Abstract
Topiramate, an anticonvulsant medication, is an efficacious treatment for alcohol dependence. To date, little is known about genetic moderators of side effects from topiramate. The objective of this study was to examine three single nucleotide polymorphisms (SNPs) of the glutamate receptor GluR5 gene (GRIK1) as predictors of topiramate-induced side effects in the context of a laboratory study of topiramate. Heavy drinkers (n = 51, 19 females), 75% of whom met criteria for an alcohol use disorder, completed a 5-week dose escalation schedule to a target dose of either 200 or 300 mg, or matched placebo. The combined medication groups were compared to placebo-treated individuals for side effects at target dose. Analyses revealed that a SNP in intron 9 of the GRIK1 gene (rs2832407) was associated with the severity of topiramate-induced side effects and with serum levels of topiramate. Genes underlying glutamatergic neurotransmission, such as the GRIK1 gene, may help predict heterogeneity in topiramate-induced side effects. Future studies in larger samples are needed to more fully establish these preliminary findings.
Introduction
The field of pharmacogenetics seeks to uncover genetic causes of heterogeneity of pharmacotherapy treatment effects, including the susceptibility to side effects. Increased knowledge about genetic moderators of medication response can be applied to treatment decision-making including dosing algorithms based on genetically-determined differences in drug metabolism (Kirchheiner et al., 2004; O'Brien, 2008). Both pharmacokinetic and pharmacodynamic genes are thought to influence medication response. Genes that influence pharmacokinetic processes are those involved in the absorption, distribution, metabolism, and excretion of drugs in the body. In contrast, genes that influence pharmacodynamic response are those directly responsible for the structure and/or functioning of the molecules or receptors targeted by the medication. These genes may also influence signaling or metabolic pathways involved in the manifestation of a disease, such that the mechanisms that affect the risk for a disorder may also underlie clinical response to its pharmacological treatment. While pharmacokinetic genes comprise the largest group of known pharmacogenetic factors, some have argued that pharmacodynamic genes have stronger effects on medication response (Goldstein, Need, Singh, & Sisodiya, 2007).
Topiramate (TOP) is an anticonvulsant medication recently identified as efficacious for the treatment of alcohol dependence (AD). Individuals with AD were randomized to TOP, in escalating dose up to 300 mg/day, reported reductions in drinking across several measures of alcohol consumption, with large magnitude effect sizes, as compared to placebo (PLAC) (Johnson et al., 2003; Johnson et al., 2007). Results of two open-label trials found similarly positive results (Raguraman, Priyadharshini, & Chandrasekaran, 2005; Rubio et al., 2004). Glutamate antagonist medications, including TOP, have also received empirical support for the treatment of alcohol withdrawal symptoms (Krupitsky et al., 2007).
TOP is a promising pharmacotherapy for AD, although TOP's side effect profile may limit its use in clinical practice given that approximately 20% of patients drop out of clinical trials for alcoholism due to TOP-induced side effects (Johnson et al., 2007). Therefore, it would be useful to identify individuals who are more likely to report greater severity of side effects to this medication. In a recent large clinical trial, the most commonly reported side effects among individuals with AD were: paresthesia (51%), taste perversion (23%), anorexia (20%), and difficulty with concentration (15%) (Johnson et al., 2007). These results are similar to those of previous reports (Johnson et al., 2003; Johnson, Swift, Addolorato, Ciraulo, & Myrick, 2005). Likewise, the literature on TOP for the treatment of epilepsy, at higher doses to those used for alcoholism treatment, has suggested that it causes frequent cognitive side effects in both clinical trials and in practice (Fritz et al., 2005; Thompson, Baxendale, Duncan, & Sander, 2000). A number of variables have been examined as predictors of cognitive adverse events to TOP (i.e., difficulty with attention, concentration, and memory) among patients treated for seizure disorders, including individual differences (e.g., gender, age at start of treatment, diagnosis) and clinical (e.g., titration, maximum dose, maintenance dose) variables, but none have successfully predicted treatment response (Goldstein et al., 2007). Alternatively, genetic differences may contribute to the heterogeneity of adverse events from TOP.
TOP has multiple neuropharmacological mechanisms of action (Johnson, 2005). These actions include the facilitation of gamma aminobutyric acid (GABA) neurotransmission and the inhibition of glutamatergic receptors. Acute dopamine (DA) release across the corticomesolimbic axis plays a critical role in determining the motivational significance of alcohol (Hyman & Malenka, 2001) and alcohol reward (Dodd, Beckmann, Davidson, & Wilce, 2000; Weiss, Lorang, Bloom, & Koob, 1993; Weiss & Porrino, 2002). Because corticomesolimbic DA release is under tonic inhibitory control via GABAergic neurons and excitatory control via glutamatergic neurons, TOP's concurrent GABAergic agonism and glutamatergic antagonism has been hypothesized to inhibit acute motivation for alcohol (i.e., craving). Importantly, TOP antagonizes the ability of kainate to activate kainate/alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) subtype of glutamate receptors, with no apparent activity on the N-methyl-D-aspartic acid (NMDA) subtype (Johnson, 2005). Glutamatergic neurotransmission, in turn, is thought to play an important role in various alcohol-related behaviors such as alcohol withdrawal symptoms (Littleton, 1998) and responses to alcohol (Lipsky & Goldman, 2003). In addition to its effects on glutamatergic transmission, TOP potentiates inhibitory GABAA receptor-mediated input, which represents another important mechanism underlying TOP's effects (Johnson, 2005; Johnson & Ait-Daoud, 2000).
Based on the putative neurobiological mechanisms of action of TOP, particularly its antagonist effects on glutamateric neurotransmission, genes coding for glutamate receptors represent plausible candidate genes as they harbor allelic variants that may underlie the pharmacodynamics of TOP. This investigation focuses on the kainate-selective glutamate receptor GluR5 gene (GRIK1), located on chromosome 21q, as a candidate pharmacogenetic predictor of TOP-induced side effects given that animal (Gryder & Rogawski, 2003) and in-vitro (Kaminski, Banerjee, & Rogawski, 2004) studies found that receptors containing the GluR5 subunit selectively bind topiramate. In this study, we selected three SNPs in the 3’ half of the GRIK1 gene (rs2832387, rs2186305, and rs2832407), given that these were the only SNPs significantly associated with AD in a recent study evaluating seven GRIK1 SNPs (Kranzler, 2007). Additionally, the last two SNPs are located in exons 9 and 17, respectively, for which there is evidence of alternative splicing (Barbon & Barlati, 2000), although their functional significance remains unclear.
The objective of this study was to examine three single nucleotide polymorphisms (SNPs) of the glutamate receptor GluR5 gene (GRIK1) as predictors of TOP-induced side effects in the context of a recent laboratory study (Miranda et al., 2008). A community sample of non-treatment seeking heavy drinkers, 75% of whom met criteria for an alcohol use disorder (AUD), completed a 5-week titration period to two target doses of TOP (TOP, 200 mg, 300 mg) or matched PLAC. Participants were stabilized on the target medication dose, or given an equivalent schedule of PLAC, before completing a laboratory protocol consisting of cue-exposure followed by alcohol administration (not reported here). This study focuses on genetic predictors of severity of side effects reported upon stabilization at the target dose of TOP versus PLAC. A secondary aim of this study is to test GRIK1 SNPs for their association with serum levels of TOP at the target dose. Consistent with the pharmacogenetics framework, it is hypothesized that genetic factors of putative influence on the targets of a given pharmacotherapy, in this case, glutamate receptors targeted by topiramate, may ultimately predict responses to this pharmacotherapy. Associations between the candidate polymorphisms and drinking behavior are also reported.
Method
Participants
Inclusion criteria were the following: consuming ≥ 18 alcoholic drinks per week (≥ 14 for females) during the 90 days before enrollment; not currently seeking treatment for alcoholism; and age between 21 and 65 years. Exclusion criteria were: clinically significant medical contraindications; use of medications that could affect mood, drinking, or TOP's effects; current or recent alcohol treatment or treatment-seeking; allergy to TOP; weight <110 or > 250 lbs; living with another participant in the study; consuming ≥ 60 drinks/week (≥ 53 for females); not using reliable birth control, if female; positive urine pregnancy screen; Beck Depression Inventory-II score > 14.
Of 78 individuals enrolled in the study, 14 withdrew from the study, 6 for discomfort from side effects, 2 for misgivings about participation in a medication trial, and 6 for miscellaneous personal reasons. Of those participants who withdrew due to side effects, four were in the 300 mg condition, one was in the 200 mg condition, and one was in the PLAC condition. No medication-related serious adverse events occurred. Three additional participants in the 300 mg group who completed the study were determined to have zero TOP levels by serum assay so their data were not used in the analyses.
A total of 61 participants completed the trial. A subset of participants (n = 51, 19 females) provided consent for DNA collection and represent the sample for this preliminary investigation of TOP pharmacogenetics (n = 32 in TOP, 19 in PLAC). The mean age of the study sample was 43.1 years (SD = 12.7), 90% were Caucasian, and the mean level of education was 13.8 years (SD = 3.0). At baseline, participants reported drinking an average of 4.8 days per week and consuming an average of 6.3 drinks (SD = 2.9) per drinking day. Diagnostically, 75.5% of participants met current criteria for an AUD at enrollment, 22.5% for alcohol abuse and 53% for alcohol dependence.
Procedure
All procedures were approved by the Brown University Institutional Review Board. A telephone screen and a face-to-face screening session assessed inclusion and exclusion criteria. A medical examination by a physician determined medical and medication-related exclusions. Upon enrollment in the study, participants were provided with their first week of medication and the MEMS compliance system and completed baseline assessments. Participants attended weekly sessions during which they were assessed for alcohol use, craving, and side effects and were given their next week's medication supply. All side effects were communicated to the study physician on a weekly basis. The physician and all staff were blind to medication conditions. The present study focuses on the assessment completed at the target dose of medication.
Medication and Compliance
Participants underwent a 32-day medication titration period that is described in detail elsewhere (Miranda et al., 2008). The target dose was maintained for up to 7-days, with a modal stabilization period of 4 days, during which participants completed a laboratory session that included alcohol administration and cue-reactivity. Doses were compounded by an independent pharmacy and all capsules appeared identical. Compliance was assessed using two methods. First, a Medication Event Monitoring System (MEMS) was used, which employs an electronic medication bottle cap to record and store the date/time that participants open the bottle. The MEMS system also prompted the participants to take the medication in the morning and evening. Second, plasma levels of TOP were quantified via blood assay. Participants provided blood samples at the target dose (i.e., immediately prior to the laboratory session) which were analyzed by an independent laboratory. Test method consisted of a fluorescence polarization immunoassay, which has a high degree of sensitivity, 2.0 mcg/ml. Only samples from individuals randomized to TOP were analyzed. Of 32 participants on TOP, 27 provided blood samples; however, samples from 9 participants hemolyzed in transit and were nonviable for analysis, leaving a total of 18 valid samples (11 in the 200 mg, 7 in the 300 mg condition).
Assessment
Alcohol use disorders were assessed through diagnostic interviews conducted by a trained Ph.D.-level psychologist using the Structured Clinical Interview for DSM-IV (SCID-Patient Version) (First, 1995). Alcohol use prior to and during the study was assessed using the Timeline Followback (Sobell, Maisto, Sobell, & Cooper, 1979), with heavy drinking days defined as 6/4 standard drinks in a day for males/females, respectively (Flannery et al., 2002). Side effect severity was the primary dependent variable in this study and consisted of a continuous measure defined as the mean ratings of the 19 side effects at target dose. Side effects were assessed weekly in a structured interview in which participants were asked to rate the severity of 38 specific potential side effects using the categories “None,” “Mild,” “Moderate,” and “Severe.” Participants were also asked open-ended questions to determine the incidence of any non-anticipated side effects, which were rated using the same severity ratings. Data gathered during days 32-37 (target dose in TOP conditions) were used. The following 19 side effects at the target medication dose (or PLAC) represent the focus of the analyses: dizziness, decrease in appetite, changes in vision, difficulty sleeping, weight loss, fatigue, difficulty with coordination or balance, difficulty with concentration or attention, tingling in finger or toes (paresthesia), word-finding difficulties, memory difficulties, tremor, constipation or diarrhea, restlessness, nervousness or anxiety, irritability, depression, confusion, and changes in sexual function.
DNA Analyses
DNA was collected following published procedures (Freeman et al., 1997; Walker et al., 1999). Participants swabbed their cheeks with three cotton swabs, followed by a rinse of the mouth with tap water. Genomic DNA was isolated from buccal cells using published procedures (Lench, Stanier, & Williamson, 1988; Spitz et al., 1996). An ABI 7300 instrument was used to conduct 5'-nuclease (TaqMan) assays of the three GRIK1 candidate SNPs using assays commercially available from Applied Biosystems. This method relies on allele-specific hybridization of oligonucleotide probes (Livak, 1999). For quality assurance purposes, 20% of the samples were genotyped twice. Allele frequencies and tests for Hardy-Weinberg Equilibrium are reported in Table 1.
Table 1.
Allele and genotype frequencies for the three candidate SNPs of the GRIK1 gene by medication condition (TOP versus PLAC) and Hardy-Weinberg Equilibrium (HWE) test
| SNP rs# Location | Allele FrequencyA | Genotype FrequencyA | TopiramateB (n = 32) | PlaceboB (n = 19) | HWCC |
|---|---|---|---|---|---|
| SNP 1: rs2186305 intron 17 | T = 0.75 C = 0.25 |
TT = 0.55 TC/CC = 0.45 |
TT = 19 TC/CC = 13 |
TT = 10 TC/CC = 9 |
χ2(1) = 0.07, ns |
| SNP 2: rs2832407 intron 9 | C = 0.62 A = 0.38 |
CC = 0.37 CA/AA = 0.63 |
CC = 11 CA/AA = 21 |
CC = 8 CA/AA = 11 |
χ2(1) = 0.08, ns |
| SNP 3: rs2832387 13 kb 3’ | G = 0.70 A = 0.30 |
GG = 0.53 GA/AA = 0.47 |
GG = 17 GA/AA = 15 |
GG = 10 GA/AA = 9 |
χ2(1) = 2.34, ns |
Allele and genotype frequencies presented in percentages
Allele and genotype frequencies for TOP and PLAC conditions are presented in terms of sample size (n) for each cell
Non-significant χ2 results indicate that the observed allele frequencies were in conformity with Hardy-Weinberg Equilibrium expectations.
Data Analysis
All data were initially examined for distribution normality and outliers. The probability level of α = .05 was selected for these exploratory hypotheses based on hypothesis-wide number of dependent variables (per Dar, Serlin, & Omer, 1994) and since Bonferroni corrections are known to overcorrect (Duggal, Gillanders, Holmes, & Bailey-Wilson, 2008). Since the gene by medication effect is the effect relevant to the hypotheses and since mean side effect severity rating is the dependent measure of side effects, there are only three tests of the first aim's hypotheses and only three tests for the second aim's hypotheses. Thus, an alpha of .05 within each separate set of hypotheses is acceptable, given the importance of detecting any effect and avoiding false negative results (Type II error).
Due to the small sample size for the analyses of genetic moderators, the two TOP groups were combined (n = 32) and compared to the PLAC condition (n = 19) in order to increase the statistical power to detect group differences. A series of t-tests and χ2 tests compared the two groups for demographics, drinking history, and number of side effects at target dose. Cronbach's alpha was used to test the internal consistency of the 19 potential side effects. A series of 2 × 2 between-groups analyses of variance (ANOVAs) were used to test the study hypotheses regarding the main effects of medication and medication × GRIK1 genotype interactions on mean severity ratings across side effects at target dose. Significant interactions were followed by planned comparisons between genes within the TOP conditions. Each candidate SNP of the GRIK1 gene was tested separately because we were not interested in the linear combination of variables (Dar et al., 1994). A 2 × 2 between-groups ANOVA was also used to address the secondary aim regarding a main effect of genotype and interaction effects with dose (200 vs. 300 mg) on TOP serum levels. Associations between the candidate SNPs and drinking at target dose were also tested using ANCOVAs, which controlled for baseline drinking measures. Linkage disequilibrium (LD) plots for individuals of European Ancestry were generated from Hapmap data using the software program Haploview v4.1 (Barrett, Fry, Maller, & Daly, 2005).
Results
Preliminary Analyses
All dependent variables were found to be adequately normally distributed, without outliers. The sample was predominantly made up of individuals of European Ancestry (i.e., 90%). GRIK1 genotype groups were compared with regard to ethnicity and results suggest that they did not differ on ethnicity (χ2 p > .30); therefore, it is highly unlikely that population stratification confounded the analyses presented herein. Univariate ANOVAs revealed no differences between the medication and genotype groups in terms of demographic and drinking variables (p > .05). In addition, GRIK1 genotype ratios did not differ significantly between medication conditions.
Linkage Disequilibrium
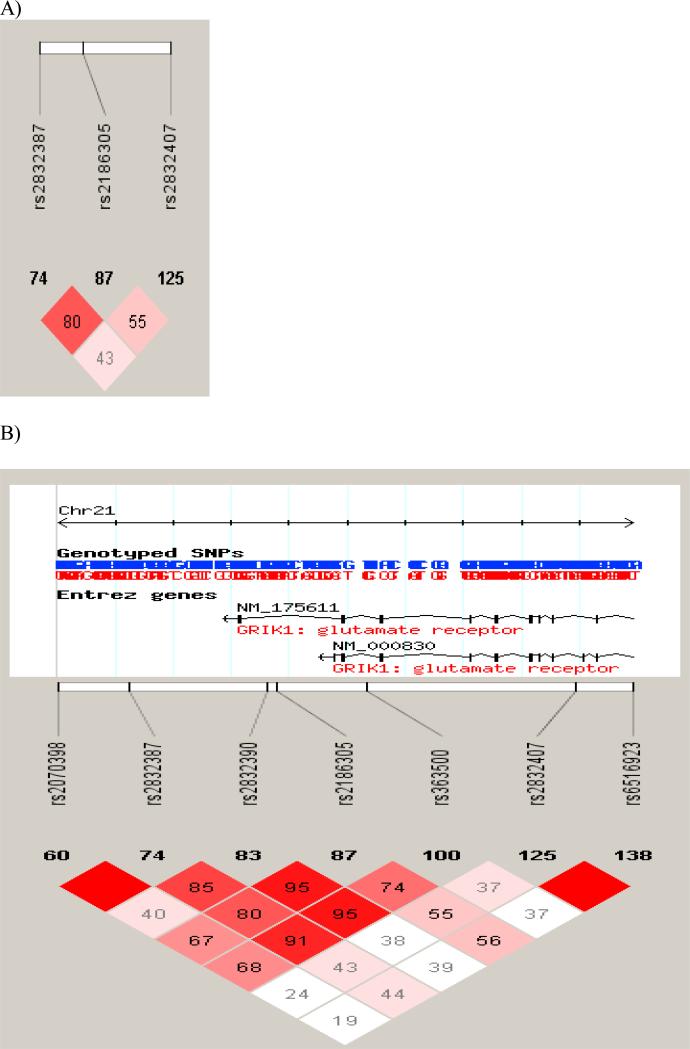
Linkage disequilibrium (LD) among the three candidate SNPs are presented in Figure 1a and among the three SNPs and the ones studied by Kranzler et al. (2007) are presented in Figure 1b. Pairwise SNP |D’| values (× 100) are presented with darkened blocks (i.e., high D’ values) indicating SNPs with limited recombination. In other words, SNPs with high D’ values indicate that markers are good surrogates for each other, likely to be transmitted together and to capture similar genetic variance. As can be seen in Figure 1a, SNPs 1 and 3 in this study had the highest D’ value, as estimated from Hapmap data for pedigrees of European Ancestry. High D’ values may indicate close topographic location on the chromosome and therefore redundancy in the information capture. Kranzler et al. (2007) built haplotype blocks for the GRIK1 SNPs and in that study SNPs 1 and 3 were in the same haplotype block, whereas SNP 2 was in block 2, suggesting greatest overlap between SNPs 1 and 3.
Figure 1.
LD plot from Haploview 4.1 for EA subjects based on Hapmap samples of individuals of European Ancestry for the three genotypes evaluated in this study (A) and for all seven SNPs evaluated by Kranzler et al. (2007) (B). Pair-wise SNP |D’| values (x100) of linkage are shown. Darkened blocks indicate SNP pairs without evidence of extensive recombination.
Compliance and Side Effects
All participants met the medication compliance criterion of 80%, with medication taken on a mean of 96.5% of days (range = 82% - 100%) according to MEMS data. The most frequently endorsed side effects in this sample were: paresthesia (33.3%), difficulty with concentration or attention (19.6%), constipation or diarrhea (17.6%), nervousness or anxiety (15.7%), memory difficulties (13.7%), decreased appetite (11.8%), and word-finding difficulties (11.8%). In the TOP condition, participants reported a mean of 2.53 (SD = 2.96) side effects, as compared to 0.84 (SD = 1.21) in the PLAC condition, t (49) = -2.85, p < .01. Side effects were generally in the mild-to-moderate range, such that 83% of the reported side effects were rated as mild, 16% as moderate, and 1% as severe. The internal consistency of the severity ratings for the 19 side effects was Cronbach's α = 0.77.
Medication and Genetic Effects on Severity of Side Effects
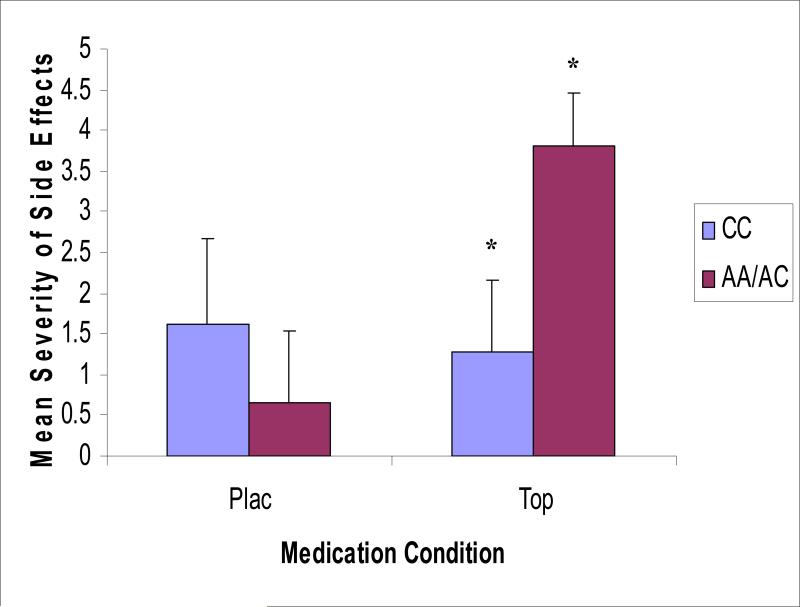
Although there was no significant main effect of medication (TOP vs. PLAC; F [1, 50] = 2.57, p = .12) or GRIK1 genotype (AC/AA vs. CC; F [1, 50] = 0.78, p = .38) on mean side effect severity, there was a significant GRIK1 SNP 2 (rs2832407; see Table 1) genotype × medication interaction. Carriers of the A allele reported greater severity of side effects on TOP versus PLAC, as compared to individuals who were homozygous for the C allele (F [1, 50] = 4.02, p = .05; see Figure 2). Planned comparisons revealed that the two genotype groups did not differ on severity of side effects on PLAC, t (18) = -1.21, p = .26. However, in the TOP condition, carriers of the A allele reported significantly higher side effect severity as compared to individuals who were homozygous for the C allele, t (31) = 2.56, p < .05. GRIK1 SNPs 1 and 3 did not have significant main effects or interaction effects with medication conditions on severity ratings of side effects.
Figure 2.
GRIK1 genotype (rs2832407) by medication interaction on mean severity of side effects at target dose (TOP vs. PLAC) along with standard error bars. Significant (p < .05) genotype differences within the TOP condition are indicated by an asterisk.
Medication and Genetic Effects on TOP Serum Levels
As a manipulation check serum TOP levels were compared between the two active-dose groups and the 300 mg group had higher serum TOP levels compared to 200 mg group (200 mg, n = 11, M = 4.25 mcg/ml; 300 mg, n = 7, M = 7.72 mcg/ml; F [1, 17] = 29.13, p < .0001). Additionally, there was a significant main effect of GRIK1 SNP2 genotype (rs2832407) such that carriers of the A allele had higher serum TOP levels as compared to individuals homozygous for the C allele (AA/AC, n = 13, M = 6.68 mcg/ml; CC, n = 5, M = 5.28 mcg/ml; F [1, 17] = 4.72, p < .05). The interaction effect between TOP dose (i.e., 200 vs. 300 mg/day) and GRIK1 genotype was not significant. GRIK1 SNPs 1 and 3 did not show significant effects on serum TOP levels. The relationship between serum TOP level and severity of side effects was not significant, r = 0.06, p > .10, with < 4% shared variance.
Medication and Genetic Effects on Drinking
At target dose (week 5), there was a main effect of GRIK1 SNP 2 (rs2832407) genotype on percent heavy drinking days (%HDD) (F [1, 46] = 5.46, p < .05) after controlling for baseline %HDD. Carriers of the A allele reported a higher mean %HDD, as compared to individuals who were homozygous for the C allele (40.4% versus 22.2%). There was no significant main effect of medication (TOP, M = 25.2 versus PLAC, M = 37.4), nor a genotype × medication interaction, on %HDD at target dose. There was no significant effect of GRIK1 SNPs, medication, or genotype × medication interaction on average drinks per drinking day at target dose. Lastly, there was no evidence of an association between severity of side effects and drinking behavior at target dose in the TOP condition, measured by %HDD (r = -0.15, p > .10) and average number of drinks per drinking day (r = 0.09, p > .10).
Discussion
Allelic variation in the GRIK1 (rs2832407) gene was associated with the severity of TOP-induced side effects in a community sample of non-treatment seeking heavy drinkers. The same SNP (rs2832407) was associated with differences in serum levels of TOP among participants in the TOP condition. Specifically, carriers of the A allele of SNP 2 of the GRIK1 gene reported greater severity of side effects when receiving TOP as compared to those homozygous for the C allele. Carriers of the A allele of SNP 2 of the GRIK1 gene also had higher serum TOP levels and higher percentage of heavy drinking days at target dose, than C-allele homozygotes. Interestingly, less than 4% of the variance was shared between serum TOP levels and severity of side effects, suggesting that mechanisms other than drug metabolism may account for the effects of genotype on TOP-induced side effect severity.
Thus, meaningful allelic variation in the gene responsible for the structure of one of the receptors targeted by TOP, the GluR5 kainate receptor, may contribute to the heterogeneity of treatment effects for TOP, especially side effect liability. These findings suggest that individuals with this genetic variant of the GRIK 1 gene may require additional medication management to keep from discontinuing treatment with TOP due to side effects, or the treating physician may consider alternative pharmacotherapies for these individuals. Studies of treatment-seeking samples and larger cell sizes are needed to more fully uncover the role of allelic variation in the GRIK1 gene, drinking behavior, and clinical response to TOP, including side effect liability. If supported, these preliminary findings may lead to a more personalized use of TOP for alcoholism.
The mechanisms by which this polymorphism may influence TOP-induced side effects and TOP serum levels remain unclear. Although there is some evidence of alternative splicing mechanisms operating at exon 9 of the GRIK1 gene (Barbon & Barlati, 2000) where SNP 2 (rs2832407) is located, the functional significance of this polymorphism remains unknown. On the basis of the pharmacogenetics framework employed in this manuscript, one may speculate that genetic factors of putative influence on the targets of a given pharmacotherapy, in this case glutamate receptors targeted by topiramate, may ultimately predict responses to this pharmacotherapy. Specifically, animal (Gryder & Rogawski, 2003) and in-vitro (Kaminski et al., 2004) studies have suggested that inhibition of GluR5 kainate receptors represents an important mechanism underlying TOP's effects. Therefore, it is plausible to hypothesize that genetic variation leading to structural and/or functional variation in the GluR5 kainate receptor, a selective target of topiramate coded by the GRIK1 gene, may in turn predict differential response to this pharmacotherapy. Similar findings have been demonstrated, for example, for the Asn40Asp allele of the μ-opioid receptor (OPRM1) gene, a functional polymorphism leading to greater binding affinity for β-endorphins (Bond et al., 1998). This SNP has been found to predict more favorable responses to naltrexone in the laboratory (Ray & Hutchison, 2007) and in clinical trials (Anton et al., 2008; Oslin et al., 2003). Much work remains to be done before similar assertions can be made for the pharmacogenetics of topiramate. Nevertheless, the rationale for this pharmacogenetic investigation is similar to that of naltrexone and largely based on the premise that genetic variation in the GRIK1 gene, which codes for GluR5 kainate receptor subunit, may be highly relevant to the pharmacogenetics of topiramate as receptors containing this subunit selectively bind topiramate.
As is the case for genetic associations to polymorphisms of unknown functional significance, linkage disequilibrium represents an important and plausible alternative explanation, whereby the signal detected at this locus in the present study may be a result of its close proximity to a functional polymorphism. Similarly, population stratification and other unmeasured third variables, both environment and genetic in nature, may account for these results. Future studies are undoubtedly needed to establish the role of glutamatergic genes, including GRIK1, to the pharmacogenetics of TOP, as well as to elucidate the neurobiological and molecular mechanisms underlying potential effects. Attention to mechanisms of action of TOP beyond the glutamate system, such as its effects on GABAergic neurotransmission, for instance, is needed to fully inform pharmacogenetic investigations. Nevertheless, this study contributes preliminary data using an empirically-driven approach to the pharmacogenetics of TOP, a promising pharmacotherapy for alcoholism (Johnson et al., 2007).
These results must be interpreted in light of the study's strengths and limitations. Strengths include the community sample composed of heavy drinkers, most of whom met criteria for an alcohol use disorder, the careful assessment and medication compliance protocols, and the collection of a biomarker of medication compliance via serum TOP levels. Limitations include the small sample size necessitating the combination of the two active medication conditions into one group, which in turn precluded the examination of dose-dependent effects. In addition, because participants were afforded some flexibility in scheduling the laboratory session in week 5, the number of possible drinking days during week 5 varied across participants (see Miranda et al., 2008). As such, replication of the observed association between genotype and percent heavy drinking days will be important. In these analyses, corrections for multiple comparisons were not implemented due to the preliminary nature of this investigation and the small sample size. On balance, these preliminary results suggest that genetic variation at the GluR5 kainate receptor may be a useful predictor of TOP-induced side effects. Future studies are needed to more fully ascertain these results as well as to examine the role of these polymorphisms as predictors of outcomes in clinical trials of topiramate.
Acknowledgments
This research was supported in part by a Career Development Award (1K23 AA014966) grants (5R01 AA7850-17; T32 AA007459) from the National Institute of Alcohol Abuse and Alcoholism, a Research Career Development Award, a Research Career Scientist Award, and a Senior Research Career Scientist Award from the Department of Veterans Affairs.
The authors wish to thank Amy Christian and John-Paul Massaro for their excellent technical assistance and Chinatsu McGeary for genotyping assistance.
References
- Anton RF, Oroszi G, O'Malley S, Couper D, Swift R, Pettinati H, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch Gen Psychiatry. 2008;65(2):135–144. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbon A, Barlati S. Genomic organization, proposed alternative splicing mechanisms, and RNA editing structure of GRIK1. Cytogenet Cell Genet. 2000;88(3-4):236–239. doi: 10.1159/000015558. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95(16):9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar R, Serlin RC, Omer H. Misuse of statistical test in three decades of psychotherapy research. J Consult Clin Psychol. 1994;62(1):75–82. doi: 10.1037//0022-006x.62.1.75. [DOI] [PubMed] [Google Scholar]
- Dodd PR, Beckmann AM, Davidson MS, Wilce PA. Glutamate-mediated transmission, alcohol, and alcoholism. Neurochem Int. 2000;37(5-6):509–533. doi: 10.1016/s0197-0186(00)00061-9. [DOI] [PubMed] [Google Scholar]
- Duggal P, Gillanders EM, Holmes TN, Bailey-Wilson JE. Establishing an adjusted p-value threshold to control the family-wide type 1 error in genome wide association studies. BMC Genomics. 2008;9:516. doi: 10.1186/1471-2164-9-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JBW. Structured Clinical Interview for DSM-IV Axis I Disorders - Patient edition (SCID-I/P, version 2.0) Biometrics Research Department, New York State Psychiatric Institute; New York: 1995. [Google Scholar]
- Flannery BA, Allen JP, Pettinati HM, Rohsenow DJ, Cisler RA, Litten RZ. Using acquired knowledge and new technologies in alcoholism treatment trials. Alcohol Clin Exp Res. 2002;26(3):423–429. [PubMed] [Google Scholar]
- Freeman B, Powell J, Ball D, Hill L, Craig I, Plomin R. DNA by mail: an inexpensive and noninvasive method for collecting DNA samples from widely dispersed populations. Behav Genet. 1997;27(3):251–257. doi: 10.1023/a:1025614231190. [DOI] [PubMed] [Google Scholar]
- Fritz N, Glogau S, Hoffmann J, Rademacher M, Elger CE, Helmstaedter C. Efficacy and cognitive side effects of tiagabine and topiramate in patients with epilepsy. Epilepsy Behav. 2005;6(3):373–381. doi: 10.1016/j.yebeh.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Goldstein DB, Need AC, Singh R, Sisodiya SM. Potential genetic causes of heterogeneity of treatment effects. Am J Med. 2007;120(4 Suppl 1):S21–25. doi: 10.1016/j.amjmed.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Gryder DS, Rogawski MA. Selective antagonism of GluR5 kainate-receptor-mediated synaptic currents by topiramate in rat basolateral amygdala neurons. J Neurosci. 2003;23(18):7069–7074. doi: 10.1523/JNEUROSCI.23-18-07069.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Malenka RC. Addiction and the brain: the neurobiology of compulsion and its persistence. Nat Rev Neurosci. 2001;2(10):695–703. doi: 10.1038/35094560. [DOI] [PubMed] [Google Scholar]
- Johnson BA. Recent advances in the development of treatments for alcohol and cocaine dependence: focus on topiramate and other modulators of GABA or glutamate function. CNS Drugs. 2005;19(10):873–896. doi: 10.2165/00023210-200519100-00005. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Ait-Daoud N. Neuropharmacological treatments for alcoholism: scientific basis and clinical findings. Psychopharmacology (Berl) 2000;149(4):327–344. doi: 10.1007/s002130000371. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Ait-Daoud N, Bowden CL, DiClemente CC, Roache JD, Lawson K, et al. Oral topiramate for treatment of alcohol dependence: a randomised controlled trial. Lancet. 2003;361(9370):1677–1685. doi: 10.1016/S0140-6736(03)13370-3. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Rosenthal N, Capece JA, Wiegand F, Mao L, Beyers K, et al. Topiramate for treating alcohol dependence: a randomized controlled trial. JAMA. 2007;298(14):1641–1651. doi: 10.1001/jama.298.14.1641. [DOI] [PubMed] [Google Scholar]
- Johnson BA, Swift RM, Addolorato G, Ciraulo DA, Myrick H. Safety and efficacy of GABAergic medications for treating alcoholism. Alcohol Clin Exp Res. 2005;29(2):248–254. doi: 10.1097/01.alc.0000153542.10188.b0. [DOI] [PubMed] [Google Scholar]
- Kaminski RM, Banerjee M, Rogawski MA. Topiramate selectively protects against seizures induced by ATPA, a GluR5 kainate receptor agonist. Neuropharmacology. 2004;46(8):1097–1104. doi: 10.1016/j.neuropharm.2004.02.010. [DOI] [PubMed] [Google Scholar]
- Kirchheiner J, Nickchen K, Bauer M, Wong ML, Licinio J, Roots I, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. 2004;9(5):442–473. doi: 10.1038/sj.mp.4001494. [DOI] [PubMed] [Google Scholar]
- Kranzler HR, Covault J, Herman A, Burian L, Arias A. Glutamate receptor gene variation and alcohol dependence risk. Alcohol Clin Exp Res. 2007;31(6):71A. [Google Scholar]
- Krupitsky EM, Rudenko AA, Burakov AM, Slavina TY, Grinenko AA, Pittman B, et al. Antiglutamatergic strategies for ethanol detoxification: comparison with placebo and diazepam. Alcohol Clin Exp Res. 2007;31(4):604–611. doi: 10.1111/j.1530-0277.2007.00344.x. [DOI] [PubMed] [Google Scholar]
- Lench N, Stanier P, Williamson R. Simple non-invasive method to obtain DNA for gene analysis. Lancet. 1988;1(8599):1356–1358. doi: 10.1016/s0140-6736(88)92178-2. [DOI] [PubMed] [Google Scholar]
- Lipsky RH, Goldman D. Genomics and variation of ionotropic glutamate receptors. Ann N Y Acad Sci. 2003;1003:22–35. doi: 10.1196/annals.1300.003. [DOI] [PubMed] [Google Scholar]
- Littleton J. Neurochemical mechanisms underlying alcohol withdrawal. Alcohol Health Res World. 1998;22(1):13–24. [PMC free article] [PubMed] [Google Scholar]
- Livak KJ. Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet Anal. 1999;14(5-6):143–149. doi: 10.1016/s1050-3862(98)00019-9. [DOI] [PubMed] [Google Scholar]
- Miranda R, Jr., MacKillop J, Monti PM, Rohsenow DJ, Tidey J, Gwaltney C, et al. Effects of topiramate on urge to drink and the subjective effects of alcohol: a preliminary laboratory study. Alcohol Clin Exp Res. 2008;32(3):489–497. doi: 10.1111/j.1530-0277.2007.00592.x. [DOI] [PubMed] [Google Scholar]
- O'Brien CP. Prospects for a genomic approach to the treatment of alcoholism. Arch Gen Psychiatry. 2008;65(2):132–133. doi: 10.1001/archgenpsychiatry.2007.32. [DOI] [PubMed] [Google Scholar]
- Oslin DW, Berrettini W, Kranzler HR, Pettinati H, Gelernter J, Volpicelli JR, et al. A functional polymorphism of the mu-opioid receptor gene is associated with naltrexone response in alcohol-dependent patients. Neuropsychopharmacology. 2003;28(8):1546–1552. doi: 10.1038/sj.npp.1300219. [DOI] [PubMed] [Google Scholar]
- Raguraman J, Priyadharshini RK, Chandrasekaran R. Effects of topiramate in alcohol dependence. Aust N Z J Psychiatry. 2005;39(8):736–737. doi: 10.1111/j.1440-1614.2005.01666_2.x. [DOI] [PubMed] [Google Scholar]
- Ray LA, Hutchison KE. Effects of naltrexone on alcohol sensitivity and genetic moderators of medication response: a double-blind placebo-controlled study. Arch Gen Psychiatry. 2007;64(9):1069–1077. doi: 10.1001/archpsyc.64.9.1069. [DOI] [PubMed] [Google Scholar]
- Rubio G, Ponce G, Jimenez-Arriero MA, Palomo T, Manzanares J, Ferre F. Effects of topiramate in the treatment of alcohol dependence. Pharmacopsychiatry. 2004;37(1):37–40. doi: 10.1055/s-2004-815473. [DOI] [PubMed] [Google Scholar]
- Sobell LC, Maisto SA, Sobell MB, Cooper AM. Reliability of alcohol abusers’ self-reports of drinking behavior. Behav Res Ther. 1979;17(2):157–160. doi: 10.1016/0005-7967(79)90025-1. [DOI] [PubMed] [Google Scholar]
- Spitz E, Moutier R, Reed T, Busnel MC, Marchaland C, Roubertoux PL, et al. Comparative diagnoses of twin zygosity by SSLP variant analysis, questionnaire, and dermatoglyphic analysis. Behav Genet. 1996;26(1):55–63. doi: 10.1007/BF02361159. [DOI] [PubMed] [Google Scholar]
- Thompson PJ, Baxendale SA, Duncan JS, Sander JW. Effects of topiramate on cognitive function. J Neurol Neurosurg Psychiatry. 2000;69(5):636–641. doi: 10.1136/jnnp.69.5.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect. 1999;107(7):517–520. doi: 10.1289/ehp.99107517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss F, Lorang MT, Bloom FE, Koob GF. Oral alcohol self-administration stimulates dopamine release in the rat nucleus accumbens: genetic and motivational determinants. J Pharmacol Exp Ther. 1993;267(1):250–258. [PubMed] [Google Scholar]
- Weiss F, Porrino LJ. Behavioral neurobiology of alcohol addiction: recent advances and challenges. J Neurosci. 2002;22(9):3332–3337. doi: 10.1523/JNEUROSCI.22-09-03332.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]