine, Porcu Geraldine, d’Ercole Claude. Dominique Figarella-Branger, Charles Raybaud, and Sylviane Confort-Gouny. MR imaging of acquired fetal brain disorders. Child’s Nervous System. 2003;19(7–8):490–500. doi: 10.1007/s00381-003-0761-x. [DOI] [PubMed] [Google Scholar]
- 84.Ozduman K, Pober BR, Barnes P, Copel JA, Ogle EA, Duncan CC, Ment LR. Fetal stroke. Pediatric Neurology. 2004 Mar;30(3):151–162. doi: 10.1016/j.pediatrneurol.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 85.Baker PN, Johnson IR, Gowland PA, Hykin J, Adams V, Mansfield P, Worthington BS. Measurement of fetal liver, brain and placental volumes with echoplanar magnetic resonance imaging. British Journal of Obstetrics and Gynaecology. 1995;102:35–39. doi: 10.1111/j.1471-0528.1995.tb09023.x. [DOI] [PubMed] [Google Scholar]
- 86.Gong QY, Roberts N, Garden AS, Whitehouse GH. Fetal and fetal brain volume estimation in the third trimester of human pregnancy using gradient echo MR imaging. Magnetic Resonance Imaging. 1998;16(3):235–420. doi: 10.1016/s0730-725x(97)00281-6. [DOI] [PubMed] [Google Scholar]
- 87.Limperopoulos C, Tworetzky W, McElhinney DB, Newburger JW, Brown DW, Robertson RL, Guizard N, McGrath E, Geva J, Annese D, Dunbar-Masterson C, Trainor B, Laussen PC, du Plessis AJ. Brain volume and metabolism in fetuses with congenital heart disease evaluation with quantitative magnetic resonance imaging and spectroscopy. Circulation. 2010;121:26–33. doi: 10.1161/CIRCULATIONAHA.109.865568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grossman R, Hoffman C, Mardor Y, Biegon Anat. Quantitative MRI measurements of human fetal brain development in utero. Neuroimage. 2006;33(2):463–470. doi: 10.1016/j.neuroimage.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 89.Hartley R, Zisserman A. Multiple View Geometry. UK: Cambridge University Press; 2000. [Google Scholar]
- 90.Shum H, Szeliski R. Construction and refinement of panoramic mosaics with global and local alignment; IEEE Int’l Conf. Computer Vision; 1998. pp. 953–958. [Google Scholar]
- 91.Capel D, Zisserman A. Automated mosaicing with super-resolutionzoom. Computer Vision and Pattern Recognition. 1998 [Google Scholar]
- 92.Wachinger C, Glocker B, Zeltner J, Paragios N, Komodakis N, Hansen MS, Navab N. Proceedings Med Image Comput Comput Assist Interv. Springer; 2008. Deformable mosaicing for whole-body MRI; pp. 113–121. [DOI] [PubMed] [Google Scholar]
- 93.Aymeric Perchant Tom Vercauteren, Malandain Gregoire, Pennec Xavier, Ayache Nicholas. Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy. Medical Image Analysis. 2006;10:673–692. doi: 10.1016/j.media.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 94.Davis J. Mosaics of scenes with moving objects. Computer Vision and Pattern Recognition. 1998 [Google Scholar]
- 95.Fei Baowei, Duerk Jeffrey L, Boll Daniel T, Lewin Jonathan S, Wilson David L. Slice-to-volume registration and its potential application to interventional MRI-guided radio-frequency thermal ablation of prostate cancer. IEEE Transactions on Medical Imaging. 2003 Apr;22(4):515–525. doi: 10.1109/TMI.2003.809078. [DOI] [PubMed] [Google Scholar]
- 96.Kim TS, Singh M, Sungkarat W, Zarow C, Chui H. Automatic registration of postmortem brain slices to MRI reference volume. IEEE Trans. Nucl. Sci. 2000 Aug;47:1607–1613. [Google Scholar]
- 97.Zhengping J, Mowforth PH. Mapping between MR brain images and a voxel model. Med. Inform. 1991;16:183–193. doi: 10.3109/14639239109012126. [DOI] [PubMed] [Google Scholar]
- 98.Kim B, Boes JL, Bland PH, Chenevert TL, Meyer CR. Motion correction in fMRI via registration of individual slices into an anatomical volume. Magn. Reson. Med. 1999;41:964–972. doi: 10.1002/(sici)1522-2594(199905)41:5<964::aid-mrm16>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 99.Rousseau F, Glenn O, Iordanova B, Rodriguez-Carranza C, Vigneron D, Barkovich J, Studholme C. Reconstructing high resolution in vivo fetal MR brain images. In: Duncan J, Gerig G, editors. Proceedings of Medical Image Computing and Computer Assisted Intervention (MICCAI) volume LLNCS 3749. Springer Verlag; 2005. pp. 548–555. [DOI] [PubMed] [Google Scholar]
- 100.Rousseau F, Glenn OA, Iordanova B, Rodriguez-Carranza CE, Vigneron DB, Barkovich AJ, Studholme C. Registration-based approach for reconstruction of high-resolution in utero MR brain images. Acad. Radiol. 2006 Sep;13(9):1072–1081. doi: 10.1016/j.acra.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 101.Lee S, Wolberg G, Shin SY. Scattered data interpolation with multilevel B-splines. IEEE Transactions on Visualisation and Computer Graphics. 1997;3:228–244. [Google Scholar]
- 102.Franke Richard. Scattered data interpolation: Tests of some method. Mathematics of Computation. 1982 Jan;38(157):181–200. [Google Scholar]
- 103.Amidror Isaac. Scattered data interpolation methods for electronic imaging systems: A survey. 2002 [Google Scholar]
- 104.Jiang S, Xue H, Glover A, Rutherford M, Rueckert D, Hajnal JV. MRI of moving subjects using multislice snapshot images with volume reconstruction (SVR): application to fetal, neonatal, and adult brain studies. IEEE Trans Med Imaging. 2007;26:967–980. doi: 10.1109/TMI.2007.895456. [DOI] [PubMed] [Google Scholar]
- 105.Gholipour A, Estroff JA, Warfield SK. Robust super-resulution volume reconstruction from slice acquisitions: Application to fetal brain mri. IEEE transactions on Medical Imaging. 2010 Oct;29(10):1739–1758. doi: 10.1109/TMI.2010.2051680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim K, Hansen MF, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Intersection-based registration of slice stacks to form 3D images of the human fetal brain; Proc. IEEE International Symposium on Biomedical Imaging: From Nano to Macro; 2008. May, pp. 1167–1170. [Google Scholar]
- 107.Kim K, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Intersection based motion correction of multislice MRI for 3-d in utero fetal brain image formation. IEEE Trans. Med. Imaging. 2010;vol. 29(1):146–158. doi: 10.1109/TMI.2009.2030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rousseau Francois, Studholme Colin, Koob Meriam, Diete-mann Jean-Luis. Proceedings MICCAI 2010, Beijing. Part II. volume 6361. Springer: 2010. On super-resolution for fetal brain MRI; pp. 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rousseau F, Kim K, Studholme C. A groupwise super-resolution approach: application to brain MRI; Proceedings IEEE International Symposium on Biomedical Imaging: From Nano to Macro; 2010. Apr, pp. 860–863. [Google Scholar]
- 110.Sled JG, Pike B. Standing-wave and RF penetration artifacts caused by elliptic geometry: An electrodynamic analysis of MRI. IEEE Transactions in Medical Imaging. 1998;17(4):653–662. doi: 10.1109/42.730409. [DOI] [PubMed] [Google Scholar]
- 111.Corbett-Detig JM, Habas PA, Scott JA, Kim K, Rajagopalan V, McQuillen PS, Barkovich AJ, Glenn OA, Studholme C. 3-D global and regional patterns of human fetal subplate growth determined in utero. Brain Struct. Funct. 2011 doi: 10.1007/s00429-010-0286-5. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Arnold JB, Liow JS, Schaper KA, Stern JJ, Sled JG, Shattuck DW, Worth AJ, Cohen MS, Leahy RM, Mazziotta JC, Rotten-berg DA. Qualitative and quantitative evaluation of six algorithms for correcting intensity nonuniformity effects. Neuroimage. 2001;13:931–943. doi: 10.1006/nimg.2001.0756. [DOI] [PubMed] [Google Scholar]
- 113.Kim Kio, Habas Piotr, Rajagopalan Vidya, Scott Julia, Corbett-Detig James, Rousseau Francois, James Barkovich Orit Glenn, Studholme Colin. Non-iterative relative bias correction for 3D reconstruction of in utero fetal brain MR imaging; Proceedings of 32nd Annual International Conference of the IEEE EMBS; 2010. pp. 879–882. [DOI] [PubMed] [Google Scholar]
- 114.Pham DL, Xu C, Prince JL. Current methods in medical image segmentation. Annual Review of Biomedical Engineering. 2000;2:315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 115.Brummer ME. Optimized intensity thresholds for volumetric analysis of magnetic resonance imaging data. Proc. SPIE Medical Imaging 1992. 1992;volume 1808:299–310. [Google Scholar]
- 116.Kundu A. Local segmentation of biomedical images. Comput. Med. Imag. Graph. 1990;14:173–183. doi: 10.1016/0895-6111(90)90057-i. [DOI] [PubMed] [Google Scholar]
- 117.Wells WM, Grimson EL, Kikinis R, Jolesz FA. Adaptive segmentation of MRI data. IEEE Trans. Med. Imag. 1996 Aug;15:429–442. doi: 10.1109/42.511747. [DOI] [PubMed] [Google Scholar]
- 118.Ashburner J, Friston KJ. Voxel-based morphometry-The methods. Neuroimage. 2000;11:805–821. doi: 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- 119.Van Leemput K, Maes F, Vandermeulen D, Suetens P. Automated model-based tissue classification of MR images of the brain. IEEE Transactions on Medical Imaging. 1999;18:897–908. doi: 10.1109/42.811270. [DOI] [PubMed] [Google Scholar]
- 120.Ashburner J, Friston KJ. Computing average shaped tissue probability templates. NeuroImage. 2009;45(2):333–341. doi: 10.1016/j.neuroimage.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 121.Yan MXH, Karp JS. An adaptive bayesian approach to three-dimensional MR brain segmentation. In: Bizais Y, Barillot C, Paola RD, editors. Proceedings of Information Processing in Medical Imaging. Kluwer Academic Dodrecht; 1995. pp. 201–213. [Google Scholar]
- 122.Kapur T, Grimson WEL, Wells WM, Kikinis R. Segmentation of brain tissue from magnetic resonance images. Med. Image Anal. 1996;1:109–127. doi: 10.1016/S1361-8415(96)80008-9. [DOI] [PubMed] [Google Scholar]
- 123.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder committee revisited. Nat. Rev. Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 124.Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Atlas-based segmentation of developing tissues in the human brain with quantitative validation in young fetuses. Human Brain Mapping. 2010 Sep;31(9):1348–1358. doi: 10.1002/hbm.20935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Murgasova M, Dyet L, Edwards AD, Rutherford M, Hajnal JV, Rueckert D. Segmentation of brain MRI in young children. Acad. Radiol. 2007;14(11):1350–1366. doi: 10.1016/j.acra.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 126.Yoon U, Fonov VS, Perusse D, Evans AC. The effect of template choice on morphometric analysis of pediatric brain data. Neuroimage. 2009;45(3):769–777. doi: 10.1016/j.neuroimage.2008.12.046. [DOI] [PubMed] [Google Scholar]
- 127.Altaye M, Holland SK, Wilke M, Gaser C. Infant brain probability templates for MRI segmentation and normalization. Neuroimage. 2008;43(4):721–730. doi: 10.1016/j.neuroimage.2008.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weisenfeld NI, Warfield SK. Automatic segmentation of newborn brain MRI. Neuroimage. 2009;47(2):564–572. doi: 10.1016/j.neuroimage.2009.04.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Prastawa M, Gilmore JH, Lin W, Gerig G. Automatic segmentation of MR images of the developing newborn brain. Med Image Anal. 2005;9:457–466. doi: 10.1016/j.media.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 130.Warfield S, Kaus M, Jolesz FA, Kikinis R. Adaptive, template moderated, spatially varying statistical classification. Med. Image Anal. 2000;4(1):43–55. doi: 10.1016/s1361-8415(00)00003-7. [DOI] [PubMed] [Google Scholar]
- 131.Alexander GR, Tompkins ME, Petersen DJ, Hulsey TC, Mor J. Discordance between LMP-based and clinically estimated gestational age: implications for research, programs, and policy. Public Health Rep. 1995;110(4):395–402. [PMC free article] [PubMed] [Google Scholar]
- 132.Habas PA, Kim K, Corbett-Detig JM, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. A spatiotemporal atlas of MR intensity, tissue probability and shape of the fetal brain with application to segmentation. Neuroimage. 2010 Nov;53(2):460–470. doi: 10.1016/j.neuroimage.2010.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Habas PA, Kim K, Chandramohan D, Rousseau F, Glenn OA, Studholme C. Statistical model of laminar structure for atlas-based segmentation of the fetal brain from in-utero MR images. Proceedings SPIE Medical Imaging 2009: Image Processing. 2009 Feb;volume 7259:17:1–17:8. [Google Scholar]
- 134.Grossman R, Hoffman C, Mardor Y, Biegon A. Quantitative MRI measurements of human fetal brain development in utero. Neuroimage. 2006;33:463–470. doi: 10.1016/j.neuroimage.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 135.Hu HH, Guo WY, Chen HY, Wang PS, Hung CI, Hsieh JC, Wu YT. Morphological regionalization using fetal magnetic resonance images of normal developing brains. Eur J Neurosci. 2009;29:1560–1567. doi: 10.1111/j.1460-9568.2009.06707.x. [DOI] [PubMed] [Google Scholar]
- 136.Kazan-Tannus JF, Dialani V, Kataoka ML, Chiang G, Feldman HA, Brown JS, Levine D. MR volumetry of brain and CSF in fetuses referred for ventriculomegaly. AJR Am J Roentgenol. 2007;189:145–151. doi: 10.2214/AJR.07.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Widjaja E, Geibprasert S, Mahmoodabadi SZ, Blaser S, Brown NE, Shannon P. Alteration of human fetal subplate layer and intermediate zone during normal development on MR and diffusion tensor imaging. Am J Neuroradiol. 2010;31:1091–1099. doi: 10.3174/ajnr.A1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gholipour A, Estroff JA, Barnewolt CE, Connolly SA, Warfield SK. Fetal brain volumetry through MRI volumetric reconstruction and segmentation. Int. J. Comput Assist Radiol Surg. 2010 Jul; doi: 10.1007/s11548-010-0512-x. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Davatzikos C, Vaillant M, Resnick SM, Prince JL, Letovsky S, Bryan RN. A computerised approach for morphological analysis of the corpus callosum. Journal of Computer Assisted Tomography. 1996;20(1):88–97. doi: 10.1097/00004728-199601000-00017. [DOI] [PubMed] [Google Scholar]
- 140.Rajagopalan V, Scott JA, Habas PA, Kim K, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Medical Image Computing and Computer Assisted Intervention. volume LNCS 6362. spinger; 2010. Sep, Measures for characterizing directionality specific volume changes in TBM of brain growth; pp. 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Garel C, Chantrel E, Brisse H, Elmaleh M, Luton D, Oury J-F, Sebag G, Hassan M. Fetal cerebral cortex: normal gestational landmarks identified using prenatal MR imaging. Am J Neuroradiol. 2001;22:184–189. [PMC free article] [PubMed] [Google Scholar]
- 142.Batchelor PG, Castellano Smith AD, Hill DLG, Hawkes DJ, Cox TCS, Dean AF. Measures of folding applied to the development of the human fetal brain. IEEE Transactions on Medical Imaging. 2002 Aug;21(8):953–965. doi: 10.1109/TMI.2002.803108. [DOI] [PubMed] [Google Scholar]
- 143.Do Carmo MP. Differential Geometry of Curves and Surfaces. Prentice Hall; 1976. [Google Scholar]
- 144.Koenderink Jan J, van Doorn Andrea J. Surface shape and curvature scales. image and vision computing. 1992;10(8) [Google Scholar]
- 145.Rodriguez-Carranza CE, Mukherjee P, Vigneron D, Barkovic AJ, Studholme C. A framework for in vivo quantification of regional brain folding in premature neonates. Neuroimage. 2008 Jun;41(2):462–478. doi: 10.1016/j.neuroimage.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Habas PA, Rajagopalan V, Scott JA, Kim K, Roosta A, Rousseau F, Barkovich AJ, Glenn OA, Studholme C. Detection and mapping of delays in early cortical folding derived from in utero MRI. SPIE Medical Imaging 2011: Image Processing. 2011 Feb [Google Scholar]
- 147.Le Bihan D, Breton E, Lallemand D, Grenier P, Cabanis E, Laval-Jeantet M. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology. 1986;161:401–407. doi: 10.1148/radiology.161.2.3763909. [DOI] [PubMed] [Google Scholar]
- 148.Le Bihan D, Mangin J-F, Poupon C, Clark CA, Pappata S, Molko N, Chabriat H. Diffusion tensor imaging: concepts and applications. Journal of Magnetic Resonance Imaging. 2001;13:534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- 149.Le Bihan D. Looking into the functional architecture of the brain with diffusion MRI. Nat. Rev., Neurosci. 2003;4:469–480. doi: 10.1038/nrn1119. [DOI] [PubMed] [Google Scholar]
- 150.Le Bihan D. Molecular diffusion, tissue microdynamics and microstructure. NMR Biomed. 1995;8:375–386. doi: 10.1002/nbm.1940080711. [DOI] [PubMed] [Google Scholar]
- 151.Prayer D, Prayer L. Diffusion-weighted magnetic resonance imaging of cerebral white matter development. Eur. J. Radiol. 2003;45:235–243. doi: 10.1016/s0720-048x(02)00312-1. [DOI] [PubMed] [Google Scholar]
- 152.Righini A, Bianchini E, Parazzini C, Gementi P, Ramenghi L, Baldoli C, Nicolini U, Mosca F, Triulzi F. Apparent diffusion coefficient determination in normal fetal brain: a prenatal MR imaging study. Am. J. Neuroradiol. 2003;24:799–804. [PMC free article] [PubMed] [Google Scholar]
- 153.Bui T, Daire JL, Chalard F, Zaccaria I, Alberti C, Elmaleh M, Garel C, Luton D, Blanc N, Sebag G. Microstructural development of human brain assessed in utero by diffusion tensor imaging. Pediatr. Radiol. 2006;36:1133–1140. doi: 10.1007/s00247-006-0266-3. [DOI] [PubMed] [Google Scholar]
- 154.Baldoli C, Righini A, Parazzini C, Scotti G, Triulzi F. Demonstration of acute ischemic lesions in the fetal brain by diffusion magnetic resonance imaging. Ann. Neurol. 2002;52:243–246. doi: 10.1002/ana.10255. [DOI] [PubMed] [Google Scholar]
- 155.Agid R, Lieberman S, Nadjari M, Gomori JM. Prenatal MR diffusion-weighted imaging in a fetus with hemimegalencephaly. Pediatr. Radiol. 2006;36:138–140. doi: 10.1007/s00247-005-0003-3. [DOI] [PubMed] [Google Scholar]
- 156.Huang Hao, Xue Rong, Zhang Jiangyang, Ren Tianbo, Richards Linda J, Yarowsky Paul, Miller Michael I, Mori Susumu. Anatomical characterization of human fetal brain development with diffusion tensor magnetic resonance imaging. The Journal of Neuroscience. 2009 Apr;29(13):4263–4273. doi: 10.1523/JNEUROSCI.2769-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kasprian Gregor, Brugger Peter C, Weber Michael, Krssak Martin, Krampl Elisabeth, Herold Christian, Prayer Daniela. In utero tractography of fetal white matter development. neuroimage. 2008;43:213–224. doi: 10.1016/j.neuroimage.2008.07.026. [DOI] [PubMed] [Google Scholar]
- 158.Jiang SZ, Xue H, Counsell S, Anjari M, Allsop J, Rutherford M, Rueckert D, Hajnal JV. Diffusion tensor imaging (DTI) of the brain in moving subjects: Application to in-utero fetal and ex-utero studies. Magn. Reson. Med. 2009;62:645–655. doi: 10.1002/mrm.22032. [DOI] [PubMed] [Google Scholar]
- 159.Kim K, Habas PA, Rousseau F, Glenn OA, Barkovich AJ, Studholme C. Reconstruction of a geometrically correct diffusion tensor image of a moving human fetal brain. Proceedings Medical Imaging 2010: Image Processing. 2010 Feb;volume 7623:I:1–I:9. [Google Scholar]
- 160.Oubel Estanislao, Koob Meriam, Studholme Colin, Dietemann Jean-Luis, Rousseau Francois. Proceedings MICCAI 2010, Beijing. Part I. volume 6361. Springer; 2010. Reconstruction of scattered data in fetal diffusion MRI; pp. 574–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ogawa S, Lee TM, Nayak AS, Glynn P. Oxygenation-sensitive contrast in magnetic resonance image of rodent brain at high magnetic fields. Magnetic Resonance in Medicine. 1990;14:68–78. doi: 10.1002/mrm.1910140108. [DOI] [PubMed] [Google Scholar]
- 162.Raichle Marcus E. Two views of brain function. Trends in Cognitive Sciences. 2010 Mar;14(4):180–190. doi: 10.1016/j.tics.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 163.Gao Wei, Zhu Hongtu, Giovanello Kelly S, Smith Keith, Shen Dinggang, Gilmore John H, Lin Weili. Evidence on the emergence of the brain’s default network from 2-week-old to 2-year-old healthy pediatric subjects. PNAS. 2009 Apr;106(16):6790–6795. doi: 10.1073/pnas.0811221106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Fransson Peter, den Ulrika A, Blennow Mats, Lagercrantz Hugo. The functional architecture of the infant brain as revealed by resting-state fMRI. Cerebral Cortex doi: 10.1093/cercor/bhq071. -:–, In Press. [DOI] [PubMed] [Google Scholar]
- 165.Jolles Dietsje D, van Buchem Mark A, Crone Eveline A, Rombouts Serge ARB. A comprehensive study of whole-brain functional connectivity in children and young adults. Cerebral Cortex doi: 10.1093/cercor/bhq104. -:–, In Press. [DOI] [PubMed] [Google Scholar]
- 166.Hykin J, Moore R, Duncan K, et al. Fetal brain activity demonstrated by functional magnetic resonance imaging. Lancet. 1999;354:645–646. doi: 10.1016/S0140-6736(99)02901-3. [DOI] [PubMed] [Google Scholar]
- 167.Fulford J, Vadeyar SH, Dodampahala SH, et al. Fetal brain activity in response to a visual stimulus. Hum Brain Mapp. 2003;20:239–245. doi: 10.1002/hbm.10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Born P, Leth H, Miranda MJ, Rostrop E, Stensgaard A, Peitersen B, Larsson HBW, Lou HC. Visual activation in infants and young children studied by functional magnetic resonance imaging. Pediatr. Res. 1998;44:578–583. doi: 10.1203/00006450-199810000-00018. [DOI] [PubMed] [Google Scholar]
- 169.Griffin LD. The intrinsic geometry of the cerebral cortex. Journal of Theoretical Biology. 1994;166:261–273. doi: 10.1006/jtbi.1994.1024. [DOI] [PubMed] [Google Scholar]
- 170.Van Essen DC DC. A tension-based theory of morphogenesis and compact wiring in the central nervous system. Nature. 1997;385:313–318. doi: 10.1038/385313a0. [DOI] [PubMed] [Google Scholar]
- 171.Hilgetag Claus C, Barbas Helen. Role of mechanical factors in the morphology of the primate cerebral cortex. PLoS Comput Biol. 2006 Mar;2(3):0146–0159. doi: 10.1371/journal.pcbi.0020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Flake AW. Surgery in the human fetus: the future. Journal of Physiology. 2003;547:45–51. doi: 10.1113/jphysiol.2002.022327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nelson BJ, Kaliakatsos IK, Abbot JJ. Microrobots for minimally invasive medicine. Annual Review of Biomedical Engineering. 2010:55–85. doi: 10.1146/annurev-bioeng-010510-103409. [DOI] [PubMed] [Google Scholar]
- 174.Flake AW, Crombleholme TM, Adzick NS. The current status and future potential of fetal intervention: image is everything. Computerized Medical Imaging and Graphics. 1999;23:51–57. doi: 10.1016/s0895-6111(98)00064-0. [DOI] [PubMed] [Google Scholar]