Abstract
The importance of epigenetic gene regulatory mechanisms in normal and cancer development is increasingly evident. Genome-wide analyses have revealed the mutation, deletion, and dysregulated expression of chromatin-modifying enzymes in a number of cancers, including hematologic malignancies. Genome-wide studies of DNA methylation and histone modifications are beginning to reveal the landscape of cancer-specific chromatin patterns. In parallel, recent genetic loss-of-function studies in murine models are demonstrating functional involvement of chromatin-modifying enzymes in malignant cell proliferation and self-renewal. Paradoxically, the same chromatin modifiers can, depending on cancer type, be either hyperactive or inactivated. Increasingly, cross talk between epigenetic pathways is being identified. Leukemias carrying MLL rearrangements are quintessential cancers driven by dysregulated epigenetic mechanisms in which fusion proteins containing N-terminal sequences of MLL require few or perhaps no additional mutations to cause human leukemia. Here, we review how recent progress in the field of epigenetics opens potential mechanism-based therapeutic avenues.
Introduction
The completion of the human genome project and the development of next-generation sequencing technologies have profoundly changed the study of cancer. Recent years have witnessed the accumulation of unprecedented amounts of genomic data, and the catalog of both genetic and epigenetic alterations in cancer has grown significantly. In parallel, the importance of chromatin modifications (or epigenetic mechanisms) in the regulation of tumor-associated gene expression is becoming better understood, prompting the development of therapeutic approaches that target these mechanisms in cancer cells. Epigenetic gene regulatory mechanisms can be broadly classified into chromatin remodeling, DNA cytosine methylation, and covalent histone modifications. Additional mechanisms such as non-coding RNA are also increasingly recognized. Large-scale sequencing projects of human cancer genomes have provided evidence that many epigenetic regulators are deleted and/or mutated in cancer,1,2 suggesting an interplay between genetics and epigenetics in cancer development. Interestingly, mutations in a given epigenetic regulator can be activating or inactivating,3-5 suggesting tumor promoting or tumor suppressor functions for the same gene product depending on cellular context. How mutations in epigenetic modifiers contribute to cellular transformation on a mechanistic level remains incompletely understood in most cancers. However, it is likely that mutations in epigenetic modifiers result in molecular vulnerabilities that can, pending more detailed mechanistic understanding, be exploited for future targeted therapies. Importantly, epigenetic modifications are potentially reversible. Many of the players involved are enzymes, which are generally considered more targetable by small-molecule drugs than other classes of molecules, such as transcription factors. Indeed, there is already precedence for efficacious clinical application of therapies directed toward epigenetic mechanisms. Hypomethylating agents such as 5-aza-2′deoxycytidine (decitabine) and 5-azacytidine (Vidaza) are approved for the treatment of myelodysplastic syndromes, and the histone deacetylase inhibitors vorinostat and romidepsin are approved for the treatment of cutaneous T-cell lymphoma. There is great interest in expanding the indications for these agents, and multiple clinical studies are ongoing. This success has spurred interest in the development of additional drugs that target epigenetic mechanisms. An emerging theme in epigenetics research is the concepts of “writers,”6 “readers”7 (or “binders”), and “erasers.”8 All three may open exciting avenues for targeted therapies as exemplified by recent studies that investigate writers such as DOT1L,9-11 readers such as the acetyl-binding BET-domain protein BRD4,12-14 or erasers such as the histone demethylase LSD1.15
Acute leukemia carrying a rearrangement of the mixed lineage leukemia (MLL) gene on chromosome band 11q23 is a prototypical cancer driven by epigenetic mechanisms. Typically, the 5′ end of the MLL gene is fused in frame to the 3′ portion of one of more than 70 known translocation partners. An alternative mechanism of transformation involves a partial tandem duplication of exons near the 5′ end of the MLL gene that may function via somewhat different mechanisms and will not be discussed here further. MLL rearrangements are observed in approximately 5% to 10% of acute myeloid leukemia (AML) and in >70% of infant acute lymphoblastic leukemia (ALL). They occur de novo or after chemotherapy exposure (typically topoisomerase 2 inhibitors such as etoposide or, less commonly, anthracyclines). The most common translocation partners are AF9 in AML and AF4 in ALL. MLL rearrangements are associated with standard risk in AML and poor prognosis in ALL. Event-free survival is approximately 50% for both MLL-rearranged de novo AML and ALL (which translates into an average-risk prognostic category for AML and a poor-risk category for pediatric ALL). These outcome statistics are unsatisfactory and illustrate the need for better therapies for these leukemias. Secondary disease arising from myelodysplastic syndrome or after exposure to topoisomerase 2 inhibitors carries a particularly dismal prognosis, even after allogeneic stem cell transplantation. Careful analysis has begun to delineate an impact of the fusion partner on outcome.16 Other risk factors such as overexpression of EVI1 have also been described.17,18
The past few years have seen a greatly increased understanding of the pathophysiology of MLL-rearranged leukemia. Here, we summarize some of the most recent findings and focus on mechanisms with therapeutic implications; we specifically discuss two aspects of MLL-rearranged leukemias: the direct effects of the leukemogenic fusion and the interaction of the MLL fusion with other epigenetic regulatory systems.
MLL proteins in gene expression
Wild-type (WT) MLL is a histone methyltransferase with specificity for lysine 4 on histone 3 (H3K4). MLL contains a catalytic SET domain also found in the position effect variegation modifier (Su(var)3-9), the Polycomb-group protein E(z), and the trithorax-group protein Trx). In humans, there are 3 types of complexes with H3K4 methylating activity: SETD1A/B-containing complexes (related to yeast COMPASS) are responsible for most cellular H3K4 trimethylation (H3K4me3).19 MLL (MLL1/KMT2A) or MLL4 (KMT2B, also referred to as MLL2)-containing complexes, which are related to Drosophila Trx-containing complexes, play an important role in HOX gene regulation. MLL3 (KMT2C) or MLL2 (KMT2D; also referred to as MLL4) are paralogues with homology to Drosophila Trx-related (trr). They have been implicated in nuclear receptor–mediated gene activation via locus-specific catalysis of H3K4me3 and more recently, enhancer regulation (see “Other members of the MLL family in cancer and leukemia”). A fifth member of the MLL family, MLL5, is more closely related to SETD5 than to other MLL genes and will not be discussed further here. The phylogeny and function of the different complexes have recently been reviewed.20 WT MLL proteins are large and contain multiple functional domains. In mice, knock-in of sequences encoding a truncated Mll protein without histone methyltransferase activity results in a milder phenotype than complete disruption of Mll function,21 and knockdown of Mll in mouse embryonic fibroblasts leads to decrease of H3K4me3 in only a minor proportion of genes.19 The Set-, Trx-, and Trr-containing complexes all contain multiple proteins (including multiple epigenetic modifiers), as do their mammalian counterparts. Studies in knockout mice have demonstrated that Mll is important for embryonic development, body patterning, and proper Hox gene expression in mice. A conditional knockout mouse has revealed that Mll is strictly required for normal adult hematopoietic stem cells.22 MLL2 (KMT2D) has recently received attention because it is mutated in several different cancers, with a particularly high incidence in lymphoma, and the related MLL3 is frequently mutated in a number of solid tumors (eg, medulloblastoma23).
Mechanisms of MLL fusion–mediated transformation
The available evidence suggests that MLL fusion proteins generated by MLL translocations function by transcriptionally upregulating approximately 100 target genes. The best characterized direct binding targets of MLL fusions are distal HoxA cluster genes and Meis1. Overexpression of HoxA9 and Meis1 is sufficient for leukemic transformation of mouse bone marrow, which suggests a critical role for HoxA cluster genes and Meis1 in MLL fusion–mediated transformation. The specific role of the other MLL fusion binding targets is incompletely understood. Forced expression of MLL-AF9 or combined forced expression of HoxA9 and Meis1 leads to in vivo leukemia arising from transduced hematopoietic stem cells. However, only MLL-AF9, but not the combination of HoxA9-Meis1 efficiently transforms committed granulocyte-macrophage progenitors, and this re-establishing of self-renewal on a non–self-renewing population (granulocyte-macrophage progenitors)24 and the development of drug resistance25 have been linked to activation of the β-catenin pathway by MLL-AF9, independent of HoxA9 and Meis1 function. How β-catenin signaling is activated by MLL-AF9 remains to be determined, although it appears that cell intrinsic, rather than niche-derived Wnt signals, plays a predominant role.26 These data suggest that modulation of β-catenin signaling may be of therapeutic value in MLL-rearranged leukemia. The individual contribution to cellular transformation by individual direct MLL-AF9 binding targets other than HoxA9 and Meis1 is less well characterized. Interestingly, some of the direct binding targets of MLL-AF9 are in fact tumor suppressors (eg, Cdkn1b).
Recruitment of MLL fusion proteins
WT MLL is recruited to chromatin via several protein-protein interactions that involve menin/LEDGF,27,28 PAF,29,30 and PHD29,31 fingers. A candidate therapeutic strategy is to interfere with recruitment of MLL fusions to chromatin. Fusion proteins invariably lose the PHD fingers, and artificial fusions, including the WT MLL PHD fingers, lose their transforming activity.32 Importantly, structural data suggest a role of the PHD finger in shutting off the WT MLL program during normal hematopoietic differentiation.33 Pharmacologic interference with the menin-MLL fusion interaction has shown promise as a therapeutic strategy for MLL-rearranged leukemia.34 Interfering with the CXXC-PAF interaction may also be a viable strategy,30 although a chemical inhibitor of this interaction has not been reported to the best of our knowledge.
Transcriptional upregulation mediated by MLL fusion proteins
The precise mechanism MLL fusions use to upregulate target genes is incompletely understood, but significant progress has been made. In leukemogenic fusions, the catalytic SET domain, which is located at the C-terminus, is invariably lost; however, MLL dimerizes, and WT MLL is required for appropriate target locus histone modifications, including H3K4me3, H3K79me2, and the transforming activity of MLL-AF9 in a murine model.35 Interestingly, the Polycomb-group protein Cbx8, traditionally implicated in transcriptional silencing via Polycomb repressive complex (PRC)1, has been shown to be required for full induction of MLL fusion target gene expression.36 This function is independent of PRC1 and instead involves the histone acetyl transferase Tip60, which is a modulator of an Myc transcriptional program found to be important in embryonic stem cells37 and MLL-rearranged leukemias.15,38,39 It is also noteworthy that MLL binds a number of protein complexes implicated in transcriptional elongation, including EAP,40 AEP,41 and SEC.42 P-TEFb (CDK9/cyclinT1) is found in these partially overlapping elongation-associated protein complexes that are important for MLL fusion–induced leukemia in mouse models.41,42 Small-molecule inhibitors of CDK9, such as the flavonoids, are in various stages of preclinical and clinical development for other malignancies and may represent a therapeutic opportunity for MLL-rearranged leukemias.
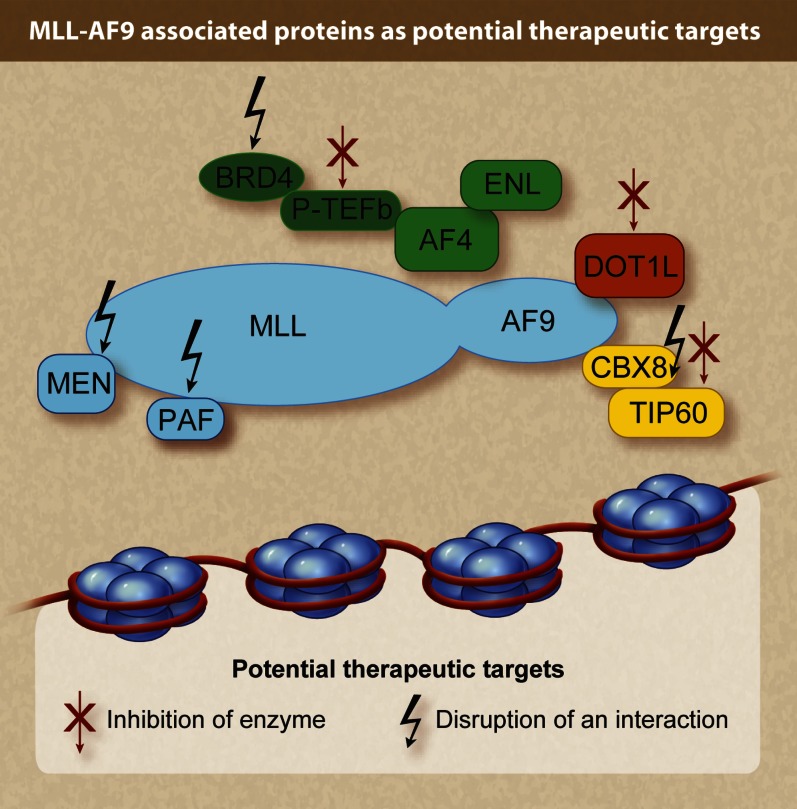
The histone methyltransferase DOT1L has been reported to be part of transcriptional elongation–associated multiprotein complexes EAP (but not AEP/SEC), and there are conflicting data regarding the simultaneous versus sequential recruitment of P-TEFb and DOT1L to chromatin. DOT1L is the only known methyltransferase for lysine 79 in histone 3 (H3K79me1/2/3) and was originally discovered as a disruptor of telomeric silencing in yeast.43,44 DOT1L was originally implicated in leukemia when it was shown to interact with the MLL fusion partner AF10 in a yeast two-hybrid screen.45 Abnormally high K79me2 signal was then found to be associated with genes directly bound by the more common MLL fusion protein MLL-AF4 in murine and human leukemias, suggesting that DOT1L may have a broader role in MLL- rearranged leukemias independent of the fusion partner.46,47 Importantly, in genetic loss-of-function mouse models, Dot1l is required for leukemia initiation and maintenance by MLL-AF99-11,48 and by 3 additional particularly aggressive fusions: MLL-AF10, CALM-AF10,49 and MLL-AF6.50 Interestingly, AF6 is a cytosolic protein that does not appear to bind any of the nuclear DOT1L-containing protein complexes but mediates dimerization of the fusion,51 a feature found to be sufficient for MLL-mediated transformation in some model systems. Genetic or pharmacologic inactivation of Dot1l leads to the collapse of an MLL fusion–driven transcriptional program that is upregulated via MLL fusion binding.9 These genetic studies coincided with the development of small-molecule DOT1L inhibitors that have been shown to selectively target MLL-rearranged leukemia cell lines.52,53 Consequently, the clinical evaluation of DOT1L inhibitors has been initiated in a phase 1 study (NCT01684150). Several important questions arise from these studies: (1) Are there resistance mechanisms to DOT1L inhibition? (2) How will cooperating mutations in genes such as N-RAS and FLT3 influence response to DOT1L inhibitors in vivo? and (3) Is there preferential activity on human leukemic stem cells over normal hematopoietic stem cells? Another important question for future preclinical studies is whether there are other malignancies sensitive to DOT1L inhibition. Improved molecular understanding of the direct action of MLL fusions, while still incomplete, has led to the discovery of several potential targeted therapies in varying stages of preclinical and clinical development (Figure 1). Given the toxicities of conventional therapeutic regimens for AML and MLL-rearranged ALL, the advent of these new areas of investigation is most welcome.
Figure 1.
Schematic of the MLL-AF9 fusion and select binding partners. Arrows indicate potential molecular therapeutic targets. Red arrows indicate targets amenable to enzymatic inhibition. Black arrows indicate targets amenable to inhibition of protein-protein interaction. (Professional illustration by Debra T. Dartez.)
Interaction of MLL fusions with other epigenetic systems
There is a growing appreciation of the interplay between different epigenetic systems. Epigenetic readers, writers, and erasers are mutated, amplified, deleted, or transcriptionally dysregulated in many cancers. We limit ourselves here to a brief discussion of DNA methylation and PRC2 in the context of MLL-fusion leukemias.
DNA cytosine methylation
DNA methylation in mammals typically occurs in the carbon 5 position of the cytosine ring, in the context of CpG dinucleotides, leading to the formation of 5-methylcytosine (5mC). There is ample evidence for dysregulation of DNA methylation in cancer, including hematologic malignancies, with important therapeutic implications.54 Here, we highlight a few points pertinent to MLL leukemia. Genome-wide analysis of DNA methylation patterns has revealed additional insights not provided by the study of the methylation status of individual gene loci (such as CDKN2A).55,56 In AML, DNA methylation programs mostly segregate with known cytogenetic groups. However, novel subgroups of AML not previously suspected on the basis of their cytogenetics have been identified on the basis of the clustering of DNA methylation patterns. There is also a set of genes aberrantly methylated in all AML. Interestingly, AML can be globally either hypo- or hypermethylated,55 demonstrating that too much or too little activity of an epigenetic pathway may contribute to leukemogenesis; this theme is also evident in the study of Polycomb proteins. Finally and importantly, DNA methylation patterns are linked to patient survival, suggesting clinical relevance to the detailed study of aberrant epigenetic changes.56 Direct binding of DNA methyltransferases (DNMTs) by prognostically relevant transcription factors such as EVI117 has been reported, and hypermethylation associated with refractory disease has been linked to overexpression of EVI1 in patients17,18 and in mouse models.57
Recent data further underscore the functional importance of aberrant DNA methylation in murine models of MLL-AF9–mediated AML: clinical samples of MLL-rearranged leukemias show pronounced global hypomethylation.55 This is mirrored by hypomethylation of murine MLL-AF9–mediated AML, with more chemosensitive progenitor-derived MLL-AF9 AML being hypomethylated compared with more resistant stem cell–derived disease.57 It is unknown whether the changes in methylation simply reflect the leukemic cell of origin or are causally related to the observed differences in latency and chemosensitivity. A more detailed functional understanding awaits whole genome bisulfite sequencing coupled with genome-wide analysis of hydroxymethylcytosine (which bisulfite sequencing cannot distinguish from methylcytosine). Of clinical interest, genetically engineered haploinsufficiency for the maintenance DNMT Dnmt1 in murine MLL-AF9 AML leads to a striking loss of self-renewal.58
These data imply that there is an optimal level of methylation for MLL-AF9 leukemia and suggest benefit to the clinical use of hypomethylating agents in MLL-rearranged AML. Another interesting finding in this study was the enrichment of derepressed genes in bivalent PRC2 targets, marked by both activating H3K4 trimethylation (placed by one of the MLL- or SET-containing complexes) and repressive H3K27 trimethylation (placed by PRC2, containing either enhancer of Zeste 2 [EZH2] or EZH1 as the active methyltransferase).58
A link between another important mechanism, the H3K9me2/3 axis, and DNA cytosine methylation has also been suggested.59,60 A recent elegant study demonstrated that forced occupation of HP1a on a transgene via a chemical inducer of dimerization can lead to H3K9 methylation. Prolonged but not short-term HP1 occupation also leads to silencing by DNA methylation, and pharmacologic inhibition of DNMTs leads to an expected reversal of DNA methylation, and also, less expectedly, to reversal of H3K9 methylation.61 More work is needed to analyze the genome-wide interplay of different epigenetic silencing mechanisms because this area clearly has therapeutic implications.
Polycomb genes
Polycomb-group genes are important developmental regulators involved in body patterning and HOX gene regulation. They are of considerable interest in cancer epigenetics. Several different Polycomb complexes have been described, and we limit ourselves here to highlighting how PRC2 intersects with MLL.
PRC2 consists of the core components embryonic ectoderm development (EED), suppressor of Zeste 12 (SUZ12), and a catalytic component, which can be an EZH2 or its less well characterized cousin, EZH1,62,63 which can partially compensate Ezh2 inactivation by preventing loss of H3K27me3 at some but not all PRC2 target gene loci.39,63 Another protein, JARID2, is involved in recruitment of the complex to target loci64-66 and is inactivated in human leukemia.67 Other factors also associate with the PRC2 complex.66 The canonical function of PRC2 is transcriptional repression of bound genes via trimethylation of histone 3 on lysine residue 27 (H3K27me3). A chromatin compacting function has been ascribed to EZH1.62 However, it should be noted that both EZH1 binding68,69 and EZH2 binding70 have recently been associated with transcriptional activation. Furthermore, at least some histone methyltransferases can methylate nonhistone substrates. This phenomenon is best characterized for SETD7.71 A function for Ezh2 in cardiac development through methylation-mediated direct inactivation of the transcription factor Gata4 has also recently been described for Ezh2.72 Thus, it is likely that EZH2 can control cell state programs and perhaps cancer development at multiple levels.
Oncogenic function of PRC2
Early studies assessing EZH2 in cancer found it to be overexpressed in cultured mantle cell lymphoma cell lines.73 EZH2 overexpression is associated with disease progression in prostate cancer,74 a function that was recently reported to be independent of PRC2/H3K27me3.75 Overexpression of EZH2 was also found in other solid tumors in follow-up studies, and forced expression of EZH2 was shown to mediate enhanced self-renewal76 and proliferation.77 A role for EZH2 in cell cycle progression of cultured cells, downstream of E2F, has also been characterized.78 In cancers of the hematopoietic system, both hyperactivity and inactivation of EZH2 have been described. There is great interest in the development of PRC2 inhibitors for use in cancer therapy, and DZNep, reportedly a PRC2 inhibitor, was recently shown to have activity in cancer models79 and specifically in leukemia.80 However, the specificity of this molecule has been called into question.81 Subsequent studies using more potent and specific EZH2 inhibitors have shown significant inhibition of cellular proliferation in lymphoma cell lines that harbor activating EZH2 mutations.82-84
We and others recently investigated the role of PRC2 in MLL-AF9–dependent AML, and found that Eed, but not Ezh2, is strictly required for MLL-AF9 AML.39,85 The inactivation of PRC2 components Ezh239,85 and Eed39,86 leads to reduction and complete loss of self-renewal, respectively, in murine MLL-AF9 leukemia. The precise mechanistic underpinnings of the role of Ezh2/PRC2 in MLL-AF9 leukemia (gene repression vs gene activation and critically important target genes) are currently unknown. Given the apparent inverse correlation between leukemogenic potency of MLL-AF9 cells and degree of Cdkn2a derepression,39 Cdkn2a, a known PRC2-repressed locus, is an important candidate locus. Double knockout of Ezh2/Eed and Cdkn2a is likely to shed more light on this issue. Normal adult hematopoietic stem cells are dependent on Ezh1,87 but not Ezh2,88 apparently via a Cdkn2a-dependent mechanism. Interestingly, a phenotype similar to the one observed after Ezh2 inactivation in MLL-AF9 leukemia, including loss in transcription of Myc targets and upregulation of PRC2 targets, was recently described for short hairpin RNA (shRNA) –mediated and pharmacologic loss-of-function studies of the histone demethylase Lsd1.15 Of note, the long noncoding RNA HOTAIR (previously implicated in PRC2 recruitment and cancer) has been described to physically link EZH2 and LSD1 proteins.89 The functional significance of this is presently unknown but may merit further investigation. MYC is an important gene in many cancers and has been validated as a potential therapeutic target in genetic models. However, direct pharmacologic interference with MYC has proven difficult. Recently, indirect modulation of MYC levels by genetic and pharmacologic interference with the epigenetic reader of acetylated histones BRD4 has been demonstrated.12-14 It appears that inhibition of epigenetic readers such as BRD4 and possibly CBX8, writers such as PRC2, and erasers such as LSD1 may open additional opportunities for targeting MYC, a long elusive therapeutic target. In summary, genetic models have demonstrated an important relationship between MLL and other epigenetic systems, pointing toward potential therapies.
Tumor suppressor function of Ezh2
Inactivating mutations in EZH2 have been described in myelodysplastic syndrome, myeloproliferative disease, and T-cell ALL and are correlated with adverse prognosis. Inactivation of Ezh2 in mice leads to T-cell ALL.90 The precise mechanisms by which EZH2 contributes to oncogenesis vs tumor suppression are unknown and deserve further study.
Other members of the MLL family in cancer and leukemia
Recurrent mutations in different epigenetic modifiers have been found in many cancers, including MLL family members. A query of the COSMIC database (performed in April 2013) reveals that there are relatively few mutations of SETD1A and SETD1B (68 and 12 unique samples, respectively, with simple mutations). There are also relatively few reported samples with simple mutations in MLL (139) and MLL4 (KMT2B; 89). However, there are substantially more frequent mutations of trr-homolog MLL family members MLL2 (KMT2D; 321) and MLL3 (370), suggesting a major tumor suppressive effect of these enzymes.23,91 What is the oncogenic mechanism of MLL2/MLL3 mutations? An only partially redundant role for MLL3 and MLL2 in the activation of p53 target genes has been reported.92 In insects, Trr functions in nuclear receptor signaling,93 and recent data from the analysis of genome-wide MLL2-binding and MLL2-inactivated cell lines support a role for MLL2 in both p53 and nuclear receptor signaling.94 Very recent data show a role for trr and Utx in enhancer function.95 Given that multiple gene products with documented or suspected involvement in enhancer function/long-range enhancer-promotor interactions (MLL2/4, CBP, UTX, CTCF, cohesin complex members) have recently been shown to be mutated in cancer, it is tempting to speculate that there may be a unifying theme. Active enhancers appear to be associated with defined chromatin modifications (H3K4me1 and H3K27Ac),96 at least in part mediated by Trr (in Drosophila)95 and p300,96 respectively. Prior to full activation, they pass through a “poised” stage (H4me1, K27me3) reminiscent of the earlier described poised promoter marked by H3K4me3 and H3K27me3. The removal of K27me3 on some enhancers is performed by Utx97 and is required for the appropriate activation of developmental programs. It may be that a similar mechanism is responsible for differentiation block in hematopoietic and other cancers, a common and widely appreciated feature of malignancy. Future studies will undoubtedly shed more light on this issue.
Concluding remarks and outlook
Epigenetic mechanisms are clearly important in cancer. New data from the past few years illustrate 2 important concepts.
First, MLL-rearranged leukemias provide a paradigm for how epigenetic dysregulation can lead to cancer through inappropriate chromatin structure with subsequent activation of target genes with oncogenic activity. An improved molecular understanding of how MLL fusions upregulate binding targets has led to the identification of a number of potential mechanism-based therapeutic vulnerabilities for this poor-prognosis malignancy (Figure 1). We have focused here on therapeutic targets related to the recruitment and transcriptional effects of MLL fusions. Other strategies such as FLT3-kinase inhibition and interference with HOXA9/MEIS1 transcriptional activities98,99 are also being explored, and their discussion here has been omitted due to space constraints. Both MLL translocations and MLL family member mutations are associated with, and probably mediate, a cellular differentiation block. We believe that future studies investigating the precise mechanisms involved are likely to uncover additional therapeutic opportunities.
Second, epigenetic pathways do not exist in a vacuum, and there is functional crosstalk. A better understanding of this crosstalk may aid in the development of more precisely targeted epigenetic therapies. For example, there may be synergies between the inhibition of PRC2 and DNA methylation. Given the frequent mutations in epigenetic modifiers, it seems likely that these mutations might sensitize the affected cancer cell to interventions that could be less toxic to cells with normal chromatin structure and regulation. Future studies will help further develop this important concept. Another understudied area concerns dynamic changes in the epigenetic makeup of cancer cells in response to external stimuli. An epigenetic drug persister phenotype in response to a targeted therapy (gefitinib) has been demonstrated.100 Deconstructing the complicated interplay between different epigenetic regulatory mechanisms and other targeted therapeutic interventions in genetically defined murine models is likely to provide the required mechanistic insight for developing the next generation of therapeutics that target epigenetic mechanisms.
Acknowledgments
The authors thank Kathrin Bernt for critically reviewing the manuscript.
T.N. was supported by grant K08CA154777 from the National Cancer Institute and by Children’s Hospital Colorado Research Institute. S.A.A. was supported by grants P01CA66996, U01CA105423, and R01CA140575 from the National Cancer Institute and by the American Cancer Society, Gabrielle’s Angel Foundation, and the Leukemia and Lymphoma Society.
Authorship
Contribution: T.N. and S.A.A. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott A. Armstrong, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: armstros@mskcc.org.
References
- 1.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Haaften G, Dalgliesh GL, Davies H, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41(5):521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107(49):20980–20985. doi: 10.1073/pnas.1012525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ernst T, Chase AJ, Score J, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 5.Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet. 2010;42(8):665–667. doi: 10.1038/ng.620. [DOI] [PubMed] [Google Scholar]
- 6.Greer EL, Shi Y. Histone methylation: a dynamic mark in health, disease and inheritance. Nat Rev Genet. 2012;13(5):343–357. doi: 10.1038/nrg3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yun M, Wu J, Workman JL, Li B. Readers of histone modifications. Cell Res. 2011;21(4):564–578. doi: 10.1038/cr.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pedersen MT, Helin K. Histone demethylases in development and disease. Trends Cell Biol. 2010;20(11):662–671. doi: 10.1016/j.tcb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 9.Bernt KM, Zhu N, Sinha AU, et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell. 2011;20(1):66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117(25):6912–6922. doi: 10.1182/blood-2011-02-334359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jo SY, Granowicz EM, Maillard I, Thomas D, Hess JL. Requirement for Dot1l in murine postnatal hematopoiesis and leukemogenesis by MLL translocation. Blood. 2011;117(18):4759–4768. doi: 10.1182/blood-2010-12-327668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson MA, Prinjha RK, Dittmann A, et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature. 2011;478(7370):529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478(7370):524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris WJ, Huang X, Lynch JT, et al. The histone demethylase KDM1A sustains the oncogenic potential of MLL-AF9 leukemia stem cells. Cancer Cell. 2012;21(4):473–487. doi: 10.1016/j.ccr.2012.03.014. [DOI] [PubMed] [Google Scholar]
- 16.Balgobind BV, Raimondi SC, Harbott J, et al. Novel prognostic subgroups in childhood 11q23/MLL-rearranged acute myeloid leukemia: results of an international retrospective study. Blood. 2009;114(12):2489–2496. doi: 10.1182/blood-2009-04-215152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lugthart S, Figueroa ME, Bindels E, et al. Aberrant DNA hypermethylation signature in acute myeloid leukemia directed by EVI1. Blood. 2011;117(1):234–241. doi: 10.1182/blood-2010-04-281337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lugthart S, van Drunen E, van Norden Y, et al. High EVI1 levels predict adverse outcome in acute myeloid leukemia: prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood. 2008;111(8):4329–4337. doi: 10.1182/blood-2007-10-119230. [DOI] [PubMed] [Google Scholar]
- 19.Wang P, Lin C, Smith ER, et al. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol. 2009;29(22):6074–6085. doi: 10.1128/MCB.00924-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shilatifard A. The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem. 2012;81:65–95. doi: 10.1146/annurev-biochem-051710-134100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terranova R, Agherbi H, Boned A, Meresse S, Djabali M. Histone and DNA methylation defects at Hox genes in mice expressing a SET domain-truncated form of Mll. Proc Natl Acad Sci USA. 2006;103(17):6629–6634. doi: 10.1073/pnas.0507425103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6(3):437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- 23.Parsons DW, Li M, Zhang X, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331(6016):435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y, Krivtsov AV, Sinha AU, et al. The Wnt/beta-catenin pathway is required for the development of leukemia stem cells in AML. Science. 2010;327(5973):1650–1653. doi: 10.1126/science.1186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung J, Esposito MT, Gandillet A, Zeisig BB, Griessinger E, Bonnet D, So CW. β-Catenin mediates the establishment and drug resistance of MLL leukemic stem cells. Cancer Cell. 2010;18(6):606–618. doi: 10.1016/j.ccr.2010.10.032. [DOI] [PubMed] [Google Scholar]
- 26.Lane SW, Wang YJ, Lo Celso C, et al. Differential niche and Wnt requirements during acute myeloid leukemia progression. Blood. 2011;118(10):2849–2856. doi: 10.1182/blood-2011-03-345165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yokoyama A, Cleary ML. Menin critically links MLL proteins with LEDGF on cancer-associated target genes. Cancer Cell. 2008;14(1):36–46. doi: 10.1016/j.ccr.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yokoyama A, Somervaille TC, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 29.Milne TA, Kim J, Wang GG, et al. Multiple interactions recruit MLL1 and MLL1 fusion proteins to the HOXA9 locus in leukemogenesis. Mol Cell. 2010;38(6):853–863. doi: 10.1016/j.molcel.2010.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muntean AG, Tan J, Sitwala K, et al. The PAF complex synergizes with MLL fusion proteins at HOX loci to promote leukemogenesis. Cancer Cell. 2010;17(6):609–621. doi: 10.1016/j.ccr.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang PY, Hom RA, Musselman CA, et al. Binding of the MLL PHD3 finger to histone H3K4me3 is required for MLL-dependent gene transcription. J Mol Biol. 2010;400(2):137–144. doi: 10.1016/j.jmb.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muntean AG, Giannola D, Udager AM, Hess JL. The PHD fingers of MLL block MLL fusion protein-mediated transformation. Blood. 2008;112(12):4690–4693. doi: 10.1182/blood-2008-01-134056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Song J, Milne TA, Wang GG, Li H, Allis CD, Patel DJ. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141(7):1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grembecka J, He S, Shi A, et al. Menin-MLL inhibitors reverse oncogenic activity of MLL fusion proteins in leukemia. Nat Chem Biol. 2012;8(3):277–284. doi: 10.1038/nchembio.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thiel AT, Blessington P, Zou T, et al. MLL-AF9-induced leukemogenesis requires coexpression of the wild-type Mll allele. Cancer Cell. 2010;17(2):148–159. doi: 10.1016/j.ccr.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan J, Jones M, Koseki H, Nakayama M, Muntean AG, Maillard I, Hess JL. CBX8, a polycomb group protein, is essential for MLL-AF9-induced leukemogenesis. Cancer Cell. 2011;20(5):563–575. doi: 10.1016/j.ccr.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Woo AJ, Chu J, et al. A Myc network accounts for similarities between embryonic stem and cancer cell transcription programs. Cell. 2010;143(2):313–324. doi: 10.1016/j.cell.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somervaille TC, Matheny CJ, Spencer GJ, et al. Hierarchical maintenance of MLL myeloid leukemia stem cells employs a transcriptional program shared with embryonic rather than adult stem cells. Cell Stem Cell. 2009;4(2):129–140. doi: 10.1016/j.stem.2008.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neff T, Sinha AU, Kluk MJ, et al. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc Natl Acad Sci USA. 2012;109(13):5028–5033. doi: 10.1073/pnas.1202258109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mueller D, Bach C, Zeisig D, et al. A role for the MLL fusion partner ENL in transcriptional elongation and chromatin modification. Blood. 2007;110(13):4445–4454. doi: 10.1182/blood-2007-05-090514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoyama A, Lin M, Naresh A, Kitabayashi I, Cleary ML. A higher-order complex containing AF4 and ENL family proteins with P-TEFb facilitates oncogenic and physiologic MLL-dependent transcription. Cancer Cell. 2010;17(2):198–212. doi: 10.1016/j.ccr.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin C, Smith ER, Takahashi H, et al. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37(3):429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singer MS, Kahana A, Wolf AJ, et al. Identification of high-copy disruptors of telomeric silencing in Saccharomyces cerevisiae. Genetics. 1998;150(2):613–632. doi: 10.1093/genetics/150.2.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Feng Q, Wang H, Ng HH, Erdjument-Bromage H, Tempst P, Struhl K, Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr Biol. 2002;12(12):1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- 45.Okada Y, Feng Q, Lin Y, et al. hDOT1L links histone methylation to leukemogenesis. Cell. 2005;121(2):167–178. doi: 10.1016/j.cell.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 46.Krivtsov AV, Feng Z, Lemieux ME, et al. H3K79 methylation profiles define murine and human MLL-AF4 leukemias. Cancer Cell. 2008;14(5):355–368. doi: 10.1016/j.ccr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guenther MG, Lawton LN, Rozovskaia T, et al. Aberrant chromatin at genes encoding stem cell regulators in human mixed-lineage leukemia. Genes Dev. 2008;22(24):3403–3408. doi: 10.1101/gad.1741408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang MJ, Wu H, Achille NJ, et al. Histone H3 lysine 79 methyltransferase Dot1 is required for immortalization by MLL oncogenes. Cancer Res. 2010;70(24):10234–10242. doi: 10.1158/0008-5472.CAN-10-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen L, Deshpande AJ, Banka D, et al. Abrogation of MLL-AF10 and CALM-AF10-mediated transformation through genetic inactivation or pharmacological inhibition of the H3K79 methyltransferase Dot1l. Leukemia. 2013;27(4):813–822. doi: 10.1038/leu.2012.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Deshpande AJ, Chen L, Fazio M, et al. Leukemic transformation by the MLL-AF6 fusion oncogene requires the H3K79 methyltransferase Dot1l. Blood. 2013;121(13):2533–2541. doi: 10.1182/blood-2012-11-465120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liedtke M, Ayton PM, Somervaille TC, Smith KS, Cleary ML. Self-association mediated by the Ras association 1 domain of AF6 activates the oncogenic potential of MLL-AF6. Blood. 2010;116(1):63–70. doi: 10.1182/blood-2009-09-243386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu W, Chory EJ, Wernimont AK, et al. Catalytic site remodelling of the DOT1L methyltransferase by selective inhibitors. Nat Commun. 2012;3:1288. doi: 10.1038/ncomms2304. [DOI] [PubMed] [Google Scholar]
- 54.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11(10):726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akalin A, Garrett-Bakelman FE, Kormaksson M, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8(6):e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Figueroa ME, Lugthart S, Li Y, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 2010;17(1):13–27. doi: 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Krivtsov AV, Figueroa ME, Sinha AU, et al. Cell of origin determines clinically relevant subtypes of MLL-rearranged AML. Leukemia. 2013;27(4):852–860. doi: 10.1038/leu.2012.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trowbridge JJ, Sinha AU, Zhu N, Li M, Armstrong SA, Orkin SH. Haploinsufficiency of Dnmt1 impairs leukemia stem cell function through derepression of bivalent chromatin domains. Genes Dev. 2012;26(4):344–349. doi: 10.1101/gad.184341.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23(1):138–149. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smallwood A, Estève PO, Pradhan S, Carey M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21(10):1169–1178. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hathaway NA, Bell O, Hodges C, Miller EL, Neel DS, Crabtree GR. Dynamics and memory of heterochromatin in living cells. Cell. 2012;149(7):1447–1460. doi: 10.1016/j.cell.2012.03.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Margueron R, Li G, Sarma K, et al. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32(4):503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen X, Liu Y, Hsu YJ, et al. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell. 2008;32(4):491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pasini D, Cloos PA, Walfridsson J, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–310. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 65.Peng JC, Valouev A, Swigut T, Zhang J, Zhao Y, Sidow A, Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen X, Kim W, Fujiwara Y, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puda A, Milosevic JD, Berg T, et al. Frequent deletions of JARID2 in leukemic transformation of chronic myeloid malignancies. Am J Hematol. 2012;87(3):245–250. doi: 10.1002/ajh.22257. [DOI] [PubMed] [Google Scholar]
- 68.Mousavi K, Zare H, Wang AH, Sartorelli V. Polycomb protein Ezh1 promotes RNA polymerase II elongation. Mol Cell. 2012;45(2):255–262. doi: 10.1016/j.molcel.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stojic L, Jasencakova Z, Prezioso C, et al. Chromatin regulated interchange between polycomb repressive complex 2 (PRC2)-Ezh2 and PRC2-Ezh1 complexes controls myogenin activation in skeletal muscle cells. Epigenetics Chromatin. 2011;4:16. doi: 10.1186/1756-8935-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee ST, Li Z, Wu Z, et al. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol Cell. 2011;43(5):798–810. doi: 10.1016/j.molcel.2011.08.011. [DOI] [PubMed] [Google Scholar]
- 71.Pradhan S, Chin HG, Estève PO, Jacobsen SE. SET7/9 mediated methylation of non-histone proteins in mammalian cells. Epigenetics. 2009;4(6):383–387. doi: 10.4161/epi.4.6.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.He A, Shen X, Ma Q, et al. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012;26(1):37–42. doi: 10.1101/gad.173930.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Visser HP, Gunster MJ, Kluin-Nelemans HC, et al. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br J Haematol. 2001;112(4):950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- 74.Varambally S, Dhanasekaran SM, Zhou M, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419(6907):624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- 75.Xu K, Wu ZJ, Groner AC, et al. EZH2 oncogenic activity in castration-resistant prostate cancer cells is Polycomb-independent. Science. 2012;338(6113):1465–1469. doi: 10.1126/science.1227604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kamminga LM, Bystrykh LV, de Boer A, et al. The Polycomb group gene Ezh2 prevents hematopoietic stem cell exhaustion. Blood. 2006;107(5):2170–2179. doi: 10.1182/blood-2005-09-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Herrera-Merchan A, Arranz L, Ligos JM, de Molina A, Dominguez O, Gonzalez S. Ectopic expression of the histone methyltransferase Ezh2 in haematopoietic stem cells causes myeloproliferative disease. Nat Commun. 2012;3:623. doi: 10.1038/ncomms1623. [DOI] [PubMed] [Google Scholar]
- 78.Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 2003;22(20):5323–5335. doi: 10.1093/emboj/cdg542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tan J, Yang X, Zhuang L, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fiskus W, Wang Y, Sreekumar A, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009;114(13):2733–2743. doi: 10.1182/blood-2009-03-213496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Miranda TB, Cortez CC, Yoo CB, et al. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8(6):1579–1588. doi: 10.1158/1535-7163.MCT-09-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.McCabe MT, Ott HM, Ganji G, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 83.Knutson SK, Wigle TJ, Warholic NM, et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat Chem Biol. 2012;8(11):890–896. doi: 10.1038/nchembio.1084. [DOI] [PubMed] [Google Scholar]
- 84.Qi W, Chan H, Teng L, et al. Selective inhibition of Ezh2 by a small molecule inhibitor blocks tumor cells proliferation. Proc Natl Acad Sci USA. 2012;109(52):21360–21365. doi: 10.1073/pnas.1210371110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tanaka S, Miyagi S, Sashida G, et al. Ezh2 augments leukemogenecity by reinforcing differentiation blockage in acute myeloid leukemia. Blood. 2012;120(5):1107–1117. doi: 10.1182/blood-2011-11-394932. [DOI] [PubMed] [Google Scholar]
- 86.Shi J, Wang E, Zuber J, et al. The Polycomb complex PRC2 supports aberrant self-renewal in a mouse model of MLL-AF9;Nras(G12D) acute myeloid leukemia. Oncogene. 2013;32(7):930–938. doi: 10.1038/onc.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hidalgo I, Herrera-Merchan A, Ligos JM, et al. Ezh1 Is Required for Hematopoietic Stem Cell Maintenance and Prevents Senescence-like Cell Cycle Arrest. Cell Stem Cell. 2012;11(5):649–662. doi: 10.1016/j.stem.2012.08.001. [DOI] [PubMed] [Google Scholar]
- 88.Mochizuki-Kashio M, Mishima Y, Miyagi S, et al. Dependency on the polycomb gene Ezh2 distinguishes fetal from adult hematopoietic stem cells. Blood. 2011;118(25):6553–6561. doi: 10.1182/blood-2011-03-340554. [DOI] [PubMed] [Google Scholar]
- 89.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Simon C, Chagraoui J, Krosl J, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26(7):651–656. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Morin RD, Mendez-Lago M, Mungall AJ, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476(7360):298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lee J, Kim DH, Lee S, et al. A tumor suppressive coactivator complex of p53 containing ASC-2 and histone H3-lysine-4 methyltransferase MLL3 or its paralogue MLL4. Proc Natl Acad Sci USA. 2009;106(21):8513–8518. doi: 10.1073/pnas.0902873106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sedkov Y, Cho E, Petruk S, et al. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426(6962):78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Guo C, Chang CC, Wortham M, et al. Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways. Proc Natl Acad Sci USA. 2012;109(43):17603–17608. doi: 10.1073/pnas.1208807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Herz HM, Mohan M, Garruss AS, et al. Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev. 2012;26(23):2604–2620. doi: 10.1101/gad.201327.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470(7333):279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee S, Lee JW, Lee SK. UTX, a histone H3-lysine 27 demethylase, acts as a critical switch to activate the cardiac developmental program. Dev Cell. 2012;22(1):25–37. doi: 10.1016/j.devcel.2011.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang Z, Smith KS, Murphy M, Piloto O, Somervaille TC, Cleary ML. Glycogen synthase kinase 3 in MLL leukaemia maintenance and targeted therapy. Nature. 2008;455(7217):1205–1209. doi: 10.1038/nature07284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang Z, Iwasaki M, Ficara F, et al. GSK-3 promotes conditional association of CREB and its coactivators with MEIS1 to facilitate HOX-mediated transcription and oncogenesis. Cancer Cell. 2010;17(6):597–608. doi: 10.1016/j.ccr.2010.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141(1):69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]