Abstract
Summary
We tested whether two genetic variants were associated with BMD at multiple clinically relevant skeletal sites in Caucasians. We found that variant rs7776725 is consistently associated with hip, spine, wrist and whole-body BMD, which highlights the potential importance of this variant or linked variants for osteoporosis.
Introduction
A recent genome-wide association study identified two single nucleotide polymorphisms (SNPs), rs7776725 and rs1721400, that were associated with bone mineral density (BMD) variation at the radius, tibia and calcaneus in a Korean population. In this study, we aimed to test whether the association of these two genetic variants can be replicated in Caucasians and whether their association with BMD can be extended to other clinically relevant skeletal sites.
Methods
We performed this study in two large cohorts of unrelated US Caucasians. Area BMD at the hip, spine, wrist (ultra-distal radius) and whole body were measured with Hologic dual-energy X-ray absorptiometer. SNPs were genotyped with Affymetrix human genome-wide genotyping arrays. Association analyses were performed using PLINK.
Results
We detected highly significant association (combined p=1.42×10−16) of rs7776725 with wrist BMD but only borderline association signal (combined p=0.017) for rs1721400 with wrist BMD. In addition, we found that rs7776725 was associated with BMD at the hip, spine and whole body. At the FAM3C gene locus where rs7776725 was located, we identified several other SNPs (rs4727922, rs1803389, rs718766 and rs7793554) that were also associated with BMD.
Conclusions
This is the first follow-up association study of rs7776725 and rs1721400 with BMD. The rs7776725 showed consistent association with BMD at multiple clinically important skeletal sites, which highlighted the potential importance of rs7776725 or linked SNPs for risk of osteoporosis. Further in-depth re-sequencing studies and functional assays are necessary to elucidate the underlying mechanisms.
Keywords: Bone mineral density, FAM3C, Osteoporosis, Single nucleotide polymorphisms
Introduction
Osteoporosis is a public health problem characterized by low bone mass as well as the deterioration of the bone structure with a consequent increased susceptibility to fractures. Osteoporosis-related fractures most likely occur at the hip, spine or wrist [1]. Osteoporosis is highly influenced by genetic factors. Bone mineral density (BMD), which is the widely used proxy measure of osteoporosis and the most important predictor for osteoporotic fractures, has a heritability of 60–90% [2].
Genome-wide association studies (GWASs) can systematically survey the whole genome for causal genetic variants underlying complex traits/diseases and are powerful tools for unlocking the genetic basis of osteoporosis [3–5]. Recently, a large-scale two-stage GWAS conducted in a Korea population identified two single nucleotide polymorphisms (SNPs), rs7776725 and rs1721400, that influence BMD at multiple skeletal sites [6]. In that study, rs7776725 which was located on chromosome 7q31 was identified to be associated with BMD at the radius (p=1.0×10−11) and tibia (p=1.6×10−6) in the discovery cohort of 8,842 Koreans, and associated with BMD at the calcaneus (p=1.9×10−10) in the replication cohort of 7,861 Koreans. The rs1721400, located on chromosome 7p14, also showed consistent associations at the tibia (p=1.4× 10−7), radius (p=2.2×10−3) and calcaneus (p=6.0×10−4) [6]. So far no replication study has been performed on the association of these two SNPs with BMD in independent Asian populations or in other ethnic groups. The validity and generality of the genetic effects of these two variants on BMD need to be evaluated.
Here, we performed a replication study to investigate the genetic effects of rs7776725 and rs1721400 on BMD in two cohorts of US Caucasians of European origin. BMD at whole body and three clinically important skeletal sites for osteoporotic fractures (hip, spine and wrist) were studied.
Methods
Study populations
This study was approved by the involved Institutional Review Board. All participants provided signed informed consents. The inclusion and exclusion criteria were the same as those detailed elsewhere [7]. Two populations of unrelated US Caucasians of European origin were involved in this study. The first population, denoted as KC-population in the following text, included 2,286 US Caucasians living in Kansas City and its surrounding areas. After exclusion of people with missing data on covariates and/or extreme phenotypic outliers, 2,279 people were included in the analysis. Area BMD (gram per square centimeter) at the lumbar spine (L2–L4), hip (femoral neck, Ward’s triangle, trochanter and intertrochanter), wrist (ultra-distal radius, UD) and whole body (WB) were measured with Hologic Discovery™ dual-energy X-ray absorptiometry (DXA) scanner (Hologic Inc., Bedford, MA, USA). The 2,279 subjects included 555 men (age range at recruitment: 19.0–89.7 years) and 1724 women (age range at recruitment: 18.4–91.5 years).
The second population, denoted as OM-population in the following text, included 1,000 US Caucasians living in Omaha, Nebraska, and its surrounding areas. DXA scans of the hip, spine and wrist UD BMD of this population were performed with Hologic QDR 4500 W (Hologic Inc., Bedford, MA, USA). These 1,000 subjects included 499 men (age range at recruitment: 19.1–87.2 years) and 501 women (age range at recruitment: 19.2–80.6 years). The detailed characteristics of these two cohorts are listed in Table 1.
Table 1.
Characteristics of the populations involved in this study
| KC-population |
OM-population |
|||||
|---|---|---|---|---|---|---|
| All | Male | Female | All | Male | Female | |
| Subjects | 2279 | 555 | 1724 | 1000 | 499 | 501 |
| Age (years) | 51.32 (13.74) | 50.61 (16.03) | 51.55 (12.91) | 50.23 (18.28) | 50.31 (18.87) | 50.15 (17.69) |
| Height (cm) | 166.35 (8.46) | 175.87 (7.24) | 163.29 (6.26) | 170.83 (9.74) | 177.83 (7.05) | 163.85 (6.51) |
| Weight (kg) | 75.28 (17.55) | 87.14 (16.75) | 71.47 (16.04) | 80.16 (17.89) | 89.02 (14.94) | 71.32 (15.92) |
| Hip BMD | 0.965 (0.145) | 1.029 (0.145) | 0.944 (0.139) | 1.032 (0.164) | 1.070 (0.169) | 0.994 (0.151) |
| Spine BMD | 1.024 (0.157) | 1.067 (0.154) | 1.010 (0.146) | 0.973 (0.156) | 1.038 (0.146) | 0.908 (0.137) |
| UD BMD | 0.437 (0.075) | 0.505 (0.069) | 0.415 (0.062) | 0.463 (0.082) | 0.507 (0.073) | 0.418 (0.065) |
| WB BMD | 1.112 (0.108) | 1.181 (0.105) | 1.090 (0.100) | – | – | – |
Data presented are unadjusted means (SD)
Genotyping and SNP calling
Genomic DNA was extracted from peripheral blood leukocytes using standard protocols. For the KC-population, SNP genotyping was performed using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA), which includes 906,600 SNPs and 940,000 copy number probes. Briefly, approximately 250 ng of genomic DNA was digested with restriction enzyme NspI and StyI, respectively. Digested DNA was adaptor-ligated and PCR-amplified for each sample. Fragment PCR products were then labeled with biotin, denatured and hybridized to the arrays. Arrays were then washed and stained using phycoerythrin on Affymetrix Fluidics Station, and scanned using the Gene-Chip Scanner 3000 7 G to quantitate fluorescence intensities. Data management and analyses were conducted using the Genotyping Command Console. SNPs were identified using Birdsuite (version 1.5.2; http://www.broad.mit.edu/mpg/birdsuite/analysis.html). A per-sample call rate threshold of 95% was used to remove subjects with unsuccessful genotyping. For all 14 SNPs analyzed in the present study, the per-SNP call rates are>99%. The rs7776725 and other 12 SNPs in vicinity to it were located in or near FAM3C locus on chromosome 7q31. The rs1721400 was mapped close to SFRP4 locus on chromosome 7p14.
The subjects in OM-population were genotyped with the Affymetrix GeneChip human mapping 500 K array set. The genotyping was performed at the Vanderbilt Microarray Shared Resource at Vanderbilt University Medical Center using the standard protocol recommended by the manufacturer (Affymetrix, Inc., USA). The detailed genotyping and quality control procedure were published elsewhere [7]. The per-SNP call rates of SNPs studied here are >99%.
Statistical analyses
The BMD measurements were normally distributed (WB BMD in the KC-population and UD BMD in the OM-population) or approximately normally distributed. Markertrait and multi-marker haplotype association analyses were performed using the quantitative trait association function in PLINK (v1.05, http://pngu.mgh.harvard.edu/purcell/plink/) [8] with untransformed BMD data. Before association analysis, age, age2, sex, age/age2-by-sex interaction, height and weight were tested for their associations with BMD, and only the significant (p≤0.05) terms were included as covariates to adjust the raw BMD values in subsequent analyses. Quantitative trait gene by environment interaction (G × E) analysis implemented in PLINK was performed to test whether the genetic effects of rs7776725 is different by genders. In the genetic–gender interaction analysis, only age, age2, height and weight were used as covariates.
Hardy–Weinberg equilibrium (HWE) test and pairwise linkage disequilibrium (LD) analyses of the SNPs as well as the LD plotting of interested genome region were conducted using the program Haploview (http://www.broad.mit.edu/mpg/haploview) [9]. All analyzed SNPs followed HWE (P>0.01).
Conservative Bonferroni corrections were applied to minimize false-positive results caused by multiple testing of SNPs. Since there was prior evidence of association for rs7776725 and rs1721400 with BMD, a p value cutoff of 0.025 (0.05/2, as two SNPs were studied) was used in this study for claiming replication of association. In the association analysis of other SNPs near rs7776725, a threshold of significance was set at a p value of <4.17×10−3 (0.05/12, as 12 SNPs were studied).
Sample-size-based weighted Z-score method was used to combine the association p values across two populations. The overall Z-scores were generated using the formula:
where Zi and Ni are the Z-score and sample size of each study to be combined, respectively. Based on the overall Z-score, a combined association p value was obtained [10].
The imputation of un-genotyped SNPs in the OM-population was carried out by using the program MACH [11, 12] (http:// www.sph.umich.edu/csg/abecasis/MACH/) on the basis of the reference sample of European ancestry derived from the 1000 Genomes Project (release of August, 2010). Associations between the phenotype and imputed SNPs were examined by MACH2QTL, in which allele dosage was used as the predictor for the phenotype in a linear regression model.
Results
Association of rs7776725 with BMD
In the KC-population, the association of rs7776725 with wrist UD BMD could be replicated, and association of rs7776725 with BMD at all other studied skeletal sites was also found. As shown in Table 2, in this population the p values of association analysis between rs7776725 and BMD at the hip, spine, UD and WB were 1.54×10−5, 5.22×10−3, 6.83×10−16 and 1.26× 10−8, respectively. It is worth noting that the extremely low p value between rs7776725 and UD BMD was comparable to that reported previously in the Korean population (p=1.0× 10−11 for radius BMD) [6]. And the direction of the allelic effects is the same, with the minor allele C of rs7776725 associated with an increased BMD. As summarized in Fig. 1, subjects with the CC or CT genotypes had significant higher BMD than those with the TT genotype. The mean dose effects per copy of the C allele on the hip, spine, UD and WB BMD were 0.016, 0.017, 0.015 and 0.018 g/cm2, respectively.
Table 2.
Association of rs7776725 and rs1721400 with BMD in two Caucasian populations
| rs7776725 |
rs1721400 |
|||||
|---|---|---|---|---|---|---|
| KC-population | OM-population | Combined p | KC-population | OM-population | Combined p | |
| Hip BMD | 1.54×10−5 | 0.286 | 2.72×10−5 | 0.393 | 0.652 | 0.336 |
| Spine BMD | 5.22×10−3 | 0.771 | 0.013 | 0.209 | 0.525 | 0.162 |
| UD BMD | 6.83×10−16 | 5.83×10−3 | 1.42×10−16 | 0.085 | 0.084 | 0.017 |
| WB BMD | 1.26×10−8 | – | 0.235 | – | ||
Boldface values represent the case of p<0.05
En dash means that item was not studied for no available WB BMD data
Fig. 1.
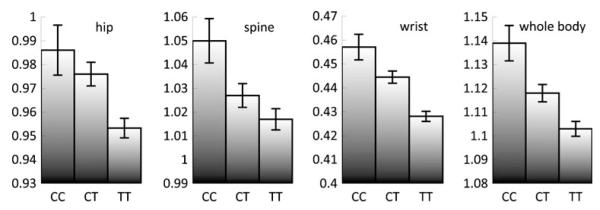
Unadjusted BMD values according to genotype at rs7776725. Data are expressed as means±SE (error bars). The adjusted BMD values in subjects with rs7776725 genotypes CC:CT:TT were as follows: 0.0206:0.0090:−0.0096 at hip; 0.0259:0.0020:−0.0055 at spine; 0.0202:0.0072:−0.0084 at wrist; and 0.0279:0.0064:−0.0089 at whole body. These are consistent with the results of the unadjusted BMD values in this figure
In the OM-population, the association of rs7776725 with BMD was only significant at the wrist (p=5.83×10−3). We could not find association between rs7776725 with hip and spine BMD in this population. Meta-analysis of the KC-population and OM-population achieved statistically significant associations for rs7776725 with hip, spine and UD BMD (Table 2).
Association of rs1721400 with BMD
Association results of rs1721400 with BMDs in the two populations are presented in Table 2. Small but statistically non-significant p values were achieved for rs1721400 with UD BMD in each of the two populations, with p=0.085 in the KC-population and p=0.084 in the OM-population. However, the combined p value of rs1721400 with UD BMD in the meta-analysis of these two populations is borderline significant (p=0.017). The minor allele T of rs1721400 was a risk factor for low UD BMD. No significant association of rs1721400 with hip, spine or WB BMD was detected in KC-population. No significant association of rs1721400 with hip or spine BMD was detected in OM-population as well as the meta-analysis of these two populations. The two studied cohorts lack the information of BMD at the tibia and calcaneus which were studied by Chao et al. [6].
Gene–gender interaction analyses
BMD has gender-specific effects. Previous studies have suggested that some genes regulating BMD might act in a gender-specific manner [13, 14]. To test if rs7776725 and rs1721400 have differential effects on BMD between two sexes, we further performed the gene–gender interaction analysis. No statistically significant gene–gender interactions were found in the two cohorts (data not shown).
Association study of other SNPs in vicinity to rs7776725
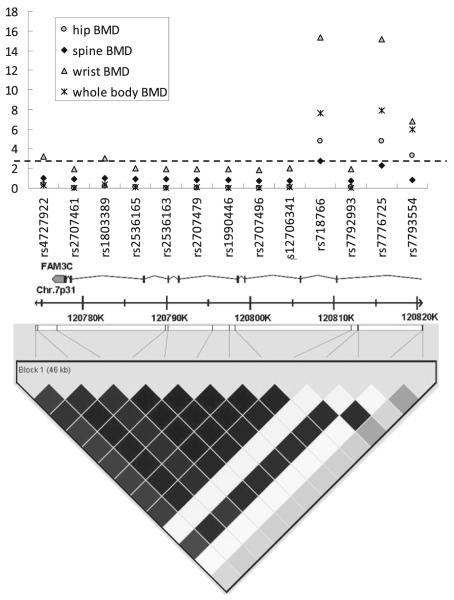
The rs7776725 was located in the first intron of the FAM3C gene. We further examined the genotyped variants within/ around the FAM3C gene in KC-population. Twelve SNPs dispersedly located from 1787 bp downstream to the first intron of FAM3C gene were tested for their association with hip, spine, UD and WB BMD in the KC population. All these 12 SNPs followed HWE (p>0.01) with MAF (Minor Allele Frequency) >5%. The basic information of these SNPs (together with rs7776725) and the LD coefficients D’ and r2 with rs7776725 are tabulated in Table 3.
Table 3.
Information of the analyzed SNPs in the FAM3C locus
| dbSNP RS ID | Physical position | Location to FAM3C | Allelea | MAFb | MAFc | D’ (r2 ) with rs7776725d | D’ (r2) with rs7776725e |
|---|---|---|---|---|---|---|---|
| rs4727922 | 120774355 | Downstream | C/T | 0.162 | 0.133 | 1 (0.049) | 0.777 (0.045) |
| rs2707461 | 120774586 | Downstream | T/C | 0.188 | 0.142 | 1 (0.053) | 0.822 (0.060) |
| rs1803389 | 120776915 | 3′ UTR | T/A | 0.172 | 0.133 | 1 (0.049) | 0.806 (0.052) |
| rs2536165 | 120789829 | Intron | C/T | 0.189 | 0.144 | 1 (0.055) | 0.815 (0.059) |
| rs2536163 | 120790146 | Intron | A/G | 0.190 | 0.142 | 1 (0.053) | 0.824 (0.061) |
| rs2707479 | 120795439 | Intron | C/T | 0.190 | 0.142 | 1 (0.053) | 0.823 (0.061) |
| rs1990446 | 120797445 | Intron | C/T | 0.190 | 0.142 | 1 (0.053) | 0.824 (0.061) |
| rs2707496 | 120798134 | Intron | T/C | 0.189 | 0.142 | 1 (0.053) | 0.832 (0.062) |
| rs12706341f | 120811944 | Intron | T/C | 0.187 | 0.142 | 1 (0.053) | 0.819 (0.059) |
| rs718766f | 120812738 | Intron | C/T | 0.269 | 0.242 | 1 (1) | 0.987 (0.937) |
| rs7792993f | 120812813 | Intron | C/A | 0.195 | 0.142 | 1 (0.053) | 0.829 (0.063) |
| rs7776725 | 120820357 | Intron | C/T | 0.276 | 0.242 | – | – |
| rs7793554f | 120820427 | Intron | C/T | 0.457 | 0.383 | 1 (0.513) | 0.996 (0.451) |
The former allele represents the minor allele of each locus
Minor allele frequency calculated in our Caucasian KC-population
Minor allele frequency reported for Caucasians in HapMap
D’ (r2 ) with rs7776725 for Caucasians in HapMap
D’ (r2 ) with rs7776725 in our Caucasian KC-population
Genotypes of these SNPs are only available in KC-population for they are not included in Affymetrix GeneChip 500 K array set
The association results of these 12 SNPs and rs7776725 are presented in Fig. 2. Four SNPs (rs4727922, rs1803389, rs718766 and rs7793554) showed significant association with BMD at one or more skeletal sites after multiple testing correction (threshold p=4.17×10−3). The significant associations were achieved for rs4727922 with wrist BMD (p=6.23×10−4 ), rs1803389 with wrist BMD (p=8.53×10−4), rs718766 with BMD at all sites analyzed (p=1.60×10−5, 1.83×10−3, 4.36× 10−16 and 2.17×10−8 for hip, spine, UD and WB BMD, respectively) and rs7793554 with hip, UD and WB BMD (p=5.42×10−4, 1.69×10−7 and 1.09×10−6 respectively). The minor alleles of rs4727922 and rs1803389 are risk factors for low BMD. The minor alleles of rs718766 and rs7793554 are associated with increased BMD.
Fig. 2.
LD plot and the association results of SNPs in FAM3C region. Upper panel: The association results of rs7776725 and other 12 SNPs. y-axis: -log10(p-value). The dashed line shows the significant threshold for the association results of these 12 SNPs. Lower panel: Pairwise LD plot for SNPs studied (rs4727922, rs2707461, rs1803389, rs2536165, rs2536163, rs2707479, rs1990446, rs2707496, rs12706341, rs718766, rs7792993, rs7776725 and rs7793554 from left to right) based on genotyping data in the KC-population. LD is colored with the correlation coefficients r2 scheme. White means r2=0. Squares are colored darker if the r2 value is higher. Black means r2=1. All the markers were shown to be grouped into a single LD block when analyzed with the “Four gamete rule” or “solid spine of LD” option in Haploview software
The associations of rs4727922 and rs1803389 with wrist BMD were not replicated in the OM-population, with p values of 0.161 and 0.167, respectively. However, in the meta-analysis of these two populations, rs4727922 and rs1803389 were significantly associated with wrist BMD (p=2.86×10−4 and 3.94×10−4 respectively). The genotyping data for rs718766 and rs7793554 were not available in the OM-population. With the 1000 Genomes Project data as reference, we imputed these two un-genotyped SNPs in the OM-population. Association analyses of the imputed data provided further supports for the role of rs718766 and rs7793554 in BMD determination (p=2.19×10−3 for rs718766 and 0.071 for rs7793554 with wrist BMD).
Haplotype association of FAM3C locus with BMD
Haplotype association analyses were performed in KC-population to test if any haplotypes in FAM3C locus could show stronger association with BMD than individual SNPs. Five haplotype windows of interest (WIN1–WIN5, as shown in Table 4)were specified mainly based on the LD relationship between SNPs. WIN1 contained all 13 FAM3C SNPs studied in KC-population. These 13 SNPs lie in a single LD block defined by the “Four gamete” or “solid spine of LD” algorithm implemented in the Haploview software. WIN2, extending from rs4727922 to rs12706341, covered nine SNPs that were highly linked to each other (average r2=0.94) but only showed very low r2 with rs7776725 (r2<0.1). WIN3 included other four SNPs that were not covered by WIN2. WIN4 was a sub-haplotype of WIN3 with SNP rs7792993 (r2<0.1 with rs7776725) excluded and thus consisted of three strongly linked SNPs (rs718766, rs7776725 and rs7793554). WIN5 only included two SNPs (rs718766 and rs7776725) that were in almost complete LD (r2≈1).
Table 4.
Haplotype-based association of FAM3C gene locus with BMD
| Window name | Haplotypes |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4727922 | rs2707461 | rs1803389 | rs2536165 | rs2536163 | rs2707479 | rs1990446 | rs2707496 | rs12706341 | rs718766 | rs7792993 | rs7776725 | |
| WIN1 | T | C | A | T | G | T | T | C | C | C | A | C |
| T | C | A | T | G | T | T | C | C | T | A | T | |
| C | T | T | C | A | C | C | T | T | T | C | T | |
| WIN2 | T | C | A | T | G | T | T | C | C | – | – | – |
| C | T | T | C | A | C | C | T | T | – | – | – | |
| WIN3 | – | – | – | – | – | – | – | – | – | C | A | C |
| – | – | – | – | – | – | – | – | – | T | A | T | |
| – | – | – | – | – | – | – | – | – | T | C | T | |
| WIN4 | – | – | – | – | – | – | – | – | – | C | – | C |
| – | – | – | – | – | – | – | – | – | T | – | T | |
| – | – | – | – | – | – | – | – | – | T | – | T | |
| WIN5 | – | – | – | – | – | – | – | – | – | C | – | C |
| – | – | – | – | – | – | – | – | – | T | – | T | |
| Window name | Haplotypes |
Hap. frequency | Hip BMD |
Spine BMD |
UD BMD |
WB BMD |
||||
|---|---|---|---|---|---|---|---|---|---|---|
| rs7793554 | BETA | p Value | BETA | p Value | BETA | p Value | BETA | p Value | ||
| WIN1 | C | 0.264 | 0.016 | 2.83×10−5 | 0.014 | 1.35×10−3 | 0.015 | 1.12×10−15 | 0.017 | 1.70×10−8 |
| T | 0.532 | −0.014 | 4.17×10−5 | −0.007 | 0.065 | −0.009 | 4.99×10−8 | −0.014 | 2.25×10−7 | |
| C | 0.151 | −0.002 | 0.571 | −0.007 | 0.245 | −0.008 | 3.54×10−4 | −0.002 | 0.582 | |
| WIN2 | – | 0.808 | −0.001 | 0.883 | 0.007 | 0.157 | 0.005 | 0.012 | 0.000 | 0.950 |
| – | 0.160 | −0.003 | 0.563 | −0.008 | 0.125 | −0.008 | 8.14×10−4 | −0.002 | 0.527 | |
| WIN3 | C | 0.266 | 0.016 | 2.24×10−5 | 0.014 | 2.20×10−3 | 0.015 | 7.76×10−16 | 0.017 | 2.09×10−8 |
| T | 0.534 | −0.013 | 1.65×10−4 | −0.007 | 0.093 | −0.009 | 5.26×10−8 | −0.014 | 4.07×10−7 | |
| C | 0.177 | −0.002 | 0.680 | −0.007 | 0.198 | −0.006 | 3.33×10−3 | −0.001 | 0.805 | |
| WIN4 | C | 0.266 | 0.016 | 2.33×10−5 | 0.014 | 2.30×10−3 | 0.015 | 7.99×10−16 | 0.017 | 2.20×10−8 |
| T | 0.543 | −0.012 | 3.65×10−4 | −0.006 | 0.124 | −0.008 | 1.37×10−7 | −0.013 | 9.35×10−7 | |
| C | 0.179 | −0.002 | 0.646 | −0.007 | 0.195 | −0.006 | 6.04×10−3 | −0.001 | 0.841 | |
| WIN5 | – | 0.266 | 0.017 | 1.77×10−5 | 0.014 | 1.83×10−3 | 0.015 | 9.36×10−16 | 0.017 | 1.21×10−8 |
| – | 0.721 | −0.016 | 1.41×10−5 | −0.012 | 4.84×10−3 | −0.015 | 4.50×10−16 | −0.017 | 1.35×10−8 | |
Boldface SNPs (rs4727922, rs1803389, rs718766, rs7776725 and rs7793554) are those identified to be associated with at least one BMD trait BETA regression coefficient
Only common haplotypes (frequency >5%) were included in the association test. The association results of FAM3C haplotypes with BMD are shown in Table 4. The haplotype– BMD association results seemed to be driven mainly by the sub-haplotype composed of SNPs rs718766 and rs7776725. In this two-SNP haplotype window (WIN5), the haplotype C-C is associated with increased BMD (p=1.77×10−5, 1.83× 10−3, 9.36×10−16 and 1.21×10−8 for or hip, spine, UD and WB BMD, respectively), and haplotype T-T is associated with lower BMD (p=1.41×10−5, 4.84×10−3, 4.50×10−16, 1.35× 10−8 for hip, spine, UD and WB BMD, respectively). Every haplotype showed to be associated with higher BMD as long as C-C sub-haplotypes for rs718766 and rs7776725 were included. In WIN2, in agreement with the association results of rs4727922 and rs1803389, the haplotype (C-T-T-C-A-C-C-T-T) which included the C allele of rs4727922 and the T allele of 1803389 was associated with lower wrist BMD (p=8.14× 10−4), and this haplotype was not associated with BMD at other sites. In general, regarding the association p value or regression coefficient, haplotype analysis only gave results of similar magnitude to those obtained from SNP association analysis.
Discussion
In this study, we examined the associations of two common variants on chromosome 7 (rs7776725 and rs1721400) with hip, spine, wrist and whole-body BMD in Caucasian populations. These two SNPs were initially identified to be associated with BMD at radius, tibia and heel in Koreans. To the best of our knowledge, the present study is the first association study in Caucasians on the effects of these two SNPs on BMD variation.
The ethnic differences in BMD have been well characterized before. For example, Caucasians and Asians had lower BMD than those of African-American and Hispanic descent [15]. The environmental and lifestyle factors as well as the genetic background should contribute to the difference of the osteoporosis risks among ethnic groups. Generally, Caucasians and Asians share the majority of osteoporosis genetics. However, genetic variants important for BMD in one population may not necessarily be important in another population [16].
In this study, the association of rs7776725 with wrist BMD was well reproduced in two Caucasian populations. Notably, the association signal of this SNP for wrist BMD in the KC-population (p=6.83×10−16) and the combined p value across two populations (p=1.42×10−16) have reached genome-wide level of significance, which is consistent with those reported in Korean population (p=1.0×10−11) [6]. This study also suggested an association of rs1721400 with wrist BMD in Caucasians since the combined p value in meta-analysis of the two studied populations was statistically significant (p= 0.017). The direction of the allelic effects of these two SNPs was the same as those detected in the initial GWAS [6]. Although further studies with larger samples of other populations are needed, variant rs7776725 is more likely to have general effects on BMD in multiple ethnic groups. Only suggestive association of rs1721400 with BMD was detected in this study; more replication studies and meta-analyses in large samples are thus required before any firm conclusions about the effect of this SNP on BMD can be drawn.
One limitation of this study is that the two studied cohorts lack the information of BMD at the tibia and calcaneus which were studied by Chao et al. [6]. That prevents direct replication at these two sites. In addition, to avoid high rate of false negatives, we did not apply multiple testing correction for the multiple correlated BMD phenotypes studied; therefore, the false discovery rate was controlled at the level of 0.05 only for each phenotype instead of all phenotypes together.
It is important to examine the relevance of the significant genetic variations to different clinically important skeletal sites. In this study, rs7776725 was associated with BMD at the hip, spine and whole body. The extremely low p value and consistent associations observed with multiple clinically important sites highlight the potential importance of rs7776725 for risk of osteoporosis.
The variant rs7776725 was located in the first intron of the FAM3C gene. FAM3C is expressed in almost all tissues [17]. It encodes a cytokine necessary for epithelial to mesenchymal transition, tumor formation and metastasis in epithelial cells [18]. FAM3C is also involved in retinal laminar formation [19]. To further explore the relationship between FAM3C gene and human BMD, we extended the association analysis to other 12 SNPs within/around the FAM3C gene locus in KC-population. After multiple-testing adjustment, rs4727922 and rs1803389 were associated with wrist BMD, and rs718766 and rs7793554 were associated with BMD at multiple sites. Notably, the magnitude of the association signal of rs718766 and rs7793554 was in well accordance with the LD levels of these two SNPs with rs7776725. Specifically, the rs718766 which is in perfect LD with rs7776725 (r2≈1) achieved almost the same level of association as those of rs7776725. The rs7793554 and rs7776725 were in moderate LD (r2≈0.5) with each other, and the association of rs7793554 with BMD was compromised.
We used the FASTSNP program (http://fastsnp.ibms.sinica.edu.tw) to analyze the potential functions of these SNPs. No known functional relevance of rs4727922, rs1803389, rs718766 and rs7776725 with bone metabolism was available. The SNP rs7793554 located in the intron of FAM3C was suggested to be an intronic enhancer. An A to G change at this locus will delete a binding site of the transcription factor CRE-BP. It is possible that these SNPs are only tags of one or more causal genetic variant(s) that contribute to the BMD variation. Haplotype analysis of FAM3C locus did not reveal stronger signals compared to single-SNP analysis. It seems more likely that rs718766, rs7776725 and rs7793554 may tag the same causal variant, while rs4727922 and 1803389 may tag another one. Fine mapping of FAM3C gene or even more extended genomic region will be helpful to identify the true functional variant(s) underlying BMD.
Conclusions
In summary, with two unrelated US Caucasian populations, this study replicated the association of rs7776725. In addition, we identified that rs7776725 was associated with BMD at hip, spine and whole body in Caucasians. In FAM3C gene locus (where rs7776725 is located), we identified several other SNPs associated with BMD variation. Our group will perform in-depth re-sequencing study of this region to pinpoint the true functional variant(s).
Acknowledgement
This work was partially supported by grants from the NIH (R01 AR050496, R21 AG027110, R01 AG026564, P50 AR055081, R01 AR057049 and R21 AA015973). The study was also supported by the National Natural Science Foundation of China (31150006, 81000363, 31000554), the Fundamental Research Funds for the Central Universities (2011JBM134), Shanghai Leading Academic Discipline Project (S30501), and startup fund from University of Shanghai for Science and Technology, Xi’an Jiaotong University, and the Ministry of Education of China.
Footnotes
Conflicts of interest None.
Contributor Information
L.-S. Zhang, College of Life Sciences and Bioengineering, School of Science, Beijing Jiaotong University, Beijing 100044, People’s Republic of China
H.-G. Hu, College of Life Sciences and Bioengineering, School of Science, Beijing Jiaotong University, Beijing 100044, People’s Republic of China
Y.-J. Liu, Center for Bioinformatics and Genomics, Department of Biostatistics and Bioinformatics, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA
J. Li, Center for Bioinformatics and Genomics, Department of Biostatistics and Bioinformatics, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA
P. Yu, Center for Bioinformatics and Genomics, Department of Biostatistics and Bioinformatics, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA
F. Zhang, The Key Laboratory of Biomedical Information Engineering, Ministry of Education and Institute of Molecular Genetics, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an 710049, People’s Republic of China
T.-L. Yang, The Key Laboratory of Biomedical Information Engineering, Ministry of Education and Institute of Molecular Genetics, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an 710049, People’s Republic of China
Q. Tian, Center for Bioinformatics and Genomics, Department of Biostatistics and Bioinformatics, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA
Y.-P. Zheng, College of Life Sciences and Bioengineering, School of Science, Beijing Jiaotong University, Beijing 100044, People’s Republic of China
Y. Guo, The Key Laboratory of Biomedical Information Engineering, Ministry of Education and Institute of Molecular Genetics, School of Life Science and Technology, Xi’an Jiaotong University, Xi’an 710049, People’s Republic of China
H.-W. Deng, College of Life Sciences and Bioengineering, School of Science, Beijing Jiaotong University, Beijing 100044, People’s Republic of China; Center for Bioinformatics and Genomics, Department of Biostatistics and Bioinformatics, Tulane University School of Public Health and Tropical Medicine, New Orleans, LA 70112, USA; Center of System Biomedical Sciences, Shanghai University of Science and Technology, Shanghai, People’s Republic of China
References
- 1.Wardlaw GM. Putting osteoporosis in perspective. J Am Diet Assoc. 1993;93(9):1000–1006. doi: 10.1016/0002-8223(93)92038-y. [DOI] [PubMed] [Google Scholar]
- 2.Tse KY, Macias BR, Meyer RS, Hargens AR. Heritability of bone density: regional and gender differences in monozygotic twins. J Orthop Res. 2009;27(2):150–154. doi: 10.1002/jor.20721. [DOI] [PubMed] [Google Scholar]
- 3.Guo Y, Tan LJ, Lei SF, Yang TL, Chen XD, Zhang F, Chen Y, Pan F, Yan H, Liu X, Tian Q, Zhang ZX, Zhou Q, Qiu C, Dong SS, Xu XH, Guo YF, Zhu XZ, Liu SL, Wang XL, Li X, Luo Y, Zhang LS, Li M, Wang JT, Wen T, Drees B, Hamilton J, Papasian CJ, Recker RR, Song XP, Cheng J, Deng HW. Genome-wide association study identifies ALDH7A1 as a novel susceptibility gene for osteoporosis. PLoS Genet. 6(1):e1000806. doi: 10.1371/journal.pgen.1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong DH, Liu XG, Guo YF, Tan LJ, Wang L, Sha BY, Tang ZH, Pan F, Yang TL, Chen XD, Lei SF, Yerges LM, Zhu XZ, Wheeler VW, Patrick AL, Bunker CH, Guo Y, Yan H, Pei YF, Zhang YP, Levy S, Papasian CJ, Xiao P, Lundberg YW, Recker RR, Liu YZ, Liu YJ, Zmuda JM, Deng HW. Genome-wide association and follow-up replication studies identified ADAMTS18 and TGFBR3 as bone mass candidate genes in different ethnic groups. Am J Hum Genet. 2009;84(3):388–398. doi: 10.1016/j.ajhg.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kung AW, Xiao SM, Cherny S, Li GH, Gao Y, Tso G, Lau KS, Luk KD, Liu JM, Cui B, Zhang MJ, Zhang ZL, He JW, Yue H, Xia WB, Luo LM, He SL, Kiel DP, Karasik D, Hsu YH, Cupples LA, Demissie S, Styrkarsdottir U, Halldorsson BV, Sigurdsson G, Thorsteinsdottir U, Stefansson K, Richards JB, Zhai G, Soranzo N, Valdes A, Spector TD, Sham PC. Association of JAG1 with bone mineral density and osteoporotic fractures: a genome-wide association study and follow-up replication studies. Am J Hum Genet. 86(2):229–239. doi: 10.1016/j.ajhg.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho YS, Go MJ, Kim YJ, Heo JY, Oh JH, Ban HJ, Yoon D, Lee MH, Kim DJ, Park M, Cha SH, Kim JW, Han BG, Min H, Ahn Y, Park MS, Han HR, Jang HY, Cho EY, Lee JE, Cho NH, Shin C, Park T, Park JW, Lee JK, Cardon L, Clarke G, McCarthy MI, Lee JY, Lee JK, Oh B, Kim HL. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat Genet. 2009;41(5):527–534. doi: 10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 7.Liu YJ, Liu XG, Wang L, Dina C, Yan H, Liu JF, Levy S, Papasian CJ, Drees BM, Hamilton JJ, Meyre D, Delplanque J, Pei YF, Zhang L, Recker RR, Froguel P, Deng HW. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum Mol Genet. 2008;17(12):1803–1813. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 10.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 34(8):816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Willer C, Sanna S, Abecasis G. Genotype imputation. Annu Rev Genomics Hum Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edderkaoui B, Baylink DJ, Beamer WG, Shultz KL, Wergedal JE, Mohan S. Genetic regulation of femoral bone mineral density: complexity of sex effect in chromosome 1 revealed by congenic sublines of mice. Bone. 2007;41(3):340–345. doi: 10.1016/j.bone.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Karasik D, Ferrari SL. Contribution of gender-specific genetic factors to osteoporosis risk. Ann Hum Genet. 2008;72(Pt 5):696–714. doi: 10.1111/j.1469-1809.2008.00447.x. [DOI] [PubMed] [Google Scholar]
- 15.Barrett-Connor E, Siris ES, Wehren LE, Miller PD, Abbott TA, Berger ML, Santora AC, Sherwood LM. Osteoporosis and fracture risk in women of different ethnic groups. J Bone Miner Res. 2005;20(2):185–194. doi: 10.1359/JBMR.041007. [DOI] [PubMed] [Google Scholar]
- 16.Lei SF, Chen Y, Xiong DH, Li LM, Deng HW. Ethnic difference in osteoporosis-related phenotypes and its potential underlying genetic determination. J Musculoskelet Neuronal Interact. 2006;6(1):36–46. [PubMed] [Google Scholar]
- 17.Zhu Y, Xu G, Patel A, McLaughlin MM, Silverman C, Knecht K, Sweitzer S, Li X, McDonnell P, Mirabile R, Zimmerman D, Boyce R, Tierney LA, Hu E, Livi GP, Wolf B, Abdel-Meguid SS, Rose GD, Aurora R, Hensley P, Briggs M, Young PR. Cloning, expression, and initial characterization of a novel cytokine-like gene family. Genomics. 2002;80(2):144–150. doi: 10.1006/geno.2002.6816. [DOI] [PubMed] [Google Scholar]
- 18.Waerner T, Alacakaptan M, Tamir I, Oberauer R, Gal A, Brabletz T, Schreiber M, Jechlinger M, Beug H. ILEI: a cytokine essential for EMT, tumor formation, and late events in metastasis in epithelial cells. Cancer Cell. 2006;10(3):227–239. doi: 10.1016/j.ccr.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 19.Katahira T, Nakagiri S, Terada K, Furukawa T. Secreted factor FAM3C (ILEI) is involved in retinal laminar formation. Biochem Biophys Res Commun. 392(3):301–306. doi: 10.1016/j.bbrc.2009.12.180. [DOI] [PubMed] [Google Scholar]