Abstract
mRNAs can be targeted to specific neuronal subcellular domains, which enables rapid changes in the local proteome through local translation. This mRNA-based mechanism links extrinsic signals to spatially restricted cellular responses and can mediate stimulus-driven adaptive responses such as dendritic plasticity. Local mRNA translation also occurs in growing axons where it can mediate directional responses to guidance signals. Recent profiling studies have revealed that both growing and mature axons possess surprisingly complex and dynamic transcriptomes, thereby suggesting that axonal mRNA localization is highly regulated and has a role in a broad range of processes, a view that is increasingly being supported by new experimental evidence. Here, we review current knowledge on the roles and regulatory mechanisms of axonal mRNA translation and discuss emerging links to axon guidance, survival, regeneration and neurological disorders.
Cell–cell signalling relies on the ability of cells to adjust their local proteome (protein abundance and complexity) with high spatiotemporal precision in response to extracellular signals. Thousands of mRNAs exhibit specific subcellular localization patterns in mammals1 and in Drosophila melanogaster2, which suggests that local mRNA translation may have a general role in controlling the local proteome. Conceptually, local protein synthesis provides several advantages over the transport of pre-existing proteins from one part of the cell to another3. First, translationally silent forms of mRNAs can be stored locally and used to make many copies of a protein when needed, thereby providing economic advantages. Second, the ectopic presence of proteins in other parts of the cell during protein transport is avoided. Third, mRNAs can be targeted to different subcellular localizations using ‘address’ information in their untranslated regions (UTRs) without changing the structure and function of the proteins they encode. Finally, properties that are unique to newly made proteins (such as minimal post-translational modification) may provide an additional layer of signalling information.
Intuition suggests that highly polarized cells like neurons would benefit greatly from local mRNA translation. Indeed, local mRNA translation is known to mediate long-lasting synaptic plasticity in dendritic spines, and studies in dendrites have provided significant mechanistic and functional insights into how extrinsic signals regulate local mRNA translation4,5. Although evidence for axonal protein synthesis in mammals dates back to the 1960s6–8, surprisingly little is known about local mRNA translation in axons. The general view of the axon as a passive transmitter of information may have contributed to this neglect. Nevertheless, axons — like dendrites — are signal-receiving compartments. Growing axons require extrinsic signals to regulate their growth, navigation and synapse formation and mature axons depend on external cues for maintenance and repair. Thus, there is clearly a conceptual need for local protein synthesis in distal axons9. Indeed, stimulus-induced responses of axons in cell culture — such as chemotropic responses10 and regeneration following injury11 — require axonal mRNA translation, and evidence suggests that some of these findings could be applicable in vivo12–14.
Thus, local mRNA translation can be regarded as a common mechanism for the regulation of local proteomic homeostasis in response to extracellular signals in axons and dendrites. Both developing15,16 and mature17,18 axons contain complex and dynamic mRNA repertoires, and recent studies have provided insights into the regulation of axonal mRNA translation. Here, we aim to provide a conceptual framework for this emerging field of active research by reviewing our understanding of the current literature and discussing future directions.
Evidence for axonal mRNA translation
The initial lack of interest in local axonal mRNA translation can be traced back to findings in the 1970s using the squid giant axon. It was suggested that protein synthesis is unlikely to occur in mature axons as little or no ribosomal RNA (as measured by their optical densities on polyacrylamide gels) was observed in the axoplasm19. This interpretation was, however, later disputed by evidence obtained using more sensitive biochemical methods showing the presence of ribosomal RNAs20, mRNAs21 and actively translating polysomes22 in squid giant axons. In mammals, ribosomes were identified by electron microscopy in embryonic cortical23 and sympathetic24 neuronal axons in cell culture, and in embryonic peripheral sensory axons in vivo25. Recent immuno-electron microscopy has shown specific immunoreactivity to the ribosomal protein S6 in the axons of cultured embryonic sympathetic and hippocampal neurons26. Notably, axonal ribosomes rarely form polysomes in vivo24,25,27, unlike in culture conditions23, which suggests that monosomal translation may predominate or that translational activation is spatiotemporally restricted in axons in normal conditions.
In an electron microscopy study using adult rats, electron-dense ribosomal rosettes — the typical ultrastructural appearance of polysomes — were limited to the axon initial segment and not detectable in the axon shaft of CNS neurons28, thus leading to speculation that axonal protein synthesis only occurs during development. Immunohistological studies, however, showed that mature PNS axons contain ribosomal proteins and RNAs, which form intermittent ribosomal plaques that are distributed unevenly in the peripheral axoplasm close to the plasma membrane29–32. These observations were consistent with an overlooked electron microscopy study from 1970 (REF. 33). The peripheral localization of ribosomes also agrees with a recent finding that a direct interaction between cell surface receptors and ribosomes spatially restricts mRNA translation to the site at which an extrinsic cue is received26. The specialized morphology and distribution of axonal ribosomes may thus have made their ultrastructural identification difficult28. Indeed, immuno-electron microscopy evidence from transgenic mice showed that an enhanced green fluorescent protein (EGFP)-tagged ribosomal protein L10a (EGFP–L10a) localizes to the nodes of Ranvier in corticospinal tract axons in vivo34. Although some EGFP–L10a may be present outside the ribosome (as it probably competes with endogenous L10a for ribosomal occupancy), this nonetheless supports the idea that mature axons contain intermittent ribosomal plaques that are not readily identifiable by conventional ultrastructural criteria29–33.
Compelling evidence for axonal protein synthesis came from metabolic labelling experiments that showed that unmyelinated axons without somas are capable of mRNA translation-dependent protein synthesis in vertebrates7,35–39 and invertebrates6,21. Another key finding was that exogenous mRNA can be translated when injected into mollusc axons without somas40. The list of axonally localizing and translatable mRNAs is rapidly growing and includes those encoding membrane-targeted proteins such as ephrin type-A receptor 2 (EPHA2)41 and κ-type opioid receptor (KOR1; also known as OPRK1)42.
Several electron microscopy studies have failed to identify rough endoplasmic reticulum (RER) and Golgi apparatus in vertebrate axons24,25,27,43, thus raising the question of whether vertebrate axons have the capacity to process proteins. However, a recent study using the severed axons of cultured peripheral sensory neurons showed that metabolically labelled, newly synthesized proteins are trafficked to the plasma membrane44, thus suggesting that the functional equivalent of RER and Golgi apparatus exists in these axons. ER-associated and Golgi-associated proteins have been detected immunocytologically in the axons of vertebrates44, but the morphology of the axonal ER is distinct from that found in the soma44,45. These findings are consistent with those seen in invertebrates. Mollusc axons are capable of local protein targeting but do not appear to possess ultrastructurally identifiable RER and Golgi apparatus46. Nevertheless, proteins residing in the ER and Golgi can be detected immunocytologically47. It will be interesting to explore whether the axonal RER and Golgi adopt specialized morphologies, perhaps similar to those of primitive parasites48. Together, these findings show that both growing and mature axons possess protein synthesis and processing machinery, but the unconventional nature of their histological structure presents a puzzle.
Function of axonal mRNA translation
Growing axons receive continuous guidance information from their environment as they travel along long stereotypical paths to reach their synaptic partners. The distal tip of a growing axon, a specialized structure known as the growth cone, must quickly process this information, often without enough time to communicate with its soma. Indeed, axons severed from their cell bodies can correctly pathfind in vivo49, and the growth cone can respond to guidance cues without the cell body in vitro10,50.
Local mRNA translation is a key mechanism in this autonomous signalling, and protein synthesis inhibitors block the ability of growth cones that are severed from their somas to respond to several guidance cues. These guidance cues include netrin 1 (REFS 51–54), semaphorin 3A (SEMA3A)10,55, SLIT2 (REF. 56), engrailed 1 and engrailed 2 (EN1 and EN2)57–59, pituitary adenylate cyclase-activating polypeptide (PACAP; also known as ADCYAP1)60, nerve growth factor (NGF)52,61, brain-derived neurotrophic factor (BDNF)54 and neurotrophin 3 (NT3)62,63. Increasing evidence shows that axonal mRNA translation continues to play roles in adulthood, particularly during plastic responses such as injury-induced axon regeneration11,13,14,64–66. These extrinsic factors rapidly induce local protein synthesis by activating the mammalian target of rapamycin (mTOR) pathway (TABLE 1).
Table 1.
Signals that regulate axonal mRNA translation
| Signal | Neuronal type | Target mRNA | Regulator | Function | Refs |
|---|---|---|---|---|---|
| Signals that increase axonal protein synthesis | |||||
| Netrin 1 | RGC* | β-actin | ZBP1 | GC turning | 53 |
| P19 | KOR1 | GRB7 | Analgesia | 164 | |
| DRG‡ | PAR3 | Unknown | Axon elongation | 52 | |
| Semaphorin 3A | DRG‡ | RHOA | Unknown | GC collapse | 55 |
| DRG‡ | Calreticulin | Unknown | Axon regeneration | 156 | |
| SLIT2B | RGC* | Cofilin | Unknown | GC collapse | 56 |
| Nerve growth factor | PC12 | Unknown | Unknown | Neurite regeneration | 114 |
| PC12 | RPL4 | Unknown | Neurite regeneration | 197 | |
| Sympathetic§ | β-actin, ADF, NF | Unknown | Axon guidance¶ | 194 | |
| Sympathetic‡ | IMPA1 | Unknown | Axon maintenance | 15 | |
| Hippocampal‡, CTX‡ | β-actin | Unknown | Axon guidance¶ | 63 | |
| DRG‡ | β-actin | Unknown | Axon regeneration | 156 | |
| DRG‡ | CREB | Unknown | Cell survival | 61 | |
| DRG‡ | PAR3 | Unknown | Axon elongation | 52 | |
| SCG‡ | COX4I1 | miR-338 | Axon maintenance | 116,178 | |
| SCG‡ | ATP5G1 | miR-338 | Axon maintenance | 198 | |
| PC12 | GAP43 | HUD | Axon guidance¶ | 103 | |
| SMN‡ | CPG15 | SMN and HUD | Axon maintenance | 12 | |
| Brain-derived neurotrophic factor | Spinal cord* | β-actin | ZBP1 | GC turning | 54 |
| Hippocampal‡, CTX‡ | β-actin | ZBP1 | Axon guidance¶ | 63 | |
| DRG‡ | β-actin | ZBP1 | Axon regeneration | 156 | |
| Spinal cord‡, SMN‡ | CPG15 | SMN and HUD | Axon maintenance | 12 | |
| Neurotrophin 3 | Hippocampal‡ | β-catenin | CPEB1 | Axon branching | 174 |
| DRG‡ | β-actin | ZBP1 | Axon regeneration | 156 | |
| DRG‡, CTX‡ | Calreticulin | Unknown | Axon guidance | 155 | |
| Pituitary adenylate cyclase-activating polypeptide |
Spinal cord* | Unknown | Unknown | GC turning | 60 |
| 5-hydroxytryptamine (serotonin) | Sensory∥ | eEF1A | Unknown | Long-term facilitation | 199 |
| Myelin-associated glycoprotein | DRG‡ | αB crystallin | Unknown | Axon regeneration | 156 |
| DRG‡, CTX‡ | Calreticulin | Unknown | Axon guidance¶ | 155 | |
| Engrailed 1 and 2 | RGC*ठ| Unknown | Unknown | Topographic mapping | 58,59 |
| RGC* | Lamin B2 | Unknown | Axon maintenance | 118 | |
| Midbrain dopaminergic‡ |
COX1 components | Unknown | Cell survival | 57 | |
| Axotomy | DRG‡ | Importin-β | Unknown | Axon regeneration | 64 |
| DRG‡ | Vimentin | Unknown | Axon regeneration | 45,65 | |
| DRG‡ | STAT3A | Unknown | Axon regeneration | 14 | |
| DRG‡ | NF-L, NF-M, NF-H | Unknown | Axon regeneration | 200 | |
| Differentiation | P19 | Tau | Unknown | Axon elongation | 201 |
| Unknown | OSN‡ | Odorant receptors | Unknown | Axon regeneration | 128 |
| Unknown | Caenorhabditis elegans | C/EBP1 | MAPK | Axon regeneration | 130 |
| Depolarization | DRG‡ | KOR1 | Unknown | Analgesia | 42 |
| Inflammation | DRG‡ | TRPV1¶ | Unknown | Hyperalgesia | 134 |
| Injury | DRG‡ | Nav1.8 | Unknown | Hyperalgesia | 135,137 |
| Signals that do not increase axonal protein synthesis | |||||
| Ephrin B | RGC* | Not applicable | Not applicable | Not applicable | 67 |
| Lysophosphatidic acid | RGC* | Not applicable | Not applicable | Not applicable | 10 |
| Signals that decrease axonal protein synthesis | |||||
| Ephrin A | RGC‡ | Unknown | ERK1/2 and TSC1/2 | Axon guidance | 152 |
ADF, actin depolymerizing factor; ATP5G1, ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9); C/EBP1, CAAT enhancer binding protein 1; COX1, cyclooxygenase 1; COX4I1, cytochrome c oxidase subunit IV isoform 1; CPEB1, cytoplasmic polyadenylation element binding protein 1; CPG15, translation of candidate plasticity-related gene 15 (also known as NRN1); CREB, cyclic AMP responsive element-binding protein; CTX, cerebral cortical; DRG, dorsal root ganglion; eEF1A, eukaryotic elongation factor 1α; ERK, extracellular signal-regulated kinase; GAP43, growth associated protein 43; GC, growth cone; GRB7, growth factor receptor-bound protein 7; HUD, Hu-antigen D (also known as ELAVL4); IMPA1, inositol monophosphatase 1; KOR1, κ-type opioid receptor; MAPK, mitogen-activated protein kinase; miR, microRNA; Nav1.8, sodium channel 1.8; NF (L, M, H), neurofilament protein (light, medium, heavy); OSN, olfactory sensory; PAR3, proteinase-activated receptor 3; RGC, retinal ganglion cell; RPL4, ribosomal protein L4; SCG, superior cervical ganglion; SMN, spinal motor neuron; STAT3A, signal transducer and activator of transcription 3A; TRPV1, transient receptor potential cation channel subfamily V member 1; TSC, tuberous sclerosis protein; ZBP1, zipcode binding protein 1.
Xenopus laevis.
Mouse or rat.
Chicken.
Aplaysia.
Indicates that the finding was not directly addressed in the study.
Chemotropic responses of growth cones
One of the first identified functions of axonal protein synthesis was its ability to mediate netrin 1-induced and sema3a-induced chemotropic responses in cultured growth cones10 (FIG. 1). Xenopus laevis retinal growth cones without somas turn towards netrin 1 gradients and away from sema3a gradients in culture10. Application of these cues to soma-less growth cones increases mTOR activity (as measured by phosphorylation of its substrate eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4e-bp1)) and axonal protein synthesis (as measured by 3H-leucine incorporation) within minutes. In addition, inhibiting mTOR activation (with rapamycin) or ribosome function (with cycloheximide or anisomycin) blocks both local protein synthesis and the chemotropic responses10. Inhibition of mTOR or protein synthesis also blocks sema3a-induced growth cone collapse (the rapid withdrawal of filopodia in response to uniformly applied repulsive cues)10. Intriguingly, a phosphoinositide 3-kinase (PI3K) inhibitor blocks the growth cone’s response to netrin 1, whereas the response to sema3a is not blocked. This suggests that different signalling pathways downstream of netrin 1 and sema3a converge on mTOR10. Lysophosphatidic acid-induced growth cone collapse is not affected by any of these inhibitors, and lysophosphatidic acid application increases neither mTOR activity nor local protein synthesis10. Other cues that do not induce protein synthesis include ephrin B67 and sphingosine 1-phosphate68.
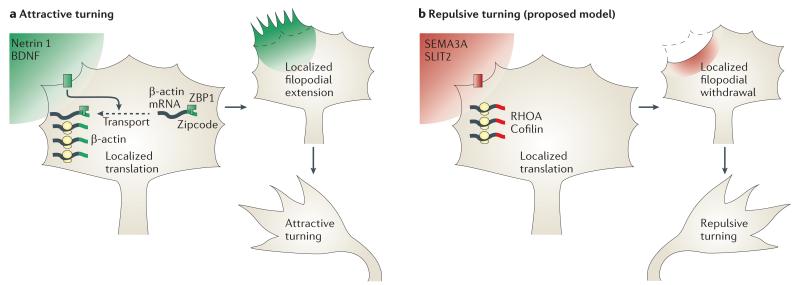
Figure 1. Growth cone turning regulated by differential mRNA translation.
Gradients of protein synthesis-inducing guidance cues commonly activate global translational activity on the side of the growth cone nearest to the gradient by activating mammalian target of rapamycin (mTOR). However, the specific mRNA translated in response to the cue differs depending on whether it is an attractive or repulsive cue and determines the direction of growth cone turning. a | Stimulation by attractive cues, such as netrin 1 and brain-derived neurotrophic factor (BDNF), leads to asymmetric synthesis of β-actin on the side near to the source of the gradient, which is mediated by β-actin mRNA transport to this region by zipcode-binding protein 1 (ZBP1)53,54. Spatially restricted synthesis of β-actin may lead to actin polymerization, cytoskeletal assembly and attractive turning of the growth cone. b | By contrast, repulsive cues, such as semaphorin 3A (SEMA3A) and SLIT2, activate the axonal translation of the actin-depolymerizing proteins RHOA55 and cofilin56 when uniformly applied in cell culture. A proposed model is shown, in which localized cytoskeletal disassembly may result in repulsive turning through polarized filopodial collapse. However, whether these molecules are translated asymmetrically in a repulsive gradient has not yet been tested.
Interestingly, SEMA3A-induced, protein synthesis-dependent growth cone collapse occurs at low, but not at high, concentrations of SEMA3A69. This agrees with the finding that growth cones lose their physiological cue selectivity when the cues are applied in high concentrations69. For example, spinal motor neuron growth cones show neuronal subtype-specific collapse responses to either SEMA3A or SEMA3F in culture, and these responses are blocked by protein synthesis inhibitors69. When these cues are applied in high concentrations, the growth cones lose both cue specificity and protein synthesis dependence69. Differences in the concentration and/or bioactivity of SEMA3A may therefore account for discrepancies in the literature regarding the protein synthesis dependence of SEMA3A-induced growth cone collapse10,55,70.
Although the detailed mechanisms vary, most protein synthesis-inducing cues seem to activate the translational machinery through the mTOR pathway10,56,58 and the direction of turning responses by growth cones is determined by the specific mRNAs that are translated. For example, stimulating growth cones with a netrin 1 or BDNF gradient results in rapid trafficking of β-actin mRNA to the side of the growth cone nearest the gradient, resulting in asymmetric β-actin synthesis that is rapamycin-sensitive53,54. Zipcode binding protein 1 (ZBP1; also known as VG1RBP), an RNA-binding protein (RBP), transports β-actin mRNA in a translationally silent state by binding to a cis-acting element zipcode in its 3′-UTR53,54,63 and repressing its translation71. Attractive growth cone turning towards netrin 1 or BDNF can be prevented by inhibiting β-actin synthesis using antisense oligonucleotides that block translation53 and by disrupting the β-actin mRNA–ZBP1 interaction with antisense zipcode oligonucleotides54 or sense zipcode oligonucleotides that competitively inhibit endogenous β-actin mRNA13. Attractive growth cone turning can also be prevented by genetically deleting ZBP1 (REF. 72); these all suggest that cue-induced β-actin synthesis is required for this process in culture. By contrast, the repulsive cues SLIT2 and SEMA3A do not elicit β-actin synthesis, but induce rapid axonal synthesis of proteins that promote actin disassembly such as cofilin56 and RHOA55. SEMA3A-induced growth cone collapse can be blocked by inhibiting axonal RHOA mRNA translation55.
These findings led to the development of the differential translation model (FIG. 1), whereby the direction of growth cone turning is determined by the localized synthesis of proteins that promote either assembly (mediating attraction) or disassembly (mediating repulsion) of the cytoskeleton73. This agrees with studies in fibroblasts that show a requirement for the localization of β-actin mRNA to the leading lamellipodia for cell polarization and motility74,75. Given that pre-existing β-actin monomers are abundant in growth cones and that local mRNA translation in fibroblast lamellipodia gives no clear temporal advantage over transporting pre-existing β-actin monomers, the primary role of de novo β-actin synthesis probably lies in the spatial bias it provides to the polarization of cytoskeletal dynamics. For example, newly synthesized β-actin that lacks post-translational modifications (such as arginylation76 and glutathionylation77) could provide a more efficient nucleation signal than pre-existing monomers and thus seed actin polymerization on the nearside of the growth cone in an attractive gradient, thus driving growth in this direction. Conversely, polarized disassembly of the cytoskeleton on the nearside of a repulsive gradient would steer axons away from the gradient; however, this idea has not yet been tested. Nevertheless, it is noteworthy that cytoskeletal-associated molecules are highly represented in the axonal transcriptome15–18 (TABLE 2).
Table 2.
Selected functional categories of axonally enriched mRNAs
| Neuronal type | Subcellular compartment |
Protein synthesis |
Mitochondrial | Cytoskeletal | Cell–cell signalling |
Ref. |
|---|---|---|---|---|---|---|
| Rat | ||||||
| Embryonic SCG | Axons | Yes | Yes | Yes | No | 64 |
| Embryonic DRG | Axons | Yes | Yes | Yes | No | 65 |
| Adult DRG | Young growth cones |
Yes | Yes | Yes | Yes | 65 |
| Neonatal cerebral cortical |
Axons (cultured to maturity) |
Yes | Yes | Yes | No | 66 |
| Xenopus laevis | ||||||
| Embryonic RGC | Young growth cones |
Yes | Yes | Yes | No | 67 |
| Embryonic RGC | Old growth cones |
Yes | Yes | Yes | Yes | 67 |
| Mouse | ||||||
| Embryonic RGC | Growth cones | Yes | Yes | Yes | No | 67 |
DRG, dorsal root ganglion; RGC, retinal ganglion cell; SCG, superior cervical ganglion.
It is not yet clear what specific aspects of axonal pathfinding in vivo correspond to these in vitro findings. Although there is no reported axon guidance defect in mice with only one functional Zbp1 allele72, the peripheral sensory neurons of these mice, as well as those of mice overexpressing zipcode competitive inhibitors, exhibit reduced axon branching in culture13. Similarly, blocking the ubiquitin–proteasome system in vivo results in cell intrinsic defects in retinal axon branching but does not affect long-range pathfinding78. However, chemotropic responses of cultured growth cones to netrin 1 are blocked either by a proteasome or translation inhibitor alone10. Therefore, growth cone chemotropic responses in vitro may reflect short-range, cue-induced events, such as axon branching. It remains to be seen which particular processes are regulated by axonal mRNA translation in vivo.
Growth cone adaptation and gradient sensing
Growth cones must sense chemical gradients in order to properly guide their axons79. Growth cone adaptation, which is crucial for gradient sensing, involves successive cycles of desensitization and resensitization of the growth cone to an extracellular signal, and the resensitization step requires axonal protein synthesis50,80. Cultured X. laevis embryonic spinal neuronal growth cones lose their ability to respond to netrin 1 or BDNF after preincubation with the cue but regain responsiveness after 60–90 minutes in a manner that is sensitive to a translational inhibitor50. This enables these growth cones to respond to a netrin 1 or BDNF gradient many times in a lengthy period of exposure, which results in a zigzag pattern of growth50. This homeostatic reset mechanism is likely to be efficient for rapidly elongating axons that need to take only intermittent bearings to steer towards a distant cue.
Adaptation can also occur on a shorter timescale80. Cultured X. laevis retinal axons briefly preincubated with a low dose of repulsive sema3a or netrin 1 (netrin 1 can be either attractive or repulsive depending on the context81,82) lose their ability to respond to a collapse-inducing dose of the same cue80. This rapid (1–2 minutes) desensitization is cue-specific and mediated by ligand-induced receptor endocytosis80. Desensitized growth cones regain responsiveness in 4 minutes in a translation-dependent manner, but a higher dose of netrin 1 or sema3a is now required to induce the same degree of growth cone collapse80. This rapid recalibration mechanism allows advancing growth cones to readjust their sensitivity quickly and to compare minute differences in a concentration gradient. Interestingly, both receptor endocytosis and translation are spatially polarized to the gradient nearside of growth cones83, which suggests that the two processes are intimately linked.
Changes at intermediate targets
Growth cones encounter different guidance cues along their pathway and change cue responsiveness while they travel. For example, X. laevis retinal axons are attracted towards netrin 1 expressed in the optic nerve head, but netrin 1 becomes repellent upon exit from the eye81,82. Such switches might be regulated by local translation in distal axons.
In chick commissural axons transfected with a fluorescent reporter protein mRNA — the translation of which is regulated by the 3′-UTR of the ephrin A receptor epha2 mRNA (BOX 1) — increased reporter expression was observed in distal axons that had passed the midline of the spinal cord41. Although it is unknown whether endogenous epha2 mRNA is localized and regulated in this manner, this suggests the possibility that intermediate targets regulate the future responsiveness of axons by stimulating local synthesis of new guidance cue receptors. The expression of robo3, a receptor for slits that repels axons at the midline, is also regulated at the translational level84. It will be interesting to determine whether mRNAs encoding this and other receptors are axonally translated and regulate axon guidance in vivo.
Box 1.
Methods used to understand axonal mRNA translation and transport
Axonal mRNA isolation and transcriptome analysis
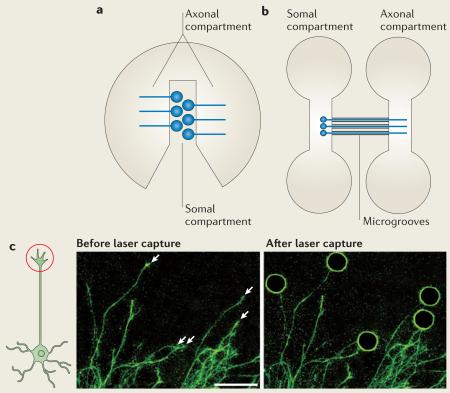
The accurate description of subcellular compartment-specific mRNA repertoires16–18 provides invaluable information regarding axonal mRNA transport and translation. The axons of cultured neurons are separated from their cell bodies using compartmentalized culture systems15,17,18 or laser-capture microdissection16. The Campenot chamber (see the figure, part a)185,189, made of a Teflon divider attached to a petri dish, has two compartments with distinct fluid environments. The proximal compartment contains cell bodies, dendrites and proximal axons, whereas the distal compartment contains distal axons. Typically, the distal compartment is supplemented with nerve growth factor (NGF), which promotes the growth of peripheral sensory61 and sympathetic neurons15. The microfluidic culture platform (see the figure, part b)18,183 has two mirror-imaged compartments. Dissociated neurons are added to the somal compartment, and axons grow into the axonal compartment through microgrooves. In laser-capture microdissection (see the figure, part c)16, cultured neurons labelled with a fluorescent lipophilic dye are fixed, and then axons or growth cones (indicated by white arrows in the figure) are microdissected individually. Because the amount of RNA obtained is minute, amplification is required before microarray analysis. This method, however, enables subcellular compartment-specific comparison, which is not possible in compartmentalized culture systems.
Quantitative immunofluorescence
The relative amount of a specific protein in cultured axons can be measured using quantitative immunofluorescence. Cultured axons without somas are treated with a protein synthesis-inducing cue with or without translational inhibitors and then fixed. The protein of interest is visualized by immunofluorescence. Background-subtracted average fluorescent intensity per unit area in axons or growth cones represents the relative protein amount10. An increase in protein quantity in axons without somas that is blocked by a translational inhibitor probably results from axonal mRNA translation.
Live imaging of translational reporter fluorescence
For mRNAs whose cis-acting translational regulatory element is known, translation can be visualized in real-time using a fluorescent reporter whose translation is regulated by the same cis-acting element. Diffusion-limited and short-lived fluorescent proteins are common translational reporters and their appearance indicates local mRNA translation61,186. Another class of reporter molecules is photoconvertible fluorescent proteins: the appearance of new fluorescent proteins after irreversible photoconversion of pre-existing proteins indicates local mRNA translation53,190.
Visualization of mRNAs and RNA-binding proteins
An mRNA can be directly visualized by delivery of in vitro-transcribed mRNAs that have incorporated fluorescent nucleotides191 into neurons by electroporation or microinjection. Alternatively, mRNAs can be visualized indirectly using the sequence-specific interaction of MS2 bacteriophage RNA hairpin and capsid protein (known as MS2 tagging)191. An mRNA fused to the MS2 binding site is co-expressed with a fluorescent MS2 protein, and the mRNA is visualized by MS2 fluorescence in axons192. MS2 tagging enables endogenous mRNAs to be labelled by generating MS2 binding site-containing knock-in mice193. RNA-binding proteins fused to a fluorescent protein can also be visualized53. Extracellular cue-induced changes in localization of mRNAs and RNA-binding proteins can be visualized in real-time or after fixation, and are commonly quantified by examining the behaviours of individual fluorescent particles or the distribution of total fluorescent particles53.
Unbiased screening of de novo proteome or translatome
De novo proteins in axons that do not have somas or glia in culture can be metabolically labelled (for example, using radioactive methionine), separated by two-dimensional electrophoresis, and identified by mass spectrometry. A challenge is to obtain sufficient amounts of axonally synthesized proteins, which are estimated to account for less than 5% of total protein synthesis in cultured sympathetic neurons194. Recently developed click chemistry enables extremely sensitive labelling of newly synthesized proteins195 and was successfully applied to visualize axonally synthesized proteins118. Alternatively, the translatome can be described196. Axonally translating mRNAs can be isolated by immunoprecipitating ribosomes and associated mRNAs specifically from axons118. For example, a tagged ribosomal protein can be specifically expressed in the eye, and ribosome–mRNA complexes can be isolated from the brain, in which the only source of a tagged ribosome is the retinal axons118.
Figure part a is modified, with permission, from REF. 185 © (2009) Macmillan Publishers Ltd. All rights reserved. Part b is modified, with permission, from REF. 183 © (2005) Macmillan Publishers Ltd. All rights reserved. Part c is reproduced, with permission from REF. 16 © (2010) Society for Neuroscience. Scale bar represents 150 μm.
Axon elongation
The short-term basal growth of axons does not require axonal protein synthesis36, and severed axons grow normally for 2–5 hours in the presence of protein synthesis inhibitors in culture10,53,85. Cue-induced axon growth promotion does, however, require axonal protein synthesis52,86. NGF and netrin 1, when applied acutely to cultured mammalian sensory and commissural neurons, respectively, promote axon outgrowth. Axonal synthesis of proteinase-activated receptor 3 (PAR3), a component of a complex that controls cytoskeletal dynamics, is required for NGF-induced and netrin 1-induced axon outgrowth52. β-thymosin, which prevents actin polymerization, is locally synthesized in cultured mollusc neurites treated with brain lysate, and inhibiting the translation of β-thymosin mRNA in isolated neurites promotes their elongation86. These studies suggest that extrinsic signals might shape the pattern of axonal outgrowth by regulating local synthesis of positive and negative regulators of cytoskeletal dynamics.
Synapse formation
Upon reaching its target, the growth cone transforms into the presynaptic terminal, which is highly branched and enriched with synaptic vesicles. Comparative subcellular mRNA profiling analysis has revealed that mRNAs encoding proteins implicated in this process (branch-promoting proteins and synaptic vesicle proteins) become specifically enriched in the growth cone as X. laevis retinal ganglion cells mature in culture16. Thus, these mRNAs are trafficked to the growth cone to coincide with target arrival and might be translated in response to target-derived cues. Indeed, presynaptic protein synthesis is required for BDNF-induced and NT3-induced potentiation of synaptic vesicle release in cultured X. laevis lower motor neurons62,87. Presynaptic protein release is also required for an increase in synaptic vesicle number with synaptic maturation in cultured mouse hippocampal neurons88.
This finding is in accordance with studies using mollusc neurons, which show that presynaptic protein synthesis89 is required for synapse formation47,90–92. It is also in agreement with a study of the Drosophila melanogaster neuromuscular junction, which shows that 3-phosphoinositide-dependent protein kinase 1 (PDK1) and ribosomal protein S6 kinase (S6K) — which are both components of the mTOR pathway — control synaptic bouton size, active zone number and synaptic function, although translation-dependency has not been tested in this case93. Furthermore, fragile X mental retardation protein (FMRP), a known translational regulator and mediator of synaptic plasticity in dendritic spines94,95, is found in axons and growth cones96–98, and its loss leads to a cell autonomous defect in the formation of presynaptic terminals in organotypic mouse hippocampal slices99. Other translational regulators such as survival of motor neuron (SMN1)100,101 and Hu-antigen D (HUD; also known as ELAVL4)102,103 also localize to axons and growth cones, thus indicating a probable broad presynaptic role of translation in aspects of synapse formation.
Transmitter biogenesis
Nerve terminals that secrete neurotransmitters remotely from their cell bodies might benefit from local mRNA translation. Indeed, a metabolic labelling study carried out in the 1960s using rat brains showed that newly (15–90 minute) synthesized proteins localize to nerve endings104. There is evidence suggesting that enzymes that regulate neurotransmitter metabolism, such as tyrosine hydroxlase (a key regulator of catecholamine biogenesis) and acetylcholinesterase (AChE; an enzyme that degrades acetylcholine), might be synthesized in nerve terminals.
A drug-induced increase in tyrosine hydroxlase activity105 and the reappearance of AChE activity after irreversible pharmacological inactivation106 occur in nerve terminals at speeds that cannot be explained by axonal transport from the cell bodies. Tyrosine hydroxlase mRNAs can be detected in the striatum by reverse transcription-polymerase chain reaction and in situ hybridization and is decreased by pharmacological lesion to the nigrostriatal pathway107.
Neuropeptides and hormones might also be synthesized in nerve terminals. Oxytocin and vasopressin mRNAs can be detected in the neurohypophysis by reverse transcription-polymerase chain reaction, in situ hybridization and northern blot108–112, and several invertebrate hormones can be locally synthesized in isolated axons47,90–92,113. However, whether these proteins are synthesized in the nerve endings in vivo and whether axonally synthesized proteins can be secreted is unknown.
Cell survival and axon maintenance
The promotion of axon survival by target-derived trophic factors can also be considered an adaptive response to extrinsic cues, and appears to involve axonal protein synthesis. For example, NGF supports the survival of cultured peripheral sensory and sympathetic neurons in a translational inhibitor-sensitive manner15,61,114. In cultured sympathetic neurons, inositol monophosphatase 1 (IMPA1) is axonally synthesized in response to NGF, and initiates an unknown signalling cascade that relays a survival signal to the nucleus15 (FIG. 2a). In cultured peripheral sensory neurons, the transcription factor cyclic AMP responsive element-binding protein (CREB) is axonally synthesized in response to NGF and retrogradely transported to the nucleus where it initiates transcription-dependent cell survival mechanisms61 (FIG. 2a). Interestingly, axonally synthesized CREB is specifically required for the NGF-induced survival signal: CREB synthesized in the cell body is unable to mediate NGF-induced cell survival61. This effect may be cell type-specific, because CREB mRNAs are found exclusively in peripheral sensory neurons17,61.
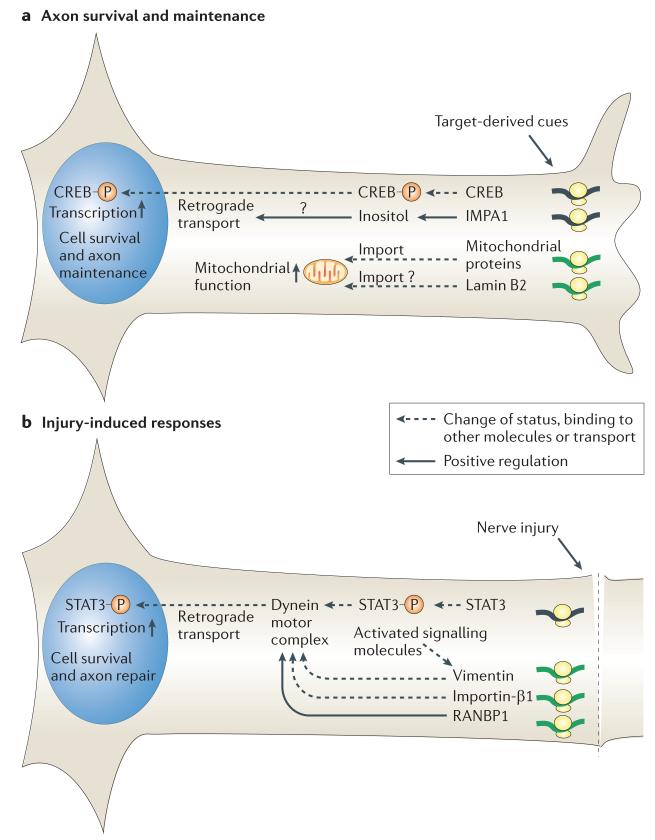
Figure 2. Axon survival, maintenance and injury-induced responses regulated by local protein synthesis.
a | Distal axons receive target-derived trophic factors. These target-derived cues activate local synthesis of mitochondrial, signalling or nuclear proteins required for axon maintenance and cell survival. Stimulation of axons with nerve growth factor (NGF) results in local synthesis of cyclic AMP responsive element-binding protein (CREB), which is then locally phosphorylated61. This active form of axonal CREB is transported into the nucleus and is required for axon survival in cultured sensory neurons61. NGF also stimulates the axonal synthesis of inositol monophosphatase 1 (IMPA1), an enzyme that regulates the inositol cycle. Axonally synthesized IMPA1 may regulate retrogradely transporting vesicles and is required for axon survival in cultured sympathetic neurons15. Distal axons also synthesize nuclear-encoded mitochondrial proteins, and sustained local synthesis and mitochondrial import of such proteins are required for axon maintenance in cultured neurons117. Lamin B2, a known nuclear envelope component, is also axonally synthesized and localized to mitochondria in distal axons, and sustained axonal synthesis of lamin B2 is required for axon maintenance in vitro and in vivo118. b | Nerve injury generates a retrograde survival and/or repair signal, and local protein synthesis is required for its generation and relay to the nucleus. The retrograde dynein motor complex is inactivated by Ran GTPase in normal conditions. Nerve injury stimulates local synthesis of Ran-specific GTPase-activating protein (RANBP1), which displaces Ran from the dynein motor66. Importin-β1 is also locally synthesized in response to nerve injury and binds to the Ran-free dynein motor complex, which results in the activated dynein motor complex64. Transcription factors, such as signal transducer and activator of transcription 3 (STAT3), are also locally synthesized and activated in injured axons and bind to the activated dynein motor complex using the nuclear localization signal14. A type III intermediate filament vimentin is also locally synthesized in injured axons and co-transported with other signalling molecules that bind to it65.
Axonally synthesized proteins can also support axon survival locally by promoting mitochondrial function. Many axons contain nuclear-transcribed mRNAs encoding mitochondrial proteins115–117 or proteins involved in mitochondrial function15–18 (TABLE 2). Axon-specific inhibition of mRNA translation or mitochondrial protein import decreases mitochondrial membrane potential in cultured sympathetic neurons117, thus suggesting that nuclear-encoded mitochondrial proteins are locally synthesized and imported to axonal mitochondria to support their function. New evidence shows that this mechanism may operate in vivo118. In cultured X. laevis retinal axons, target-derived cues such as en1 stimulate the local synthesis of lamin B2, a component of the nuclear lamina, which, surprisingly, then localizes to axonal mitochondria118 (FIG. 2a). Sustained axonal translation of lamin B2 mRNA is required for mitochondrial function in vitro and axon maintenance in vivo118. Interestingly, en1 also induces the synthesis of nuclear-encoded mitochondrial proteins in midbrain dopaminergic neurons in mice and supports their survival57. Therefore, stimulating the local synthesis of proteins involved in mitochondrial function might be a common mechanism by which target-derived cues support axon maintenance. Intriguingly, mutations in either mitochondrial proteins119 or lamins120 can cause Charcot–Marie–Tooth disease type 2, a neuropathic disorder that is characterized by chronic axonal degeneration. Together, these suggest links among mitochondria, lamins and axon maintenance.
Response to nerve injury and axon regeneration
As local mRNA translation mediates adaptive responses to extracellular signals, it is not surprising that mRNA translation can occur even in mature axons, especially during plastic responses such as regeneration. Generally, the ability of axons to synthesize proteins decreases as they mature, which has been associated with a coincident reduction in their ability to re-grow after axotomy121,122. Indeed, axons preconditioned with nerve injury in vivo show increased local protein synthesis11,38,39,43,64 and regeneration11 in vitro, and further increasing mTOR activity at the injury site (by locally applying phosphatase and tensin homologue (PTEN) short interfering RNA (siRNA) or the PTEN inhibitor bisperoxovanadium) promotes regeneration in vivo123. Intriguingly, there is evidence suggesting that axonal protein synthesis might be augmented after nerve injury by an unconventional mechanism of transceullar ribosomal delivery from glial cells124,125. Critically, blocking mRNA translation in severed axons inhibits the regeneration of new growth cones in culture11, and mice with reduced activity of ZBP1 exhibit attenuated axon regeneration after nerve injury in vivo13. These findings suggest that the ability of axons to locally synthesize proteins is related to their ability to regenerate. Indeed, olfactory sensory neurons, which naturally regenerate their axons, contain multiple axonal mRNAs126,127, some of which associate with axonal polysomes in vivo128.
In peripheral sensory neurons, nerve injury generates a cascade of retrograde signalling events that ultimately activates transcription of genes required for axon survival and regeneration129 (FIG. 2b). Local synthesis of transcription factors, such as signal transducer and activator of transcription 3 (STAT3), at the injury site is required for generating this signal14. Newly synthesized STAT3 is locally phosphorylated and retrogradely transported in an active state14. This is similar to CREB, which is locally synthesized and activated by NGF58. Intriguingly, the retrograde transport itself is also regulated by local mRNA translation. Ran GTPase, a regulator of nuclear transport, localizes to axons in a GTP-bound state in normal conditions, preventing cargo proteins from binding to the importin-α–dynein motor complex that is involved in retrograde transport66. Nerve injury induces local synthesis of Ran-specific GTPase-activating protein (RANBP1)66 and importin-β1 (REF. 64), which displaces Ran GTPase and directly binds to importin-α, respectively. The importin-α–importin-β1–dynein motor complex binds to transcription factors bearing nuclear localization signals, thus enabling their retrograde transport. Vimentin, which is also locally synthesized in response to injury, also directly binds to importin-β1 and other signalling molecules such as extracellular signal-regulated kinase 1 and 2 (ERK1/2), thereby facilitating the relay of complex information to the cell body65.
Surprisingly, local application of membrane-permeable nuclear localization signal peptides — which prevent axonally synthesized transcription factors from binding to importin-β1 — at the time of nerve injury abolishes the positive effect of preconditioning lesion to axon regeneration in vitro64 and neuronal survival in vivo14. Local translation of transcription factors might be an evolutionarily conserved mechanism to protect axons, because axonal translation of CAAT enhancer binding protein 1 (cebp-1) mRNA is required for axon regeneration after axotomy in Caenorhabditis elegans130.
In the CNS, axons lose their ability to regenerate in adulthood. Intriguingly, increasing protein synthesis might restore their regenerative potential131. In mice, global translational activity (as measured by the phosphorylation of the ribosomal protein S6) in retinal ganglion cells decreases with age132,133. In contrast to peripheral axons11, injury to postnatal retinal axons downregulates global translational activity in retinal ganglion cells in vivo131. Intriguingly, genetic deletion of the negative regulators of mTOR, PTEN and tuberous sclerosis protein 1 (TSC1), prevents this decrease in translational activity131. Remarkably, these rejuvenated neurons can regenerate axons, although whether axonal or somal protein synthesis plays a more important role in this effect is unknown131. Thus, different translational responses to nerve injury might account for the different abilities of CNS and PNS neurons to regenerate axons.
Receptor expression and pain regulation
Some evidence suggests that local mRNA translation may regulate pain sensitivity. mRNAs encoding transient receptor potential cation channel subfamily V member 1 (TRPV1), a capsaicin-gated and high temperature-gated ion channel, are detected in the axons of peripheral nociceptors in adult mice134. Interestingly, inflammatory insults increase the axonal transport of Trpv1 mRNA to central endings in the spinal cord, and intrathecal delivery of Trpv1 antisense oligodeoxynucleotides reduces the inflammation-induced TRPV1 hypersensitivity in spinal cord slices134. Nerve injury also induces specific accumulation of tetrodotoxin-resistant voltage-gated sodium channel 1.8 (Nav1.8) mRNA in injured axons in vivo135. Subcutaneous injection of Nav1.8 siRNA into areas in which peripheral endings terminate specifically reduces Nav1.8 mRNA in the axon but not in the cell body, and reduction of axonal Nav1.8 mRNA correlates with the reversal of neuropathic pain in mice135. Furthermore, delivery of inhibitors of protein synthesis either intrathecally (rapamycin136 and cycloheximide134) or intraplantarly (rapamycin137 and anisomycin138) alleviates pathological pain in mice, although these effects may indirectly result from the decreased production and release of pro-inflammatory cytokines from neighbouring cells. The axons of cultured peripheral sensory neurons also contain mRNAs encoding KOR1, a GPCR that generates an antinociceptive signal, and the axonal localization of Kor1 mRNA is enhanced by depolarization in culture42,139. Direct evidence showing that these receptors are trafficked to the plasma membrane is still lacking, although these axons have functional machinery to target proteins to the cell surface44 and mollusc axons can target locally synthesized conopressin receptor to the plasma membrane46.
Neurodevelopmental and neurodegenerative disorders
Numerous neurodevelopmental disorders, such as fragile X mental retardation and autism spectrum disorders, seem to share dysregulated protein synthesis as an underlying cause of clinical pathologies140. Although dendrites are well-documented sites of synaptic pathology140, there is less evidence for axons. Recent evidence, however, shows that translational regulators such as FMRP, also localize to axons and regulate presynaptic function99. Therefore, dysregulated axonal mRNA translation may, in future, be found to contribute to clinical pathologies of neurodevelopmental disorders141.
Recent evidence links axonal mRNA translation to neurodegenerative diseases, such as spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS). SMA is characterized by lower motor neuron degeneration and muscular atrophy and is caused by deletions in SMN1 (REF. 142). SMN1 associates with small nuclear ribonucleoproteins (snRNPs) in the nucleus to regulate pre-mRNA splicing143 but also localizes to axons and growth cones144,145. In the cytoplasm, SMN1 regulates mRNA translation by forming RNA granules with FMRP146, thereby suggesting that defective axonal mRNA translation may contribute to SMA. Indeed, SMN1 positively regulates the translation of candidate plasticity-related gene 15 (cpg15; also known as NRN1) mRNA in axons by interacting with HUD, and, remarkably, overexpressing cpg15 partially rescues SMN1 loss-of-function phenotype in zebrafish12. Therefore, the regulation of axonal mRNA translation by SMN1 may be required for the maintenance of these axons. ALS is the most common motor neuron disease in adults147 and is associated with mutations in genes encoding RBPs such as TAR DNA-binding protein 43 (TDP43), RNA-binding protein FUS (FUS), and angiogenin148,149. These mutations commonly increase the numbers of stress granules, which reversibly sequester mRNAs and repress their translation150. Because sustained translation of axonal mRNAs is required for axon maintenance12,117,118, decreased axonal mRNA translation caused by these mutations might contribute to axon degeneration in ALS.
Regulation of axonal mRNA translation
Global translational activity
The rate of protein synthesis in cells is tightly coupled to their nutritional status: cells increase protein synthesis when they receive growth-promoting signals. This cellular homeostasis is regulated by mTOR, which activates translational initiation by phosphorylating its two major substrates 4E-BP1 and S6K132,133. Accumulating evidence shows that local protein synthesis in axons and growth cones is also regulated through mTOR. Protein synthesis-inducing cues activate mTOR in a concentration-dependent manner, which results in the asymmetric activation of the protein synthesis machinery within the growth cone10,53,56,151.
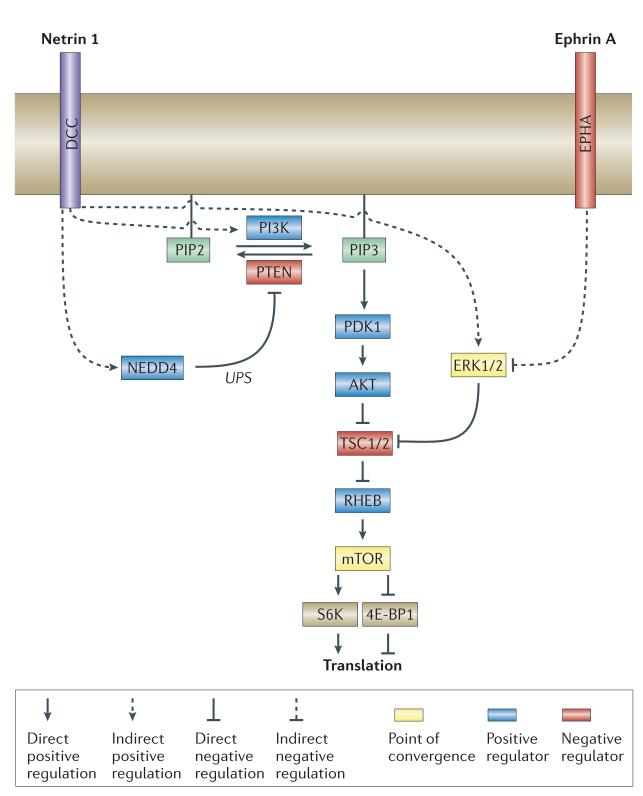
mTOR activity is positively regulated by a GTP-bound form of RHEB GTPase, which is inactivated by its GTPase-activating proteins TSC1 and TSC2 (REFS 132,133). TSC1 and TSC2 are negatively regulated by AKT and ERK1/2 (REFS 132,133). Protein synthesis-inducing cues can activate translation through the PI3K–AKT–mTOR pathway by increasing PI3K or decreasing PTEN activities. For example, netrin 1 activates PI3K and targets PTEN for ubiquitin-mediated proteolysis by activating the E3 ubiquitin-protein ligase NEDD4 (REF. 78), but also through the mitogen-activated protein kinase (MAPK)–TSC2–RHEB–mTOR pathway by activating MAPK ERK1/2 (in cases of netrin 1 (REF. 151), BDNF50 and SEMA3A151). By contrast, ephrin A reduces the activity of ERK1/2, which results in a decreased inhibition of TSC2 and a decreased mTOR activity152. This raises the interesting possibility that translational activity in growth cones could be regulated through spatial and temporal integration of diverse receptor-mediated signalling pathways converging on mTOR (FIG. 3). Cell contact-dependent changes in the cytoskeletal network may also regulate translational activity, as the cytoskeleton directly associates with the protein synthetic machinery and regulates mTOR activity153.
Figure 3. Regulation of global translational activity through mTOR.
Cues that induce and inhibit protein synthesis antagonistically regulate the activity of mammalian target of rapamycin (mTOR), which regulates cap-dependent mRNA translation by phosphorylating its two major targets: eukaryotic translation initiation factor 4E (eIF4E)-binding protein 1 (4E-BP1) and ribosomal protein S6 kinase (S6K). Protein synthesis-inducing cues, such as netrin 1 or brain-derived neurotrophic factor (BDNF), may increase mTOR activity through the AKT–mTOR pathway by activating phosphoinositide 3-kinase (PI3K)10 or by promoting ubiquitin–proteasome system (UPS)-mediated degradation of phosphatase and tensin homologue (PTEN)78, or through the mitogen-activated protein kinase (MAPK)–mTOR pathway by activating MAPK extracellular signal-regulated kinase 1 and 2 (ERK1/2)50,151 that inhibits the mTOR negative regulators tuberous sclerosis protein 1 (TSC1) and TSC2. Some cues, such as ephrin A, can inhibit protein synthesis by inhibiting ERK1/2 leading to TSC1/2 activation and mTOR inhibition152. Spatiotemporal summation of cue-induced signals converging on mTOR might lead to asymmetric translational activity. DCC, deleted in colorectal carcinoma (netrin 1 receptor); PDK1, 3-phosphoinositide-dependent protein kinase 1; PIP2, phosphatidylinositol-4,5-bisphosphate (also known as PtdIns(4,5)P2); PIP3, phosphatidylinositol-3,4,5-triphosphate (also known as PtdIns(3,4,5)P3).
In addition to regulating mTOR, guidance cues may directly regulate ribosomes26. DCC (deleted in colorectal carcinoma), a netrin 1 receptor, directly binds to ribosomal protein L5, a component of the 60S ribosomal subunit30. Binding of netrin 1 to DCC activates translational initiation and subsequently releases the ribosome–mRNA complex from DCC, thereby allowing more ribosomes to form polysomes in the vicinity of receptor activation. This provides a crucial mechanism for the localized control of mRNA translation near the site of signal activation and may also prevent unnecessary translation in the basal state by sequestering ribosomes. It will be of interest to determine whether other receptors show similar interactions with the translational machinery.
mRNA transport and axonal mRNA repertoire
Axonal transcriptome analyses from diverse neurons — including microarrays studies in X. laevis, mouse embryonic retinal ganglion cells16, rat embryonic and perinatal cortical and hippocampal neurons aged to maturity in culture18 and rat embryonic and adult peripheral sensory neurons17, and a sequential analysis of gene expression study in rat perinatal sympathetic neurons15 — identified thousands of different mRNAs in their axons. Notably, these axons contain common mRNAs, mainly those encoding protein synthesis machinery, mitochondrial proteins and components of the cytoskeleton (TABLE 2). However, some mRNAs were enriched in axons of specific cell types (for example, Impa1 mRNA is found in peripheral15,17 but not central16,18 neuronal axons, and Creb mRNAs are found exclusively in peripheral sensory neuronal axons17,61). Comparative bioinformatics analyses revealed that specific anterograde transport is likely to account for mRNA enrichment in axons15 and growth cones16. Axonal transcriptomes change dynamically during development, even in the same axons16, which suggests that axons may recruit specific and biologically relevant mRNAs using active transport.
The cis-acting localization elements that target mRNAs to axons have been identified only for a few mRNAs, such as β-actin mRNA154,155. Recent evidence shows that alternative transcription termination produces mRNAs that are targeted to distinct neuronal subcellular compartments (FIG. 4). Notably, a longer 3′-UTR targets Ranbp1 mRNA to adult sciatic nerve66, and a distinct 3′-UTR element targets Impa1 mRNA into developing sympathetic axons in response to NGF stimulation15. Furthermore, cis-acting elements can be differentially regulated by extrinsic cues. In cultured adult peripheral sensory neurons, for example, NGF, BDNF, NT3, SEMA3A and myelin-associated glycoprotein (MAG) stimulation results in cue-specific mRNA axonal repertoires, thereby suggesting that extrinsic stimuli recruit specific mRNAs156. RBPs directly associate with target mRNAs to form transport ribonucleo-protein particles (RNPs), the components of which may directly associate with molecular motors to mediate bidirectional axonal transport on microtubules157. For example, the RBP La binds and promotes translation of mRNAs containing the 5′-terminal oligopyrimidine tract (TOP) sequence in their 5′-UTRs158,159. In axons, unmodified La interacts with the kinesin motors and mediates anterograde transport of RNPs, but La switches to interact with the retrograde dynein motor upon sumoylation158. Short-range RNP movements are likely to be mediated by myosin on actin filaments53,54,160 and can be regulated by extracellular signals53,54,156. RNA granules may also switch between microtubule and microfilament tracks141. mRNAs are translationally repressed during transport and remain silent in RNA granules until released for translation161. Co-translational transport mechanisms might regulate subcellular localization of mRNAs according to the properties of proteins they encode, as seen in budding yeasts, in which actively translating ABP140 mRNA is tethered to actin filaments162. Upon completion of translation, mRNAs can again associate with different RNA granules, such as transport RNPs, stress granules that sequester non-essential mRNAs upon cellular stress, and processing bodies (P bodies) where mRNAs are stored or degraded163.
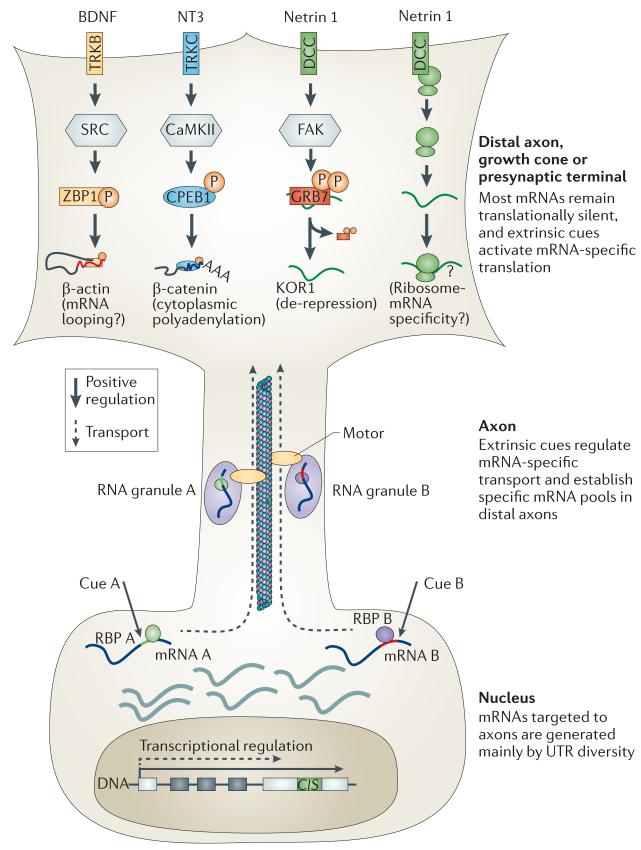
Figure 4. RNA-specific transport and translation.
Axonal targeting of mRNAs is directed by cis-acting elements that are mainly localized to the 3′-untranslated regions (UTRs) of mRNAs. Retention of these axon-targeting cis-acting elements is commonly regulated by the use of different transcriptional termination sites15,66. Extrinsic cues influence axonal mRNA repertoires by promoting transport of specific mRNAs156. Axonally targeted mRNAs are recruited to RNA granules (transport ribonuceloproteins (RNPs)) by specific RNA-binding proteins (RBPs) and are transported along microtubules probably by kinesin motors157. mRNAs remain translationally silent during transport21. Extracellular signals activate the translation of specific mRNAs mainly by regulating RBPs. For example, neurotrophins and guidance cues activate the kinases SRC71, calcium/calmodulin-dependent protein kinase II (CaMKII)174 and focal adhesion kinase (FAK)164, which phosphorylate the RBPs, zipcode binding protein 1 (ZBP1), cytoplasmic polyadenylation element binding protein (CPEB1), and growth factor receptor-bound protein 7 (GRB7), respectively. Cell surface receptors might regulate mRNA-specific translation by directly regulating ribosomes. For example, unstimulated netrin receptor DCC directly binds to ribosomes and inhibits translation26, and ribosome composition influences mRNA selectivity181. Different receptors may bind to ribosomes that are pre-tuned to specific mRNAs, and ligand stimulation might release such ribosomes and result in mRNA-specific translation. BDNF, brain-derived neurotrophic factor; KOR1, κ-type opioid receptor; NT3, neurotrophin 3; TRK, tyrosine kinase receptor.
mRNA-specific translation
Protein synthesis-inducing cues increase global translational activity but initiate translation of only a subset of mRNAs. For example, netrin 1 and bdnf, but not en1, induce axonal translation of β-actin mRNA in X. laevis retinal axons, whereas all three cues increase global translational activity53,54,118. Furthermore, cues that increase global protein synthesis can also repress translation of some mRNAs. For example, en1 increases overall axonal protein synthesis in cultured X. laevis retinal axons but decreases the translation of heat shock protein 70 (hsp70) mRNA118. How different extracellular signals activate the translation of functionally coherent subsets of mRNAs is largely unknown.
Most known mRNA cis-acting elements reside in 3′-UTRs, including those in β-actin, RHOA, EPHA2, cytochrome c oxidase subunit IV isoform 1 (COX4I1) and IMPA1 mRNAs15,23,41,116,154, but they can also be localized to 5′-UTRs (such as that of KOR1 mRNA)164, protein coding regions (such as that of ROBO3 mRNA)84 and potentially to introns (as found in some dendritically targeted RNAs)165. Intron-retaining mRNAs in axons (such as ROBO3 mRNA)166 may be prone to nonsense-mediated decay in P bodies, a mechanism that is also regulated by extracellular cues167.
Trans-acting molecules regulate mRNA transport, stability and translation by reversibly associating with specific mRNAs to form RNA granules168. Components of RNA granules, such as RBPs and microRNAs, localize to axons and growth cones, and can be regulated by extrinsic cues. For example, SRC-dependent phosphorylation of ZBP1 (REF. 71) is required for BDNF-induced β-actin synthesis and growth cone turning169. Conformational changes induced by ZBP1 binding170 or translational de-repression from ZBP1 (REF. 71) may promote the translation of β-actin mRNA.
FMRP is an RBP that recognizes target mRNAs based on their secondary structure171 and represses their translation. The translational repression by FMRP is relieved by its dephosphorylation by protein phosphatase 2A, which can be activated by neuronal activity172. FMRP localizes to axons and growth cones and regulates cue-responsive local mRNA translation in axons96,97.
Cytoplasmic polyadenylation element binding proteins (CPEB1, CPEB2, CPEB3 and CPEB4) are translational repressors that recognize a specific sequence element173. CPEB1 is required for NT3-induced axonal translation of β-catenin mRNA and axon branching in cultured rat hippocampal neurons174. Blocking CPEB function by overexpressing a dominant negative mutant impairs elongation of X. laevis retinal axons in vivo, but knocking down CPEB1 expression does not, suggesting tissue-specific function of CPEB isoforms175. These findings are consistent with a recent study using transgenic mice, which showed that CPEB1 inhibits the translation of Pten mRNA176.
MicroRNAs are another class of trans-acting elements that could regulate mRNA translation in a sequence-specific manner. Axons and growth cones contain functional RNA interference machinery55, and microRNAs repress the translation of their target mRNA in RNA granules until stimulated177. Recent evidence indicates that a single microRNA can regulate the translation of functionally related mRNAs, as miR-338 inhibits axonal translation of mRNAs encoding COX4I1 (REF. 178) and ATP5G1 (ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9))117, the components of mitochondrial complexes IV and V, respectively. These translational regulators are likely to interact, as FMRP associates with ribosomes179, microRNAs180, and other RBPs such as SMN1 (REF. 146).
Finally, receptor–ribosome interactions could potentially regulate mRNA-specific translation. Expression of individual ribosomal proteins shows tissue specificity181, which suggests that variable compositions of ribosomes may exist. Intriguingly, each composition may preferentially translate specific mRNAs, as mutant mice that are hypomorphic for the ribosomal protein L22 show specific defects in translation of related Hox mRNAs181. Considering that guidance cue receptors may regulate mRNA translation by sequestering ribosomes26, it is tantalizing to speculate that different extracellular signals might release different ribosomes that are pre-tuned to translate a specific subset of mRNAs. It is intriguing that axons and growth cones contain mRNAs encoding most ribosomal proteins15–18,156, which themselves are regulated by mTOR through their 5′-TOP sequences159, and there is evidence that at least some ribosomal proteins can be exchanged in the cytoplasm182. Thus, axons might be able to fine-tune mRNA selectivity by dynamically modifying ribosomal compositions using locally synthesized ribosomal proteins.
Summary and future directions
Local mRNA translation is widely used to maintain subcellular autonomy in axons and dendrites. Here, we have focused our attention on various stimulus-driven axonal responses, such as growth cone chemotropic responses and axon regeneration, but evidence also suggests that local mRNA translation might contribute to the maintenance of the steady state of the local proteome in the distal axons9. Axonal mRNA translation occurs during development and in adulthood, but the ability of axons to activate translational machinery is closely associated with their ability to regenerate. Axons store neuronal subtype-specific and age-specific repertoires of local mRNAs, and translation of a specific subset of mRNAs is likely to determine the nature of extrinsic cue-induced axonal responses (FIG. 5).
Figure 5. Local mRNA translation as a mediator of stimulus-induced axonal responses.
A proposed model for the function and mechanism of axonal mRNA translation. Neuronal axons contain a complex and dynamic transcriptome, and many mRNAs remain translationally silent. Various extrinsic cues stimulate translation of a distinct subset of mRNAs during development and in adulthood. For example, guidance cues induce local synthesis of cytoskeletal proteins in growing axons and regulate axon guidance and branching. Target-derived trophic factors promote local synthesis of proteins required for mitochondrial function and support the survival of distal axons. Nerve injury in adulthood stimulates local synthesis of nuclear factors that activate repair mechanisms.
Recent advances in compartmentalized cell culture techniques36,183–185 (BOX 1) have provided tools to study local mRNA translation in axons, and several candidate-based approaches have revealed crucial insights into the role of axonal mRNA translation. The unexpected complexity of axonal transcriptomes15–18 indicates that many biological pathways are post-transcriptionally regulated in axons. In order to understand the roles that local mRNA translation may have in vivo, it will be important to investigate which of these axonally localized mRNAs are translated. It is possible to isolate axonally translating mRNAs by affinity purification and axonally synthesized proteins by metabolic labelling (BOX 1), although the minute amounts of these mRNAs and proteins makes it technically challenging. Genome-wide correlative analyses of axonal transcriptomes and translatomes in the context of extrinsic cue stimulation would provide insights into both axonal mRNA translation and cue-mRNA specificity.
Another question to investigate is whether the complex axonal mRNA repertoire is a property of individual axons or a collective property of diverse axons. Transcriptome analyses have shown that axons of similar origins (such as dorsal root ganglia neurons, sympathetic ganglia neurons or retinal ganglion cells) contain thousands of different mRNAs, whereas immunocytological and ultrastructural evidence indicates that few polysomes are present in individual axons. Information regarding the number and distribution of mRNAs and protein synthesis machinery at the single axon level is needed to improve our basic understanding of how mRNA translation is regulated in axons and to gain insights into the puzzling paucity of polysomes and the apparent unconventional nature of the axonal ER and Golgi apparatus.
Although candidate studies have identified several cis-acting elements that regulate axonal mRNA localization, our knowledge about how specific mRNAs are selected for axonal transport is limited. Most screening studies have used microarray techniques to identify axonally localizing mRNAs, followed by rapid amplification of cDNA ends of individual candidates to explore whether UTR diversity mediates selective axonal mRNA transport. Recent advances in RNA sequencing technologies will provide new opportunities for unbiased genome-wide screens for novel regulatory cis-acting elements that are not limited to the UTRs.
Recent evidence using transgenic mice bearing a local translational reporter (BOX 1) demonstrated that axonal mRNA translation occurs in vivo186, and the studies using genetically modified mice with compromised functions of axonally localizing translational regulators12,13,72,99,174 revealed potential roles that axonal mRNA translation may play in vivo. However, direct evidence linking axonal mRNA translation to a specific function in vivo is still scarce, because it is technically challenging to inhibit mRNA translation specifically in axons in intact animals. One approach to address this point in candidate genes is to introduce targeted mutations to known axon-localization elements in the UTRs, which would result in normal protein expression but the specific loss of axonal mRNA localization and translation. This is analogous to an in vitro study, in which an axon-targeted isoform of Impa1 mRNA was selectively targeted by siRNA15. Another candidate approach involves delivering caged translation-blocking antisense oligonucleotides, which can be activated by local light stimulation in axons187, to specific neurons. These methods may provide ways to inhibit the translation of a specific mRNA in axons in live animals. Intriguingly, a novel method to inhibit translation of all mRNAs in presynaptic terminals in mouse brain slices was recently developed: an inactive chemically inducible, genetically encoded translational inhibitor was expressed in a specific subset of neurons and the activating chemical was locally applied to their axons62,188. Imaginative new approaches along these lines that facilitate axon-specific inhibition of mRNA translation in vivo are needed to fully uncover the role of local synthesis in nervous system assembly, maintenance and repair.
Acknowledgements
We apologize to the authors of papers we could not include in this Review owing to space limitations. We thank J. Hu for critical reading of the manuscript. This work was supported by a Wellcome Trust Programme Grant (085314/Z/08/Z) to C.E.H.
Glossary
- Polysomes
Strings of 80S ribosomes bound to mRNA molecules.
- Ribosomes
Large RNA–protein complexes (80S) at which mRNA translation occurs. They contain 4 rRNAs and more than 79 proteins and are composed of a large (60S) subunit and a small (40S) subunit.
- RNA-binding protein (RBP)
A protein that binds RNAs. Most RBPs have modular structures containing specific RNA-binding domains, catalytic domains and/or protein-binding domains.
- Small nuclear ribonucleoproteins (snRNPs)
Complexes that are composed of a small nuclear RNA and a specific set of proteins.
- RNA granules
Intermediate RNA–protein complexes that regulate RNA transport, translation and degradation. RNA granules include transport ribonucleoproteins, stress granules and processing bodies.
- Stress granules
Dense cytosolic proteins and RNA aggregations that appear under conditions of cellular stress. The RNA molecules are thought to be stalled translation pre-initiation complexes.
- MicroRNAs
Non-coding RNA molecules of 21–24 nucleotides in length that inhibit mRNA expression.
Footnotes
Competing interests statement The authors declare no competing financial interests.
FURTHER INFORMATION Christine E. Holt’s homepage: http://www.pdn.cam.ac.uk/staff/holt
References
- 1.Mili S, Moissoglu K, Macara IG. Genome-wide screen reveals APC-associated RNAs enriched in cell protrusions. Nature. 2008;453:115–119. doi: 10.1038/nature06888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lecuyer E, et al. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 3.Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 5.Wang DO, Martin KC, Zukin RS. Spatially restricting gene expression by local translation at synapses. Trends Neurosci. 2010;33:173–182. doi: 10.1016/j.tins.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giuditta A, Dettbarn WD, Brzin M. Protein synthesis in the isolated giant axon of the squid. Proc. Natl Acad. Sci. USA. 1968;59:1284–1287. doi: 10.1073/pnas.59.4.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koenig E. Synthetic mechanisms in the axon. IV. In vitro incorporation of [3H]precursors into axonal protein and RNA. J. Neurochem. 1967;14:437–446. doi: 10.1111/j.1471-4159.1967.tb09542.x. Together with reference 6, landmark studies that showed evidence for axonal protein synthesis. Using metabolic labelling, these studies showed that vertebrate and invertebrate axons without somas are capable of translation-dependent protein synthesis.
- 8.Edstrom A, Sjostrand J. Protein synthesis in the isolated Mauthner nerve fibre of goldfish. J. Neurochem. 1969;16:67–81. doi: 10.1111/j.1471-4159.1969.tb10344.x. [DOI] [PubMed] [Google Scholar]
- 9.Alvarez J, Giuditta A, Koenig E. Protein synthesis in axons and terminals: significance for maintenance, plasticity and regulation of phenotype. With a critique of slow transport theory. Prog. Neurobiol. 2000;62:1–62. doi: 10.1016/s0301-0082(99)00062-3. [DOI] [PubMed] [Google Scholar]
- 10.Campbell DS, Holt CE. Chemotropic responses of retinal growth cones mediated by rapid local protein synthesis and degradation. Neuron. 2001;32:1013–1026. doi: 10.1016/s0896-6273(01)00551-7. First demonstration of a functional role for axonal mRNA translation in mediating chemotropic responses of growth cones to guidance cue gradients. Netrin 1 and SEMA3A increase global translational activity in cultured growth cones by activating mTOR. This study also showed that proteasomal degradation and translation are intricately linked in cue-stimulated axonal responses.
- 11.Verma P, et al. Axonal protein synthesis and degradation are necessary for efficient growth cone regeneration. J. Neurosci. 2005;25:331–342. doi: 10.1523/JNEUROSCI.3073-04.2005. Key evidence showing that axonal protein synthesis is required for axon regeneration. Comparing regeneration of embryonic and adult, CNS and PNS neuronal axons in culture, with or without translation inhibitors, led to this conclusion.
- 12.Akten B, et al. Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits. Proc. Natl Acad. Sci. USA. 2011;108:10337–10342. doi: 10.1073/pnas.1104928108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donnelly CJ, et al. Limited availability of ZBP1 restricts axonal mRNA localization and nerve regeneration capacity. EMBO J. 2011;30:4665–4677. doi: 10.1038/emboj.2011.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ben-Yaakov K, et al. Axonal transcription factors signal retrogradely in lesioned peripheral nerve. EMBO J. 2012 Jan 13; doi: 10.1038/emboj.2011.494. (doi:10.1038/emboj.2011.494) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andreassi C, et al. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nature Neurosci. 2010;13:291–301. doi: 10.1038/nn.2486. Using sequential analysis of gene expression analysis, this study identified more axonal mRNAs (>11,000 sequence tags), among which IMPA1 mRNA was most abundant. A novel 3′-UTR element mediates axonal transport of IMPA1 mRNA, the axonal translation of which is required for NGF-mediated cell survival.
- 16.Zivraj KH, et al. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 2010;30:15464–15478. doi: 10.1523/JNEUROSCI.1800-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gumy LF, et al. Transcriptome analysis of embryonic and adult sensory axons reveals changes in mRNA repertoire localization. RNA. 2011;17:85–98. doi: 10.1261/rna.2386111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor AM, et al. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J. Neurosci. 2009;29:4697–4707. doi: 10.1523/JNEUROSCI.6130-08.2009. References 15–18 were key axonal transcriptome studies. Using compartmentalized culture systems and laser-capture microdissection, these studies provided comprehensive information on the complex and dynamic nature of axonal mRNA repertoires in embryonic and adult, growing and mature PNS and CNS neurons. Additionally, reference 16 showed that the growth cone of embryonic neurons has a translatome distinct from that of the axon shaft.
- 19.Lasek RJ, Dabrowski C, Nordlander R. Analysis of axoplasmic RNA from invertebrate giant axons. Nature New Biol. 1973;244:162–165. doi: 10.1038/newbio244162a0. [DOI] [PubMed] [Google Scholar]
- 20.Giuditta A, Cupello A, Lazzarini G. Ribosomal RNA in the axoplasm of the squid giant axon. J. Neurochem. 1980;34:1757–1760. doi: 10.1111/j.1471-4159.1980.tb11271.x. [DOI] [PubMed] [Google Scholar]
- 21.Giuditta A, Hunt T, Santella L. Messenger RNA in squid axoplasm. Neurochem. Int. 1986;8:435–442. doi: 10.1016/0197-0186(86)90019-7. [DOI] [PubMed] [Google Scholar]
- 22.Giuditta A, et al. Active polysomes in the axoplasm of the squid giant axon. J. Neurosci. Res. 1991;28:18–28. doi: 10.1002/jnr.490280103. [DOI] [PubMed] [Google Scholar]
- 23.Bassell GJ, et al. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. Together with reference 154, this showed evidence for an isoform-specific axonal transport of β-actin mRNAs that is regulated by extrinsic cues. Binding of ZBP1 to the zipcode in the β-actin 3′-UTR mediates this transport, which is enhanced by NT3 and necessary for growth cone motility.
- 24.Bunge MB. Fine structure of nerve fibers and growth cones of isolated sympathetic neurons in culture. J. Cell Biol. 1973;56:713–735. doi: 10.1083/jcb.56.3.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tennyson VM. The fine structure of the axon and growth cone of the dorsal root neuroblast of the rabbit embryo. J. Cell Biol. 1970;44:62–79. doi: 10.1083/jcb.44.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tcherkezian J, Brittis PA, Thomas F, Roux PP, Flanagan JG. Transmembrane receptor DCC associates with protein synthesis machinery and regulates translation. Cell. 2010;141:632–644. doi: 10.1016/j.cell.2010.04.008. First evidence that guidance cue receptors can directly regulate ribosome activity. DCC inhibits translation by sequestering ribosomes and netrin 1 binding releases ribosomes from DCC, thus providing a novel mechanism to localize mRNA translation in the vicinity of receptor activation.
- 27.Yamada KM, Spooner BS, Wessells NK. Ultrastructure and function of growth cones and axons of cultured nerve cells. J. Cell Biol. 1971;49:614–635. doi: 10.1083/jcb.49.3.614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steward O, Ribak CE. Polyribosomes associated with synaptic specializations on axon initial segments: localization of protein-synthetic machinery at inhibitory synapses. J. Neurosci. 1986;6:3079–3085. doi: 10.1523/JNEUROSCI.06-10-03079.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koenig E, Martin R. Cortical plaque-like structures identify ribosome-containing domains in the Mauthner cell axon. J. Neurosci. 1996;16:1400–1411. doi: 10.1523/JNEUROSCI.16-04-01400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koenig E, Martin R, Titmus M, Sotelo-Silveira JR. Cryptic peripheral ribosomal domains distributed intermittently along mammalian myelinated axons. J. Neurosci. 2000;20:8390–8400. doi: 10.1523/JNEUROSCI.20-22-08390.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kun A, Otero L, Sotelo-Silveira JR, Sotelo JR. Ribosomal distributions in axons of mammalian myelinated fibers. J. Neurosci. Res. 2007;85:2087–2098. doi: 10.1002/jnr.21340. [DOI] [PubMed] [Google Scholar]
- 32.Li YC, et al. Subsurface cisterna-lined axonal invaginations and double-walled vesicles at the axonal–myelin sheath interface. Neurosci. Res. 2005;53:298–303. doi: 10.1016/j.neures.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Zelena J. Ribosome-like particles in myelinated axons of the rat. Brain Res. 1970;24:359–363. doi: 10.1016/0006-8993(70)90120-4. [DOI] [PubMed] [Google Scholar]
- 34.Walker BA, et al. Reprogramming axonal behavior by axon-specific viral transduction. Gene Ther. 2012 Jan 26; doi: 10.1038/gt.2011.217. (doi:10.1038/gt.2011.217) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koenig E, Adams P. Local protein synthesizing activity in axonal fields regenerating in vitro. J. Neurochem. 1982;39:386–400. doi: 10.1111/j.1471-4159.1982.tb03960.x. [DOI] [PubMed] [Google Scholar]
- 36.Eng H, Lund K, Campenot RB. Synthesis of β-tubulin, actin, and other proteins in axons of sympathetic neurons in compartmented cultures. J. Neurosci. 1999;19:1–9. doi: 10.1523/JNEUROSCI.19-01-00001.1999. Using a highly efficient compartmentalized culture system, now known as the Campenot chamber, this study showed that local protein synthesis occurs and that it is not required for the basal rate of axon growth.
- 37.Koenig E. Evaluation of local synthesis of axonal proteins in the goldfish Mauthner cell axon and axons of dorsal and ventral roots of the rat in vitro. Mol. Cell Neurosci. 1991;2:384–394. doi: 10.1016/1044-7431(91)90025-j. [DOI] [PubMed] [Google Scholar]
- 38.Tobias GS, Koenig E. Influence of nerve cell body and neurolemma cell on local axonal protein synthesis following neurotomy. Exp. Neurol. 1975;49:235–245. doi: 10.1016/0014-4886(75)90207-1. [DOI] [PubMed] [Google Scholar]
- 39.Tobias GS, Koenig E. Axonal protein synthesizing activity during the early outgrowth period following neurotomy. Exp. Neurol. 1975;49:221–234. doi: 10.1016/0014-4886(75)90206-x. [DOI] [PubMed] [Google Scholar]
- 40.Van Minnen J, et al. De novo protein synthesis in isolated axons of identified neurons. Neuroscience. 1997;80:1–7. doi: 10.1016/s0306-4522(97)00137-1. [DOI] [PubMed] [Google Scholar]
- 41.Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. First evidence that extrinsic cues may regulate translation of guidance cue receptor mRNAs in growing axons. The authors suggested an intriguing mechanism by which intermediate targets regulate future responsiveness of pathfinding axons by stimulating local synthesis of new guidance cue receptors needed for the next part of the journey.
- 42.Bi J, Tsai NP, Lin YP, Loh HH, Wei LN. Axonal mRNA transport and localized translational regulation of κ-opioid receptor in primary neurons of dorsal root ganglia. Proc. Natl Acad. Sci. USA. 2006;103:19919–19924. doi: 10.1073/pnas.0607394104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng JQ, et al. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. J. Neurosci. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. Landmark study that showed evidence for involvement of axonal protein synthesis in axon regeneration. Adult peripheral sensory neurons can locally synthesize proteins in vitro, and this ability is enhanced by preconditioning nerve injury in vivo.
- 44.Merianda TT, et al. A functional equivalent of endoplasmic reticulum and Golgi in axons for secretion of locally synthesized proteins. Mol. Cell Neurosci. 2009;40:128–142. doi: 10.1016/j.mcn.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Willis D, et al. Differential transport and local translation of cytoskeletal, injury-response, and neurodegeneration protein mRNAs in axons. J. Neurosci. 2005;25:778–791. doi: 10.1523/JNEUROSCI.4235-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spencer GE, et al. Synthesis and functional integration of a neurotransmitter receptor in isolated invertebrate axons. J. Neurobiol. 2000;44:72–81. doi: 10.1002/1097-4695(200007)44:1<72::aid-neu7>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 47.Lyles V, Zhao Y, Martin KC. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 2006;49:349–356. doi: 10.1016/j.neuron.2005.12.029. [DOI] [PubMed] [Google Scholar]
- 48.Lujan HD, et al. Developmental induction of Golgi structure and function in the primitive eukaryote Giardia lamblia. J. Biol. Chem. 1995;270:4612–4618. doi: 10.1074/jbc.270.9.4612. [DOI] [PubMed] [Google Scholar]
- 49.Harris WA, Holt CE, Bonhoeffer F. Retinal axons with and without their somata, growing to and arborizing in the tectum of Xenopus embryos: a time-lapse video study of single fibres in vivo. Development. 1987;101:123–133. doi: 10.1242/dev.101.1.123. [DOI] [PubMed] [Google Scholar]
- 50.Ming GL, et al. Adaptation in the chemotactic guidance of nerve growth cones. Nature. 2002;417:411–418. doi: 10.1038/nature745. First evidence that local mRNA translation may be necessary for the adaptation of growth cones to extracellular signals. Cultured growth cones are desensitized to continuously applied guidance cues but later resensitized to the same cue, and resensitization is blocked by inhibitors of MAPKs and ribosome function.
- 51.Campbell DS, et al. Semaphorin 3A elicits stage-dependent collapse, turning, and branching in Xenopus retinal growth cones. J. Neurosci. 2001;21:8538–8547. doi: 10.1523/JNEUROSCI.21-21-08538.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nature Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leung KM, et al. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nature Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yao J, Sasaki Y, Wen Z, Bassell GJ, Zheng JQ. An essential role for β-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nature Neurosci. 2006;9:1265–1273. doi: 10.1038/nn1773. Together with reference 53, this provided the first evidence that asymmetric mRNA translation mediates chemotropic growth cone turning. Netrin 1 and BDNF gradients activate asymmetric β-actin synthesis by increasing its transport and translation via ZBP1, a process that is required for attractive growth cone turning towards the sources of gradients.
- 55.Wu KY, et al. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piper M, et al. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. Together with reference 55, these were the first studies to suggest a mechanism for translation-dependent growth cone repulsion. Repulsive cues SEMA3A and SLIT2 increase global translational activity but activate translation of selective mRNAs that encode cytoskeletal-disassembling molecules, such as RHOA and cofilin, respectively.
- 57.Alvarez-Fischer D, et al. Engrailed protects mouse midbrain dopaminergic neurons against mitochondrial complex I insults. Nature Neurosci. 2011;14:1260–1266. doi: 10.1038/nn.2916. [DOI] [PubMed] [Google Scholar]
- 58.Brunet I, et al. The transcription factor Engrailed-2 guides retinal axons. Nature. 2005;438:94–98. doi: 10.1038/nature04110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wizenmann A, et al. Extracellular Engrailed participates in the topographic guidance of retinal axons in vivo. Neuron. 2009;64:355–366. doi: 10.1016/j.neuron.2009.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guirland C, Buck KB, Gibney JA, DiCicco-Bloom E, Zheng JQ. Direct cAMP signaling through G-protein-coupled receptors mediates growth cone attraction induced by pituitary adenylate cyclase-activating polypeptide. J. Neurosci. 2003;23:2274–2283. doi: 10.1523/JNEUROSCI.23-06-02274.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nature Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. Together with references 64 and 130, provided key evidence that axonally synthesized proteins generate retrograde signaling to the nucleus. Axonally synthesized importin-β1 mediates retrograde transport of transcription factors to the cell body, a process essential for axon regeneration after injury in vivo. Axonally synthesized transcription factors CREB and C/EBP1 mediate cell survival in vitro and in vivo, respectively.
- 62.Je HS, et al. Presynaptic protein synthesis required for NT-3-induced long-term synaptic modulation. Mol. Brain. 2011;4:1. doi: 10.1186/1756-6606-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang HL, Singer RH, Bassell GJ. Neurotrophin regulation of β-actin mRNA and protein localization within growth cones. J. Cell Biol. 1999;147:59–70. doi: 10.1083/jcb.147.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanz S, et al. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 65.Perlson E, et al. Vimentin-dependent spatial translocation of an activated MAP kinase in injured nerve. Neuron. 2005;45:715–726. doi: 10.1016/j.neuron.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 66.Yudin D, et al. Localized regulation of axonal RanGTPase controls retrograde injury signaling in peripheral nerve. Neuron. 2008;59:241–252. doi: 10.1016/j.neuron.2008.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mann F, Miranda E, Weinl C, Harmer E, Holt CE. B-type Eph receptors and ephrins induce growth cone collapse through distinct intracellular pathways. J. Neurobiol. 2003;57:323–336. doi: 10.1002/neu.10303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strochlic L, Dwivedy A, van Horck FP, Falk J, Holt CE. A role for S1P signalling in axon guidance in the Xenopus visual system. Development. 2008;135:333–342. doi: 10.1242/dev.009563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nedelec S, et al. Concentration-dependent requirement for local protein synthesis in motor neuron subtype-specific response to axon guidance cues. J. Neurosci. 2012;32:1496–1506. doi: 10.1523/JNEUROSCI.4176-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roche FK, Marsick BM, Letourneau PC. Protein synthesis in distal axons is not required for growth cone responses to guidance cues. J. Neurosci. 2009;29:638–652. doi: 10.1523/JNEUROSCI.3845-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huttelmaier S, et al. Spatial regulation of β-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 72.Welshhans K, Bassell GJ. Netrin-1-induced local β-actin synthesis and growth cone guidance requires zipcode binding protein 1. J. Neurosci. 2011;31:9800–9813. doi: 10.1523/JNEUROSCI.0166-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin AC, Holt CE. Local translation and directional steering in axons. EMBO J. 2007;26:3729–3736. doi: 10.1038/sj.emboj.7601808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kislauskis EH, Zhu X, Singer RH. β-actin messenger RNA localization and protein synthesis augment cell motility. J. Cell Biol. 1997;136:1263–1270. doi: 10.1083/jcb.136.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shestakova EA, Singer RH, Condeelis J. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl Acad. Sci. USA. 2001;98:7045–7050. doi: 10.1073/pnas.121146098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Karakozova M, et al. Arginylation of β-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- 77.Wang J, et al. Reversible glutathionylation regulates actin polymerization in A431 cells. J. Biol. Chem. 2001;276:47763–47766. doi: 10.1074/jbc.C100415200. [DOI] [PubMed] [Google Scholar]
- 78.Drinjakovic J, et al. E3 ligase Nedd4 promotes axon branching by downregulating PTEN. Neuron. 2010;65:341–357. doi: 10.1016/j.neuron.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.von Philipsborn A, Bastmeyer M. Mechanisms of gradient detection: a comparison of axon pathfinding with eukaryotic cell migration. Int. Rev. Cytol. 2007;263:1–62. doi: 10.1016/S0074-7696(07)63001-0. [DOI] [PubMed] [Google Scholar]
- 80.Piper M, Salih S, Weinl C, Holt CE, Harris WA. Endocytosis-dependent desensitization and protein synthesis-dependent resensitization in retinal growth cone adaptation. Nature Neurosci. 2005;8:179–186. doi: 10.1038/nn1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hopker VH, Shewan D, Tessier-Lavigne M, Poo M, Holt C. Growth-cone attraction to netrin-1 is converted to repulsion by laminin-1. Nature. 1999;401:69–73. doi: 10.1038/43441. [DOI] [PubMed] [Google Scholar]
- 82.Shewan D, Dwivedy A, Anderson R, Holt CE. Age-related changes underlie switch in netrin-1 responsiveness as growth cones advance along visual pathway. Nature Neurosci. 2002;5:955–962. doi: 10.1038/nn919. [DOI] [PubMed] [Google Scholar]
- 83.Kamiguchi H, Yoshihara F. The role of endocytic L1 trafficking in polarized adhesion and migration of nerve growth cones. J. Neurosci. 2001;21:9194–9203. doi: 10.1523/JNEUROSCI.21-23-09194.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kuwako K, et al. Neural RNA-binding protein Musashi1 controls midline crossing of precerebellar neurons through posttranscriptional regulation of Robo3/Rig-1 expression. Neuron. 2010;67:407–421. doi: 10.1016/j.neuron.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 85.Shaw G, Bray D. Movement and extension of isolated growth cones. Exp. Cell Res. 1977;104:55–62. doi: 10.1016/0014-4827(77)90068-4. [DOI] [PubMed] [Google Scholar]
- 86.van Kesteren RE, et al. Local synthesis of actin-binding protein β-thymosin regulates neurite outgrowth. J. Neurosci. 2006;26:152–157. doi: 10.1523/JNEUROSCI.4164-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- 88.Sebeo J, et al. Requirement for protein synthesis at developing synapses. J. Neurosci. 2009;29:9778–9793. doi: 10.1523/JNEUROSCI.2613-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 89.Crispino M, et al. Active polysomes are present in the large presynaptic endings of the synaptosomal fraction from squid brain. J. Neurosci. 1997;17:7694–7702. doi: 10.1523/JNEUROSCI.17-20-07694.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hu JY, Meng X, Schacher S. Target interaction regulates distribution and stability of specific mRNAs. J. Neurosci. 2002;22:2669–2678. doi: 10.1523/JNEUROSCI.22-07-02669.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schacher S, Wu F, Panyko JD, Sun ZY, Wang D. Expression and branch-specific export of mRNA are regulated by synapse formation and interaction with specific postsynaptic targets. J. Neurosci. 1999;19:6338–6347. doi: 10.1523/JNEUROSCI.19-15-06338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee W, Jones AM, Ono JK, Wayne NL. Regional differences in processing of locally translated prohormone in peptidergic neurons of Aplysia californica. J. Neurochem. 2002;83:1423–1430. doi: 10.1046/j.1471-4159.2002.01252.x. [DOI] [PubMed] [Google Scholar]
- 93.Cheng L, Locke C, Davis GW. S6 kinase localizes to the presynaptic active zone and functions with PDK1 to control synapse development. J. Cell Biol. 2011;194:921–935. doi: 10.1083/jcb.201101042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ronesi JA, Huber KM. Metabotropic glutamate receptors and fragile X mental retardation protein: partners in translational regulation at the synapse. Sci. Signal. 2008;1:pe6. doi: 10.1126/stke.15pe6. [DOI] [PubMed] [Google Scholar]
- 95.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Antar LN, Li C, Zhang H, Carroll RC, Bassell GJ. Local functions for FMRP in axon growth cone motility and activity-dependent regulation of filopodia and spine synapses. Mol. Cell. Neurosci. 2006;32:37–48. doi: 10.1016/j.mcn.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 97.Li C, Bassell GJ, Sasaki Y. Fragile X mental retardation protein is involved in protein synthesis-dependent collapse of growth cones induced by semaphorin-3A. Front. Neural Circuits. 2009;3:11. doi: 10.3389/neuro.04.011.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Christie SB, Akins MR, Schwob JE, Fallon JR. The FXG: a presynaptic fragile X granule expressed in a subset of developing brain circuits. J. Neurosci. 2009;29:1514–1524. doi: 10.1523/JNEUROSCI.3937-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hanson JE, Madison DV. Presynaptic Fmr1 genotype influences the degree of synaptic connectivity in a mosaic mouse model of fragile X syndrome. J. Neurosci. 2007;27:4014–4018. doi: 10.1523/JNEUROSCI.4717-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fallini C, et al. The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons. J. Neurosci. 2011;31:3914–3925. doi: 10.1523/JNEUROSCI.3631-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang H, et al. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J. Neurosci. 2006;26:8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J. Cell Sci. 2002;115:3817–3827. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- 103.Smith CL, et al. GAP-43 mRNA in growth cones is associated with HuD and ribosomes. J. Neurobiol. 2004;61:222–235. doi: 10.1002/neu.20038. [DOI] [PubMed] [Google Scholar]
- 104.Droz B, Barondes SM. Nerve endings: rapid appearance of labeled protein shown by electron microscope radioautography. Science. 1969;165:1131–1133. doi: 10.1126/science.165.3898.1131. [DOI] [PubMed] [Google Scholar]
- 105.Thoenen H, Mueller RA, Axelrod J. Phase difference in the induction of tyrosine hydroxylase in cell body and nerve terminals of sympathetic neurones. Proc. Natl Acad. Sci. USA. 1970;65:58–62. doi: 10.1073/pnas.65.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Koenig E, Koelle GB. Acetylcholinesterase regeneration in peripheral nerve after irreversible inactivation. Science. 1960;132:1249–1250. doi: 10.1126/science.132.3435.1249. [DOI] [PubMed] [Google Scholar]
- 107.Melia KR, Trembleau A, Oddi R, Sanna PP, Bloom FE. Detection and regulation of tyrosine hydroxylase mRNA in catecholaminergic terminal fields: possible axonal compartmentalization. Exp. Neurol. 1994;130:394–406. doi: 10.1006/exnr.1994.1219. [DOI] [PubMed] [Google Scholar]
- 108.Jirikowski GF, Sanna PP, Bloom FE. mRNA coding for oxytocin is present in axons of the hypothalamo–neurohypophysial tract. Proc. Natl Acad. Sci. USA. 1990;87:7400–7404. doi: 10.1073/pnas.87.19.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trembleau A, Melia KR, Bloom FE. BC1 RNA and vasopressin mRNA in rat neurohypophysis: axonal compartmentalization and differential regulation during dehydration and rehydration. Eur. J. Neurosci. 1995;7:2249–2260. doi: 10.1111/j.1460-9568.1995.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 110.Trembleau A, Morales M, Bloom FE. Differential compartmentalization of vasopressin messenger RNA and neuropeptide within the rat hypothalamo–neurohypophysial axonal tracts: light and electron microscopic evidence. Neuroscience. 1996;70:113–125. doi: 10.1016/0306-4522(95)00328-g. [DOI] [PubMed] [Google Scholar]
- 111.Mohr E, Fehr S, Richter D. Axonal transport of neuropeptide encoding mRNAs within the hypothalamo–hypophyseal tract of rats. EMBO J. 1991;10:2419–2424. doi: 10.1002/j.1460-2075.1991.tb07781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mohr E, Richter D. Diversity of mRNAs in the axonal compartment of peptidergic neurons in the rat. Eur. J. Neurosci. 1992;4:870–876. doi: 10.1111/j.1460-9568.1992.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 113.Dirks RW, et al. Ultrastructural evidence for the axonal localization of caudodorsal cell hormone mRNA in the central nervous system of the mollusc Lymnaea stagnalis. Microsc. Res. Tech. 1993;25:12–18. doi: 10.1002/jemt.1070250104. [DOI] [PubMed] [Google Scholar]
- 114.Twiss JL, Shooter EM. Nerve growth factor promotes neurite regeneration in PC12 cells by translational control. J. Neurochem. 1995;64:550–557. doi: 10.1046/j.1471-4159.1995.64020550.x. [DOI] [PubMed] [Google Scholar]
- 115.Gioio AE, et al. Local synthesis of nuclear-encoded mitochondrial proteins in the presynaptic nerve terminal. J. Neurosci. Res. 2001;64:447–453. doi: 10.1002/jnr.1096. [DOI] [PubMed] [Google Scholar]
- 116.Aschrafi A, Natera-Naranjo O, Gioio AE, Kaplan BB. Regulation of axonal trafficking of cytochrome c oxidase IV mRNA. Mol. Cell Neurosci. 2010;43:422–430. doi: 10.1016/j.mcn.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hillefors M, Gioio AE, Mameza MG, Kaplan BB. Axon viability and mitochondrial function are dependent on local protein synthesis in sympathetic neurons. Cell. Mol. Neurobiol. 2007;27:701–716. doi: 10.1007/s10571-007-9148-y. Together with reference 118, this showed that mitochondria in distal axons are maintained by locally synthesized nuclear-encoded proteins. Blocking either local protein synthesis or protein import in distal axons leads to mitochondrial dysfunction and axon degeneration.
- 118.Yoon BC, et al. Local translation of extranuclear lamin B promotes axon maintenance. Cell. 2012;148:1–13. doi: 10.1016/j.cell.2011.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot–Marie–Tooth disease. Lancet Neurol. 2009;8:654–667. doi: 10.1016/S1474-4422(09)70110-3. [DOI] [PubMed] [Google Scholar]
- 120.Capell BC, Collins FS. Human laminopathies: nuclei gone genetically awry. Nature Rev. Genet. 2006;7:940–952. doi: 10.1038/nrg1906. [DOI] [PubMed] [Google Scholar]
- 121.Wang W, van Niekerk E, Willis DE, Twiss JL. RNA transport and localized protein synthesis in neurological disorders and neural repair. Dev. Neurobiol. 2007;67:1166–1182. doi: 10.1002/dneu.20511. [DOI] [PubMed] [Google Scholar]
- 122.Gumy LF, Tan CL, Fawcett JW. The role of local protein synthesis and degradation in axon regeneration. Exp. Neurol. 2010;223:28–37. doi: 10.1016/j.expneurol.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Christie KJ, Webber CA, Martinez JA, Singh B, Zochodne DW. PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J. Neurosci. 2010;30:9306–9315. doi: 10.1523/JNEUROSCI.6271-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Court FA, Hendriks WT, MacGillavry HD, Alvarez J, van Minnen J. Schwann cell to axon transfer of ribosomes: toward a novel understanding of the role of glia in the nervous system. J. Neurosci. 2008;28:11024–11029. doi: 10.1523/JNEUROSCI.2429-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Court FA, et al. Morphological evidence for a transport of ribosomes from Schwann cells to regenerating axons. Glia. 2011;59:1529–1539. doi: 10.1002/glia.21196. [DOI] [PubMed] [Google Scholar]
- 126.Vassar R, et al. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- 127.Wensley CH, et al. Olfactory marker protein mRNA is found in axons of olfactory receptor neurons. J. Neurosci. 1995;15:4827–4837. doi: 10.1523/JNEUROSCI.15-07-04827.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Dubacq C, Jamet S, Trembleau A. Evidence for developmentally regulated local translation of odorant receptor mRNAs in the axons of olfactory sensory neurons. J. Neurosci. 2009;29:10184–10190. doi: 10.1523/JNEUROSCI.2443-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Michaelevski I, et al. Signaling to transcription networks in the neuronal retrograde injury response. Sci. Signal. 2011;3:ra53. doi: 10.1126/scisignal.2000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yan D, Wu Z, Chisholm AD, Jin Y. The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138:1005–1018. doi: 10.1016/j.cell.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Park KK, et al. Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322:963–966. doi: 10.1126/science.1161566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol. Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 134.Tohda C, et al. Axonal transport of VR1 capsaicin receptor mRNA in primary afferents and its participation in inflammation-induced increase in capsaicin sensitivity. J. Neurochem. 2001;76:1628–1635. doi: 10.1046/j.1471-4159.2001.00193.x. [DOI] [PubMed] [Google Scholar]
- 135.Ruangsri S, et al. Relationship of axonal voltage-gated sodium channel 1.8 (NaV1.8) mRNA accumulation to sciatic nerve injury-induced painful neuropathy in rats. J. Biol. Chem. 2011;286:39836–39847. doi: 10.1074/jbc.M111.261701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Geranton SM, et al. A rapamycin-sensitive signaling pathway is essential for the full expression of persistent pain states. J. Neurosci. 2009;29:15017–15027. doi: 10.1523/JNEUROSCI.3451-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jimenez-Diaz L, et al. Local translation in primary afferent fibers regulates nociception. PLoS One. 2008;3:e1961. doi: 10.1371/journal.pone.0001961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Melemedjian OK, et al. IL-6- and NGF-induced rapid control of protein synthesis and nociceptive plasticity via convergent signaling to the eIF4F complex. J. Neurosci. 2010;30:15113–15123. doi: 10.1523/JNEUROSCI.3947-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bi J, Hu X, Loh HH, Wei LN. Mouse κ-opioid receptor mRNA differential transport in neurons. Mol. Pharmacol. 2003;64:594–599. doi: 10.1124/mol.64.3.594. [DOI] [PubMed] [Google Scholar]
- 140.Bear MF, Dolen G, Osterweil E, Nagarajan N. Fragile X: translation in action. Neuropsychopharmacology. 2008;33:84–87. doi: 10.1038/sj.npp.1301610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Liu-Yesucevitz L, et al. Local RNA translation at the synapse and in disease. J. Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 143.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. A novel function for SMN, the spinal muscular atrophy disease gene product, in pre-mRNA splicing. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 144.Sharma A, et al. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp. Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 145.Rossoll W, et al. SMN, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of β-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Piazzon N, et al. In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein. J. Biol. Chem. 2008;283:5598–5610. doi: 10.1074/jbc.M707304200. [DOI] [PubMed] [Google Scholar]
- 147.Cleveland DW, Rothstein JD. From Charcot to Lou Gehrig: deciphering selective motor neuron death in ALS. Nature Rev. Neurosci. 2001;2:806–819. doi: 10.1038/35097565. [DOI] [PubMed] [Google Scholar]
- 148.Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nature Rev. Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Greenway MJ, et al. ANG mutations segregate with familial and ‘sporadic’ amyotrophic lateral sclerosis. Nature Genet. 2006;38:411–413. doi: 10.1038/ng1742. [DOI] [PubMed] [Google Scholar]
- 150.Emara MM, et al. Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 2010;285:10959–10968. doi: 10.1074/jbc.M109.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Campbell DS, Holt CE. Apoptotic pathway and MAPKs differentially regulate chemotropic responses of retinal growth cones. Neuron. 2003;37:939–952. doi: 10.1016/s0896-6273(03)00158-2. [DOI] [PubMed] [Google Scholar]
- 152.Nie D, et al. Tsc2-Rheb signaling regulates EphA-mediated axon guidance. Nature Neurosci. 2010;13:163–172. doi: 10.1038/nn.2477. First evidence showing that some guidance cues can repress mRNA translation. Ephrin A represses mTOR by inhibiting MAPK ERK1/2, providing a mechanism by which multiple cues regulate local protein synthesis in axons and growth cones by modulating diverse pathways that converge on mTOR.
- 153.Kim S, Coulombe PA. Emerging role for the cytoskeleton as an organizer and regulator of translation. Nature Rev. Mol. Cell Biol. 2010;11:75–81. doi: 10.1038/nrm2818. [DOI] [PubMed] [Google Scholar]
- 154.Zhang HL, et al. Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 155.Vuppalanchi D, et al. Conserved 3′-untranslated region sequences direct subcellular localization of chaperone protein mRNAs in neurons. J. Biol. Chem. 2010;285:18025–18038. doi: 10.1074/jbc.M109.061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Willis DE, et al. Extracellular stimuli specifically regulate localized levels of individual neuronal mRNAs. J. Cell Biol. 2007;178:965–980. doi: 10.1083/jcb.200703209. Comprehensive profiling study that showed extrinsic cues can influence the axonal transcriptome. Systematic analysis of axons treated with NGF, BDNF, NT3, MAG and SEMA3A showed that these cues regulate axonal transport of 50 candidate mRNAs in culture.
- 157.Sotelo-Silveira JR, Calliari A, Kun A, Koenig E, Sotelo JR. RNA trafficking in axons. Traffic. 2006;7:508–515. doi: 10.1111/j.1600-0854.2006.00405.x. [DOI] [PubMed] [Google Scholar]
- 158.van Niekerk EA, et al. Sumoylation in axons triggers retrograde transport of the RNA-binding protein La. Proc. Natl Acad. Sci. USA. 2007;104:12913–12918. doi: 10.1073/pnas.0611562104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 160.Vuppalanchi D, Willis DE, Twiss JL. Regulation of mRNA transport and translation in axons. Results Probl. Cell Differ. 2009;48:193–224. doi: 10.1007/400_2009_16. [DOI] [PubMed] [Google Scholar]
- 161.Krichevsky AM, Kosik KS. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 2001;32:683–696. doi: 10.1016/s0896-6273(01)00508-6. [DOI] [PubMed] [Google Scholar]
- 162.Kilchert C, Spang A. Cotranslational transport of ABP140 mRNA to the distal pole of S. cerevisiae. EMBO J. 2011;30:3567–3580. doi: 10.1038/emboj.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol. Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 164.Tsai NP, Bi J, Wei LN. The adaptor Grb7 links netrin-1 signaling to regulation of mRNA translation. EMBO J. 2007;26:1522–1531. doi: 10.1038/sj.emboj.7601598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Buckley PT, et al. Cytoplasmic intron sequence-retaining transcripts can be dendritically targeted via ID element retrotransposons. Neuron. 2011;69:877–884. doi: 10.1016/j.neuron.2011.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Chen Z, Gore BB, Long H, Ma L, Tessier-Lavigne M. Alternative splicing of the Robo3 axon guidance receptor governs the midline switch from attraction to repulsion. Neuron. 2008;58:325–332. doi: 10.1016/j.neuron.2008.02.016. [DOI] [PubMed] [Google Scholar]
- 167.Giorgi C, et al. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 168.Kiebler MA, Bassell GJ. Neuronal RNA granules: movers and makers. Neuron. 2006;51:685–690. doi: 10.1016/j.neuron.2006.08.021. [DOI] [PubMed] [Google Scholar]
- 169.Sasaki Y, et al. Phosphorylation of zipcode binding protein 1 is required for brain-derived neurotrophic factor signaling of local β-actin synthesis and growth cone turning. J. Neurosci. 2010;30:9349–9358. doi: 10.1523/JNEUROSCI.0499-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chao JA, et al. ZBP1 recognition of β-actin zipcode induces RNA looping. Genes Dev. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Melko M, Bardoni B. The role of G-quadruplex in RNA metabolism: involvement of FMRP and FMR2P. Biochimie. 2010;92:919–926. doi: 10.1016/j.biochi.2010.05.018. [DOI] [PubMed] [Google Scholar]
- 172.Narayanan U, et al. S6K1 phosphorylates and regulates fragile X mental retardation protein (FMRP) with the neuronal protein synthesis-dependent mammalian target of rapamycin (mTOR) signaling cascade. J. Biol. Chem. 2008;283:18478–18482. doi: 10.1074/jbc.C800055200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Richter JD. Cytoplasmic polyadenylation in development and beyond. Microbiol. Mol. Biol. Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kundel M, Jones KJ, Shin CY, Wells DG. Cytoplasmic polyadenylation element-binding protein regulates neurotrophin-3-dependent β-catenin mRNA translation in developing hippocampal neurons. J. Neurosci. 2009;29:13630–13639. doi: 10.1523/JNEUROSCI.2910-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lin AC, et al. Cytoplasmic polyadenylation and cytoplasmic polyadenylation element-dependent mRNA regulation are involved in Xenopus retinal axon development. Neural Dev. 2009;4:8. doi: 10.1186/1749-8104-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Alexandrov IM, et al. Cytoplasmic polyadenylation element binding protein deficiency stimulates PTEN and Stat3 mRNA translation and induces hepatic insulin resistance. PLoS Genet. 2012;8:e1002457. doi: 10.1371/journal.pgen.1002457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Schratt GM, et al. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- 178.Aschrafi A, et al. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J. Neurosci. 2008;28:12581–12590. doi: 10.1523/JNEUROSCI.3338-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 180.Jin P, et al. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nature Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- 181.Kondrashov N, et al. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Tsurugi K, Ogata K. Evidence for the exchangeability of acidic ribosomal proteins on cytoplasmic ribosomes in regenerating rat liver. J. Biochem. 1985;98:1427–1431. doi: 10.1093/oxfordjournals.jbchem.a135410. [DOI] [PubMed] [Google Scholar]
- 183.Taylor AM, et al. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nature Methods. 2005;2:599–605. doi: 10.1038/nmeth777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Park JW, Vahidi B, Taylor AM, Rhee SW, Jeon NL. Microfluidic culture platform for neuroscience research. Nature Protoc. 2006;1:2128–2136. doi: 10.1038/nprot.2006.316. [DOI] [PubMed] [Google Scholar]
- 185.Campenot RB, Lund K, Mok SA. Production of compartmented cultures of rat sympathetic neurons. Nature Protoc. 2009;4:1869–1887. doi: 10.1038/nprot.2009.210. [DOI] [PubMed] [Google Scholar]
- 186.Willis DE, et al. Axonal localization of transgene mRNA in mature PNS and CNS neurons. J. Neurosci. 2011;31:14481–14487. doi: 10.1523/JNEUROSCI.2950-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Dmochowski IJ, Tang X. Taking control of gene expression with light-activated oligonucleotides. Biotechniques. 2007;43:161–171. doi: 10.2144/000112519. [DOI] [PubMed] [Google Scholar]
- 188.Je HS, et al. Chemically inducible inactivation of protein synthesis in genetically targeted neurons. J. Neurosci. 2009;29:6761–6766. doi: 10.1523/JNEUROSCI.1280-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Campenot RB. NGF and the local control of nerve terminal growth. J. Neurobiol. 1994;25:599–611. doi: 10.1002/neu.480250603. [DOI] [PubMed] [Google Scholar]
- 190.Leung KM, Holt CE. Live visualization of protein synthesis in axonal growth cones by microinjection of photoconvertible Kaede into Xenopus embryos. Nature Protoc. 2008;3:1318–1327. doi: 10.1038/nprot.2008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Park HY, Buxbaum AR, Singer RH. Single mRNA tracking in live cells. Methods Enzymol. 2010;472:387–406. doi: 10.1016/S0076-6879(10)72003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bi J, Tsai NP, Lu HY, Loh HH, Wei LN. Copb1-facilitated axonal transport and translation of κ opioid-receptor mRNA. Proc. Natl Acad. Sci. USA. 2007;104:13810–13815. doi: 10.1073/pnas.0703805104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Lionnet T, et al. A transgenic mouse for in vivo detection of endogenous labeled mRNA. Nature Methods. 2011;8:165–170. doi: 10.1038/nmeth.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Lee SK, Hollenbeck PJ. Organization and translation of mRNA in sympathetic axons. J. Cell Sci. 2003;116:4467–4478. doi: 10.1242/jcs.00745. [DOI] [PubMed] [Google Scholar]
- 195.Dieterich DC, et al. In situ visualization and dynamics of newly synthesized proteins in rat hippocampal neurons. Nature Neurosci. 2010;13:897–905. doi: 10.1038/nn.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Twiss JL, Smith DS, Chang B, Shooter EM. Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration. Neurobiol. Dis. 2000;7:416–428. doi: 10.1006/nbdi.2000.0293. [DOI] [PubMed] [Google Scholar]
- 198.Natera-Naranjo O, et al. Local translation of ATP synthase subunit 9 mRNA alters ATP levels and the production of ROS in the axon. Mol. Cell Neurosci. 2012;49:263–270. doi: 10.1016/j.mcn.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 199.Giustetto M, et al. Axonal transport of eukaryotic translation elongation factor 1α mRNA couples transcription in the nucleus to long-term facilitation at the synapse. Proc. Natl Acad. Sci. USA. 2003;100:13680–13685. doi: 10.1073/pnas.1835674100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sotelo-Silveira JR, et al. Neurofilament mRNAs are present and translated in the normal and severed sciatic nerve. J. Neurosci. Res. 2000;62:65–74. doi: 10.1002/1097-4547(20001001)62:1<65::AID-JNR7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 201.Aronov S, Aranda G, Behar L, Ginzburg I. Axonal tau mRNA localization coincides with tau protein in living neuronal cells and depends on axonal targeting signal. J. Neurosci. 2001;21:6577–6587. doi: 10.1523/JNEUROSCI.21-17-06577.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]