Abstract
High molecular weight thioredoxin reductases (TRs) catalyze the reduction of the redoxactive disulfide bond of thioredoxin, but an important difference in the TR family is the sequence of the C-terminal redox-active tetrapeptide that interacts directly with thioredoxin, especially the presence or absence of a selenocysteine (Sec) residue in this tetrapeptide. In this study we have employed protein engineering techniques to investigate the C-terminal redox active tetrapeptides of three different TRs: mouse mitochondrial TR (mTR3), Drosophila melanogaster TR (DmTR), and the mitochondrial TR from C. elegans (CeTR2), which have C-terminal tetrapeptide sequences of Gly-Cys-Sec-Gly, Ser-Cys-Cys-Ser, and Gly-Cys-Cys-Gly, respectively.
Three different types of mutations and chemical modifications were performed in this study: insertion of alanine residues in between the cysteine residues of the Cys-Cys or Cys-Sec dyads, modification of the charge at the C-terminus, and altering the position of the Sec residue in the mammalian enzyme. The results show that mTR3 is quite accommodating to insertion of alanine residues into the Cys-Sec dyad, with only a 4-6 fold drop in catalytic activity. In contrast, both DmTR and CeTR2 lost 100-300 fold activity when alanine residues were inserted into the Cys-Cys dyad. We have tested the importance of a salt bridge between the C-terminus and a basic residue that was proposed for orienting the Cys-Sec dyad of mTR3 for proper catalytic position by changing the C-terminal carboxylate to a carboxamide. The result is an enzyme with twice the activity as the wild-type mammalian enzyme. A similar result was achieved when the C-terminal carboxylate of DmTR was converted to a hydroxamic acid or a thiocarboxylate. Lastly, reversing the positions of the Cys and Sec residues in the catalytic dyad resulted in a 100 fold loss in catalytic activity. Taken together, the results support our previous model of Sec as the leaving group during reduction of the C-terminus during the catalytic cycle.
Thioredoxin reductases (TR)1 from higher eukaryotes are members of the glutathione reductase (GR) family of pyridine nucleotide-disulfide oxidoreductases (1). These enzymes are homodimeric with each monomer constructed from a three-domain architecture and catalyze the reduction of a cognate substrate by thiol-disulfide exchange. Electrons supplied by NADPH are transferred to the enzyme active site disulfide via a bound FAD. TRs use the same general mechanism as GR except that a second thiol-disulfide exchange step is added to the enzymatic cycle prior to reduction of its cognate substrate, thioredoxin (Trx). This second thiol-disulfide exchange step utilizes a 16 amino acid C-terminal extension, which contains an additional disulfide redox center (1-3). Upon reduction of this C-terminal disulfide by the N-terminal redox center (on the opposite chain), reduction of the substrate, Trx, can commence.
The C-terminal redox center of most of the high Mr TRs are notable because they contain a vicinal Cys-Cys dyad that forms an 8-membered ring when oxidized. This type of disulfide bond is rare in protein structures and is almost always found as part of a type VIII β-turn, which is thought to help stabilize high energy turns (4, 5). In TR, the 8-membered ring structure is not part of a β-turn, but part of a redox center that cycles between reduced and oxidized states. Though the Cys-Cys dyad is also found in methanol dehydrogenases (6), it is thought to play a structural role, thus TR is the only known example where this dyad is involved in redox cycling. In contrast, the substrate Trx has a very common redox center, the CXXC motif, which is found in many proteins with thioredoxin folds (7). Why TRs use this rare type of disulfide bond in redox cycling remains an unanswered question.
Mammalian TRs are distinguished from other TRs by the presence of the rare amino acid selenocysteine (Sec, U), which has been shown to be essential for Trx reduction, as part of the C-terminal motif Gly-Cys-Sec-Gly (GCUG) (8). Thus the mammalian enzymes have the interesting feature of containing a selenium atom in the 8-membered ring. The selenium atom is apparently only required in mammalian TRs (9), since several other homologues, notably the TR from Drosophila melanogaster (DmTR) (2) and the mitochondrial TR from C. elegans (CeTR2) (10), can catalyze the same reaction with relatively high efficiency with a standard cysteine (Cys) residue.
A generalized reaction mechanism for the reaction catalyzed by high Mr TRs is shown in Figure 1A. In the mammalian enzyme it is thought that the attacking nucleophile on the disulfide bond of Trx is the selenolate of the Sec residue (11). This inference is based upon the mammalian enzyme’s ability to reduce H2O2 (12), which is abolished upon mutation of Sec to Cys. It is thus likely that the selenolate is the attacking nucleophile in the Trx reductase reaction as well. Since the Sec residue occupies the C-terminal position of the redox-dyad in the mammalian enzyme, the C-terminal Cys of the Cys-Cys dyad of DmTR and CeTR2 should act as the attacking nucleophile for their respective cognate Trx’s as well. The adjacent Cys residue in the N-terminal position of the redox-dyad would therefore act to resolve the mixed selenylsulfide or mixed disulfide between TR and Trx (Figure 1B). The subsequent release of substrate reforms the 8-membered ring, which then must be reduced by the N-terminal redox center for the enzymatic cycle to continue (Figure 1C).
Figure 1.
(A) Proposed pathway for transfer of electrons to Trx by TR. The Cys residue that interacts with the flavin cofactor is labeled as “CT” (charge-transfer), while the Cys residue that acts as the interchange residue is labeled “IC”. Once the interchange Cys becomes reduced by the flavin cofactor, it initiates attack on the 8-membered ring formed by either adjacent Cys residues (DmTR and CeTR2) or adjacent Cys and Sec residues (mTR3) on the C-terminus on the opposing subunit. We refer to this step in the reaction mechanism as the “ring opening” step and is highlighted in red in the diagram. Once this ring is reduced, the attacking nucleophile (either a sulfur atom or a selenium atom – labeled as X), initiates attack on the disulfide bond of Trx. The N-terminal Cys of the dyad would then “resolve” the mixed disulfide formed between TR and Trx and is thus labeled as “Res”. The “prime” designation indicates residues that are on the adjacent subunit. (B) Diagram of the TR/Trx complex formed when the attacking nucleophile (either S or Se) attacks the disulfide bond of Trx. The adjacent Cys residue then attacks to resolve this complex, releasing product and forming the oxidized, 8-membered ring. (C) The 8-membered ring is then reduced by CysIC. CysIC can attack the N-terminal sulfur atom (pathway 1) or the C-terminal atom (S or Se, pathway 2). We have argued here and previously (17) for pathway 1). Termaa is the C-terminal amino acid (either Gly or Ser).
To test the importance of this 8-membered ring structure to the catalytic cycle, we have constructed mutant enzymes in which we have enlarged the ring by inserting alanine residues in between the Cys-Cys or Cys-Sec dyad for three different TRs. These TRs are DmTR, CeTR2 and the mouse mitochondrial TR (mTR3). We have chosen these TRs because they represent both Cys-containing TRs (DmTR and CeTR2) and Sec-TR-containing TRs (mTR3). While DmTR and CeTR2 are both Cys-containing TRs, they have different flanking residues on either side of the Cys-Cys dyad, glycine residues in the case of CeTR2 (GCCG) and serine residues in the case of DmTR (SCCS). This is important because it has been proposed that the flanking serine residues in DmTR participate in catalysis by acting as acid/base catalysts, thereby eliminating the need for Sec (13).
We also have performed additional structure-function analysis on these enzymes using intein-mediated peptide ligation (semisynthesis), which allowed us to construct several mutants that are not possible using routine site-directed mutagenesis methodologies. These experiments allowed us to modify the C-terminal carboxylate to a carboxamide, a hydroxamic acid, and a thiocarboxylate. This was done to test the hypothesis that the C-terminal carboxylate forms a salt bridge with Lys29. This salt bridge was hypothesized to be important for positioning the C-terminal redox center in the proper position for catalysis (14). Lastly in the mammalian enzyme, we tested the importance of the position of Sec within the Cys-Sec dyad by switching the position of these two redox active residues using our semisynthetic technique. Our results are reported herein.
MATERIALS AND METHODS
Materials
Chitin agarose beads were purchased from New England Biolabs (Ipswich, MA). DTNB (5,5′dithio-bis(2-nitrobenzoic acid)), NADPH, bovine pancreatic insulin and N-methylmercaptoacetamide (NMA) was purchased from Sigma-Aldrich (St. Louis, MO). Hydrogen peroxide (30% solution) was purchased from J.T Baker (Phillipsburg, NJ). All other chemicals were purchased from Fisher Scientific (Fair Lawn, NJ) and were of reagent grade or better. All restriction endonucleases, Vent DNA polymerase, and T4 DNA ligase were purchased from New England Biolabs (Ipswich, MA) and used with the supplied buffers according to manufacturers guidelines.
Peptide Synthesis
Sec-containing peptides were produced by Fmoc solid phase synthesis as previously described (15). Peptides were purified by preparative HPLC from the Shimadzu Corporation (Kyoto, Japan) and verified by matrix assisted laser desorption ionization time-of-flight mass spectrometry on a Voyager DE PRO Workstation from Applied Biosystems (Framingham, MA). Synthesis of the Cys-Sec-Gly-NH2 (CUG) tripeptide with C-terminal carboxyamide utilized a similar protocol with the exception of substitution of Fmoc-PAL resin from PE Biosystems (Hamburg, Germany) to yield the carboxamide upon cleavage from the resin.
Production of Semisynthetic Mouse Thioredoxin Reductase
The general method for producing mouse TR3 by semisynthesis has been previously described (16). Briefly, the full length Sec489Cys mutant as well as the truncated mTR3 missing the C-terminal CUG tripeptide sequence is expressed as a TR-intein-chitin binding domain fusion protein in E. coli. E. coli cell lysate supernatant is applied to chitin agarose beads to affinity purify the truncated TR. Cleavage of TR from the intein was performed on-resin using 70 mM NMA in the presence or absence of peptide. The TR mutant in which tripeptide UUG (Sec-Sec-Gly) was to be incorporated used 140 mM NMA instead. Final purification of TR was performed by hydrophobic interaction and anionic exchange chromatography as previously described (16). The efficiency of peptide incorporation for semisynthetic mTR3 was determined by inductively coupled plasma mass spectrometry (ICP-MS) calculated to the molar amount of protein analyzed. The yield of semisynthetic TR was 20-30 mg per 6 L of culture.
Cloning and Expression of DmTR
We have previously reported the expression and purification of DmTR as an intein fusion protein (17). We also have reported the conditions for cloning and constructing the plasmids via PCR necessary for expression of this fusion protein (17). In this study we have constructed two new mutants using identical PCR and cloning conditions to our earlier report. For construction of DmTR with mutant C-terminal redox center Ser-Cys-Ala-Cys-Ser (SCACS488-492) we used downstream primer 5′-ACAGCCGGTACCCTTGGCAAAGCAGCTGCAGGCGCAGCTGGCCGGCGTGGG-3′. For construction of the DmTR mutant with C-terminal sequence Ser-Cys-Ala-Ala-Cys-Ser (SCAACS488-493) we used the downstream primer 5′-ACAGCCGGTACCCTTGGCAAAGCAGCTGCAGGCGGCGCAGCTGGCCGGCGTGGG-3′. The sequence of the upstream primer was the same as previously reported (17). Sequencing of the DNA of the resultant plasmids was done at the University of Vermont DNA Sequencing Facility using an ABI 3100-Avant Genetic Analyzer.
Production of C-terminal Variants of DmTR
Each of the TR mutants from Drosophila is expressed as a TR-intein-chitin binding domain fusion protein and affinity purified from chitin agarose as described for mTR3 (16). E. coli ER2566 cells were used for production of recombinant WT and mutant enzymes. The cells were transformed with the plasmid, plated on LB-ampicillin plates containing 200 μg/mL ampicillin, and incubated at 37 °C overnight. A single colony was used to grow a 100 mL inoculum culture of LB (200 μg/mL ampicillin). This culture was incubated overnight at 37 °C with shaking. An inoculum culture of 10 mL was added to a 1 L baffled Pyrex Fernbach flask containing TB media (12 g/L tryptone, 24 g/L yeast extract, 4 mL/L glycerol, 16 mM potassium phosphate monobasic and 72 mM potassium phosphate dibasic at pH 7.0) containing 200 μg/mL ampicillin. The cells were incubated at 37 °C with shaking (100 rpm) until the O.D. at 600 nm reached 1.0. The cells were then chilled on ice until the temperature decreased to 20 °C and then induced by adding IPTG to a to final concentration of 0.5 mM. The induced cells were incubated at 20 °C overnight with shaking (100 rpm). The cells were harvested by centrifugation and stored at −20 °C then thawed on ice, homogenized in MOPS buffer, and lysed by probe-sonication as previously described (16). The WT enzyme having C-terminal sequence SCCS, as well as mutants with C-terminal sequences of SCACS and SCAACS are released from the intein, while bound on chitin agarose, by the addition of NMA.
To produce DmTR mutants in which the carboxylate group was changed to another functional group by addition of small molecules to the column buffer, four parallel experiments were performed. Chitin resin with bound fusion protein was separated evenly into four columns with 20 mL of chitin resin in each. Column 1 contained buffer only (no cleavage reagent) to evaluate non-specific hydrolysis from the intein with subsequent release of the fusion protein. Column 2 contained 100 mM NMA. Column 3 contained 100 mM (NH4)2S to produce the thiocarboxylic acid derivative (18). Column 4 contained 100 mM hydroxylamine to produce the hydroxamic acid derivative. Each column was equilibrated with three column volumes of cleavage reagent. The column buffer containing 50 mM MOPS buffer, 300 mM NaCl, and 100 mM of the cleavage reagent was then adjusted to pH 7.5. The equilibrated resin was transferred to a 50 mL conical tube and brought to a final volume of 45 mL. Cleavage was allowed to proceed for 24 hr at 4 °C followed by 4 hr at 25 °C, after which the column was drained of cleavage buffer and 100 mL of fresh eluant was added and then collected. The reactions were evaluated by 12% SDS-PAGE with the final purification of each DmTR enzyme by hydrophobic interaction chromatography followed by ion exchange chromatography as previously described (16).
Cloning of His-Tagged CeTR2
Plasmid pCeTR2 (10) was used as a template to clone the CeTR2 gene into plasmid pET45b(+) (Novagen) using an upstream primer containing a “hexaHis-tag”. For this construct, an upstream primer of sequence 5′-ACAGCCCCGGATCCCTTCTCATCAAATAAATTTGATCTG-3′ was used with downstream primer, 5′-ACAGCCAAGCTTGTCGACTCATCCACAGCATCCCTGAGTTCTTGG-3′ in the amplification process to produce the WT enzyme with C-terminal sequence Gly-Cys-Cys-Gly. C-terminal mutants were produced using downstream primers 5′-ACAGCCAAGCTTGTCGACTCATCCACATGCGCATCCCTGAGTTCTTGG-3′ (Gly-Cys-Ala-Cys-Gly mutant), and 5′-ACAGCCAAGCTTGTCGACTCATCCACATGCTGCGCATCCCTGAGTTCTTGG-3β (Gly-Cys-Ala-Ala-Cys-Gly mutant). The resulting plasmids (pHisCeTR2) encode a protein with a N-terminal hexa-histidine tag followed by an enterokinase cleavage site and the CeTR2 protein. Each PCR reaction was identical to the one as described for DmTR. The amplification reaction was monitored by 0.8% analytical agarose gel electrophoresis. The DNA from the PCR was then purified using the Qiagen QIAquick PCR Purification Kit.
The PCR product and pET45b(+) plasmid were then treated with restriction enzymes Bam HI and Sal I for 2 h at 37°C. Digests were purified using the Qiagen QIAquick PCR purification kit. Ligation reactions contained 200 ng of plasmid and 400 ng of PCR product as described for DmTR. The ligation reaction was then incubated at 37 °C with 5U Asc I for 2 h to enhance the amount of positive clones as Asc I will only cut plasmids that have not been ligated with the PCR insert. Positive clones were screened by restriction analysis using Bam HI and Sal I, analyzed by analytical agarose gel electrophoresis, and verified by sequencing of the DNA coding region.
Purification of WT and Mutant CeTR2 Enzymes
For expression of recombinant WT CeTR2 and mutant enzymes in E. coli, the same protocol was used as was described for DmTR except the TB media was supplemented with 20 mg/L each of niacinamide, pyridoxine, and riboflavin. The frozen cell pellets were thawed on ice and homogenized in 100 mL of 50 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole, 10% glycerol, 20 mM βME, pH 8.0, until homogeneous. Lysozyme was added to the homogenate at a concentration of 1 mg/mL and allowed to stir at 4 °C for 1 h. The homogenate was then briefly sonicated (5 × 30 sec). The sample was then centrifuged with a Beckman J21B centrifuge (JA-14 rotor) at 7000 ×g for 90 min at 4 °C. The supernatant was gravity loaded onto a 5 mL column of Ni-NTA resin with a flow rate of 0.5–1.0 mL/min. The column was washed with the buffer until the absorbance at 280 nm reached 0.05. This was followed by a wash with buffer containing 20 mM imidazole to eliminate nonspecific protein binding after which the protein was eluted in a buffer containing 250 mM imidazole
The protein was dialyzed against modified TE buffer (10 mM Tris, 1 mM EDTA, 10 mM NaCl, pH 8.0, 2 × 4L). The dialyzed protein was then reduced by addition of 20 mM βME and then loaded on to a 2′,5′-ADP Sepharose column (20 mL, Amersham) equilibrated with 10 mM Tris, 1 mM EDTA, 10 mM NaCl, 20 mM βME, pH 8.0 and washed until the A280 was below 0.05. Protein was eluted in buffer containing 3 M NaCl and analyzed by 12% SDS-PAGE. Fractions were pooled and concentrated by ultrafiltration using an Amicon Ultra 30,000 molecular weight cut off (MWCO) (Millipore) concentrator to a final volume of 1–2 mL. The concentrated protein was gravity loaded onto a Sephacryl S-200 gel filtration column (Pharmacia) of dimensions 3.8 cm × 97 cm. The column was equilibrated with 50 mM potassium phosphate, 300 mM NaCl, and 1 mM EDTA at pH 8.0. Fractions were collected (4 mL each) and then analyzed by UV–Vis spectrophotometry at 280 and 460 nm and examined for purity by 12 % SDS–PAGE. Fractions exhibiting the 55 kD band were pooled and concentrated by ultrafiltration.
Production of Thioredoxin
The clone containing the gene for TrxA from E. coli was a gift from Ronald T. Raines (19) and was produced as previously described (20). E. coli BL21(DE3) cells made competent with CaCl2 and transformed with 50 ng of DNA and plated onto LB agar supplemented with 200 Qg/mL ampicillin. Single colonies were used to inoculate 100 mL ampicillin-containing LB media and allowed to shake overnight at 37 °C.
Six liters of LB media (10 g/L tryptone, 5 g/L yeast extract, and 10 g/L NaCl) supplemented with 200 Qg/mL ampicillin was inoculated with 10 mL (each liter) of inoculum culture and grown to an OD of 0.6 at 600 nm while shaking at 37 °C in a C25KC shaker incubator (New Brunswick Scientific). The cells were induced for 3 h at 37 °C with 0.5 mM IPTG, then harvested by a 10 min centrifugation at 10,000 rpm in a JA-14 rotor using a Model J-21B centrifuge from Beckman. The resulting cell pellets were frozen at −20 °C. The cells were then thawed on ice, homogenized in 20 mM Tris, pH 8.4 containing 1 mM EDTA, and lysed by probe sonication using a Branson Sonifier. The lysate was centrifuged as described above for 90 min and the resulting supernatant was loaded onto a 70 mL column of DEAE sephacel (Sigma-Aldrich) equilibrated with the lysis buffer. After loading, the column was washed with buffer until the A280 of the effluent was 0.05. The sample was eluted using two 400 mL gradients containing NaCl (0 mM to 100 mM and 100 mM to 250 mM). Fractions were collected (3 mL) and evaluated by absorbance at 280 nm and further analyzed by 15% SDS-PAGE. Fractions containing Trx were pooled and then adjusted to 60% ammonium sulfate saturation and centrifuged as above for 60 min. The pellet was solubilized in a minimal volume of 20 mM Tris pH 8.4, 250 mM NaCl, 1 mM EDTA.
A 2 mL sample was loaded onto a Sephacryl S-100 HR, 98 cm × 3.3 cm, gel filtration column from Pharmacia-Amersham Biosciences (Uppsala, Sweden) equilibrated with sample buffer. Collected fractions (1 mL) were evaluated by absorbance at 280 nm and SDS-PAGE. The fractions containing Trx were pooled, concentrated using an Amicon Ultra with 5000 MWCO from Millipore. Purity was > 95% as judged by 15% SDS-PAGE. The Trx concentration was calculated using the extinction coefficient at 280 nm of 13,700 M−1•cm−1 (21).
Enzymatic Characterization of Thioredoxin Reductase
The TR mutants were assayed for activity towards DTNB, Trx, and hydrogen peroxide as described by Arner (22). All assays were performed on a Cary 50 UV/VIS spectrophotometer from Varian (Walnut Creek, CA) at 25 °C, pH 7.0, and were initiated by addition of enzyme. Spectral properties of purified TR were evaluated at 275 nm, 370 nm, 460 nm (summarized in Table S1 in the Supporting Information) as previously described (11) and the concentration of homodimeric TR was determined using the flavin extinction coefficient of 22.6 mM−1•cm−1 (22). Activity was monitored over 2 min with Vo determined from the linear fit. Plots of Vo/ET vs. substrate concentration were fit by the Michaelis-Menten equation using KaleidaGraph 4.02 from Synergy Software (Reading, PA) and activities reported as moles of NADPH consumed per min per mole of TR dimer.
The DTNB assay contained 0.2 mM NADPH and 10 mM EDTA in 100 mM potassium phosphate. For each concentration of DTNB, activity was corrected for background by addition of buffer only. Activity was measured by the increase in absorbance at 412 nm, calculated using the extinction coefficient for TNB-S−(5-thio-2-nitrobenzoate, 13.6 mM−1•cm−1), and divided by two to account for the production of 2TNB-S− per NADPH consumed. The concentration of mTR3 in the assay was 2 nM, 5 nM for DmTR, and 4 nM for CeTR2.
The Trx assay contained 0.15 mM NADPH, 1 mM EDTA, and 10 mg/mL insulin in 50 mM potassium phosphate in a volume of 500 μL. Activity was background corrected for each concentration of Trx by addition of buffer only as well as with enzyme in the absence of substrate. Activity was measured by the decrease in absorbance at 340 nm for the consumption of NADPH and calculated using the extinction coefficient of 6200 M−1•cm−1. The concentration of WT mTR3 in the assay was 2 nM, 25 nM for WT DmTR, and 40 nM for WT CeTR2. The concentration of each mutant TR was adjusted to achieve a similar change in absorbance at 340 nm as that of the respective WT TR. Mutants with poor Trx activity required 0.4 μM to 2.5 μM homodimeric TR in the assay.
The hydrogen peroxide assay for semisynthetic mTR3 is formulated similar to that of the DTNB assay with activity measured at 340nm and background corrected as with the previous assays. The concentration of WT mTR3 in the assay was 50 nM and the concentration of each mutant TR was adjusted to achieve a similar change in absorbance at 340 nm.
pH Optima of Trx Reduction
Activity towards Trx was tested as a function of pH for each construct. Due to the insolubility of insulin below pH 7.0, each assay utilized 500 μM Trx, which is approximately ten times greater than the estimated Km. To avoid differences in ionic strength, each assay contained buffer with a final concentration of 30 mM citrate/ 30 mM Tris/ 30 mM phosphate adjusted from pH 5.5 to 10.5. The concentration of TR in the assay was the same as used for the Trx Michaelis-Menten profile. Each assay contained 0.15 mM NADPH and 1 mM EDTA. The activity was measured at 340nm and background corrected. The data were collected in duplicate and then normalized to the percent of maximal activity for each given mutant and plotted as percent activity vs. pH.
RESULTS
Production of TRs with Modified C-termini
Because the last steps of catalysis occur near the C-terminus of TR and the C-terminal carboxylate has been proposed to be involved in a salt bridge that is important for catalytic function (14, 23), we are able to use an intein to modify the C-terminus in ways that would be impossible with other protein engineering techniques. In this system, the use of “intein-mediated engineering” allows us to convert the C-terminal carboxylate of DmTR into either a thiocarboxylate or a hydroxamic acid as illustrated in Figure 2. The thiocarboxylate form of TR is produced by adding (NH4)2S to the column buffer of the chitin affinity column, with bound TR-intein fusion protein. The sulfide anion causes cleavage of the fusion protein resulting in a thiocarboxylate (18). Similarly, addition of NH2OH to the column buffer will result in cleavage of the fusion protein to produce a protein with a hydroxamic acid functionalized C-terminus. Cleavage of DmTR from the intein using NMA yielded 35 mg of TR while cleavage with (NH4)2S and NH2OH yielded 31 mg and 41 mg of enzyme, respectively. If no cleavage reagent is added to the column buffer, the fusion protein will hydrolyze to a small extent. This control reaction yielded 1.4 mg of TR as a combination of TR and the fusion protein. Based on the above yields, this represents 2.5 – 4.0 % non-specific release or hydrolysis. A 12% SDS-PAGE gel demonstrating the cleavage efficiencies of the various reagents is available in the Supporting Information (Supporting Information Figure S1).
Figure 2.
The use of “intein engineering” to incorporate changes at the C-terminus of DmTR. DmTR is produced as a fusion protein with the VMA1 intein. The fusion protein exists in equilibrium between amide and thioester forms and the thioester form can be liberated from the intein by the addition of small molecules added to the column buffer. Addition of a thiol to the buffer results in a thioester-tagged protein. This thioester will hydrolyze to form the free carboxylic acid. Addition of hydroxylamine (NH2OH) liberates DmTR with the C-terminus being functionalized to a hydroxamic acid, while addition of ammonium sulfide ((NH4)2S) results in the formation of a thiocarboxylate at the C-terminus.
Activity of Semisynthetic Mouse TR Towards Thioredoxin
In order to overcome the barriers to heterologous production of selenoproteins in E. coli (24) we have utilized intein-mediated peptide ligation to join residues 1-487 of mTR3 with the tripeptide Cys-Sec-Gly, to produce a fully functional enzyme (16). Using this technique we have made a number of mutations to the C-terminal tetrapeptide of mTR3. Because these mutants contain an atom of selenium, we were able to determine the efficiency of ligation using ICP-MS to determine the mol % of selenium in each mutant then dividing the molar concentration of selenium by the molar concentration of TR in the sample (16). The efficiency of peptide incorporation is much higher when Cys is in the N-terminal position of the attacking peptide. Presumably, when Sec is in the N-terminal position, the resulting selenoester is much more prone to hydrolysis, resulting in a lower ligation efficiency. The ligation efficiency for each mutant enzyme is summarized in Table 1 along with non-corrected catalytic parameters for the reduction of E. coli Trx.
Table 1.
Thioredoxin reductase activity of semisynthetic mTR3 enzymesa
| Enzyme | kcat (min−1) | Km (μM) | % Peptide Incorporation |
|---|---|---|---|
| b,cTR-G-COO- | No activity | No activity | NA |
| b,dTR-GCCG-COO- | 4.1 ± 0.11 | 49.1 ± 3.2 | NA |
| b,eTR-GCUG-COO- | 2220 ± 78 | 67.6 ± 6 | 91 |
| fTR-GCUG-CONH2 | 3010 ± 351 | 41.6 ± 5.0 | 63 |
| TR-GUCG-COO- | 8.3 ± 0.1 | 36.1 ± 1.3 | 32 |
| TR-GUUG-COO- | 1.2 ± 0.1 | 64.8 ± 16.1 | 10 |
| TR-GCAUG-COO- | 350 ± 14 | 20.8 ± 3.5 | 100 |
| TR-GCAAUG-COO- | 501 ± 41 | 34.9 ± 10.0 | 100 |
| grat TR1 | 3000 | 35 | NA |
Spectral properties of these enzymes are listed in Table S1.
Reported previously in (16).
The truncated form of the enzyme missing the C-terminal tripeptide Cys-Sec-Gly.
The full length mutant Sec489Cys.
The WT enzyme produced by semisynthesis.
The WT enzyme produced by semisynthesis with a C-terminal carboxamide.
The data in Table 1 demonstrates that changing the C-terminal carboxylate of mTR3 to a carboxamide increases the activity of the enzyme. This increase in activity is not limited to the reduction of Trx, this mutant enzyme also has an increased hydrogen peroxidase activity (Table 2) and enhanced DTNB reductase activity as well (Table 3). If the selenium content is normalized for both samples, the carboxamide mutant has nearly twice the activity as the semisynthetic WT-carboxylate enzyme. The position of the Sec residue in mTR3 is important for catalysis as demonstrated by the mutant in the which Sec and Cys residues are “switched” (TR-GUCG)2, showing activity similar to the Sec489Cys mutant (TR-GCCG), 8.3 and 4.1 min−1 respectively. Replacement of the Cys-Sec dyad with a Sec-Sec dyad (TR-GUUG) results in a mutant enzyme with very low catalytic activity. Even if the kcat values were normalized for selenium content, the TR-GUCG and TR-GUUG mutants have 90 fold and 185 fold lower catalytic activity than the WT enzyme. The mutants in which alanine residues were inserted to increase the ring size (TR-GCAUG and TR-GCAAUG) show only a modest decrease in catalytic activity, 6 and 4 fold, respectively.
Table 2.
Hydrogen peroxidase activity of semisynthetic mTR3 enzymes
| Enzyme | kcat (min−1) | Km (mM) | kcat / Km (min−1•mM−1) |
|---|---|---|---|
| bTR-G-COO- | No activity | No activity | NA |
| cTR-GCCG-COO- | 19.6 ± 1.9 | 233 ± 36 | 0.084 |
| dTR-GCUG-COO- | 1753 ± 257 | 259 ± 46 | 6.77 (5.30) a |
| eTR-GCUG-CONH2 | 3204 ± 351 | 167 ± 24 | 19.18 (13.46) a |
| TR-GUCG-COO- | 19.7 ± 0.8 | 9.5 ± 1.2 | 2.07 |
| TR-GUUG-COO- | 6.0 ± 0.7 | 47 ± 10 | 0.128 |
| TR-GCAUG-COO- | 284 ± 3.9 | 14 ± 0.7 | 20.29 |
| TR-GCAAUG-COO- | 125 ± 6.4 | 6.8 ± 1.2 | 18.38 |
The numbers in parentheses are the second order rate constant determined from the linear fit due to poor saturation.
The truncated form of the enzyme missing the C-terminal tripeptide Cys-Sec-Gly.
The full length mutant Sec489Cys.
The WT enzyme produced by semisynthesis.
The WT enzyme produced by semisynthesis with a C-terminal carboxamide.
Table 3.
DTNB reductase activity for semisynthetic mTR3
| Enzyme | kcat (min−1) | Km (mM) | kcat / Km (min−1•mM−1) |
|---|---|---|---|
| aTR-G-COO- | 856 ± 43 | 2.72 ± 0.43 | 314 |
| bTR-GCCG-COO- | 794 ± 78 | 1.75 ± 0.41 | 454 |
| cTR-GCUG-COO- | 1251 ± 71 | 0.46 ± 0.09 | 2720 |
| dTR-GCUG-CONH2 | 3284 ± 133 | 0.13 ± 0.02 | 25260 |
| TR-GUCG-COO- | 751 ± 51 | 1.61 ± 0.34 | 466 |
| TR-GUUG-COO- | 914 ± 18 | 1.84 ± 0.12 | 497 |
| TR-GCAUG-COO- | 1010 ± 26 | 0.26 ± 0.03 | 3885 |
| TR-GCAAUG-COO- | 999 ± 44 | 0.43 ± 0.08 | 2323 |
The truncated form of the enzyme missing the C-terminal tripeptide Cys-Sec-Gly.
The full length mutant Sec489Cys .
The WT enzyme produced by semisynthesis.
The WT enzyme produced by semisynthesis with a C-terminal carboxamide.
pH Rate Profiles of Semisynthetic mTR3 Enzymes
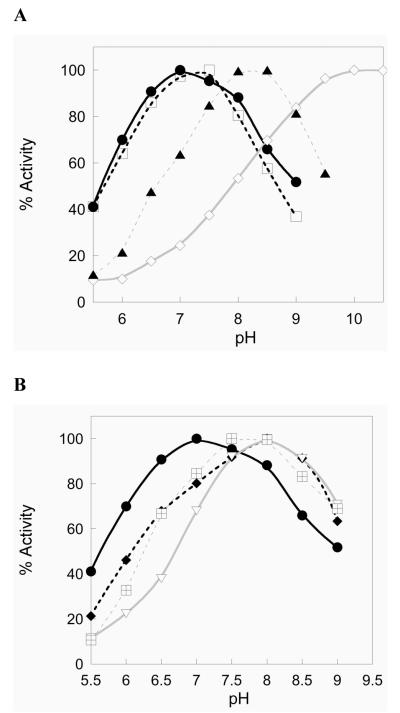
It has been well documented that mutation of the catalytic Sec residue to Cys in the mammalian enzyme results in a shift of the maximal activity towards higher pH (11). As such, part of our mechanistic evaluation of the semisynthetic mutants included an evaluation of TR activity as a function of pH. We also observe a shift from pH 7.0 to pH 8.0 for the Sec to Cys mutant (Figure 3A). The carboxamide mutant (showing higher activity than WT) shows an identical profile to that of the WT-carboxylate enzyme. However, the di-Sec mutant shows a pronounced shift towards pH 10.0 for maximal activity. In contrast to the large pH shift observed for the di-Sec mutant, the TR-GUCG mutant shows only a modest alkaline shift in pH optima (0.5 – 1.0 units higher than the WT enzyme, Figure 3B). The alanine insertion mutants also display an alkaline shift near pH 8.0 in the Trx reductase assay (Figure 3B).
Figure 3.
Activity towards Trx as a function of pH for semisynthetic mTR3. Panel A shows the WT enzyme with the naturally occurring carboxylic acid (closed circle), the WT amino acid sequence produced as a C-terminal carboxamide (open square), the Sec489Cys mutant mutant (closed triangle), and the GUUG mutant (open diamond). Panel B shows the WT-carboxylate enzyme (closed circle) for reference, the GUCG mutant (checkered box), the GCAUG mutant (closed diamond), and the GCAAUG mutant (inverted, open triangle).
Peroxidase Activity of Semisynthetic Mouse TR
Hydrogen peroxidase activity is a characteristic that is unique to the mammalian TR due to the presence of the Sec residue (8, 11, 12), and thus this activity is absent in DmTR and CeTR2. However, H2O2 is a poor substrate for mTR3 as indicated by the high Km values (low mM) and may not be a physiologically relevant activity for the enzyme. Peroxidase activity plots for mammalian TRs reported previously show poor saturation but have been limited to concentrations less than 5 mM of substrate (11, 16). In this study, we have expanded the profile to 70 mM peroxide. The results demonstrate poor saturation for WT-carboxylate and carboxamide forms (Supporting Information Figure S2). We previously reported an activity of 71 ± 7 min−1 with a Km of 6.6 ± 0.5 mM (16) when limiting the analysis to substrate concentrations equivalent to that reported in the literature (11). After repeated extended analysis of the WT-carboxylate enzyme we observe an activity of 1753 ± 257 min−1 with a Km of 259 ± 33 mM using 50 nM of enzyme, while the carboxamide mutant has an activity of 3204 ± 351 min−1 with a Km of 167 ± 17 mM using 20 nM of enzyme (Table 2).
However, as shown in Table 2, the Km values for mutant enzymes TR-GCAUG, TR-GCAAUG, and TR-GUCG are dramatically lower. The interpretation of this data is that, while the WT enzyme displays significant peroxidase activity, the kinetics are more typical of a second order reaction due to minimal binding of the substrate by the enzyme. In contrast the mutants mentioned above display saturation kinetics, typical of enzymes that behave in a Michaelis-Menten fashion. The saturation curves for these mutants can be seen in Figure 4 and are dramatically different than the WT enzyme. One interpretation of this result is that insertion of alanine residues into the dyad creates a binding site for H2O2 that is not present in the WT enzyme. This would explain the saturation behavior of the mutants and the high Km of the WT enzyme.
Figure 4.

Michaelis-Menten plot of the hydrogen peroxidase activity of semisynthetic mouse enzymes. Shown are WT mTR3 (closed circles), the GCAUG mutant (open squares), and the GCAAUG mutant (closed triangles). The dramatic shift in Km is apparent.
We also tested the impact of the concentration of the co-substrate NADPH on the saturation profile since TR has a ping-pong bi-bi mechanism (25). This analysis was done for the WT enzyme and for the TR-GCAUG mutant. The kinetic plots are shown in the Supporting Information as Figure S3A (WT enzyme) and Figure S3B (GCAUG mutant). Detailed information about these plots is provided in Supporting Information Table S2. For each enzyme the saturation curve are similar from 25 μM to 500 μM NADPH. In both cases, progress curves remained linear over the 2 min time frame of the assay with the exception of the assay using 25 μM NADPH and 70 mM peroxide, where linearity was maintained to 90 sec due to NADPH depletion. Thus, the change in Km observed in the mutants is not due to an altered affinity for NADPH.
Thioredoxin Reductase Activity for DmTR and CeTR
The mutants produced for DmTR can be subdivided into those that have higher or lower activity compared to WT DmTR (Table 4). In the case of the mutant enzymes that have increased activity, the results are analogous to mTR3. When the C-terminus of DmTR is changed from a carboxylate to either a thiocarboxylate or a hydroxamic acid, the result is a mutant enzyme with ~1.7 fold increase in activity. The data shows that, like mTR3, either neutralizing the charge at the C-terminus (hydroxamic acid), or changing the charge distribution (thiocarboxylate) results in an increase in the observed activity. Spectral characteristics of WT DmTR and mutant enzymes are reported in the Supporting Information as Table S3.
Table 4.
DmTR thioredoxin reductase activitya
| Enzyme | kcat (min−1) | Km (μM) |
|---|---|---|
| aTR-S-COO- | No activity | No activity |
| bTR-SCCS-COO- | 299.4 ± 7.4 | 173.3 ± 8.1 |
| cTR-SCCS-CONHOH | 513.3 ± 33.3 | 172.5 ± 21.7 |
| dTR-SCCS-COS- | 491.9 ± 19.6 | 67.8 ± 7.3 |
| TR-SCACS-COO- | 2.12 ± 0.3 | 298.3 ± 58.0 |
| TR-SCAACS-COO- | 0.91 ± 0.2 | 166 ± 58.0 |
Spectral properties are listed in Table S2.
The truncated form of the enzyme missing the C-terminal tripeptide Cys-Cys-Ser.
The WT enzyme.
The WT enzyme produced as the C-terminal hydroxamic acid.
The WT enzyme produced as the C-terminal thiocarboxylate.
In contrast to mTR3 where increasing ring size of the Cys-Sec dyad by inserting alanine residues had modest effect on activity (4-6 fold lower), alanine insertion mutants (DmTR-SCACS and DmTR-SCAACS) had greatly reduced activity compared to WT DmTR. Increasing the ring size of the Cys-Cys dyad resulted in a 150 to 300 fold loss in kcat, while the Km was little affected. Similar results are observed for CeTR2, which shows a 145 fold decrease for the single insertion and 90 fold decrease for the double insertion respectively (Supporting Information Table S4). The pH optima for the WT DmTR enzyme is ~7.5 with no change observed with either of the carboxyl variants and an alkaline shift for both of the Ala insertion mutants (Supporting Information Figure S4). These trends are identical to that observed for mTR3. These results indicate that the non-selenium enzymes have a greater dependence on a vicinal disulfide than does the mammalian enzyme for a vicinal selenylsulfide.
Comparison of DTNB Reductase Activities
The small molecule disulfide DTNB has long been used for quantification of free thiols in proteins (26) as well as to evaluate thiol-disulfide exchange reactions (27), as is the process catalyzed by TR. As a substrate of TR, DTNB displays Michaelis-Menten kinetics, but it is not a physiologically relevant substrate, which is reflected by the high Km values that have been previously reported for this substrate (2, 11, 28). It does, however, provide a substrate suitable for evaluating the activity of TR mutants that show little or no activity towards Trx since both the N-terminal and C-terminal disulfide redox centers are capable of reducing DTNB as has been been reported by us (16), and others (2, 28). A good model for either redox center interacting with DTNB has been previously presented (29).
For our truncated form of mTR3 (Table 3), we observe a kcat of 856 ± 43 min−1. This is 68% of the semisynthetic WT enzyme, which has a kcat of 1251 ± 71 min−1. The WT does however, have a significantly lower Km. This is different from what is observed for the C-terminal mutant of rat TR1 (11) or the truncated form of human TR (30), which show poor activity. However, this result is similar to a C-terminal mutant of Plasmodium falciparum TR (PfTR), which had 64% of WT DTNB reductase activity (28). The alanine insertion mutants (TR-GCAUG and TR-GCAAUG) have slightly elevated activities compared to the truncated enzyme, but less than the semisynthetic WT enzyme. The C-terminal carboxamide mutant merits special note as the catalytic efficiency of this enzyme (kcat/Km) is ca. 25,000 min−1•mM−1. This is > 10 fold higher than the WT enzyme when the 28% lower selenium content of this mutant is taken into consideration.
DTNB reductase activity of DmTR is also increased when the negative charge at the C-terminus is either neutralized by converting the carboxylate to a neutral hydroxamic acid, or modulated by conversion to a thiocarboxylate (Table 5). Similar to the Sec-containing mammalian enzyme, the truncated DmTR mutant also shows very high DTNB reductase activity, as do the alanine insertion mutants (TR-SCACS and TR-SCAACS). This same effect is observed in CeTR2, where alanine insertion between the redox-active Cys residues of the C-terminal redox center has very little effect on DTNB reductase activity (Supporting Information Table S5). The Cys-containing DmTR and CeTR2 enzymes have ~12% of the DTNB reductase activity of the Sec-containing mTR3 (kcat of 157 min−1 for DmTR and 134 min−1 for CeTR2, respectively), while their catalytic efficiencies are only 3-fold and 8-fold lower than mTR3, respectively.
Table 5.
DTNB reductase activity for DmTR
| Enzyme | kcat (min−1) | Km (mM) | kcat / Km (min−1•mM−1) |
|---|---|---|---|
| aTR-S-COO- | 178.0 ± 7.0 | 0.75 ± 0.09 | 237 |
| bTR-SCCS-COO- | 157 ± 12.4 | 0.22 ± 0.07 | 713 |
| cTR-SCCS-CONHOH | 230.9 ± 8.6 | 0.14 ± 0.02 | 1650 |
| dTR-SCCS-COS- | 337.2 ± 19.9 | 0.45 ± 0.06 | 748 |
| TR-SCACS-COO- | 187.5 ± 17 | 0.20 ± 0.06 | 935 |
| TR-SCAACS-COO- | 94.9 ± 4.1 | 0.25 ± 0.03 | 380 |
The truncated form of the enzyme missing the C-terminal tripeptide Cys-Cys-Ser.
The WT enzyme.
The WT enzyme produced as the C-terminal hydroxamic acid.
The WT enzyme produced as the C-terminal thiocarboxylate.
DISCUSSION
TRs from higher eukaryotes share a common structure with the flavoprotein GR with the exception of a 16 amino acid C-terminal extension (1). This C-terminal tail contains a unique redox center as it consists of either adjacent Cys residues or or adjacent Cys and Sec residues that either form a vicinal disulfide or vicinal selenylsulfide bond (analogous to a disulfide bond – Supporting Information Figure S5). In this study we have used a combination of protein engineering techniques to investigate the C-terminal redox center of Sec and Cys-containing TRs. This investigation is divided into three areas: ring size, the role of charge at the C-terminus, and requirement for the position of the Sec residue in the mammalian enzyme. These areas are discussed below.
Ring Size
When a disulfide bond forms between adjacent residues, the result is the formation of an 8-membered ring. This type of disulfide is a rare occurrence and has been found in only 32 structures out of 28,000 structures in the Protein Data Bank3 (4, 5). Based upon NMR studies of small proteins where the Cys-Cys(ox) dyad is found, we believe that this unit in TR may be undergoing conformational switching between cis and trans forms of the ring (Figure S5). This is apparently the case in human and bass hepcidin, which also contains this unique ring structure (31, 32). A comparison can be made between this peptidyl 8-membered ring and cyclooctene. Cyclooctene exists in a cis configuration as there is ~10 kcal/mol difference in stability between the cis and trans isomers of cyclooctene (33). By analogy, the peptidic 8- membered ring having partial double bond character (40 %) that restricts rotation about the peptide bond, could also relieve ring strain by adopting a cis geometry. Interestingly, insertion of a selenium atom into this ring should increase the overall ring size and decrease the strain of the ring.
Insertion of either one or two alanine residues in between residues of the Cys-Sec or Cys-Cys dyad of TR results in a larger, less strained ring of either 11 or 14 atoms instead of 8. The kinetic data in Table 1 indicates that these insertions have only a modest effect on the kcat (4-6 fold) of mTR3 when assayed with Trx and minimal impact on Km. In contrast, the data indicates for DmTR that, the same alanine insertion mutants are greatly affected as shown by the large decrease in kcat (150-300 fold, Table 4), while the kcat of CeTR2 (also a Cys-containing TR) is decreased by 90 – 145 fold. Taken together, the data shows that for Sec-containing mTR3, the catalytic impact of ring size is minimal, while for the Cys-containing TRs, the catalytic impact of increasing ring size is significant. Therefore relief of ring strain may not significantly contribute to the catalytic function of mammalian TR.
When the 8-membered ring of the Cys-Cys(ox) dyad is opened, one of the sulfur atoms will be attacked by the interchange Cys and the other sulfur atom will be the leaving group (Figure 1C). In examining the crystal structure of DmTR, we recently proposed that Cys490′ (the C-terminal Cys) was in the leaving group position, while Cys489′ was the site of nucleophilic attack (17). We argued that the intervening peptide bond of this dyad was in a cis conformation and this conformation allowed for proton transfer from His464′ to the thiolate of Cys490′. In contrast, we argued that in the mammalian enzyme, the intervening peptide bond of the Cys-Sec(ox) dyad was in a trans conformation and this placed the selenolate in a position where it could not accept a proton from the general acid (His). However, the low pKa of the selenolate would obviate the need for proton transfer. If the model is correct, then insertion of alanine residues into the dyad would disrupt proton transfer from the catalytic His and result in a loss in activity in the absence of Sec, which is observed experimentally.
We see different results with respect to ring size for the non-physiological substrates H2O2 and DTNB. Enlarging the ring in mTR3 does significantly improve the Km for H2O2 and these mutant enzymes behave with typical Michaelis-Menten kinetics unlike the WT enzyme with this substrate (Table 2, Figure 4). While we see that ring size does significantly impact Trx reductase activity of DmTR and CeTR2, enlarging the ring does not effect DTNB reductase activity to an appreciable extent. The fact that all of the mutants in this study (including the truncated forms of TR) exhibit high DTNB reductase activity indicates the importance of the N-terminal redox center in the reduction of DTNB for these three enzymes.
Comment on DTNB Reductase Activity of the TRs in this Study
In comparing the DTNB reductase activity for the enzymes in this study (Tables 3, 5, and S5), three important conclusions can be made. First, the C-terminal active site is not required for DTNB reductase activity but can affect catalytic efficiency. Secondly, it offers a plausible direct correlation to the results for the reduction of Trx and H2O2 by mTR3. This is accomplished by dividing the results into groups based on catalytic efficiencies (kcat/Km). The C-terminal carboxamide variant has the highest catalytic efficiency for DTNB and the highest Trx activity, while the WT-carboxylate enzyme and Ala insertion mutants have moderate catalytic efficiency for DTNB. The mutants with poor Trx reductase activity have the lowest catalytic efficiency for DTNB and are similar to truncated mTR3, indicating that the DTNB reductase activity from these mutants can be attributed to the N-terminal redox center. The decrease in efficiency is due to the increase in Km, as has been reported for mutants of DmTR (2) and PfTR (28).
We offer an explanation for the varying activities of mutant TRs for DTNB reported in the literature. DTNB has two carboxylate groups as well as a low pKa thiol expected to be unprotonated upon reduction (34, 35). Therefore, the difference in activities between mutant forms of different TRs can be explained by comparing differences in the active site electrostatic surface potentials (shown in Supporting Information Figure S6). The least electronegative potential is for mTR3 (36) (highest DTNB reductase activity) while rat TR1 (14) is the most electronegative (poor DTNB reductase activity (11)). A similar interpretation was made for the poor activity of truncated human TR toward oxidized diglutathione (30).
Role of Charge at the C-terminus
Our use of inteins has allowed us to explore the role of the C-terminal carboxylate in the catalytic mechanism of TR, which is fortuitously placed close to the site of catalysis. In the crystal structure of rat TR1 (14), it was proposed that a salt bridge interaction between the C-terminal carboxylate of mammalian TR and Lys29 (Lys26 in DmTR) was important for maintaining the position of the Cys-Sec(ox) dyad as it approached the N-terminal redox center prior to reduction. The same role was suggested for Arg351 in a modeling study for complex formation between TR and Trx (23). Another role suggested for Lys29 is that of a general acid (W. Brandt, personal communication). In this role, Lys29 would donate a proton to the thiolate of the N-terminal Cys of the Cys-Sec dyad in mammalian TR or the thiolate of the N-terminal Cys of the Cys-Cys dyad in DmTR. If a conventional site-directed mutagenesis experiment were performed on Lys29, then these two roles could not be distinguished if the Lys mutant lost significant activity. In addition, any time a mutant is made there is the additional complication of altering the structure of the protein, making interpretation of the result difficult in such cases.
In this study, we have made the most conservative change possible by creating a semisynthetic mammalian TR that has a C-terminal carboxamide in place of the usual carboxylate. The carboxamide is isosteric with the carboxylate, but is neutral, allowing for an uncomplicated analysis of the resultant mutation. In this case, eliminating the charge at the C-terminus increases the activity of the enzyme over a broad range of substrates (Trx, H2O2, and DTNB). In all of the substrates tested, there is an increase in activity of two fold. Similar results are also observed for DmTR when the C-terminal carboxylate is converted to a hydroxamic acid ( pKa = 8.88) (37) or thiocarboxylate. If the proposed salt bridge were important for aligning the Cys-Sec or Cys-Cys dyad for catalysis, one would expect a significant decrease in activity, which is not observed. Therefore, it seems highly unlikely that the C-terminal carboxylate is involved in an important catalytic electrostatic interaction but could contribute to product release. A hydrogen bond, however, cannot be ruled out.
Position of the Sec Residue in Mammalian TR
Our semisynthetic technique for production of mTR3 has also allowed us to make several other unique changes to the C-terminal redox-active tetrapeptide. In DmTR, there are two Cys residues whose roles in catalysis are difficult to discern using conventional protein engineering techniques. In the mammalian enzyme however, the two residues in this dyad are distinguishable since one position of the dyad is occupied by Cys and the other by Sec. Here we have investigated the importance of the position of Sec in the dyad by creating a Sec-Sec dyad (TR-GUUG) and a switch mutant (TR-GUCG).
The redox potential of the dyad decreases from −326 mV to −385 mV by changing it from a Cys-Sec dyad to a Sec-Sec dyad (38, 39). The very low redox potential of the diselenide bond makes it very difficult to open by the thiol-disulfide exchange chemistry. This mutation resulted in an enzyme with greatly impaired catalytic function (185 fold loss in Trx reductase activity, Table 1). This loss in activity is most likely explained by a large decrease in the ring opening step. The difficulty in opening up this diselenide bond is shown by the very large shift in pH optimum for this mutant. The Sec-Sec mutant has a pH optimum for reduction of Trx near 10, while the WT enzyme is centered on pH 7 (Figure 3A and 3B). The most likely explanation for this dramatic shift in pH optimum for this mutant is that the thiolate (Cys52) that is responsible for initial nucleophilic attack on the dyad must be fully ionized for this to occur, which our data suggests occurs near pH 10 in this mutant (Figure 3A).
It is thought that the Sec residue is the attacking nucleophile in the reduction of Trx. This is so because the selenolate of a Sec residue is much more nucleophilic than a thiolate of a Cys residue. Indirect evidence for this hypothesis is given by the hydrogen peroxidase activity of the enzyme (8, 12), which is abolished upon mutation of Sec to Cys (11). There are two consequences to switching the positions of sulfur and selenium atoms in the dyad. First, the interchange Cys residue (CysIC in Figure 1C) would have to attack a more electronegative atom, which is unfavorable, thus leading to a decrease in rate. This switch would also place the Cys residue in the leaving group position during ring opening, which would also significantly decrease the activity towards all three substrates in this study, due to the higher pKa of the thiolate compared to a selenolate as we have argued previously (17).
The kinetic profile of the hydrogen peroxidase activity of the WT enzyme is more like that of a chemical reaction (2nd order kinetics – Figure 4) rather than an enzymatic reaction. Small organic molecules with a selenium atom also have non-enzymatic peroxidase activity (40). Therefore one would expect the position of Sec to have little impact on peroxidase activity. However, switching positions of the two atoms (S and Se) results in a 30 fold decrease in peroxidase activity when selenium content is normalized – Table 2. The two reasons for the poor activity of the switch mutant in the reduction of Trx given above apply to the hydrogen peroxidase reaction as well. Thus we argue that pathway 1 in Figure 1C should be correct. This model is further supported by the data for the Ala insertion mutants, as these mutants would maintain Cys in the interchange position and Sec in the leaving group position in the ring opening step.
Finally, the results of the pH profiles for Trx reduction are also supportive of this hypothesis. If the ring opening step is responsible for the change in pH optimum, then the interpretation that we have put forth previously, namely that protonation of the leaving group in the ring opening step is partially rate limiting (17), can be used to explain the shift towards basic pH for the switch mutant since the leaving group ability of the thiolate of Cys increases at more basic pH. The data presented here (while not definitive) is supportive of our model for Sec as the leaving group in the ring opening step.
Supplementary Material
Footnotes
These studies were supported by National Institutes of Health Grant GM070742 to RJH.
ABBREVIATIONS ADP, adenosine diphosphate; (NH4)2S, ammonium sulfide; βME, β-mercaptoethanol; CeTR2, mitochondrial TR from Caenorhabditis elegans; CXXC, the tetrapeptide motif Cys-Xaa-Xaa-Cys; CUG, the tripeptide Cys-Sec-Gly; DEAE, diethylaminoethyl; DTNB, 5,5′dithio-bis(2-nitrobenzoic acid); DmTR, TR from Drosophila melanogaster; EDTA, ethylaminediamine tetraacetic acid; Fmoc, 9-fluoroenylmethoxycarbonyl; HPLC, high pressure liquid chromatography; H2O2, hydrogen peroxide; NH2OH, hydroxylamine; ICP-MS, inductively coupled plasma mass spectrometry; IPTG, isopropyl-β-D-thiogalactopyranoside; LB, Luria-Bertani media; Mr, molecular ratio; MOPS, 3-(N-morpholino) propane sulfonic acid; mTR3, mouse mitochondrial thioredoxin reductase; NADPH, β-nicotinamide adenine dinucleotide phosphate, reduced; Ni-NTA, nickel nitrilotriacetic acid; NMA, N-methylmercaptoacetamide; PCR, polymerase chain reaction; PfTR, Plasmodium falciparum TR; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; Sec, selenocysteine; TB, terrific broth; TNB- , thiobis(2-nitrobenzoic acid) anion; TR, thioredoxin reductase; Tris, tris-(hydroxymethyl)aminomethane; Trx, thioredoxin; U, the one letter code for selenocysteine; UUG, the tripeptide Sec-Sec-Gly.
Throughout this study, we use the nomenclature (TR-aa1aa2aa3aa4) as an abbreviation to show the changes we have made to the C-terminal tetrapeptide. In the Tables, we add the designation COO-, CONH2, or COS- to show what changes we have made to the C-terminal carboxyl group.
The study by Perczel and coworkers is the most current study on the number of vicinal disulfide bonds in the PDB. At the time of the publication of (4, 5), there were ca. 28,000 structures in the PDB. There may be more structures and more proteins containing vicinal disulfide bonds, but this comment is limited to known structures deposited in the PDB.
SUPPORTING INFORMATION AVAILABLE There are five tables in the Supporting Information that give spectral properties of mTR3 mutant enzymes, spectral properties of DmTR mutant enzymes, and kinetic data for CeTR2. There are also six figures available. Figure 1 shows a 12% SDS-PAGE gel of the cleavage experiment with DmTR and various cleavage reagents. Figure 2 shows a Michaelis-Menten plot of the hydrogen peroxidase activity of semisynthetic mTR3 enzymes. Figure 3 shows the effect of titrating NADPH on the Km for hydrogen peroxide on mTR3 enzymes. Figure 4 shows the activity dependence on pH for the DmTR mutants. Figure 5 shows a potential conformational switching mechanism for the Cys-Cys dyad. Figure 6 shows the electrostatic potential of tetrapeptide binding pocket of various TRs. This material is available free of charge via the Internet at http://pubs.acs.org
REFERENCES
- 1.Arscott LD, Gromer S, Schirmer RH, Becker K, Williams CH., Jr. The mechanism of thioredoxin reductase from human placenta is similar to the mechanisms of lipoamide dehydrogenase and glutathione reductase and is distinct from the mechanism of thioredoxin reductase from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3621–3626. doi: 10.1073/pnas.94.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer H, Massey V, Arscott LD, Schirmer RH, Ballou DP, Williams CH., Jr. The mechanism of high Mr thioredoxin reductase from Drosophila melanogaster. J. Biol. Chem. 2003;278:33020–33028. doi: 10.1074/jbc.M303762200. [DOI] [PubMed] [Google Scholar]
- 3.Wang PF, Arscott LD, Gilberger TW, Muller S, Williams CH., Jr. Thioredoxin reductase from Plasmodium falciparum: evidence for interaction between the C-terminal cysteine residues and the active site disulfide-dithiol. Biochemistry. 1999;38:3187–3196. doi: 10.1021/bi982674g. [DOI] [PubMed] [Google Scholar]
- 4.Hudaky I, Gaspari Z, Carugo O, Cemazar M, Pongor S, Perczel A. Vicinal disulfide bridge conformers by experimental methods and by ab initio and DFT molecular computations. Proteins. 2004;55:152–168. doi: 10.1002/prot.10581. [DOI] [PubMed] [Google Scholar]
- 5.Carugo O, Cemazar M, Zahariev S, Hudaky I, Gaspari Z, Perczel A, Pongor S. Vicinal disulfide turns. Protein Eng. 2003;16:637–639. doi: 10.1093/protein/gzg088. [DOI] [PubMed] [Google Scholar]
- 6.Blake CC, Ghosh M, Harlos K, Avezoux A, Anthony C. The active site of methanol dehydrogenase contains a disulphide bridge between adjacent cysteine residues. Nat. Struct. Biol. 1994;1:102–105. doi: 10.1038/nsb0294-102. [DOI] [PubMed] [Google Scholar]
- 7.Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. doi: 10.1021/bi9628580. [DOI] [PubMed] [Google Scholar]
- 8.Gladyshev VN, Jeang KT, Stadtman TC. Selenocysteine, identified as the penultimate C-terminal residue in human T-cell thioredoxin reductase, corresponds to TGA in the human placental gene. Proc. Natl. Acad. Sci. U. S. A. 1996;93:6146–6151. doi: 10.1073/pnas.93.12.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanzok SM, Fechner A, Bauer H, Ulschmid JK, Muller HM, Botella-Munoz J, Schneuwly S, Schirmer R, Becker K. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–646. doi: 10.1126/science.291.5504.643. [DOI] [PubMed] [Google Scholar]
- 10.Lacey BM, Hondal RJ. Characterization of mitochondrial thioredoxin reductase from C. elegans. Biochem. Biophys. Res. Commun. 2006;346:629–636. doi: 10.1016/j.bbrc.2006.05.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J. Biol. Chem. 2000;275:18121–18128. doi: 10.1074/jbc.M000690200. [DOI] [PubMed] [Google Scholar]
- 12.Zhong L, Arner ES, Holmgren A. Structure and mechanism of mammalian thioredoxin reductase: the active site is a redox-active selenolthiol/selenenylsulfide formed from the conserved cysteine-selenocysteine sequence. Proc. Natl. Acad. Sci. U. S. A. 2000;97:5854–5859. doi: 10.1073/pnas.100114897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gromer S, Johansson L, Bauer H, Arscott LD, Rauch S, Ballou DP, Williams CH, Jr., Schirmer RH, Arner ES. Active sites of thioredoxin reductases: why selenoproteins? Proc. Natl. Acad. Sci. U. S. A. 2003;100:12618–12623. doi: 10.1073/pnas.2134510100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sandalova T, Zhong L, Lindqvist Y, Holmgren A, Schneider G. Three-dimensional structure of a mammalian thioredoxin reductase: implications for mechanism and evolution of a selenocysteine-dependent enzyme. Proc. Natl. Acad. Sci. U. S. A. 2001;98:9533–9538. doi: 10.1073/pnas.171178698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris KM, Flemer S, Jr., Hondal RJ. Studies on deprotection of cysteine and selenocysteine side-chain protecting groups. J Pept Sci. 2007;13:81–93. doi: 10.1002/psc.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eckenroth B, Harris K, Turanov AA, Gladyshev VN, Raines RT, Hondal RJ. Semisynthesis and characterization of mammalian thioredoxin reductase. Biochemistry. 2006;45:5158–5170. doi: 10.1021/bi0517887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckenroth BE, Rould MA, Hondal RJ, Everse SJ. Structural and Biochemical Studies Reveal Differences in the Catalytic Mechanisms of Mammalian and Drosophila melanogaster Thioredoxin Reductases. Biochemistry. 2007;46:4694–4705. doi: 10.1021/bi602394p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinsland C, Taylor SV, Kelleher NL, McLafferty FW, Begley TP. Overexpression of recombinant proteins with a C-terminal thiocarboxylate: implications for protein semisynthesis and thiamin biosynthesis. Protein Sci. 1998;7:1839–1842. doi: 10.1002/pro.5560070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–14991. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 20.Williams CH, Jr., Zanetti G, Arscott LD, McAllister JK. Lipoamide dehydrogenase, glutathione reductase, thioredoxin reductase, and thioredoxin. J. Biol. Chem. 1967;242:5226–5231. [PubMed] [Google Scholar]
- 21.Holmgren A, Reichard P. Thioredoxin 2: cleavage with cyanogen bromide. Eur. J. Biochem. 1967;2:187–196. doi: 10.1111/j.1432-1033.1967.tb00125.x. [DOI] [PubMed] [Google Scholar]
- 22.Arner ES, Zhong L, Holmgren A. Preparation and assay of mammalian thioredoxin and thioredoxin reductase. Methods Enzymol. 1999;300:226–239. doi: 10.1016/s0076-6879(99)00129-9. [DOI] [PubMed] [Google Scholar]
- 23.Brandt W, Wessjohann LA. The functional role of selenocysteine (Sec) in the catalysis mechanism of large thioredoxin reductases: proposition of a swapping catalytic triad including a Sec-His-Glu state. Chembiochem. 2005;6:386–394. doi: 10.1002/cbic.200400276. [DOI] [PubMed] [Google Scholar]
- 24.Tormay P, Bock A. Barriers to heterologous expression of a selenoprotein gene in bacteria. J. Bacteriol. 1997;179:576–582. doi: 10.1128/jb.179.3.576-582.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gromer S, Arscott LD, Williams CH, Jr., Schirmer RH, Becker K. Human placenta thioredoxin reductase. Isolation of the selenoenzyme, steady state kinetics, and inhibition by therapeutic gold compounds. J. Biol. Chem. 1998;273:20096–20101. doi: 10.1074/jbc.273.32.20096. [DOI] [PubMed] [Google Scholar]
- 26.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 27.Wilson JM, Bayer RJ, Hupe DJ. Structure-Reactivity Correlations for the Thiol-Disulfide Interchange Reaction. J. Am. Chem. Soc. 1977;99:7922–7926. [Google Scholar]
- 28.Krnajski Z, Gilberger TW, Walter RD, Muller S. Intersubunit interactions in Plasmodium falciparum thioredoxin reductase. J. Biol. Chem. 2000;275:40874–40878. doi: 10.1074/jbc.M008443200. [DOI] [PubMed] [Google Scholar]
- 29.Fujiwara N, Fujii T, Fujii J, Taniguchi N. Roles of N-terminal active cysteines and C-terminal cysteine-selenocysteine in the catalytic mechanism of mammalian thioredoxin reductase. J. Biochem., (Tokyo) 2001;129:803–812. doi: 10.1093/oxfordjournals.jbchem.a002923. [DOI] [PubMed] [Google Scholar]
- 30.Urig S, Lieske J, Fritz-Wolf K, Irmler A, Becker K. Truncated mutants of human thioredoxin reductase 1 do not exhibit glutathione reductase activity. FEBS Lett. 2006;580:3595–3600. doi: 10.1016/j.febslet.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 31.Hunter HN, Fulton DB, Ganz T, Vogel HJ. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 2002;277:37597–37603. doi: 10.1074/jbc.M205305200. [DOI] [PubMed] [Google Scholar]
- 32.Lauth X, Babon JJ, Stannard JA, Singh S, Nizet V, Carlberg JM, Ostland VE, Pennington MW, Norton RS, Westerman ME. Bass hepcidin synthesis, solution structure, antimicrobial activities and synergism, and in vivo hepatic response to bacterial infections. J. Biol. Chem. 2005;280:9272–9282. doi: 10.1074/jbc.M411154200. [DOI] [PubMed] [Google Scholar]
- 33.McMurray J. Organic Chemistry. 2nd Edition 1988. p. 173. [Google Scholar]
- 34.Arscott LD, Veine DM, Williams CH., Jr. Mixed disulfide with glutathione as an intermediate in the reaction catalyzed by glutathione reductase from yeast and as a major form of the enzyme in the cell. Biochemistry. 2000;39:4711–4721. doi: 10.1021/bi9926431. [DOI] [PubMed] [Google Scholar]
- 35.Wong KK, Vanoni MA, Blanchard JS. Glutathione reductase: solvent equilibrium and kinetic isotope effects. Biochemistry. 1988;27:7091–7096. doi: 10.1021/bi00418a063. [DOI] [PubMed] [Google Scholar]
- 36.Biterova EI, Turanov AA, Gladyshev VN, Barycki JJ. Crystal structures of oxidized and reduced mitochondrial thioredoxin reductase provide molecular details of the reaction mechanism. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15018–15023. doi: 10.1073/pnas.0504218102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monzyk B, Crumbliss AL. Acid Dissociation Constants (Ka) and Their Temperature Dependencies (delta Ha, delta Sa) for a Series of Carbon- and Nitrogen-Substituted Hydroxamic Acids in Aqueous Solution. J. Org. Chem. 1980;45:4670–4675. [Google Scholar]
- 38.Besse D, Moroder L. Synthesis of selenocysteine peptides and their oxidation to diselenide-bridged compounds. J Pept Sci. 1997;3:442–453. doi: 10.1002/(SICI)1099-1387(199711)3:6%3C442::AID-PSC122%3E3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Beld J, Woycechowsky KJ, Hilvert D. Selenoglutathione: efficient oxidative protein folding by a diselenide. Biochemistry. 2007;46:5382–5390. doi: 10.1021/bi700124p. [DOI] [PubMed] [Google Scholar]
- 40.Muller A, Cadenas E, Graf P, Sies H. A novel biologically active seleno-organic compound--I. Glutathione peroxidase-like activity in vitro and antioxidant capacity of PZ 51 (Ebselen) Biochem. Pharmacol. 1984;33:3235–3239. doi: 10.1016/0006-2952(84)90083-2. [DOI] [PubMed] [Google Scholar]
- 41.Luthman M, Holmgren A. Rat liver thioredoxin and thioredoxin reductase: purification and characterization. Biochemistry. 1982;21:6628–6633. doi: 10.1021/bi00269a003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.