Abstract
Zinc finger homeodomain enhancer-binding protein (Zfhep/Zfhx1a) is a transcription factor essential for immune system development, skeletal patterning, and life. Regulation of the interleukin 2 gene in T cells has been suggested to depend on post-translational processing of Zfhep, however, no modifications of Zfhep are known. Here we demonstrate that Zfhep is present in both hyperphosphorylated and hypophosphorylated forms. Western analysis demonstrates two forms of Zfhep with different mobilities. Differences in phosphorylation are sufficient to explain the difference in mobilities. Zfhep is primarily phosphorylated on Ser and Thr residues since PP2A dephosphorylates the slower mobility band. Treatment of nuclear extract with O-GlcNAcase did not detect O-linked sugar. Importantly, post-translational processing is cell-specific. Doublets of Zfhep were detected in five cell lines, whereas 6 cell lines contain only, or predominantly, non-phosphorylated Zfhep, and Saos-2 cells contain predominantly the phosphorylated form. These data provide the first demonstration that Zfhep is post-translationally modified.
Keywords: ZEB, deltaEF1, AREB6, Zfhx1a, phosphatase 2A, vomitoxin, COS, C2C12, CHO
INTRODUCTION
The activity of many transcription factors is physiologically regulated by post-transcriptional modifications, including phosphorylation and O-linked glycosylation. For example, phosphorylation of CREB is essential for transcriptional activation (1). Even nuclear receptors, which are ligand-dependent transcription factors, are ligand-independently activated by phosphorylation. In many cases, extracellular signals regulate the activity of kinases and phosphatases that modify transcription factors (2). Phosphorylation of transcription factors can regulate various activities, including DNA-binding, as observed with Dlx3 (3), CREB (4), GATA-4 (5, 6), Nur-77 (7), and c-Myb (8). Transcription factors can also be dynamically glycosylated with O-linked N-acetylglucosamine (O-GlcNAc) on Ser or Thr residues. GlcNAcylation can occur in the nucleus, and regulate the activity of transcription factors, such as Sp1 and c-Myc (9–11).
Zfhep (Zfhx1a/Nil-2a/AREB6/ZEB/δEF-1/BZP) is a member of an unusual family of transcription factors having multiple zinc fingers and also having one or more homeodomains (12–21). Zfhep is essential for differentiation or proliferation of a very early T cell precursor in the embryo (22), as well as craniofacial development, and skeletal patterning (23). Zfhep has been suggested to be a repressor of myogenesis (24) since cotransfection of myoD and Zfhep cDNAs shows that Zfhep effectively blocks myoD-induced myogenesis or transcriptional activation in vitro (25, 26). We recently demonstrated that the expression pattern of Zfhep is consistent with a role in development of the brain and proliferation of brain progenitor cells (27).
In addition to its role in early differentiation, Zfhep participates in regulation of interleukin-2 (IL-2) in mature T cells. Prior to T cell activation, Zfhep binds a negative regulatory element (NRE-A) in the IL-2 gene and represses transcription (21). Activation of T cells increases the binding of transcriptional activators (such as NF-κB and AP1) to the IL-2 gene, and decreases Zfhep binding to NRE-A (28). The mycotoxin deoxynivalenol induces the IL-2 gene in part by inhibition of I-κB (28), and inhibition of Zfhep/ZEB binding to NRE-A (29). Deoxynivalenol decreases binding to NRE-A without blocking expression of Zfhep, indicating a post-translational mechanism. However, no modifications of Zfhep have been described to date.
While transfection of exogenous Zfhep has been extensively used to study the role of Zfhep, nothing is known about the regulation of Zfhep protein activity. Zfhep is a repressor of transcription at certain target genes (17, 19, 20, 25, 26, 30–33), and yet is reported to activate transcription of other target genes (34–36). Activation of the vitamin D receptor gene by Zfhep/ZEB is cell-specific (35). The molecular mechanisms underlying the choice between activation or repression by Zfhep are unknown, but may include cell-specific differences in post-translational modification. We demonstrate that the Zfhep protein is expressed in both a hyperphosphorylated, and a hypophosphorylated form. These two forms are differentially expressed in different cell types.
MATERIALS AND METHODS
Cell lines
COS-7 and CV-1 (green monkey kidney lines), U138 MG (human glioblastoma), JEG-3 (human choriocarcinoma), C2C12 (mouse myoblast), Saos-2 (human osteosarcoma), Neuro2a (mouse neuroblastoma), F9 (mouse embryonal carcinoma), SK-N-SH (human neuroblastoma), GH3, GH4C1, and αT3-1 (rat pituitary tumor lines), PC12 (pheochromocytoma), CHO (Chinese hamster ovary), and HEPM (human embryonic palatal mesenchyme) cells were obtained from American Type Culture Collection (ATCC, Rockville, MD), Dr. William Young (CHO), or Dr. P. L. Mellon (αT3-1). Cells were propagated at 37°C in 5% CO2 with Dulbecco’s modified Eagle’s medium (DMEM, Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum, 100 U/ml penicillin and 100 μg/ml streptomycin, or alpha minimum essential medium (αMEM, Invitrogen) containing 10 μg/ml ribonucleosides and deoxyribonucleosides and antibiotics.
Nuclear extracts
Nuclear extracts were prepared by the method of Schrieber et al. (37) with modifications. Cells grown in T150 flasks to 90–100% confluency were washed with 10 ml ice-cold 10 mM Tris pH 7.4, 150 mM NaCl (TBS) and scraped in TBS with 1 mM PMSF. Cells were pelleted by centrifuging 1500 × g at 4°C for 5 minutes and washed in ice cold TBS. The cells were resuspended in 800 μl ice cold Buffer A (10 mM Hepes pH 7.9, 10 mM KCl, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 1 × protease inhibitors having 10 μg/ml each of the following: antipain, aprotinin, chymostatin, leupeptin and pepstain A (ICN Pharmaceuticals, Costa Mesa, CA)) and allowed to swell on ice for 15 minutes then lysed with 0.625% NP-40. After vortexing vigorously for 10 seconds, the lysate was centrifuged for 45 seconds at 4°C and 16,000 × g. To extract the nuclear protein, the nuclear pellet was resuspended in 200 μl ice-cold Buffer C (20 mM Hepes pH 7.9, 400 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1 mM DTT, 1 mM PMSF, 1 × protease inhibitors). The tubes were mixed on a rotator 30 minutes at 4°C, then centrifuged for 15 minutes at 4°C and 16,000 × g. The supernatant was dialyzed against 250 volumes dialysis buffer (20 mM Hepes, 50 mM KCl, 20% glycerol, 0.5 mM DTT, 0.5 mM PMSF) overnight. The dialyzed extract was centrifuged for 30 minutes at 4°C and 16,000 × g. The soluble supernatant was either used immediately or frozen in aliquots at −80°C.
Enzymatic treatment of Zfhep
Zfhep was dephosphorylated to determine whether it was post-translationally modified by phosphorylation in certain cell types (38). U138MG nuclear extract (50 μg) was brought to 50 mM Hepes pH 7.9, 1 mM MgCl2, and 1 × protease inhibitor mix and pre-incubated for 10 minutes at 30°C. 20 U of Calf Intestinal alkaline Phosphatase (CIP, Roche Molecular Biochemicals, Indianapolis, IN), or 0.3 U of human purified protein phosphatase 2A (PP2A, Upstate Biotechnology, Inc., Lake Placid, NY) was then added and the sample incubated an additional 15 minutes at 30°C. To demonstrate that the changes in electrophoretic mobility of Zfhep were due to the dephosphorylating activity of the CIP enzyme, and not to contaminating proteases or glycosidases, samples were incubated with CIP in the presence of 100 mM sodium phosphate, an inhibitor of CIP activity. Dephosphorylation reactions were terminated by the addition of 20 μl of 4 × SDS sample buffer followed by boiling for 5 minutes. Changes in the electrophoretic migration of Zfhep in mock-treated and phosphatase-treated samples were visualized by immunoblotting. The presence of O-linked glycosylation was tested by incubation for 18 hours at 37°C with N-acetyl-D glucosaminidase (O-GlcNAcase; Roche) in 0.1% SDS, 50 mM citric phosphate pH 4.
Immunoblotting
Codons for amino acids 557–663 encompassing the Zfhep homeodomain (HD) were cloned into pGEX-2TK (Amersham Pharmacia, Piscataway, NJ). Bacterially expressed peptide was purified and injected into rabbits to raise antibodies to rat Zfhep protein (27, 39). Zfhep protein was visualized by western blotting. Protein samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride membrane. The blot was blocked in 0.5% Tween 20 and 5% non-fat dried milk in TTBS (20 mM Tris pH 7.5, 500 mM NaCl, 0.05% Tween 20) and washed in TTBS. Primary antibody was diluted 1:6,000 in TTBS and incubated with the blot overnight. The blot was then washed extensively in TTBS. Goat anti-rabbit horseradish peroxidase conjugated secondary antibody was diluted 1:10,000 in TTBS and incubated with the blot for 1 hour. The blot was then rinsed twice and washed for 30 minutes, 15 minutes, and four times at 5 minutes in TTBS. The blot was reacted with the chemiluminescent substrate Luminol (Pierce, Rockford, IL) for 10 minutes and exposed to Biomax MR film (Kodak, New Haven, CT).
RESULTS
Western blot of nuclear proteins separated on an 8% polyacrylamide gel decorated protein that is present in COS-7 and CHO (Figure 1a), with a molecular mass of approximately 180 kDa. This is consistent with earlier reports with the chicken (18, 26), mouse and hamster (17) orthologues of Zfhep, and is significantly higher than the calculated molecular mass (124 kDa). Preimmune serum did not label protein in nuclear extracts (Figure 1a). We used a lower percentage acrylamide SDS-PAGE to further define the size of Zfhep. Analysis of COS-7 cell nuclear extracts on 4% SDS-PAGE demonstrated the presence of two separate immunoreactive proteins. The upper band has a relative mobility of 185 kDa whereas the lower band is 180 kDa (Figure 1b). The doublet was observed in multiple, independent COS-7 nuclear extracts, including different preparations of protease inhibitors. The doublet was present in COS-7 cells from either low density or confluent cultures, and was also observed in nuclear extracts from U138 cells.
Figure 1.
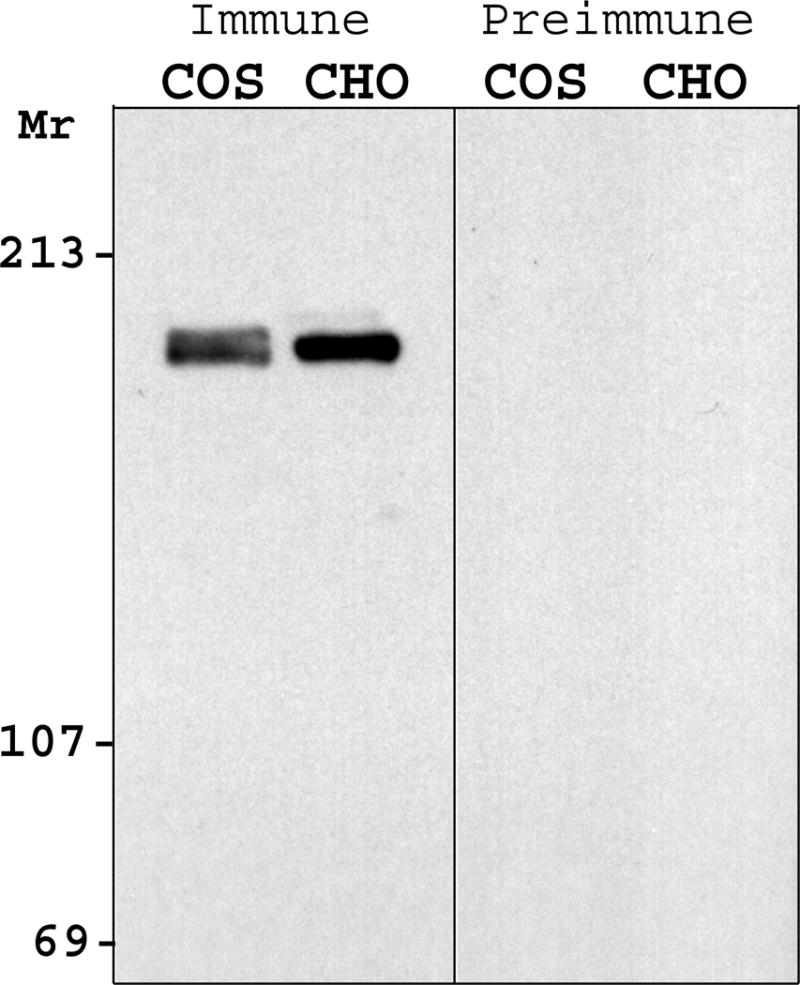
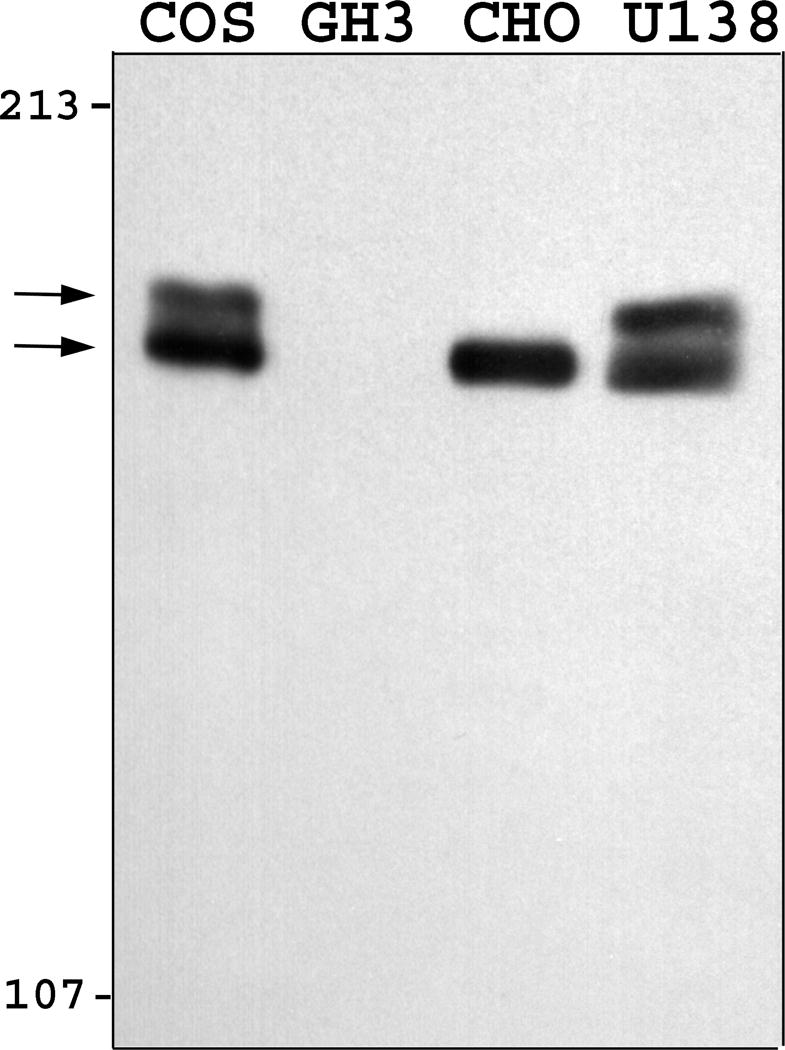
Zfhep is expressed as a doublet. Nuclear extracts were separated on SDS-PAGE with either (A) 10% polyacrylamide, or (B) 4% polyacrylamide gels. Western analysis was done with either the anti-Zfhep antiserum, or preimmune serum. Zfhep protein appears as a doublet in COS-7 and U138MG cells (but not CHO cells) only with the 4% gel.
To investigate the cause of the Zfhep doublet, we determined whether dephosphorylation or deglycosylation alters the mobility of the bands. Nuclear extract from U138MG cells was incubated with, or without, calf intestinal phosphatase to remove phosphate groups. Mock incubation did not alter the ratio of the two bands in the Zfhep doublet, however, addition of CIP caused a complete disappearance of the upper band, and an increased intensity of the lower band (Figure 2a). Incubation with CIP in the presence of phosphate buffer (to block phosphatase activity) demonstrated that the loss of the upper band was specifically due to the phosphatase activity of CIP, not any contaminating protease (Figure 2a). CIP treatment of COS-7 nuclear extracts also collapsed the Zfhep doublet into a single band (Figure 2b). CIP treatment of CHO nuclear extract does not change the alignment of the single band on the immunoblots (not shown). In an independent set of experiments, treatment of COS nuclear extract with protein phosphatase 2A also diminished the upper band (Figure 3). PP2A is active at phospho-Ser and phospho-Thr, but not phospho-Tyr. These data demonstrate that the slower mobility anti-Zfhep-reactive protein is phosphorylated, whereas the higher mobility form of Zfhep is not. Therefore, we tested whether the presence of the hypophosphorylated band was an artifact of dephosphorylation by endogenous phosphatases during cell lysis. To address this question, nuclear extracts were isolated in the presence of 100 mM phosphate buffer to block endogenous phosphatase activity, however, this did not shift the ratio of the two bands (not shown). Many nuclear proteins have O-linked GlcNAc moieties, therefore, we tested whether deglycosylation also altered the mobility of the Zfhep bands. Treatment of COS-7 nuclear extract with O-GlcNAcase had no effect on either the mobility or the ratio of the two Zfhep bands (Figure 4). In summary, differences in phosphorylation are sufficient to explain the different mobilities of the two forms of Zfhep.
Figure 2.
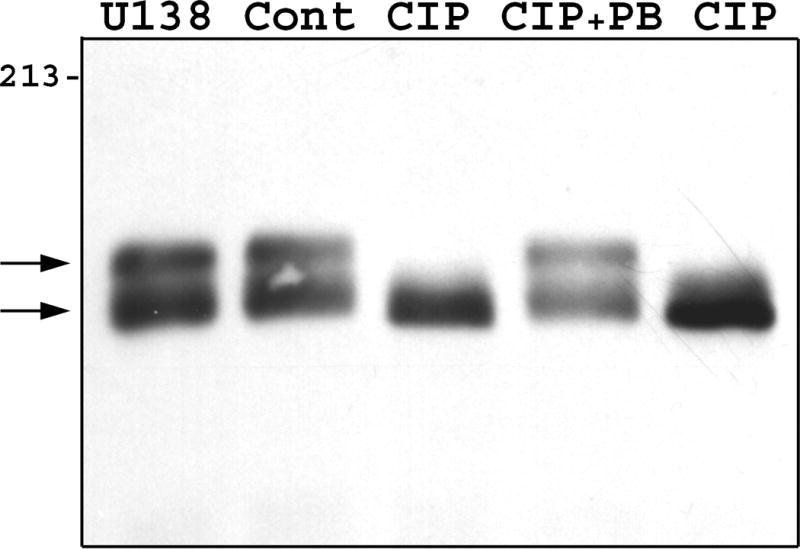
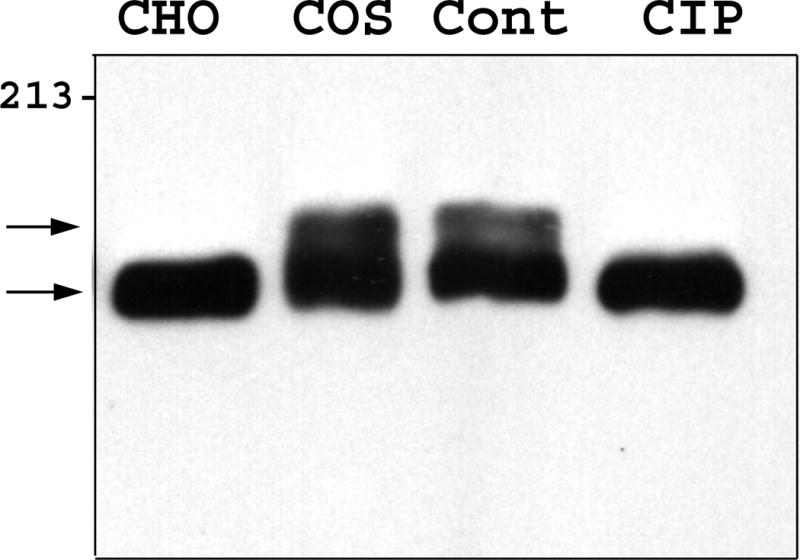
The difference in mobility of the two Zfhep bands is caused by phosphorylation. A.) U138MG nuclear extract was incubated with or without CIP prior to western analysis with anti-Zfhep antiserum. Lane 1: untreated U138 proteins, Lane 2: control incubation, Lane 3: CIP, Lane 4: CIP in 100 mM phosphate buffer, Lane 5: CIP in HEPES buffer. B.) Nuclear extract from COS-7 cells was pretreated as described prior to immunoblotting with anti-Zfhep. Lane 1: untreated CHO nuclear extract, Lane 2: untreated COS-7 nuclear extract, Lane 3: control incubation of COS extract, Lane 4: CIP incubation of COS extract.
Figure 3.
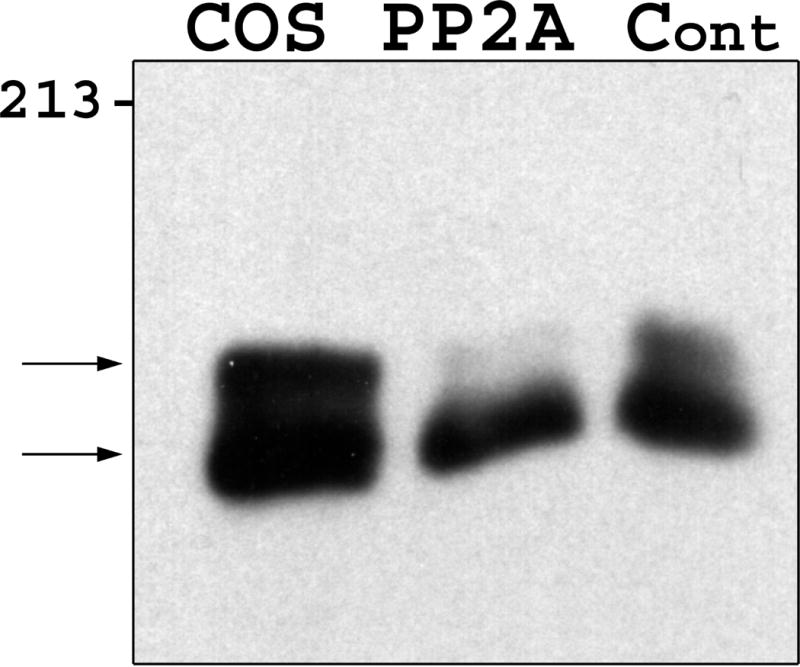
Zfhep is phosphorylated on Ser/Thr residues. Nuclear extract from COS-7 was incubated with or without PP2A prior to immunoblotting. PP2A dephosphorylates pSer or pThr residues, but not pTyr. Lane 1: untreated COS nuclear extract, Lane 2: PP2A incubation, Lane 3: control incubation.
Figure 4.
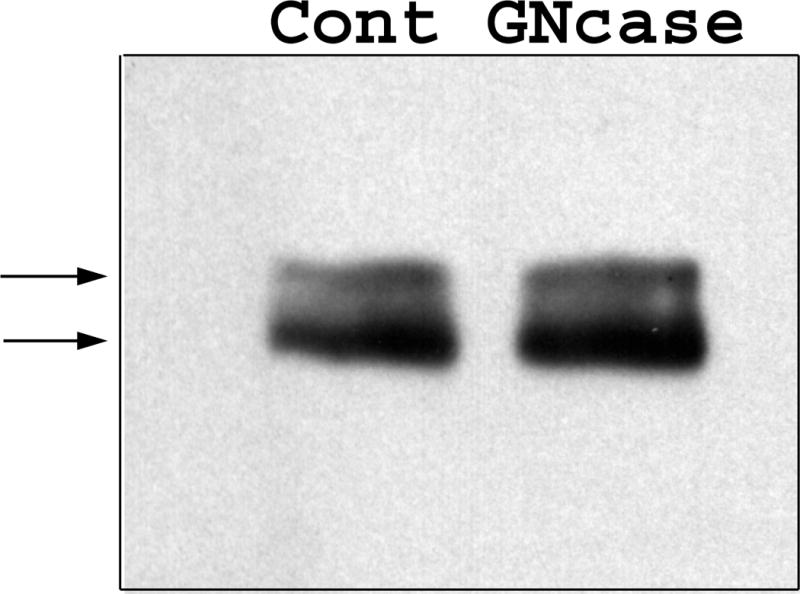
O-GlcNAcylation does not alter the mobility of Zfhep. Nuclear extract from COS-7 were incubated with or without O-GlcNAcase (Gncase) prior to western analysis for Zfhep.
We investigated whether Zfhep is present as a doublet in various cell types. Nuclear extracts were prepared from cultures of 15 cell types. We found a doublet of Zfhep immuno-reactive bands in HEPM, COS-7, U138 MG, CV1, and SK-N-SH cells, but no expression in GH3, JEG3, and PC12 cells (Figure 5). Only, or predominantly, the hypophosphorylated band was expressed in F9, GH4C1, Neuro2a, αT3-1, CHO, and C2C12 cells, as opposed to Saos-2 cells which produce predominantly the upper band. Very long exposures of westerns with CHO nuclear extracts failed to show any evidence of phosphorylated Zfhep. The absence of the slower mobility phosphorylated Zfhep protein in certain cell types indicates that the post-translational processing of Zfhep is cell-specific.
Figure 5.
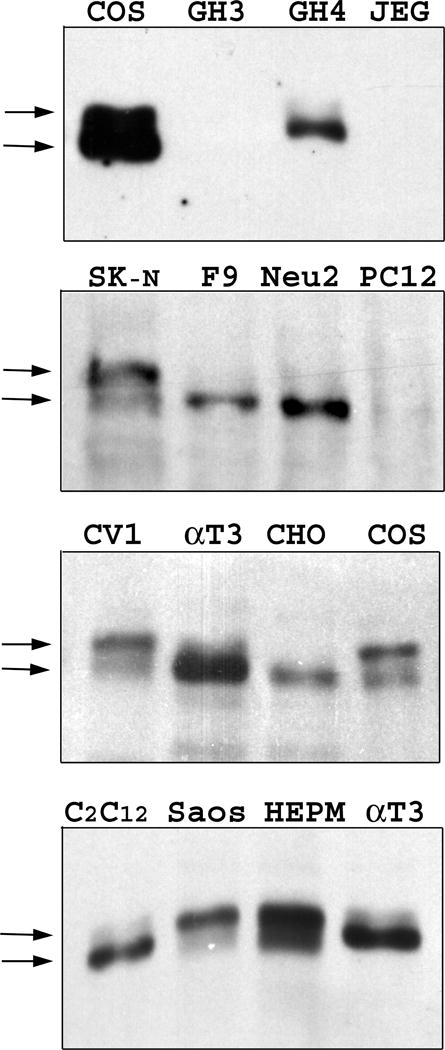
Phosphorylation of Zfhep is cell-specific. Expression of Zfhep in untreated nuclear extracts from different cell types was analyzed by western blot of 4% acrylamide gels. The full names of the cell lines are given in the Results section.
We also investigated whether nuclear localization was controlled by phosphorylation of Zfhep. Given the presence of both forms of Zfhep in COS-7 cells, immunofluorescence with the anti-Zfhep primary antiserum and FITC-conjugated anti-rabbit IgG secondary antibody (Chemicon, Temecula, CA) was used to determine if either form was localized outside the nucleus. Immunocytochemical assay of COS-7 cells detected Zfhep only in the nucleus, indicating that both isoforms are nuclear (not shown). In support of this, no Zfhep was detected in western analysis of cytosol from COS-7, CHO, or GH3 cells (not shown). Also, immunocytochemistry did not detect cytosolic or nuclear Zfhep in JEG-3 cells, consistent with the western analysis in Figure 5.
DISCUSSION
We have made the novel observation that Zfhep is present as two forms. The increased acuity provided by a 4% acrylamide gel in western analysis shows that Zfhep is expressed as a doublet of proteins with relative mobilities of approximately 180 kDa and 185 kDa in several cell types. The presence of two separate forms of Zfhep on western analysis allows direct assay of the modifications that are responsible for the different mobilities. We have shown that the 185 kDa form of Zfhep is phosphorylated, whereas the 180 kDa Zfhep is hypophosphorylated, if at all. The lower Zfhep band apparently contains few or no phosphate groups since CIP caused no change of mobility in these experiments. The difference of mobility of 5,000 Da is similar to that seen with phosphorylation of protein kinase C epsilon (40). The shift to a slower mobility likely reflects decreased binding of SDS anions due to the negatively charged phosphates.
The presence of the phosphorylated form of Zfhep is cell-specific, which may contribute to different activities of Zfhep in different cell types. For example, transcriptional activation of the VDR gene by Zfhep is known to be cell-type specific (35). The number of potential Ser/Thr phosphorylation sites in rat Zfhep was determined by analysis with PhosphoBase (http://www.cbs.dtu.dk/databases/PhosphoBase/) (41). PhosphoBase identified 61 consensus phosphorylation sequences conserved in rat and human Zfhep, representing 8 different kinases. The locations of potential phosphorylation sites suggest possible roles for phosphorylation. Zfhep contains three PLD/NLS repeats that are binding sites for the corepressor CtBP (30, 32). These residues are not phosphorylated, however, clusters of phosphorylation sequences are located around and between the PLD/NLS sequences. Phosphorylation at these sites may modify the ability of CtBP to bind Zfhep. Similarly, clusters of phosphorylation sites flanking the acidic activation domain may influence the activity of this domain, providing a mechanism for regulating transcriptional activation by Zfhep.
Zfhep/ZEB is a physiologically important repressor of the interleukin 2 gene in T cells. Yang et al. (2002) recently found that superinduction of IL-2 by deoxynivalenol is due in part to loss of the DNA-binding activity of Zfhep, apparently due to post-translational modification of Zfhep (29). Our data provide the first demonstration that Zfhep is post-translationally modified, suggesting the possibility that changes in phosphorylation in response to deoxynivalenol are responsible for the loss of DNA-binding activity of Zfhep.
Acknowledgments
Supported by NIH DE 13614, the University of Louisville School of Medicine, and the Jewish Hospital Foundation.
Abbreviations used
- Zfhep
zinc finger homeodomain enhancer-binding protein
- IL-2
interleukin 2
- PP2A
protein phosphatase 2A
- O-GlcNAc
O-linked N-acetylglucosamine
- CIP
calf intestinal phosphatase
- O-GlcNAcase
N-acetyl-D glucosaminidase
- CtBP
C-terminal binding protein
- NRE-A
negative regulatory element-A
- NF-κB
nuclear factor kappa B
- I-κB
inhibitor of κB
- DTT
dithiothreitol
- GST
glutathione-S-transferase
- NP-40
Nonidet-P40
References
- 1.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 2.Hazzalin CA, Mahadevan LC. MAPK-regulated transcription: a continuously variable gene switch? Nat Rev Mol Cell Biol. 2002;3:30–40. doi: 10.1038/nrm715. [DOI] [PubMed] [Google Scholar]
- 3.Park GT, Denning MF, Morasso MI. Phosphorylation of murine homeodomain protein Dlx3 by protein kinase C. FEBS Lett. 2001;496:60–65. doi: 10.1016/s0014-5793(01)02398-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grimes CA, Jope RS. CREB DNA binding activity is inhibited by glycogen synthase kinase-3 beta and facilitated by lithium. J Neurochem. 2001;78:1219–1232. doi: 10.1046/j.1471-4159.2001.00495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kitta K, Clement SA, Remeika J, Blumberg JB, Suzuki YJ. Endothelin-1 induces phosphorylation of GATA-4 transcription factor in the HL-1 atrial-muscle cell line. Biochem J. 2001;359:375–380. doi: 10.1042/0264-6021:3590375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang Q, Wiese RJ, Bueno OF, Dai YS, Markham BE, Molkentin JD. The transcription factor GATA4 is activated by extracellular signal-regulated kinase 1- and 2-mediated phosphorylation of serine 105 in cardiomyocytes. Mol Cell Biol. 2001;21:7460–7469. doi: 10.1128/MCB.21.21.7460-7469.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masuyama N, Oishi K, Mori Y, Ueno T, Takahama Y, Gotoh Y. Akt inhibits the orphan nuclear receptor Nur77 and T-cell apoptosis. J Biol Chem. 2001;276:32799–32805. doi: 10.1074/jbc.M105431200. [DOI] [PubMed] [Google Scholar]
- 8.Cures A, House C, Kanei-Ishii C, Kemp B, Ramsay RG. Constitutive c-Myb amino-terminal phosphorylation and DNA binding activity uncoupled during entry and passage through the cell cycle. Oncogene. 2001;20:1784–1792. doi: 10.1038/sj.onc.1204345. [DOI] [PubMed] [Google Scholar]
- 9.Comer FI, Hart GW. O-GlcNAc and the control of gene expression. Biochim Biophys Acta. 1999;1473:161–171. doi: 10.1016/s0304-4165(99)00176-2. [DOI] [PubMed] [Google Scholar]
- 10.Comer FI, Hart GW. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem. 2000;275:29179–29182. doi: 10.1074/jbc.R000010200. [DOI] [PubMed] [Google Scholar]
- 11.Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto T, Nakano Y, Moringa T, Tamaoki T. A new family of homeobox genes encoding multiple homeodomain and zinc finger motifs. Mech Dev. 1992;39:125–126. doi: 10.1016/0925-4773(92)90031-e. [DOI] [PubMed] [Google Scholar]
- 13.Cabanillas AM, Darling DS. Alternative splicing gives rise to two isoforms of Zfhep, a zinc finger/homeodomain protein that binds T3-response elements. DNA and Cell Biology. 1996;15:643–651. doi: 10.1089/dna.1996.15.643. [DOI] [PubMed] [Google Scholar]
- 14.Darling DS, Gaur NK, Zhu B. A zinc finger homeodomain transcription factor binds specific thyroid hormone response elements. Mol Cell Endocrinol. 1998;139:25–35. doi: 10.1016/s0303-7207(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 15.Genetta T, Kadesch T. Cloning of a cDNA encoding a mouse transcriptional repressor displaying striking sequence conservation across vertebrates. Gene. 1996;169:289–290. doi: 10.1016/0378-1119(95)00824-1. [DOI] [PubMed] [Google Scholar]
- 16.Sekido R, Takagi T, Okanami M, Moribe H, Yamamura M, Higashi Y, Kondoh H. Organization of the gene encoding transcriptional repressor deltaEF1 and cross-species conservation of its domains. Gene. 1996;173:227–232. doi: 10.1016/0378-1119(96)00185-0. [DOI] [PubMed] [Google Scholar]
- 17.Franklin AJ, Jetton TL, Shelton KD, Magnuson MA. BZP, a novel serum-responsive zinc finger protein that inhibits gene transcription. Mol Cell Biol. 1994;14:6773–6788. doi: 10.1128/mcb.14.10.6773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funahashi J, Sekido R, Murai K, Kamachi Y, Kondoh H. Delta-crystallin enhancer binding protein delta EF1 is a zinc finger-homeodomain protein implicated in postgastrulation embryogenesis. Development. 1993;119:433–446. doi: 10.1242/dev.119.2.433. [DOI] [PubMed] [Google Scholar]
- 19.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins: implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–6163. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watanabe Y, Kawakami K, Hirayama Y, Nagano K. Transcription factors positively and negatively regulating the Na,K-ATPase alpha 1 subunit gene. J Biol Chem. 1993;114:849–855. doi: 10.1093/oxfordjournals.jbchem.a124267. [DOI] [PubMed] [Google Scholar]
- 21.Williams TM, Moolten D, Burlein J, Romano J, Bhaerman R, Godillot A, Mellon M, Rauscher FJ, III, Kant JA. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 22.Higashi Y, Moribe H, Takagi T, Sekido R, Kawakami K, Kikutani H, Kondoh H. Impairment of T cell development in deltaEF1 mutant mice. J Exp Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takagi T, Moribe H, Kondoh H, Higashi Y. δEF1, a zinc finger and homeodomain transcription factor, is required for skeleton patterning in multiple lineages. Development. 1998;125:21–31. doi: 10.1242/dev.125.1.21. [DOI] [PubMed] [Google Scholar]
- 24.Krempler A, Brenig B. Zinc finger proteins: watchdogs in muscle development. Mol Gen Genet. 1999;261:209–215. doi: 10.1007/s004380050959. [DOI] [PubMed] [Google Scholar]
- 25.Postigo AA, Dean DC. ZEB, a vertebrate homolog of Drosophila Zfh-1, is a negative regulator of muscle differentiation. EMBO J. 1997;16:3935–3943. doi: 10.1093/emboj/16.13.3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sekido R, Murai K, Funahashi J, Kamachi Y, Fujisawa-Sehara A, Nabeshima Y, Kondoh H. The delta-crystallin enhancer-binding protein delta EF1 is a repressor of E2-box-mediated gene activation. Mol Cell Biol. 1994;14:5692–5700. doi: 10.1128/mcb.14.9.5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yen PM, Croci A, Dowling A, Zhang S, Zoeller RT, Darling DS. Developmental and functional evidence of a role for Zfhep in neural cell development. Brain Res Mol Brain Res. 2001;96(1–2):57–69. doi: 10.1016/s0169-328x(01)00267-4. [DOI] [PubMed] [Google Scholar]
- 28.Ouyang YL, Li S, Pestka JJ. Effects of vomitoxin (deoxynivalenol) on transcription factor NF-kappa B/Rel binding activity in murine EL-4 thymoma and primary CD4+ T cells. Toxicol Appl Pharmacol. 1996;140:328–336. doi: 10.1006/taap.1996.0228. [DOI] [PubMed] [Google Scholar]
- 29.Yang GH, Pestka JJ. Vomitoxin (deoxynivalenol)-mediated inhibition of nuclear protein binding to NRE-A, an IL-2 promoter negative regulatory element, in EL-4 cells. Toxicology. 2002;172:169–179. doi: 10.1016/s0300-483x(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 30.Furusawa T, Moribe H, Kondoh H, Higashi Y. Identification of CtBP1 and CtBP2 as corepressors of zinc finger-homeodomain factor deltaEF1. Mol Cell Biol. 1999;19:8581–8590. doi: 10.1128/mcb.19.12.8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikeda K, Halle JP, Stelzer G, Meisterernst M, Kawakami K. Involvement of negative cofactor NC2 in active repression by zinc finger-homeodomain transcription factor AREB6. Mol Cell Biol. 1998;18:10–18. doi: 10.1128/mcb.18.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postigo AA, Dean DC. ZEB represses transcription through interaction with the corepressor CtBP. Proc Natl Acad Sci USA. 1999;96:6683–6688. doi: 10.1073/pnas.96.12.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekido R, Murai K, Kamachi Y, Kondoh H. Two mechanisms in the action of repressor deltaEF1: binding site competition with an activator and active repression. Genes Cells. 1997;2:771–783. doi: 10.1046/j.1365-2443.1997.1570355.x. [DOI] [PubMed] [Google Scholar]
- 34.Chamberlain EM, Sanders MM. Identification of the novel player deltaEF1 in estrogen transcriptional cascades. Mol Cell Biol. 1999;19:3600–3606. doi: 10.1128/mcb.19.5.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lazarova DL, Bordonaro M, Sartorelli AC. Transcriptional regulation of the vitamin D(3) receptor gene by ZEB. Cell Growth & Differ. 2001;12:319–326. [PubMed] [Google Scholar]
- 36.Ikeda K, Kawakami K. DNA binding through distinct domains of zinc-finger-homeodomain protein AREB6 has different effects on gene transcription. Eur J Biochem. 1995;233:73–82. doi: 10.1111/j.1432-1033.1995.073_1.x. [DOI] [PubMed] [Google Scholar]
- 37.Schreiber E, Matthias P, Muller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore DD, Sefton BS. Analysis of Protein Phosphorylation. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. John Wiley and Sons, Inc.; New York: 1999. pp. 18.0–18.9. [Google Scholar]
- 39.Cabanillas AM, Smith GE, Darling DS. T3-activation of the rat growth hormone gene is inhibited by a zinc finger/homeodomain protein. Mol Cell Endocrinol. 2001;181:131–137. doi: 10.1016/s0303-7207(01)00531-7. [DOI] [PubMed] [Google Scholar]
- 40.Cenni V, Doppler H, Sonnenburg ED, Maraldi N, Newton AC, Toker A. Regulation of novel protein kinase C epsilon by phosphorylation. Biochem J. 2002;363:537–545. doi: 10.1042/0264-6021:3630537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreegipuu A, Blom N, Brunak S. PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res. 1999;27:237–239. doi: 10.1093/nar/27.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]


