Abstract
Fruit ripening is a highly coordinated developmental process that coincides with seed maturation. The ripening process is regulated by thousands of genes that control progressive softening and/or lignification of pericarp layers, accumulation of sugars, acids, pigments, and release of volatiles. Key to crop improvement is a deeper understanding of the processes underlying fruit ripening. In tomato, mutations blocking the transition to ripe fruits have provided insights into the role of ethylene and its associated molecular networks involved in the control of ripening. However, the role of other plant hormones is still poorly understood. In this review, we describe how plant hormones, transcription factors, and epigenetic changes are intimately related to provide a tight control of the ripening process. Recent findings from comparative genomics and system biology approaches are discussed.
Keywords: climacteric fruit, non-climacteric fruit, hormones, ripening, regulation
Fruits are a distinctive characteristic of Angiosperms. They occur today in a wide variety of forms and types. The ancestral fruit, dry and dehiscent, probably emerged in the early Cretaceous period; fleshy fruits appeared later in the Cretaceous or early Tertiary (Eriksson et al., 2000). The diversification of fruits from a dry dehiscent form to a fleshy drupe or berry, correlated with the rise of vertebrates, main agents of seed dispersal (Knapp, 2002). The maturation of fruits is a complex and highly coordinated developmental process. In fleshy fruits, ripening results in the production of succulent, flavorful, and soft pericarp that attract animals and facilitate seed dispersal (Giovannoni, 2001). In addition to softening, fruits normally exhibit increased accumulation of sugars, acids, pigments, and volatiles that increase interest and palatability to animals. Moreover, fruits are an important source of supplementary diet, providing minerals, vitamins, fibers, and antioxidants for humans. From an agronomical point of view, nutritional value, flavor, processing qualities, and shelf-life determine the quality of fruits.
The main changes associated with ripening include color (loss of green color and increase in non-photosynthetic pigments that vary depending on species and cultivar), firmness (softening by cell wall degrading activities and alterations in cuticle properties), taste (increase in sugar and decline in organic acids), and flavor (production of volatile compounds providing the characteristic aroma).
Analytical tools that allow comprehensive phenotyping at the level of transcriptome (Alba et al., 2005; Vriezen et al., 2008; Matas et al., 2011; Rohrmann et al., 2011), proteome (Lee et al., 2004; Rose et al., 2004; Saravanan and Rose, 2004), and metabolome (Fait et al., 2008; Lombardo et al., 2011) facilitate an overview of the metabolic network (Carrari et al., 2006; Deluc et al., 2007; Grimplet et al., 2007; Enfissi et al., 2010; Zamboni et al., 2010; Osorio et al., 2011, 2012; Rohrmann et al., 2011; Lee et al., 2012; Pan et al., 2013); whilst network analysis is beginning to yield a detailed understanding of the systems regulation underlying fruit development.
HORMONAL AND TRANSCRIPTIONAL REGULATION DURING RIPENING
Fruits are generally classified into two physiological groups, climacteric and non-climacteric, according to their respiratory activity and associated ethylene biosynthesis profiles during ripening. Ethylene synthesis in climacteric fruits such as tomato, apple, and banana, is essential for normal fruit ripening and blocking either synthesis or perception of this hormone prevents ripening (Hamilton et al., 1990; Oeller et al., 1991; Rottmann et al., 1991; Barry et al., 1996).
Efforts to uncover the transcriptional regulation underlying carpel and fruit development were first focused on the dry dehiscent siliques of the model plant Arabidopsis (Liljegren et al., 2000; Dinneny et al., 2005). These studies clarified the role of several MADS-box transcription factors in tissue specification and mechanism of dehiscence. Among these, the redundant SHATTERPROOF 1/2 genes (SHP, members of the AGAMOUS subfamily) specified valve margin identity in the silique: when mutated, fruits became indehiscent. However, despite the striking anatomical differences between dry and fleshy fruits, subsequent studies, primarily focused on tomato, have shown the involvement in ripening regulation of several orthologs of those MADS-box genes previously characterized in Arabidopsis (Pnueli et al., 1994; Itkin et al., 2009; Vrebalov et al., 2009; Giménez et al., 2010; Bemer et al., 2012). It is now clear that a part of the regulatory networks underlying fruit development have been conserved during the evolution of fleshy fruits (Smaczniak et al., 2012; Seymour et al., 2013).
A number of important advances in our understanding of mechanisms that regulate ripening have also come from the characterization of monogenic tomato mutants, including ripening-inhibitor (rin), non-ripening (nor), colorless non-ripening (Cnr), green-ripe (Gr), green flesh (gf), high pigment 1 (hp1), high pigment 2 (hp2), and never-ripe (Nr; Lanahan et al., 1994; Mustilli et al., 1999; Vrebalov et al., 2002; Liu et al., 2004; Barry and Giovannoni, 2006; Manning et al., 2006; Barry et al., 2008). The rin mutant encodes a partially deleted MADS-box protein of the SEPALLATA clade (SEP4; Hileman et al., 2006), whereas Cnr is an epigenetic change which alters the promoter methylation of SQUAMOSA promoter binding (SPB) protein. NOR is a member of the NAC-domain transcription factor family (Giovannoni, 2007). A recent study in which the transcriptome, proteome, and targeted metabolite analysis were combined during development and ripening of nor and rin mutants, has helped to refine the ethylene-regulated expression of downstream genes and added to our knowledge the role of this hormone in both protein- and metabolite regulation in tomato ripening (Osorio et al., 2011). This data supported the view that nor and rin act together in a cascade to control ripening (Giovannoni et al., 1995; Thompson et al., 1999) and also suggested that nor has a more global effect on ethylene/ripening-related gene expression than rin, which indicates that nor likely operates upstream of rin. Recently, using a combined approach based on chromatin immunoprecipitation and transcriptome analysis, it was provided evidence that RIN interacts with the promoters of more than 200 genes, modulating the expression of its targets by activation or repression. RIN target genes are major regulators of ripening control, such as CNR and NOR, or belong (Martel et al., 2011) to well-known pathways active during the transition from green to ripe fruits (e.g., carotenoid accumulation, chlorophyll breakdown, ethylene synthesis and perception; Fujisawa et al., 2013).
Fruits such as strawberry, citrus, and grape have been classified as non-climacteric, based on the lack of the respiratory burst and on the low endogenous production of ethylene compared to standard climacteric fruits (Perkins-Veazie, 1995). In pepper fruits, some cultivars seem to be ethylene-insensitive, while others pepper cultivars treated with exogenous ethylene were able to stimulate the expression of ripening-specific genes (Armitage, 1989; Ferrarese et al., 1995; Harpster et al., 1997; El-Kereamy et al., 2003).
In strawberry, which has emerged as a prime model of non-climacteric fruit ripening, ethylene is relatively high in green fruits, decreases in white fruits, and finally increases again at the red stage of ripening (Perkins-Veazie et al., 1996; Iannetta et al., 2006). Interestingly, this last increase is accompanied by an enhanced respiration rate that resembles the one that occurs in climacteric fruits at the onset of ripening (Iannetta et al., 2006). For better understanding the function of ethylene during strawberry ripening, different approaches have been used. External application of ethylene caused the down-regulation of several cell wall-related genes, such as β-galactosidase, pectin methylesterase, or β-xylosidase (Trainotti et al., 2001; Castillejo et al., 2004; Bustamante et al., 2009), while the expression of other genes such as expansin, FaEXP2 (Civello et al., 1999) was ethylene-insensitive. Recent studies at transcriptomic and metabolomic levels in transgenic strawberry fruits with decreased ethylene sensitivity indicates that ethylene action is required for normal fruit development, acting differently in the two parts of strawberry fruit, achenes and receptacle (Merchante et al., unpublished data). These results show that, although not as relevant as in climacteric fruits, ethylene may nevertheless play a role in strawberry fruit ripening.
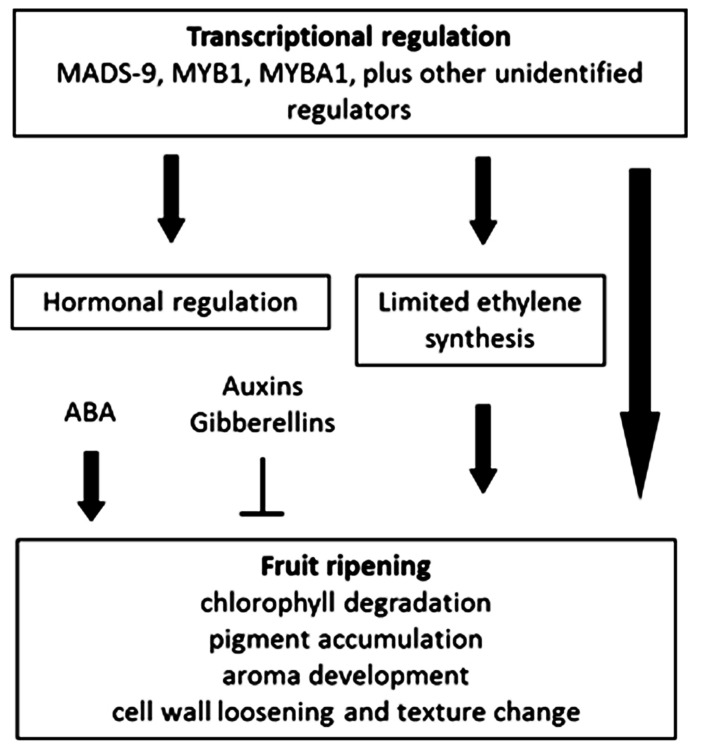
Recent comparative transcriptome and metabolome studies during the maturation processes of climacteric and non-climacteric fruits (tomato and pepper, respectively) suggest that both species have similar ethylene-mediated signaling components. In pepper, the regulation of these genes is, however, clearly different and may reflect altered ethylene sensitivity or regulators other than ethylene than in tomato (Osorio et al., 2012). Unlike the situation described in tomato the ethylene biosynthesis genes, aminocyclopropane-1-carboxylic acid (ACC) synthase, and ACC oxidase, are not induced in pepper. However, genes downstream of ethylene perception, such as cell wall-related genes, ethylene response factor 3 (ERF3), and carotenoid biosynthesis genes, are up-regulated during pepper fruit ripening (Osorio et al., 2012). Other commonly regulated genes between climacteric and non-climacteric fruits have been described. In strawberry, a SEPALLATA gene (SEP1/2; MADS-box) is needed for normal development and ripening (Seymour et al., 2011). Similarly, in banana, which is classified as a climacteric fruit, the MADS-box SEP3 gene also displays ripening-related expression (Elitzur et al., 2010). In apple, MADS2 gene expression is also associated with fruit firmness (Cevik et al., 2010), whereas in bilberry fruit, the SQUAMOSA MADS-box ortholog of the TDR4 gene in tomato, has a role in regulation of anthocyanin biosynthesis (Jaakola et al., 2010; see Figure 1).
FIGURE 1.
Overview of ripening regulation in non-climacteric fruits.
Current knowledge about the role of hormones – other than ethylene – in the development and ripening of climacteric and non-climacteric fruits is limited. In tomato, pepper, banana, muskmelon, and strawberry, the most abundant free auxin, indole-3-acetic acid (IAA), has been reported to decline prior to the onset of ripening; this reduction was accompanied by an increase of its conjugated form (IAA-Asp; Bottcher et al., 2010). The conjugation reaction is catalyzed by the IAA-amino synthase gene (GH3). In tomato, 15 members of GH3 gene family have been described, but only for two of them is the pattern of expression associated with ripening (Kumar et al., 2012). Tomato fruits overexpressing the pepper GH3 gene show anticipation of ripening (Liu et al., 2005), which is in agreement with the view that the ratio between IAA and its AA-conjugated form, rather than the level of IAA itself, may contribute to the temporal regulation of ripening (Bottcher et al., 2010). In non-climacteric fruits, no single growth regulator appears to play a positive role analogous to that played by ethylene, but it has been observed that auxin can negatively control the ripening of some non-climacteric fruits. In strawberry, it has been shown that the expression of many ripening-specific genes can be down-regulated by treatments with an exogenous auxin. Also in grape auxin seems to play a negative role in the regulation of ripening with synthetic auxin treatments delaying the expression of a number of ripening-related genes (Davies et al., 1997).
As a consequence of the prominent role of auxin in the development and ripening of some non-climacteric fruits, little attention has been paid to possible roles of other plant hormones, such as gibberellins (GAs). However, in strawberry, it has been reported that external application of GA3 to ripening fruits caused a significant delay in the development of the red color (Martinez et al., 1996) and modified the expression of genes involved in cell enlargement (de la Fuente et al., 2006) and cell wall disassembly (Bustamante et al., 2009).
In plants, the phytohormone abscisic acid (ABA) is known to be involved in various aspects of plant growth, development, and responses to environmental stresses (Leung and Giraudat, 1998; Finkelstein and Rock, 2002; Himmelbach et al., 2003; Hirayama and Shinozaki, 2007). ABA promotes sugar accumulation in fleshy fruits (Yamaki and Asakura, 1991; Kobashi et al., 1999; Richings et al., 2000; Pan et al., 2005) and plays a role in the regulation of climacteric and non-climacteric fruit ripening (Coombe, 1992; Davies et al., 1997; Giovannoni, 2001; Rodrigo et al., 2003; Zhang et al., 2009; Sun et al., 2012). In tomato, the suppression of the gene that catalyzes the first step in ABA biosynthesis (NCED1, 9-cis-epoxycarotenoid dioxygenase), results in the down-regulation of some ripening-related cell wall genes, such as polygalacturonase and pectinmethylesterase, as well as an increase in firmness and longer shelf-life (Sun et al., 2012). Similarly, reduction of NCED expression correlates with retardation of ripening in strawberry (Jia et al., 2011). ABA is considered a ripening-inducer in strawberry and grape fruits (Chai et al., 2011; Jia et al., 2011). The mechanisms of ABA signaling is not known, however, in grape, analysis of the GH3 promoter identified ABRE-like elements, which may indicate that the ABA/auxin content ratio is related to the initiation of ripening (Perkins-Veazie, 1995; Jiang and Joyce, 2003; Bottcher et al., 2010).
In recent years, the level of understanding of the molecular events at the transcriptional, biochemical, hormonal, and metabolite levels underlying ripening in climacteric and non-climacteric fruits has increased considerable (see Figures 1 and 2). However, we still poorly understand the developmental switch that occurs in hormone responsiveness during the transition from immature to ripe fruits. To date, most published studies of transcriptional and metabolic regulation are of relatively low resolution at both spatial and temporal levels and are furthermore restricted in coverage of various cell molecular entities. However, new emerging technologies as well as improved statistical tools (Klie et al., 2011) allow us to further refine our analytical ability in order to cope with issues as subcellular compartmentation and contrasting behavior of different cell types (Caldana et al., 2012). Additionally, the availability of high quality fruit genome sequence data (Jaillon et al., 2007; Shulaev et al., 2011; Tomato Genome Consortium, 2012) will aid our understanding about the genetic regulation of fruit development and ripening.
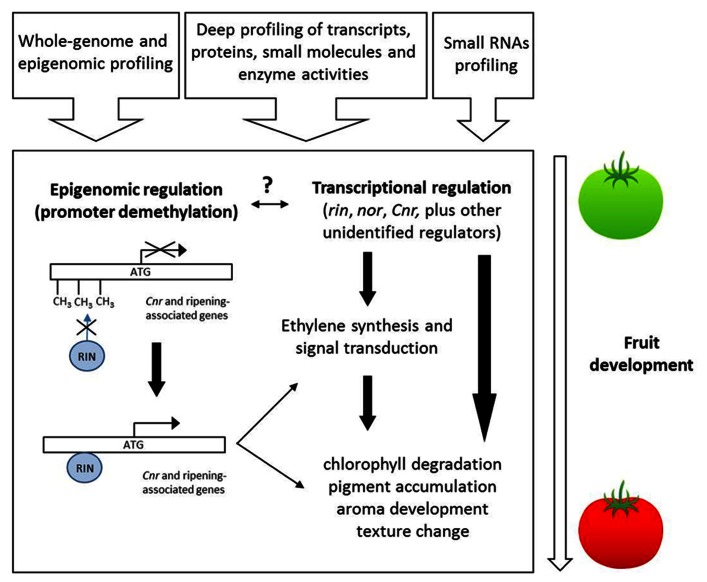
FIGURE 2.
Overview of ripening regulation in climacteric fruits. The contribution of systems profiling approaches (shown at the top) will help identify novel regulatory genes and elucidate the interplay between epigenomic remodeling and transcriptional regulation involved during the ripening process.
EPIGENETIC REMODELING DURING RIPENING
Epigenetic regulation of gene expression (inheritance without an alteration in the primary DNA sequence) is increasingly recognized as mechanism for modulating genome activity. Naturally occurring epigenetic changes at a single gene locus in plants can result in heritable morphological variation without alteration of the underlying DNA sequence (Patterson et al., 1993; Cubas et al., 1999; Manning et al., 2006). DNA methylation is one form of epigenetic regulation. It is involved in transcriptional regulation, stress responses and furthermore plays a major role in protecting the genome integrity against the activity of transposable elements (TEs) and other repetitive sequence (Chan et al., 2005).
In plants, DNA methylation occurs at cytosine residues in three different sequences (CG, CHG, and CHH, where H = A, C or T; Cokus et al., 2008) and is set in place and maintained by different factors (Law and Jacobsen, 2010).
Analysis of epigenetic variation in Arabidopsis revealed that at least one-third of expressed genes are methylated in their coding region, and only 5% of genes are methylated within promoter regions (Zhang et al., 2006; Vaughn et al., 2007). However, the promoter-methylated genes have a higher degree of tissue-specific expression (Zhang et al., 2006; Zilberman and Henikoff, 2007).
The first survey of the frequency and distribution of cytosine methylation sites in tomato dates back to more than 20 years ago, when it was found that polymorphisms in cytosine methylation between two tomato species were relatively abundant and that methylation patterns were stably inherited, from parents to offspring, segregating in a Mendelian fashion. The presence of tissue-specific methylation patterns and the overall decrease of 5-mC frequency in developing tissues also led the authors to postulate variation of methylation status of selected alleles during plant development (Messeguer et al., 1991).
More recently, the impact of cytosine methylation on tomato fruit ripening has strikingly emerged in the definition of the molecular nature of the colorless non-ripening phenotype. The tomato non-ripening Cnr mutant fails to produce ripe berries; fruits exhibit green pericarps and do not respond to external applications of ethylene. The gene at the Cnr locus was identified as a SPB protein-like using positional cloning, but the non-ripening phenotype could not be attributed to any alteration in the coding gene sequence. Bisulfite sequencing of the Cnr mutant allele showed instead hypermethylation of cytosine in the region upstream the predicted ATG start site. This hypermethylation state correlated with a drastic reduction of Cnr gene expression (Manning et al., 2006). Therefore, the non-ripening phenotype was due to the heritable cytosine hypermethylation pattern of the region including the Cnr gene promoter. Additionally, in normal tomato fruit (cv. Liberto) development, the promoter of Cnr appears to be demethylated in a specific region just prior to the onset of ripening. This lead to the hypothesis that DNA methylation contributes to the regulation of fruit ripening (Seymour et al., 2008).
Recent work by Zhong et al. (2013) provides genome-wide insights into the link between the genetic program of fruit ripening and DNA methylation state. On the basis of the previous results on the nature of the Cnr (epi) mutation, the authors injected a chemical inhibitor of cytosine methylation, 5-azacytidine, directly in the locular spaces and columella of developing tomato fruits. The methylation inhibitor induced the formation of local ripe areas, red in appearance, where the expression of typical ripening-related genes (phytoene synthase 1 and polygalacturonase) was anticipated. Moreover, the Cnr promoter region was demethylated in red sectors with respect to green parts of the fruits, pointing at the demethylation of Cnr as the epigenetic signal sufficient to induce ripening. The authors then extended their views on the role of cytosine methylation reporting the full tomato methylome sequences of leaves, immature and ripe fruits, including the ripening-impaired mutants Cnr and rin. The sequencing of the entire epigenome revealed at least three important results: (i) in wild-type fruits, the degree of methylation of regions upstream the transcription start sites (TSS) decreased gradually along fruit development; (ii) this general decline was not observed for the fruits of the ripening-impaired mutants Cnr and rin, whose CG methylation levels were constantly higher at TSS and, for Cnr, also comparable to those observed in leaves; (iii) the promoters of typical ripening-related genes were gradually demethylated during development of wild-type fruits. Further evidences about the link between ripening and cytosine methylation came from the ChIP-Seq mapping of RIN binding sites during fruit development. The set of RIN targets included 292 genes with a known role in ripening. RIN binding sites were found to be adjacent or overlapping the methylation “hotspots” upstream the TSS. The analysis of methylation status of these regions showed that they were progressively demethylated during the transition from green to red ripe fruits; and this lower level of methylation correlated with higher transcript levels of RIN target genes. A previous study showed that the binding of RIN to a limited set of promoters was inhibited in the Cnr epimutant, indicating that promoter hypermethylation may prevent RIN binding (Martel et al., 2011). These three main findings, i.e., that: (i) local treatment of immature fruits with a DNA demethylating chemical accelerates ripening; (ii) promoters of ripening genes, which contain RIN binding sites, are gradually demethylated during ripening but remain stably hypermethylated in ripening-deficient mutants; (iii) RIN does not bind hypermethylated Cnr gene promoters (and, possibly, all hypermethylated promoters of its target genes), taken together, assign a key role to the epigenome structure and developmental dynamics in coordinating tomato fruit ripening. The global scenario presented so far also suggests that progressive demethylation of ripening-related gene promoters may be the necessary condition for binding of transcriptional regulators, thus triggering the accumulation of ripening-related transcripts. In normal fruit development, however, the mechanism inducing demethylation of promoters remains elusive, and further efforts are needed in this direction to uncover the “missing link.” Given the growing importance of epigenetic modifications in impacting fruit phenotypes, we envisage that, in the future, high-throughput sequencing technologies will allow routine screening of crop epigenomes, accelerating detection of epigenetic variation. We anticipate that screening epigenome structure and dynamics will coexist with the analysis of conventional genetic variation in future plant breeding strategies. Epigenetic-based crop improvement approaches may radically impact fruit quality traits, especially for those traits whose allelic variation has been reduced during domestication or recent intensive breeding pressure. As such future modeling work aimed at integrating epigenomic profiling and small RNA profiling alongside the more frequently used transcript, protein, enzyme, and metabolite profiling (as suggested in Figure 2) will allow far greater understanding of the complex dynamics underlying this tightly regulated biological process.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by grants from the Max-Planck-Gesellschaft (to Sonia Osorio and Alisdair R. Fernie), and by Ministerio de Ciencia e Innovación, Spain (Ramón and Cajal contract). Federico Scossa acknowledges the support of CRA-Young Investigator Program.
REFERENCES
- Alba R., Payton P., Fei Z. J., McQuinn R., Debbie P., Martin G. B., et al. (2005). Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17 2954–2965 10.1105/tpc.105.036053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armitage A. M. (1989). Promotion of fruit ripening of ornamental peppers by ethephon. HortScience 24 962–964 [Google Scholar]
- Barry C. S., Blume B., Bouzayen M., Cooper W., Hamilton A. J., Grierson D. (1996). Differential expression of the 1-amino-cyclopropane-1-carboxylate oxidase gene family of tomato. Plant J. 9 525–535 10.1046/j.1365-313X.1996.09040525.x [DOI] [PubMed] [Google Scholar]
- Barry C. S., Giovannoni J. J. (2006). Ripening in the tomato green-ripe mutant is inhibited by ectopic expression of a protein that disrupts ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 103 7923–7928 10.1073/pnas.0602319103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry C. S., McQuinn R. P., Chung M. Y., Besuden A., Giovannoni J. J. (2008). Amino acid substitutions in homologs of the STAY-GREEN protein are responsible for the green-flesh and chlorophyll retainer mutations of tomato and pepper. Plant Physiol. 147 179–187 10.1104/pp.108.118430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M., Karlova R., Ballester A. R., Tikunov Y. M., Bovy A. G., Wolters-Arts M., et al. (2012). The tomato FRUITFULL homologs TDR4/FUL1 and MBP7/FUL2 regulate ethylene-independent aspects of fruit ripening. Plant Cell 24 4437–4451 10.1105/tpc.112.103283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottcher C., Keyzers R. A., Boss P. K., Davies C. (2010). Sequestration of auxin by the indole-3-acetic acid-amino synthase GH3-1 in grape berry (Vitis vinifera L.) and the proposed role of auxin conjugation during ripening. J. Exp. Bot. 61 3615–3625 10.1093/jxb/erq174 [DOI] [PubMed] [Google Scholar]
- Bustamante C. A., Civello P. M., Martinez G. A. (2009). Cloning of the promoter region of beta-xylosidase (FaXyl1) gene and effect of plant growth regulators on the expression of FaXyl1 in strawberry fruit. Plant Sci. 177 49–56 10.1016/j.plantsci.2009.03.015 [DOI] [Google Scholar]
- Caldana C., Fernie A. R., Willmitzer L., Steinhauser D. (2012). Unraveling retrograde signaling pathways: finding candidate signaling molecules via metabolomics and systems biology driven approaches. Front. Plant Sci. 3: 267. 10.3389/fpls.2012.00267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrari F., Baxter C., Usadel B., Urbanczyk-Wochniak E., Zanor M. I., Nunes-Nesi A., et al. (2006). Integrated analysis of metabolite and transcript levels reveals the metabolic shifts that underlie tomato fruit development and highlight regulatory aspects of metabolic network behavior. Plant Physiol. 142 1380–1396 10.1104/pp.106.088534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillejo C., de la Fuente J. I., Iannetta P., Botella M. A., Valpuesta V. (2004). Pectin esterase gene family in strawberry fruit: study of FaPE1, a ripening-specific isoform. J. Exp. Bot. 55 909–918 10.1093/jxb/erh102 [DOI] [PubMed] [Google Scholar]
- Cevik V., Ryder C. D., Popovich A., Manning K., King J. K., Seymour G. B. (2010). A FRUITFULL-like gene is associated with genetic variation for fruit flesh firmness in apple (Malus domestica Borkh.). Tree Genet. Genomes 6 271–279 10.1007/s11295-009-0247-4 [DOI] [Google Scholar]
- Chai Y. M., Jia H. F., Li C. L., Dong Q. H., Shen Y. Y. (2011). FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 62 5079–5089 10.1093/jxb/err207 [DOI] [PubMed] [Google Scholar]
- Chan S. W., Henderson I. R., Jacobsen S. E. (2005). Gardening the genome: DNA methylation in Arabidopsis thaliana. Genetics 6 351–360 10.1038/nrg1601 [DOI] [PubMed] [Google Scholar]
- Civello P. M., Powell A. L., Sabehat A., Bennett A. B. (1999). An expansin gene expressed in ripening strawberry fruit. Plant Physiol. 121 1273–1280 10.1104/pp.121.4.1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cokus S. J., Feng S., Zhang X., Chen Z., Merriman B., Haudenschild C. D., et al. (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452 215–219 10.1038/nature06745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombe B. G. (1992). Research on development and ripening of the grape berry. Am. J. Enol. Vitic. 43 101–111 10.1111/j.1755-0238.2000.tb00171.x [DOI] [Google Scholar]
- Cubas P., Vincent C., Coen E. (1999). An epigenetic mutation responsible for natural variation in floral symmetry. Nature 401 157–161 10.1038/43657 [DOI] [PubMed] [Google Scholar]
- Davies C., Boss P. K., Robinson S. P. (1997). Treatment of grape berries, a nonclimacteric fruit with a synthetic auxin, retards ripening and alters the expression of developmentally regulated genes. Plant Physiol. 115 1155–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente J. I., Amaya I., Castillejo C., Sanchez-Sevilla J. F., Quesada M. A., Botella M. A., et al. (2006). The strawberry gene FaGAST affects plant growth through inhibition of cell elongation. J. Exp. Bot. 57 2401–2411 10.1093/jxb/erj213 [DOI] [PubMed] [Google Scholar]
- Deluc L. G., Grimplet J., Wheatley M. D., Tillett R. L., Quilici D. R., Osborne C., et al. (2007). Transcriptomic and metabolite analyses of Cabernet Sauvignon grape berry development. BMC Genomics 8:429 10.1186/1471-2164-8-429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinneny J. R., Weigel D., Yanofsky M. F. (2005). A genetic framework for fruit patterning in Arabidopsis thaliana. Development 132 4687–4697 10.1242/dev.02062 [DOI] [PubMed] [Google Scholar]
- Elitzur T., Vrebalov J., Giovannoni J. J., Goldschmidt E. E., Friedman H. (2010). The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. J. Exp. Bot. 61 1523–1535 10.1093/jxb/erq017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Kereamy A., Chervin C., Roustan J. P., Cheynier V., Souquet J. M., Moutounet M., et al. (2003). Exogenous ethylene stimulates the long-term expression of genes related to anthocyanin biosynthesis in grape berries. Physiol. Plant. 119 175–182 10.1034/j.1399-3054.2003.00165.x [DOI] [Google Scholar]
- Enfissi E. M., Barneche F., Ahmed I., Lichtle C., Gerrish C., McQuinn R. P., et al. (2010). Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 22 1190–1215 10.1105/tpc.110.073866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson O., Friis E. M., Lofgren P. (2000). Seed size, fruit size, and dispersal systems in angiosperms from the early cretaceous to the late tertiary. Am. Nat. 156 47–58 10.1086/303367 [DOI] [PubMed] [Google Scholar]
- Fait A., Hanhineva K., Beleggia R., Dai N., Rogachev I., Nikiforova V. J., et al. (2008). Reconfiguration of the achene and receptacle metabolic networks during strawberry fruit development. Plant Physiol. 148 730–750 10.1104/pp.108.120691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarese L., Trainotti L., Moretto P., deLaureto P. P., Rascio N., Casadoro G. (1995). Differential ethylene-inducible expression of cellulase in pepper plants. Plant Mol. Biol. 29 735–747 10.1007/BF00041164 [DOI] [PubMed] [Google Scholar]
- Finkelstein R. R., Rock C. D. (2002). Abscisic acid biosynthesis and response. Arabidopsis Book 1:e0058 10.1199/tab.0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M., Nakano T., Shima Y., Ito Y. (2013). A large-scale identification of direct targets of the tomato MADS Box transcription factor RIPENING INHIBITOR reveals the regulation of fruit ripening. Plant Cell 25 371–386 10.1105/tpc.112.108118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez E., Pineda B., Capel J., Anton M. T., Atares A., Perez-Martin F., et al. (2010). Functional analysis of the Arlequin mutant corroborates the essential role of the Arlequin/TAGL1 gene during reproductive development of tomato. PLoS ONE 5:e14427 10.1371/journal.pone.0014427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52 725–749 10.1146/annurev.arplant.52.1.725 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. J. (2007). Fruit ripening mutants yield insights into ripening control. Curr. Opin. Plant Biol. 10 283–289 10.1016/j.pbi.2007.04.008 [DOI] [PubMed] [Google Scholar]
- Giovannoni J. J., Noensie E. N., Ruezinsky D. M., Lu X., Tracy S. L., Ganal M. W., et al. (1995). Molecular genetic analysis of the ripening-inhibitor and non-ripening loci of tomato: a first step in genetic map-based cloning of fruit ripening genes. Mol. Gen. Genet. 248 195–206 10.1007/BF02190801 [DOI] [PubMed] [Google Scholar]
- Grimplet J., Deluc L. G., Tillett R. L., Wheatley M. D., Schlauch K. A., Cramer G. R., et al. (2007). Tissue-specific mRNA expression profiling in grape berry tissues. BMC Genomics 8:187 10.1186/1471-2164-8-187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton A. J., Lycett G. W., Grierson D. (1990). Antisense gene that inhibits synthesis of the hormone ethylene in transgenic plants. Nature 346 284–287 10.1038/346284a0 [DOI] [Google Scholar]
- Harpster M. H., Lee K. Y., Dunsmuir P. (1997). Isolation and characterization of a gene encoding endo-beta-1,4-glucanase from pepper (Capsicum annuum L). Plant Mol. Biol. 33 47–59 10.1023/A:1005795028489 [DOI] [PubMed] [Google Scholar]
- Hileman L. C., Sundstrom J. F., Litt A., Chen M., Shumba T., Irish V. F. (2006). Molecular and phylogenetic analyses of the MADS-box gene family in tomato. Mol. Biol. Evol. 23 2245–2258 10.1093/molbev/msl095 [DOI] [PubMed] [Google Scholar]
- Himmelbach A., Yang Y., Grill E. (2003). Relay and control of abscisic acid signaling. Curr. Opin. Plant Biol. 6 470–479 10.1016/S1369-5266(03)00090-6 [DOI] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2007). Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 12 343–351 10.1016/j.tplants.2007.06.013 [DOI] [PubMed] [Google Scholar]
- Iannetta P. P. M., Laarhovenb L. J., Medina-Escobar N., James E. K., McManuse M. T., Davies H. V., et al. (2006). Ethylene and carbon dioxide production by developing strawberries show a correlative pattern that is indicative of ripening climacteric fruit. Physiol. Plant. 127 247–259 10.1111/j.1399-3054.2006.00656.x [DOI] [Google Scholar]
- Itkin M., Seybold H., Breitel D., Rogachev I., Meir S., Aharoni A. (2009). TOMATO AGAMOUS-LIKE 1 is a component of the fruit ripening regulatory network. Plant J. 60 1081–1095 10.1111/j.1365-313X.2009.04064.x [DOI] [PubMed] [Google Scholar]
- Jaakola L., Poole M., Jones M. O., Kämäräinen-Karppinen T., Koskimäki J. J., Hohtola A., et al. (2010). A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 153 1619–1629 10.1104/pp.110.158279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J. M., Noel B., Policriti A., Clepet C., Casagrande A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449 463–465 10.1038/nature06148 [DOI] [PubMed] [Google Scholar]
- Jia H. F., Chai Y. M., Li C. L., Lu D., Luo J. J., Qin L., et al. (2011). Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 157 188–199 10.1104/pp.111.177311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. M., Joyce D. C. (2003). ABA effects on ethylene production, PAL activity, anthocyanin and phenolic contents of strawberry fruit. Plant Growth Regul. 39 171–174 10.1023/A:1022539901044 [DOI] [Google Scholar]
- Klie S., Krueger S., Krall L., Giavalisco P., Flügge U. I., Willmitzer L., et al. (2011). Analysis of the compartmentalized metabolome – a validation of the non-aqueous fractionation technique. Front. Plant Sci. 2:55 10.3389/fpls.2011.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp S. (2002). Tobacco to tomatoes: a phylogenetic perspective on fruit diversity in the Solanaceae. J. Exp. Bot. 53 2001–2011 10.1093/jxb/erf068 [DOI] [PubMed] [Google Scholar]
- Kobashi K., Gemma H., Iwahori S. (1999). Sugar accumulation in peach fruit as affected by abscisic acid treatment in relation to some sugar metabolizing enzymes. J. Jpn. Soc. Hortic. Sci. 68 465–470 10.2503/jjshs.68.465 [DOI] [Google Scholar]
- Kumar R., Agarwal P., Tyagi A. K., Sharma A. K. (2012). Genome-wide investigation and expression analysis suggest diverse roles of auxin-responsive GH3 genes during development and response to different stimuli in tomato (Solanum lycopersicum). Mol. Genet. Genomics 287 221–235 10.1007/s00438-011-0672-6 [DOI] [PubMed] [Google Scholar]
- Lanahan M. B., Yen H. C., Giovannoni J. J., Klee H. J. (1994). The never ripe mutation blocks ethylene perception in tomato. Plant Cell 6 521–530 10.1105/tpc.6.4.521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law J. A., Jacobsen S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nature 11 204–220 10.1038/nrg2719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M., Joung J. G., McQuinn R., Chung M. Y., Fei Z., Tieman D., et al. (2012). Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays and important role in ripening and carotenoid accumulation. Plant J. 70 191–204 10.1111/j.1365-313X.2011.04863.x [DOI] [PubMed] [Google Scholar]
- Lee S. J., Saravanan R. S., Damasceno C. M. B., Yamane H., Kim B. D, Rose J. K. C. (2004). Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 42 979–988 10.1016/j.plaphy.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Leung J., Giraudat J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49 199–222 10.1146/annurev.arplant.49.1.199 [DOI] [PubMed] [Google Scholar]
- Liljegren S. J., Ditta G. S., Eshed Y., Savidge B., Bowman J. L., Yanofsky M. F. (2000). SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 404 766–770 10.1038/35008089 [DOI] [PubMed] [Google Scholar]
- Liu K., Kang B. C., Jiang H., Moore S. L., Li H., Watkins C. B., et al. (2005). A GH3-like gene, CcGH3, isolated from Capsicum chinense L. fruit is regulated by auxin and ethylene. Plant Mol. Biol. 58 447–464 10.1007/s11103-005-6505-4 [DOI] [PubMed] [Google Scholar]
- Liu Y., Roof S., Ye Z., Barry C., van Tuinen A., Vrebalov J., et al. (2004). Manipulation of light signal transduction as a means of modifying fruit nutritional quality in tomato. Proc. Natl. Acad. Sci. U.S.A. 101 9897–9902 10.1073/pnas.0400935101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo V. A., Osorio S., Borsani J., Lauxmann M. A., Bustamante C. A., Budde C. O., et al. (2011). Metabolic profiling during peach fruit development and ripening reveals the metabolic networks that underpin each developmental stage. Plant Physiol. 157 1696–1710 10.1104/pp.111.186064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning K., Tor M., Poole M., Hong Y., Thompson A. J., King G. J., et al. (2006). A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 38 948–952 10.1038/ng1841 [DOI] [PubMed] [Google Scholar]
- Martel C., Vrebalov J., Tafelmeyer P., Giovannoni J. J. (2011). The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiol. 157 1568–1579 10.1104/pp.111.181107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G. A., Chaves A. R., Anon M. C. (1996). Effect of exogenous application of gibberellic acid on color change and phenylalanine ammonia-lyase, chlorophyllase, and peroxidase activities during ripening of strawberry fruit (Fragaria × ananassa Duch). J. Plant Growth Regul. 15 139–146 10.1007/BF00198929 [DOI] [Google Scholar]
- Matas A. J., Yeats T. H., Buda G. J., Zheng Y., Chatterjee S., Tohge T., et al. (2011). Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23 3893–3910 10.1105/tpc.111.091173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messeguer R., Ganal M. W., Steffens J. C., Tanksley S. D. (1991). Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol. Biol. 16 753–770 10.1007/BF00015069 [DOI] [PubMed] [Google Scholar]
- Mustilli A. C., Fenzi F., Ciliento R., Alfano F., Bowler C. (1999). Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 11 145–157 10.1105/tpc.11.2.145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeller P. W., Wong L. M., Taylor L. P., Pike D. A., Theologis A. (1991). Reversible inhibition of tomato fruit senescence by antisense RNA. Science 254 437–439 10.1126/science.1925603 [DOI] [PubMed] [Google Scholar]
- Osorio S., Alba R., Damasceno C. M. B., Lopez-Casado G., Lohse M., Zanor M. I., et al. (2011). Systems biology of tomato fruit development: combined transcript, protein and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 157 405–425 10.1104/pp.111.175463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S., Alba R., Nikoloski Z., Kochevenko A., Fernie A. R., Giovannoni J. J. (2012). Integrative comparative analyses of transcript and metabolite profiles from pepper and tomato ripening and development stages uncovers species-specific patterns of network regulatory behavior. Plant Physiol. 159 1713–1729 10.1104/pp.112.199711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q. H., Li M. J., Peng C. C., Zhang N., Zou X., Zou K. Q., et al. (2005). Abscisic acid activates acid invertases in developing grape berry. Physiol. Plant. 125 157–170 10.1111/j.1399-3054.2005.00552.x [DOI] [Google Scholar]
- Pan Y., Bradley G., Pyke K., Ball G., Lu C., Fray R., et al. (2013). Network inference analysis identifies an APRR2-like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 161 1476–1485 10.1104/pp.112.212654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson G. I., Thorpe C. J., Chandler V. L. (1993). Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics 135 881–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins-Veazie P. M. (1995). Growth and ripening of strawberry fruit. Hortic. Rev. 17 267–297 10.1002/9780470650585.ch8 [DOI] [Google Scholar]
- Perkins-Veazie P. M., Huber D. J., Brecht J. K. (1996). In vitro growth and ripening of strawberry fruit in presence of ACC, STS or propylene. Ann. Appl. Biol. 128 105–116 10.1111/j.1744-7348.1996.tb07094.x [DOI] [Google Scholar]
- Pnueli L., Hareven D., Rounsley S., Yanofsky M. F., Lifschitz E. (1994). Isolation of the tomato AGAMOUS gene TAG1 and analysis of its homeotic role in transgenic plants. Plant Cell 6 163–173 10.1105/tpc.6.2.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richings E. W., Cripps R. F., Cowan A. K. (2000). Factors affecting ‘Hass’ avocado fruit size: carbohydrate, abscisic acid and isoprenoid metabolism in normal and phenotypically small fruit. Physiol. Plant. 109 81–89 10.1034/j.1399-3054.2000.100112.x [DOI] [Google Scholar]
- Rodrigo M. J., Marcos J. F., Alferez F., Mallent M. D., Zacarias L. (2003). Characterization of Pinalate, a novel Citrus sinensis mutant with a fruit-specific alteration that results in yellow pigmentation and decreased ABA content. J. Exp. Bot. 54 727–738 10.1093/jxb/erg083 [DOI] [PubMed] [Google Scholar]
- Rohrmann J., Tohge T., Alba R., Osorio S., Caldana C., McQuinn R., et al. (2011). Combined transcription factor profiling, microarray analysis and metabolite profiling reveals the transcriptional control of metabolic shifts occurring during tomato fruit development. Plant J. 68 999–1013 10.1111/j.1365-313X.2011.04750.x [DOI] [PubMed] [Google Scholar]
- Rose J. K. C., Bashir S., Giovannoni J. J., Jahn M. M., Saravanan R. S. (2004). Tackling the plant proteome: practical approaches, hurdles and experimental tools. Plant J. 39 715–733 10.1111/j.1365-313X.2004.02182.x [DOI] [PubMed] [Google Scholar]
- Rottmann W. H., Peter G. F., Oeller P. W., Keller J. A., Shen N. F., Nagy B. P., et al. (1991). 1-Aminocyclopropane-1-carboxylate synthase in tomato is encoded by a multigene family whose transcription is induced during fruit and floral senescence. J. Mol. Biol. 222 937–961 10.1016/0022-2836(91)90587-V [DOI] [PubMed] [Google Scholar]
- Saravanan R. S, Rose J. K. C. (2004). A critical evaluation of sample extraction techniques for enhanced proteomic analysis of recalcitrant plant tissues. Proteomics 4 2522–2532 10.1002/pmic.200300789 [DOI] [PubMed] [Google Scholar]
- Seymour G., Poole M., Manning K., King G. J. (2008). Genetics and epigenetics of fruit development and ripening. Curr. Opin. Plant Biol. 11 58–63 10.1016/j.pbi.2007.09.003 [DOI] [PubMed] [Google Scholar]
- Seymour G. B., Ostergaard L., Chapman N. H., Knapp S., Martin C. (2013). Fruit development and ripening. Annu. Rev. Plant Biol. 64 219–241 10.1146/annurev-arplant-050312-120057 [DOI] [PubMed] [Google Scholar]
- Seymour G. B., Ryder C. D., Cevik V., Hammond J. P., Popovich A., King G. J., et al. (2011). A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch. fruit, a non-climacteric tissue. J. Exp. Bot. 62 1179–1188 10.1093/jxb/erq360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V., Sargent D. J., Crowhurst R. N., Mockler T. C., Folkerts O., Delcher A. L., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43 109–116 10.1038/ng.740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smaczniak C., Immink R. G., Angenent G. C., Kaufmann K. (2012). Developmental and evolutionary diversity of plant MADS-domain factors: insights from recent studies. Development 139 3081–3098 10.1242/dev.074674 [DOI] [PubMed] [Google Scholar]
- Sun L., Sun Y., Zhang M., Wang L., Ren J., Cui M., et al. (2012). Suppression of 9-cis-epoxycarotenoid dioxygenase, which encodes a key enzyme in abscisic acid biosynthesis, alters fruit texture in transgenic tomato. Plant Physiol. 158 283 298 10.1104/pp.111.186866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A. J., Tor M., Barry C. S., Vrebalov J., Orfila C., Jarvis M. C., et al. (1999). Molecular and genetic characterization of a novel pleiotropic tomato-ripening mutant. Plant Physiol. 120 383–390 10.1104/pp.120.2.383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 635–641 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainotti L., Spinello R., Piovan A., Spolaore S., Casadoro G. (2001). beta-Galactosidases with a lectin-like domain are expressed in strawberry. J. Exp. Bot. 52 1635–1645 10.1093/jexbot/52.361.1635 [DOI] [PubMed] [Google Scholar]
- Vaughn M. W., Tanurdzic M., Lippman Z., Jiang H., Carrasquillo R., Rabinowicz P. D., et al. (2007). Epigenetic natural variation in Arabidopsis thaliana. PLoS Biol. 5:e174 10.1371/journal.pbio.0050174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Pan I. L., Arroyo A. J., McQuinn R., Chung M., Poole M., et al. (2009). Fleshy fruit expansion and ripening are regulated by the Tomato SHATTERPROOF gene TAGL1. Plant Cell 21 3041–3062 10.1105/tpc.109.066936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrebalov J., Ruezinsky D., Padmanabhan V., White R., Medrano D., Drake R., et al. (2002). A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 296 343–346 10.1126/science.1068181 [DOI] [PubMed] [Google Scholar]
- Vriezen W. H., Feron R., Maretto F., Keijman J., Mariani C. (2008). Changes in tomato ovary transcriptome demonstrate complex hormonal regulation of fruit set. New Phytol. 177 60–76 10.1111/j.1469-8137.2007.02254.x [DOI] [PubMed] [Google Scholar]
- Yamaki S., Asakura T. (1991). Stimulation of the uptake of sorbitol into vacuoles from apple fruit flesh by abscisic acid and into protoplasts by indoleacetic acid. Plant Cell Physiol. 32 315–318 [Google Scholar]
- Zamboni A., Di Carli M., Guzzo F., Stocchero M., Zenoni S., Ferrarini A., et al. (2010). Identification of putative stage-specific grapevine berry biomarkers and omics data integration into networks. Plant Physiol. 154 1439–1459 10.1104/pp.110.160275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Leng P., Zhang G., Li X. (2009). Cloning and functional analysis of 9-cis-epoxycarotenoid dioxygenase (NCED) genes encoding a key enzyme during abscisic acid biosynthesis from peach and grape fruits. J. Plant Physiol. 166 1241–1252 10.1016/j.jplph.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Zhang X., Yazaki J., Sundaresan A., Cokus S., Chan S. W., Chen H., et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126 1189–1201 10.1016/j.cell.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Zhong S., Fei Z., Chen Y. R., Zheng Y., Huang M., Vrebalov J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31 154–159 10.1038/nbt.2462 [DOI] [PubMed] [Google Scholar]
- Zilberman D., Henikoff S. (2007). Genome-wide analysis of DNA methylation patterns. Development 134 3959–3965 10.1242/dev.001131 [DOI] [PubMed] [Google Scholar]