SUMMARY
With rapid developments in DNA and protein sequencing technologies, combined with powerful bioinformatics tools, a continued acceleration of gene identification in parasitic helminths is predicted, potentially leading to discovery of new drug and vaccine targets, enhanced diagnostics and insights into the complex biology underlying host-parasite interactions. For the schistosome blood flukes, with the recent completion of genome sequencing and comprehensive transcriptomic datasets, there has accumulated massive amounts of gene sequence data, for which, in the vast majority of cases, little is known about actual functions within the intact organism. In this review we attempt to bring together traditional in vitro cultivation approaches and recent emergent technologies of molecular genomics, transcriptomics and genetic manipulation to illustrate the considerable progress made in our understanding of trematode gene expression and function during development of the intramolluscan larval stages. Using several prominent trematode families (Schistosomatidae, Fasciolidae, Echinostomatidae), we have focused on the current status of in vitro larval isolation/cultivation as a source of valuable raw material supporting gene discovery efforts in model digeneans that include whole genome sequencing, transcript and protein expression profiling during larval development, and progress made in the in vitro manipulation of genes and their expression in larval trematodes using transgenic and RNA interference (RNAi) approaches.
Keywords: Trematoda, larval stages, Schistosoma, in vitro culture, genome, transcriptome, gene manipulation, transgenesis, RNA interference
INTRODUCTION
A well-recognized and highly conserved aspect of the life cycles of digenetic trematodes is their use of molluscs, mainly snails, as first intermediate hosts, and within which all species undergo a complex process of asexual reproduction. Infections of the molluscan host are initiated by entry of the ciliated miracidial stage where, once inside the host, they typically shed their ciliated epidermal plates, transforming to the primary sporocyst. Germinal cells within sporocysts then begin to divide forming multicellular embryos, each of which are destined to become either secondary (daughter) sporocysts or rediae (depending on the trematode species), thus constituting the next larval generation. This asexual sporocystogenic or redial-generating reproductive cycle may repeat itself several times, but eventually gives rise to the cercarial stage, which represents the next free-living stage responsible for transmitting the infection to the next host, either a second intermediate or a definitive host. Clearly the molluscan developmental phase of trematode life cycles is critically important, serving to greatly amplify the infective cercarial population that, in turn, dramatically increases the probability of successful transmission to the next host, even if the prevalence of infection within a given snail population is low (Basch, 1991).
As alluded to above, the intramolluscan phase of development is complex and, currently, little information is available regarding the underlying mechanisms regulating successful transition of the free-swimming miracidium to the parasitic primary sporocyst stage, or the biochemical and physiological requirements for establishing and maintaining an ongoing parasite infection. In general, digeneans exhibit very narrow ranges of snail host specificity, suggesting that the physiological parameters dictating larval survival may be quite different between parasite species or their hosts. In the present review we attempt to summarize a growing body of research that focuses on the in vitro manipulation of early intramolluscan trematode stages, emphasizing the blood flukes Schistosoma spp. Topics include the current status of in vitro culture as an investigative tool, gene discovery in model digeneans, gene and protein expression profiling during in vitro larval development, and the development and application of larval transgenesis and RNA interference (RNAi) as functional genomic complements to gene discovery efforts. The results of these studies not only give important insights into fundamental questions about how host specificity is regulated and/or what physiological factors dictate infection success, but also provide a basis for the identification and possible targeting of critical molecular or biochemical pathways for disruption by chemical or biological interventions leading to termination of infections within the molluscan host and thus, transmission to humans or domestic animals.
In vitro cultivation – essential tool for gene profiling and functional genomic investigations
Earlier studies on the human blood flukes, Schistosoma mansoni (Yoshino and Laursen, 1995) and S. japonicum (Coustau et al. 1997; Coustau and Yoshino, 2000), revealed that exposure of freshly-hatched miracidia to snail isotonic saline alone, e.g. Chernin’s balanced salt solution (CBSS; Chernin, 1963) would trigger the shedding of ciliated miracidial epidermal plates, and the concomitant formation of the primary (=mother) sporocyst tegument under in vitro conditions (Fig. 1). Thus, it appeared that the change in environmental osmolarity (from ~10 mOs/L in freshwater to ~140 mOs/L in snail saline=osmolarity of snail hemolymph) was sufficient to mimic the initial penetration and early larval development in the snail intermediate host. Other trematode species including echinostomes (Echinostoma paraensei, E. caproni) (Loker et al. 1992; Ataev et al. 1998), and the common liver fluke Fasciola hepatica (Gourbal et al. 2008) also appear to share in part this osmotic ‘transformation signal’, although under in vitro conditions, transformation may not involve the release or ‘shedding’ of ciliated epidermal plates typical of S. mansoni and S. japonicum miracidia. In fact, E. caproni cultivated in vitro for 24 h or more in serum-free medium or snail cell-conditioned medium, although ceasing swimming activity and losing some cilia from epidermal plates (Ataev et al. 1998), do not ‘shed’ their plates. Thus, it appears that the physico-chemical cues required by E. caproni, and possibly other Echinostoma spp. or fasciolids, for fully transforming to sporocysts in vitro differ from the ‘simple’ osmotic cues of schistosomes.
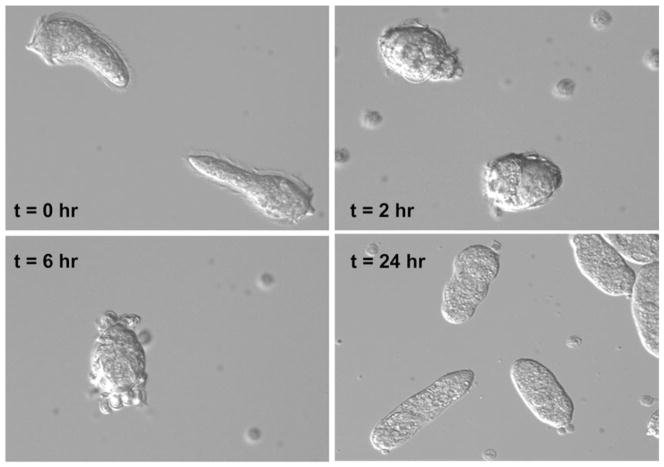
Fig. 1.
Schistosoma mansoni miracidia (t=0 h) as they undergo morphological transformation to the primary sporocyst stage (t=24 h). Miracidia, cultured in isotonic snail saline (Chernin’s balanced salt solution, CBSS; Chernin, 1963), first cease swimming, assume a rounded shape (t=2 h) as expansion of intercellular ridges force ciliated epidermal plates to round up and detach from the larval surface (t=6 h). ‘t’ indicate the approximate time elapsed from initial introduction of miracidia into culture.
The deer liver fluke Fascioloides magna provides an informative case to illustrate the complexities of miracidial transformation signals. Miracidia of F. magna, like E. caproni, do not spontaneously transform to primary sporocysts when incubated in snail ringers (CBSS) or serum-containing medium (Laursen and Yoshino, 1999). Although miracidia stop swimming within a few hours in culture, ciliated plates remained firmly attached to the majority of larvae at 24 h post-cultivation (Fig. 2A). However, when F. magna miracidia were cultured in CBSS that had been preconditioned by incubation with the Biomphalaria glabrata embryonic (Bge) cell line, miracidium-to-sporocyst transformation was readily induced (Fig. 2B, C). Moreover, treatment of the Bge cell-conditioned medium with heat (56 °C for 30 min; 100 °C for 10 min) or proteinase K significantly reduced the transformation-stimulating activity of the conditioned medium (Laursen and Yoshino, 1999). Based on these findings it appears that F. magna miracidia require an additional signal(s), likely a protein or protein-associated factor, from the snail host in addition to or in place of an osmotic signal in order to trigger transformation. Supporting these findings, Campbell and Todd (1955) reported that F. magna miracidial transformation in vitro (i.e. shedding of epidermal plates) only took place after miracidia had made physical contact with the snail, attempting to penetrate but failing, implying the requirement of a snail host molecular signal to initiate this process.
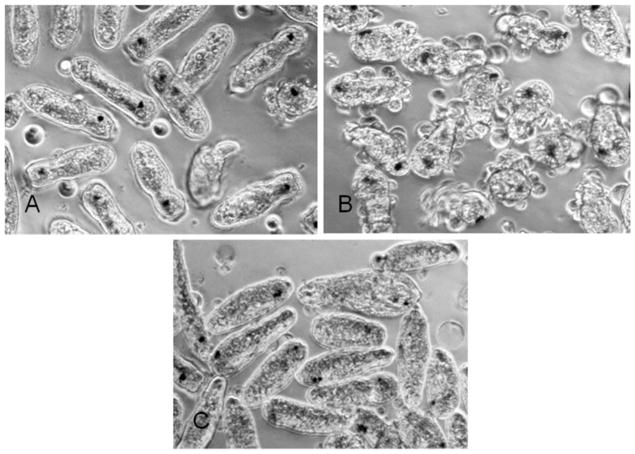
Fig. 2.
In vitro cultivation of Fascioloides magna (Fasiciolidae) miracidia in untreated CBSS (Chernin’s balanced salt solution; Chernin, 1963). Within 24 h, most miracidia cease swimming activity but the majority will not fully transform (i.e. will retain their ciliated epidermal plates (A). However, miracidia cultured in CBSS, pre-conditioned by cultivation with snail Biomphalaria glabrata embryonic (Bge) cells, are induced to transform (rounding and shedding of ciliated epidermal plates (B) to become fully-transformed primary sporocysts (C).
Clearly, the signals involved in early miracidium-to-sporocyst development are complex and undoubtedly differ between trematode groups. However, in vitro culture systems, while not mimicking precisely the environment within the natural host, do provide a means of tracking changes in gene expression that accompany early larval development that may provide critical insights into the role of specific genes or gene families in the establishment and maintenance of infections within the snail host. This is particularly true for in vitro systems capable of supporting transitions to the first intramolluscan parasitic stage (primary or mother sporocyst) and, in the case of schistosome species and F. magna, advanced development to subsequent daughter sporocyst (Yoshino and Laursen, 1995; Coustau et al. 1997; Ivanchenko et al. 1999) or redial (Laursen and Yoshino, 1999) stages. With the rapid expansion of gene/protein sequence databases for schistosomes and other trematode species, the incorporation of in vitro culture systems into experimental designs represents an invaluable tool for evaluating molecular changes linked to parasite development or for exploring gene function through manipulating transcript expression.
GENOMICS, TRANSCRIPTOMICS, AND PROTEOMICS OF LARVAL TREMATODES – SO MUCH DATA, SO LITTLE INFORMATION
Because of the recognized global importance of schistosomiasis as a significant public health concern, it is not surprising that the majority of molecular research has focused on Schistosoma spp.. In part due to the limited success of traditional methods in controlling schistosomiasis, the scientific community joined efforts in 1993 to begin generating genomic sequence data from Schistosoma mansoni (Franco et al. 1995) with funding support from the WHO/TDR(UNICEF-UNDOWorld Bank-WHO Special Programme for Research and Training in Tropical Diseases) (Oliveira et al. 2008). It was envisioned at the time that the genomic data generated would promote the acceleration of gene identification strategies and data analysis, which in turn could then be translated into new tools for fighting infection and disease including discovery of new drug and vaccine targets, improved diagnostics and clinical management approaches, and, importantly, tools for dissecting the basic biological underpinning of the complex host-parasite interactions (Franco et al. 2000; El-Sayed et al. 2004; Wilson et al. 2006; Brindley and Pearce, 2007; Hokke et al. 2007).
By means of whole-genome shotgun sequencing approaches the entire assembled genomes for S. mansoni and S. japonicum recently have been completed, revealing estimates of genome sizes of approximately 363 MB for S. mansoni and 397 MB per haploid genome for S. japonicum, and for both species, ~13 000 expressed transcripts, encoding ~11 000 genes (Haas et al. 2007; Berriman et al. 2009; The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium, 2009). Genes are inserted into 7 pairs of autosomes and one pair of sex chromosomes (female=ZW, male=ZZ), each chromosome ranging in size from 18 to 73 MB (Simpson et al. 1982; LoVerde et al. 2004; http://lifecenter.sgst.cn/schistosoma/en/genomeProject.do; www.chgc.sh.cn/japonicum). Moreover, several transcriptomic analyses have been reported for S. mansoni (Franco et al. 1995, 1997; Santos et al. 1999; Verjovski-Almeida et al. 2003; Oliveira et al. 2004), and S. japonicum (Hu et al. 2003; Peng et al. 2003; McManus et al. 2004), contributing to a total of 205 892 expressed sequence tags (ESTs) from S. mansoni and 103 725 for S. japonicum in the dbEST database (dbEST; release 071009 – July 10, 2009), the vast majority of which are still unannotated. Given that schistosomes, like all other digenean species, exhibit complex life cycles involving multiple developmental stages, it is expected that at least 1000 genes would be uniquely expressed in a stage-specific manner (Wilson et al. 2006), although this estimate has yet to be experimentally verified. In terms of identifying genes expressed in the intramolluscan larval stages, transcriptomic analysis of S. mansoni has produced the most comprehensive datasets on gene expression in which 45 367 of 163 000 open-reading frame ESTs (ORESTs) were derived from miracidia, sporocysts (germballs) and cercariae (Verjovski-Almeida et al. 2003). For S. japonicum, 84 499 total ESTs were generated, but unfortunately only 569 ESTs were derived from miracidia (433) and cercariae (136), with the rest originating from non-molluscan parasite stages including adult worms and eggs (Peng et al. 2003; Liu et al. 2006).
With the exception of S. mansoni and S. japonicum, the accumulated genomic and transcript sequence data on other schistosome species or nonschistosome flukes are sparce indeed. For the third major human-infecting species of blood fluke, S. haematobium, dbEST lists only 6 ESTs, while GenBank entries (NCBI) total 127, with 21 sequences from cercariae and the rest from adult worms or unknown sources. Fasciola hepatica (common liver fluke) and F. gigantica (intestinal fluke), causative agents of fascioliasis in sheep and cattle (occasionally humans), are distributed worldwide and represent significant sources of economic losses in the livestock industry (McManus and Dalton, 2006). To date, there are only 324 and 149 nucleotide sequences in the NCBI gene database and 242 and 91 protein sequence entries in the NCBI protein database for F. hepatica and F. gigantica, respectively, and most of these originated from adult worm sources. Another model trematode that has received considerable attention are the echinostomes, which currently lists ~400 ESTs for Echinostoma spp. in the NCBI dbEST. Most of these are from the miracidium and primary sporocyst, but very few have been annotated.
Although there is an impressive amount of gene sequence data for schistosomes and databases for other non-schistosome species continue to grow, those genes specifically expressed during larval stage development, with the exception of cercariae, have been largely under-represented or unidentified in large genome or transcriptome investigations compared to adult worm-, schistosomula- and egg-derived sequences (Verjovski-Almeida et al. 2003; Peng et al. 2003; Liu et al. 2006). Bioinformatic searches of the NCBI’s UNIGENE database revealed 10 219 and 9395 sequence clusters for S. mansoni and S. japonicum, respectively. Of total S. mansoni clusters, 3392 contain identified miracidial sequences, 3128 sporocyst cDNA sequences, and 2206 cercarial sequences, although many of the raw larval sequences are unknown/unannotated. For S. japonicum, only 352 and 137 sequence clusters contain cDNA sequences (transcripts) for miracidia and cercariae, respectively, and none for sporocysts (last update in the Unigene database 09/04/2008). Similarly few intramolluscan larval sequences are specifically identified from other non-schistosome trematode species.
Despite the considerable progress made in completing the sequencing and assembly of the S. mansoni and S. japonicum genomes, gene identification and annotation are far from complete. During annotation of the S. mansoni and S. japonicum transcriptomes approximately 55% and 35%, respectively (Verjovski-Almeida et al. 2003; Hu et al. 2003) of sequences shared no similarity with known genes. Given that approximately half of putative genes are classified as unknown or predict hypothetical proteins based on possession of apparent ORFs and/or conserved domains, there is a critical need for continued efforts in identifying and annotating these ‘unknown’ genes with the future goal of assessing their expression and functional roles throughout parasite development. Because the intramolluscan larval stages, with the exception of cercariae, have generally been neglected as specific subjects of detailed gene analyses, an updating of gene sequence identification of, and expression within, the miracidium, primary sporocysts and secondary sporocysts are needed. Another limitation of the current schistosome transcriptomes is that expression profiles represent a composite of the whole organism and not genes expressed in specific tissues/organs within these complex multi-cellular animals. Consequently, although assumed functions of certain transcripts using computational analyses may provide hints as to possible cellular localization, the actual tissue in which specific genes are expressed cannot be predicted with accuracy, rendering the data much less useful in terms of aggregated information (Dillon et al. 2007).
Finally, protein analyses (proteomics) of early developing larvae are just beginning and amino acid sequence data are slowly accumulating in the NCBI database. Currently there are only ~46 entries for S. mansoni sporocysts in the schistosome protein db, and of those, 36 are mucins (~83% of the total) reported by Roger et al. (2008a). The same pattern is observed for other larval stages like miracidia and cercariae representing all schistosome species, in which only 13 miracidial and 62 cercarial sequences have been deposited to date. In S. japonicum, approximately 900 protein sequences were reported in miracidia by Liu et al. (2006). Overall, however, protein sequence data for molluscan schistosome larval forms is highly under-represented, further emphasizing the need for continued efforts to explore the larval proteome if we are to achieve a better understanding of the biology of these and other trematode groups. As detailed below, recent proteomic analyses of in vitro-cultured S. mansoni primary sporocysts and proteins released during miracidial transformation in culture represent significant additions to the larval protein database.
Regardless of the difficulties of manipulating and analysing the vast amounts of genomic and transcriptomic data in schistosomes, the mining of these data has stimulated the conduct of many spin-off investigations centering on intramolluscan larval stages including the study of SNPs and micro-satellites (Valentim et al. 2009), large-scale profiling of larval gene expression by microarray analysis (Vermeire et al. 2006) and SAGE (Williams et al. 2007; Taft et al. 2009), stage-associated proteome analyses (Curwen et al. 2004, 2006; Knudsen et al. 2005; Roger et al. 2008b; Wu et al. 2009) and assessments of the larval glycome (Robijn et al. 2005; Lehr et al. 2008; Peterson et al. 2009). An update on the status of gene and protein expression during larval trematode development using stage-specific profiling approaches is discussed below.
GENE AND PROTEIN PROFILING DURING INTRAMOLLUSCAN LARVAL DEVELOPMENT
An important first step in evaluating the functional significance of gene expression is to identify genes whose expression is associated with specific developmental stages and localize the encoded protein products within parasite cells/tissues to help infer potential biological activities or roles. For early intramolluscan developmental stages (miracidia, sporocysts, germballs, cercariae) such gene expression profiling has been accomplished using several approaches. In each of these cases, in vitro cultivation systems have provided a critical means of identifying stage-associated expression of various molecules, from genes to carbohydrates, which may have functional significance for that particular stage of development.
In recent years, investigations of gene and protein expression in different schistosome intramolluscan larval stages have become increasingly common as more sophisticated questions are being asked regarding the regulation of larval development, growth and asexual reproduction. A number of molecular approaches, such as cDNA subtraction (Sargent and Dawid, 1983), suppression subtractive hybridization (SSH, Diatchenko et al. 1996) and mRNA differential display (Liang and Pardee, 1992), have been used in other systems to compare gene expression repertoires between different populations, and have now been applied to larval trematodes (e.g. Adema et al. 2000; Coppin et al. 2002; Nowak and Loker, 2005). These techniques principally evaluate transcript presence vs. absence and as such, generally reveal little in the way of quantification of gene expression between populations under study. Now with the accumulation of robust databases of gene sequences, especially for schistosome species, high-throughput methods for both qualitative and quantitative profiling of multiple transcript expression have made it possible to evaluate ‘global’ gene expression in stage-associated comparative analyses. Two such approaches, microarray analysis and serial analysis of gene expression (SAGE), illustrate how quantitative, multi-gene expression profiling can provide valuable insights into the regulation of early developing intramolluscan larval stages.
Microarray analyses
The cDNA/oligonucleotide microarray has recently served as an important tool for the rapid (high-throughput) quantitative analyses of gene expression patterns in the intramolluscan stages of schistosome development including miracidia, sporocysts and cercariae. Based on mRNA hybridization to its complementary cDNA template, two different cDNA populations (targets) are synthesized, usually with Cy-dye fluorescent-labeled nucleotides (cDNA populations labeled with different dyes), from RNA samples of interest. Recently, dye-labeled target cDNAs can be created from very small cell/tissue samples by incorporating high-fidelity amplified RNA (aRNA) (Petalidis et al. 2003; Tang et al. 2009), which has greatly facilitated gene expression comparisons between scarce or low-abundance tissues or between parasite stages. Typically, pair-wise comparisons of relative transcript abundances are made by simultaneous hybridization of labeled cDNA populations to an array of pre-selected known genes affixed or spotted to a glass slide or silicon chip, allowing for the quantification of relative gene expression patterns by detection and measurement of the intensity of the fluorescent labels hybridizing to each spotted gene. In the last several years, microarray techniques have been used to identify stage-specific expression patterns of transcripts in the different S. mansoni and S. japonicum intramolluscan larval stages including miracidia, primary sporocysts (Vermeire et al. 2006; Gobert et al. 2009), secondary sporocysts (germballs) (Jolly et al. 2007), cercariae (Fitzpatrick et al. 2005; Jolly et al. 2007; Gobert et al. 2009) and even gender-associated transcript expression pattern in S. mansoni cercariae (Fitzpatrick et al. 2008). A subset of the reported up-regulated transcripts is summarized in Supplemental Table 1, organized by sequence-derived biological functions based on gene ontology (GO) annotation. Although this table is only a qualitative compilation of gene undergoing expression changes, quantitative comparisons are presented in the referenced citations. Taken together, the changes in relative transcript levels associated with specific larval stages are interpreted to represent the parasite’s response to the snail host environment, as well as its own changing needs for further growth and development. Indeed, there is a predominance of up-regulated transcripts related to specific biological functions for each of the intramolluscan larval stages (Fig. 3). For example S. mansoni miracidia show enrichment in transcripts encoding proteins related to functions like refolding/chaperoning (p40, HSP40, HSP70, HSP90), energy production (triosephosphate isomerase, phosphoenolpyruvate carboxykinase PEPCK), motility and calcium binding (SME16, calcineurin and Ca binding dynein-like protein), primary sporocysts have higher gene expression levels for anti-oxidants (e.g. peroxiredoxins, GST26/28 and Cu/Zn SOD), proteases (cathepsin, elastase, preprocathepsin and haemoglobinase) and protein synthesis/degradation (40 S and L37a ribosomal proteins, ubiquitins), while cercariae are enriched in proteins associated with energy production (ATP-synthase, lipid-binding protein, NADH dehydrogenase 4/5/7, ATPase subunit 6) and motility transcripts (tubulin, actin, myosin light and heavy chain).
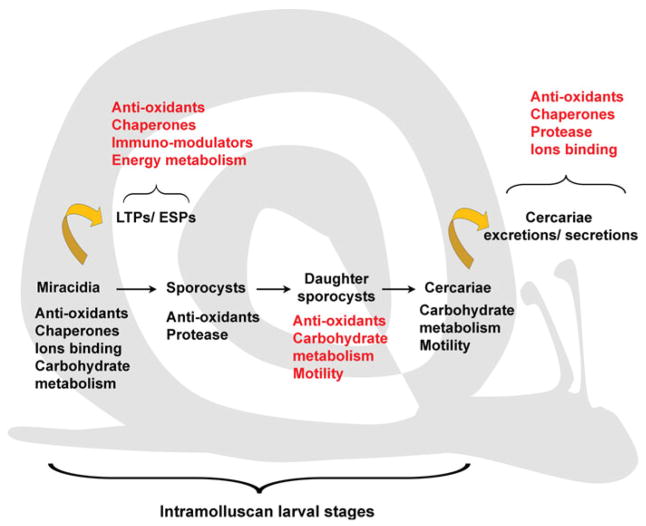
Fig. 3.
Schematic representation of Schistosoma intramolluscan larval stages; miracidia, primary (mother) sporocysts, secondary (daughter) sporocysts and cercariae. For each larval stage, specific biological functions corresponding to predominant up-regulated transcripts (black lettering) or proteins (red lettering) are described. Functional categories of differentially-expressed excretory-secretory proteins/products (ESPs) and larval transformation proteins (LTPs) released inside the molluscan intermediate host during miracidium-to-sporocysts transformation, and cercarial ESPs release during and following emergence from the snail host are also listed (red lettering).
These latter findings of higher transcript levels involving motility and energy production transcripts in both schistosome miracidia and cercariae were not unexpected considering that both are highly motile with accompanying high energy requirements. Similarly, although anti-oxidant transcripts are observed throughout the different intramolluscan larval stages, their expression is higher in miracidia and both primary and secondary sporocysts, as these are the first larval stages to confront a potentially hostile oxidative haemolymph environment of the snail host (Bender et al. 2002), as well as possible encounter with the snail’s immune defence system. In response to host reactive oxygen species (ROS), elevated anti-oxidant levels seen in early larvae suggest a potential role in protection against oxidative stress generated by the snail host, in addition to the need for larvae to maintain their own internal redox balance (Zelck and Von Janowsky, 2004; Vermeire and Yoshino, 2007). A similar transcript pattern is observed for Sm16, an anti-inflammatory glycoprotein (Ramaswamy et al. 1995), previously found in the cercarial secretory glands and believed to be involved in penetration into the mammalian host, as well as induction of apoptosis in responding immune cells (Curwen et al. 2006). Elevated steady-state Sj16 transcript levels in S. japonicum miracidia and sporocysts might suggest a similar role for this protein in host entry and/or defence against snail immune cells (Gobert et al. 2009). Sporocyst stages also appear to be significantly up-regulated in protease transcripts (cathepsin, elastase) and ribosomal genes. Proteases are believed to play an important role in the degrading of host tissue proteins, while elevated ribosomal proteins are indicative of high protein synthetic activity expected in these rapidly growing larval stages. These first multigene transcriptome analyses of different S. mansoni and S. japonicum intramolluscan stages provide valuable information supporting numerous hypotheses of the involvement of selected genes or gene families in parasite development, growth and survival within the snail host.
Gene expression in schistosome cercariae, although representing the ‘end-product’ of intramolluscan development, is of particular interest because sex-differentiation is presumed to occur during cercarial development within daughter sporocysts. Fitzpatrick et al. (2008) used a microarray referenced with genomic DNA to attempt a gender characterization of morphologically identical S. mansoni cercariae. By analyzing specific transcripts known to be gender-related, these authors successfully established specific transcriptional patterns distinguishing ‘male’ and ‘female’ cercariae. As would be expected, transcripts related to egg biology or to male germ cell development and reproduction were highly expressed in female and male cercariae, respectively. Moreover, genes encoding proteins involved in glucose transport, protein degradation (proteases) and immune response modulation dominate expression in ‘male’ cercariae, supporting the existing hypothesis that males are more capable when it comes to infection and survival within the mammalian host. The microarrays performed in S. mansoni and S. japonicum allowed the discovery of distinct patterns of gene expression that are assumed to reflect major biological functions for each of the specific intramolluscan larval stages. This suggests that regulation of transcriptional activity is highly dependent, not only on preset genetic programmes (e.g. those associated with sex-determination and activity), but certainly on the environment presented by the host. Microarray analyses allow a rapid, quantitative and simultaneous assessment of multiple gene expression in different populations at a specific developmental time points, or parasites maintained under different experimental conditions. However, one of the drawbacks of this technique is that construction of microarrays or the gene ‘chips’ themselves require prior knowledge of and sequence information for the genes of interest, thus limiting the study to the spectrum of genes for which sequence information is known.
Serial analysis of gene expression (SAGE)
An alternative to microarray analysis as an expression profiling tool is SAGE or serial analysis of gene expression (Velculescu et al. 1995). Unlike microarray, SAGE can be performed without prior pre-selection of specific transcripts, and allows for greater flexibility and potential transcripts coverage in its quantitative analysis of expressed transcripts. SAGE is based on similar principles as EST sequencing, with the additional notion that short tags (10–14 mers) of specific locations are sufficient to identify a specific transcript. In LongSAGE methods, tags are usually ~21 nt in length, a sequence length that is predicted through theoretical modelling to have a >99·8% chance of matching only once in a genome the size of humans (Saha et al. 2002). To create SAGE libraries, copies of cDNA are synthesized from target mRNA previously trapped on polyT-coated beads. Once synthesized, bead-immobilized cDNAs undergo a series of specific digestions, resulting in short nucleotide sequences (tags) that, in turn, are bound end-to-end, generating concatemers of multiple tags that are finally cloned and sequenced. Given the availability of the complete transcriptome of the organism of interest, this analysis permits the identification of expressed transcripts (genes), and by enumerating the SAGE tags for a given transcript, a quantitative comparison of specific gene expression between multiple sample libraries also can be ascertained (Williams et al. 2007). Verification that differentially expressed tags in fact represent differential expression of the genes of interest can be determined by real-time q-PCR or semi-quantitative reverse transcriptase PCR. In addition, SAGE allows for the identification of anti-sense transcripts, which have been hypothesized to function in post-transcriptional gene regulation in vertebrate cells (Luther et al. 1998; Hastings et al. 2000). The inverse-proportionality of sense-to-antisense tag counts in larval S. mansoni, and overall high representation of antisense tags (35%) (Taft et al. 2009) is consistent with high anti-sense transcript content reported in other parasites (Patankar et al. 2001; Radke et al. 2005), and suggests a potentially novel mechanism for post-translational gene regulation in larval schistosomes (Gunasekera et al. 2004).
To date, only two LongSAGE analyses in S. mansoni have included intramolluscan larval stages; the first, by Williams et al. (2007), produced an average of 60 000 tags from each of the 10 stage-specific SAGE libraries, and more recently, a detailed follow-up analysis of 21 440 unique SAGE tags from 5 intramolluscan larval libraries reported by Taft et al. (2009). In both studies, 5 groups of S. mansoni larvae were analyzed; 1 miracidial library, and 4 different primary sporocyst libraries from larvae maintained in in vitro culture for 6 or 20 days (representing early and late developing primary sporocysts, respectively) in 2 separate media supplemented with or without Biomphalaria glabrata embryonic (Bge) cell pre-conditioned medium. Larval cultivation in the presence/absence of conditioned Bge cell medium was an attempt to mimic the snail’s internal environment, and any resulting alterations in the parasite transcriptomic response. Although both SAGE studies identified numerous relevant transcripts specifically-associated with miracidial and sporocyst stages, the resulting transcript repertoire identified proteins that were highly consistent with previous microarray findings: namely a predominance of proteins/enzymes related to anti-oxidants, chaperones, egg antigens, carbohydrate metabolism and calcium-binding functions in miracidia, and anti-oxidant-, protease-, and ribosomal protein (protein synthesis)-related transcripts highly expressed in sporocysts. In general, this set of transcripts exhibiting enhanced expression appears to reflect parasite stages that are adjusting to abrupt physiological changes associated with the transition from a free-living to parasitic existence. By the same token, elevated steady-state transcript levels seen in sporocysts at 6 and 20 days of culture also were rich in growth and development-related genes, especially those involved in protein synthesis/metabolism (60 S acidic ribosomal protein P, 40 S ribosomal protein S19, elongation factor 1α, Golgi membrane protein, ubiquitin, HSP90 chaparonin) and energy production (ATP synthase B, ATPase) (Taft et al. 2009). Although not providing information as to the precise roles being played by the specific genes identified in these profiles, these studies can give an excellent impression of the major types of activities that are presumed to be important to larval stages at a particular point in their development.
Proteomic analyses
More information regarding the identity, functionality and origins of intramolluscan larval proteins would permit a better understanding of important physiological processes, in particular those associated with parasite defence and potential immune evasion mechanisms. A series of proteomic studies was recently performed focusing on: (1) miracidium-to-sporocyst transformation excretory-secretory (ES) proteins (Guillou et al. 2007; Gourbal et al. 2008), also termed larval transformation proteins (Wu et al. 2009); (2) protein differential expression between compatible and incompatible S. mansoni sporocysts (Roger et al. 2008a, b) and (3) proteins contained within or released by schistosome cercariae (Knudsen et al. 2005; Liu et al. 2006; Yang et al. 2009).
Using mass spectrometry (MS) techniques, recent studies have focused on identifying proteins released/secreted during in vitro miracidium-to-sporocyst transformation in several trematode species: S. mansoni (Guillou et al. 2007; Wu et al. 2009), E. caproni (Guillou et al. 2007) and F. hepatica (Gourbal et al. 2008). In their proteomic study, Wu et al. (2009) suggested a change in terminology from ‘excretory-secretory proteins/products’ (ESPs), commonly used to characterize proteins released during larval transformation in culture medium, to ‘larval transformation proteins’ (LTPs) in characterizing this group of molecules. The reason for the suggested change was based on their finding that approximatively 60% of the proteins identified as ‘ES’ transformation proteins lacked identifiable signal peptides or characteristics of non-classically-secreted proteins (Nickel, 2005), strongly suggesting that many proteins are being released by leakage or breakdown of ciliary epidermal plates during in vitro miracidia-to-sporocyst transformation. Interestingly, however, despite the fact that these studies focused on miracidia-to-sporocyst transformation in three different species and used different methods for preparing samples prior to and during MS analyses, they all identified proteins with similar functions. For example, as in the previously described transcriptomic studies, chaperones/stress proteins, anti-oxidants, and calcium-binding proteins were well represented in the proteome of these digenean species. In addition, the presence of various proteases and protease inhibitors are indicative of active digestive/nutritional processes or, combined with the presence of anti-oxidants and immune modulators, suggest LTP involvement in the protection of the parasite against ROS generated from plasma or circulating immune cells (haemocytes). The presence of functional antioxidant enzymes and their activity (Vermeire and Yoshino, 2007; Wu et al. 2009) and proteases/protease inhibitors that are shared between invading miracidia and the snail host suggest at least two types of LTP-mediated functions; (1) aggressive counter-measures to the host’s hostile environment or haemocyte responses or (2) more passive mechanisms whereby the parasite avoids host recognition and immune elimination by disrupting immune recognition or mimicking host molecular structures (Guillou et al. 2007; Wu et al. 2009).
Recent studies by Roger and colleagues (2008a, b, c), further support a passive interference hypothesis of host-parasite compatibility, as they identified differentially expressed sporocyst proteins between incompatible (IC, Guadeloupian) and compatible (C, Brazilian) S. mansoni strains. After clearly showing an antigenic variation between C and IC larvae, their differential proteomic study revealed an increase of a few proteins in C larval strain and the presence of 9 structurally-related, but highly polymorphic mucin proteins (Sm poMuc), differentially expressed between the 2 strains. Mucin-like proteins are believed to be involved in host-parasite interaction in various helminth species (Theodoropoulos et al. 2001), and are speculated to function as a potential smoke-screen or decoy against host defence (Roger et al. 2008c). In addition to differential expression poMuc in C and IC larvae, their presence in apical glands and in LTPs further support a potential protective role of these proteins in the intramolluscan stages of S. mansoni. As suggested earlier by Yoshino and Boswell (1986), the sharing of schistosome-specific glycotopes (Lehr et al. 2008; Peterson et al. 2009) also represents another type of mimetic mechanism in which larval stages express host-like molecules, thus avoiding recognition.
Acetabular gland contents and ESPs from S. mansoni cercariae and LTPs from transforming miracidia also exhibit similar expression patterns, with proteins belonging to the same predicted functional groups described previously (Appendix A; Supplementary Table 1). However, a few proteins appeared specific to cercarial ES, such as elastase, the 20·8/21·7 kDa Ca-binding proteins and HSP60, whereas S. mansoni LTPs were found strongly enriched in a variety of venom allergen-like proteins, suspected of being potentially involved in immuno-modulation and larval infection. Both miracidial LTPs and cercarial ES proteins (released upon simulated invasion) clearly show a similar pattern in functionality, which can be explained by the fact that these two larval stages are responsible for active penetration and host entry, migration/movement within host tissues, maintaining energy production, and aggressive counter-measures against oxidative stress and immune reactivity. A similar case can be made for S. japonicum carcariae, in which UV treatment-induced abrogation of proteins involved in motility, energy metabolism and protein transport (chaparones) were shown to be important for parasite infection and survival (Yang et al. 2009). Indeed as shown by Liu et al. (2006), like S. mansoni, cercariae and miracidia of S. japonicum share a common proteome when it comes to expression of proteins involved in host entry (calpain, calreticulin), motility (α- and β-tubulins, dynein light chain, myosin heavy chain, actin), energy metabolism (ATP synthase, ADP/ATP carrier protein, MDH, GAPDH, PepCK, fructose bisphosphate aldolase) and stress responses (several HSP homologues, thioredoxin peroxidase).
Although cercarial ES proteins are believed to potentially originate from the secretory glands and cytosolic component of the tegument (Knudsen et al. 2005), the origins of miracidial LTPs have only recently been explored. Polyclonal antibodies generated against S. mansoni LTPs and poMuc in two different studies (Wu et al. 2009; Roger et al. 2008b, respectively), revealed differential immunolocalization patterns. Anti-LTPs antibodies showed no specific vesicle or gland cross-reactivity, but strong reactivity to ciliated epidermal plates of miracida and the sporocyst tegument. This supports the hypothesis that, similar to cercarial ES proteins, a subset of miracidial LTPs probably originate from the tegument. Anti-epidermal plate reactivity likely is a result of proteins released into LTP from plates undergoing lysis following larval detachment. Anti-bodies against poMuc, however, immunolocalized to miracidial and sporocyst apical glands, which are believed to be involved in the production of secretions essential to snail host invasion. Regardless of their specific function or localization, both the miracidium and the cercaria release an impressive variety of proteins whose main functions are likely targeted larval survival, defence, and/or development.
Over the past decade, there has been an impressive collection and archiving of gene and protein sequence data focused mainly on two schistosome species, S. mansoni and S. japonicum. Due to our current ability to culture the early intramolluscan stages of these digeneans we can now qualitatively and/or quantitatively assess protein and transcript patterns across the intramolluscan larval stages, and as a result, are beginning to have a glimpse of the complex mechanisms used by trematode larvae during invasion, survival, and development within its intermediate host. With the next generation 454 pyrosequencing technology (Nordstrom et al. 2001; Vera et al. 2008) it is now possible to simultaneously evaluate multi-transcriptomic samples both qualitatively (gene discovery) and quantitatively (gene expression), and with the completion of genome sequencing, assembly and annotation of both S. mansoni and its snail intermediate host, Biomphalaria, ultra-sequencing using pyrosequencing methodologies will provide valuable insights into snail host-larval gene interactions (‘interactome’) (Barakat et al. 2009). Despite the creation of these ever-growing gene/protein databases, a full understanding of the functional role played by identified genes and gene products ultimately will depend on our ability to manipulate their expression within the living, intact parasite, and this represents the next critical challenge facing molecular helminthology investigators.
IN VITRO MANIPULATION OF GENE EXPRESSION AND FUNCTION IN INTRAMOLLUSCAN LARVAL STAGES
As amply demonstrated above, gene and protein discovery efforts have resulted in an ever-growing accumulation of molecular and biochemical data cataloguing numerous genes/gene products and their expression in various digenetic trematodes species, most notably the schistosomes, fasciolids and echinostomes. Moreover, for two of the major human fluke pathogens, S. mansoni and S. japonicum, life cycle stage-associated gene and protein expression profiling provides important clues as to the potential involvement of identified proteins or putative protein homologues in the myriad of molecular activities driving the development and/or maintenance of a given developmental stage. Unfortunately, this kind of information provides only limited understanding or insight into the actual functional role(s) of specifically-expressed genes or their protein products within the intact organism. Thus, effective approaches for predictably manipulating endogenous expression of specific genes within trematodes still represent a formidable barrier to exploring gene functions as they relate to the biology of these complex, multi-stage parasites and their interactions with their intermediate and definitive hosts.
Gene manipulation in Trematoda: development of methodologies
Methodologies for manipulating genes and their expression in flukes are still in its infancy, although significant progress is being made in the areas of transgenesis (i.e. introduction and expression of heterologous or homologous gene constructs using DNA plastid vectors, DNA transposons, retrotransposons or replication-defective retroviruses) and RNA interference (RNAi). The work involving trematode transgenesis has focused almost exclusively on the schistosome blood flukes and has emphasized both snail and mammalian host stages of development as experimental models. Likewise, functional gene studies using RNAi also have emphasized the schistosomes, with a focus mainly on the mammalian stages of infection. Recent excellent reviews have summarized in detail the technologies underlying the construction, delivery and expression analyses of the variety of gene constructs used in demonstrating transient transfection and viral transformation within the Schistosoma spp. (Grevelding, 2006; Beckmann et al. 2007; Brindley and Pearce, 2007; Kalinna and Brindley, 2007; Mann et al. 2008; Han et al. 2009), and gene-specific knockdown in flukes by RNAi (Skelly, 2006; Geldhof et al. 2007; Kalinna and Brindley, 2007; Pearce and Freitas, 2008). In keeping with the theme of this review, the following discussion will focus on how these technologies have been applied specifically to evaluating gene function in the intramolluscan developmental stages of trematodes and the critical role being played by in vitro culture systems in the manipulation of specific larval genes.
Manipulation of gene expression in larval trematodes
Methods for the in vitro cultivation of intramolluscan larval stages, from the free-living miracidium to multi-generation sporocysts or rediae, have now evolved to the point where specific larval stages can be produced in vitro for use as potential targets of genetic manipulation, making it possible to begin addressing the role of specific genes during larval development. As pointed out previously, an added feature of transgenic approaches (silencing or over-expressing genes of interest) in the miracidium or sporocyst stages is the likelihood of effecting changes within embryonic germ cells, which then have the potential of being passed on to progeny (assuming expression is not deleterious) through the normal asexual reproductive process. Live, genetically-transformed miracidia or sporocysts carrying altered genes can then be transferred to compatible uninfected snails through natural infection or surgical transplantation, respectively. Although RNAi provides only transient knockdown effects, this method currently represents the only approach in which specific endogenous genes can be manipulated and larvae assessed for potential functional defects. Advances in double-stranded RNA delivery by transgenic approaches, as well as precautions in the set-up, execution and interpretation of findings in RNAi-type experiments also will be discussed.
Transgenesis in larval trematodes
Transgenesis, or the introduction and induction of expression of exogenous or foreign genes, has enjoyed considerable success in its application to trematode larval stages, particularly in the schistosomes. Although all stages of schistosomes to date have been subjects of attempted transfection, we will mainly focus this review on approaches involving the intramolluscan larval stages. Arguably, these stages represent the most desirable targets for transfection due to their ease of in vitro manipulation, the abundance of germinal cells readily accessible to transgene targeting while carrying the possibility for germline introduction and genomic integration, and finally the ability to propagate transgene-carrying progeny by natural infection or surgical transfer of transfected asexually-reproducing stages (miracidia or sporocysts) into suitable snail hosts (Kapp et al. 2003; Grevelding, 2006; Kalinna and Brindley, 2007).
The miracidium and in vitro-cultured primary sporocyst have attracted considerable attention as targets of transgenesis for reasons mentioned above. The earliest attempts to successfully deliver and express exogenous transcripts or transgenes utilized particle bombardment (biolistics) in which the gold particles coated with DNA encoding the green fluorescent protein (GFP) ORF flanked by S. mansoni HSP70 promoter and terminator sequences, were introduced into live S. mansoni sporocysts with a particle accelerator or ‘gene gun’ (Wippersteg et al. 2002a). The presence of GFP transcripts (by RT-PCR) in sporocyst extracts and GFP protein expression (by confocal fluorescent microscopy) provided evidence for transient larval transfection. Follow-up experiments showed that biolistic introduction of GFP flanked with promoter/termination sequences of the cysteine proteinase ER60, localized and expressed in lateral glands and cytons of the protonephridia, tissues with putative proteinase activity and known excretory-secretory function (Wippersteg et al. 2002b, 2003). These seminal studies showed the tractability of introducing and expressing foreign DNA in living sporocysts and opened up the possibility of tissue-specific targeting of transgene expression. The next important break-through in larval transgenesis was the demonstration that transgenes introduced into miracidia were expressed in developing sporocysts following natural infection of snails (Heyers et al. 2003). Biolistics was used to deliver an enhanced GFP (eGFP) construct flanked by an HSP70 promoter/terminator into S. mansoni miracidia, followed by infection of B. glabrata snails. Histological localization of gold particles near germinal cells in primary sporocysts of snails with 14-day-old infections and the finding of eGFP mRNA expression by nested PCR in infected, but not uninfected, snail tissues demonstrated the feasibility of transgene delivery and expression in miracidia, and importantly, the possibility of continuing propagation of germline-altered progeny (sporocysts, cercariae) and establishment of transgenic schistosome lines. This latter senario recently was eloquently demonstrated by Beckmann et al. (2007), who introduced plastid GFP constructs driven by 1·5 kb promoter fragments of the S. mansoni actin gene (Sm Act1) into miracidia (F0 generation) using biolistics, and detected GFP transcript expression through the F0 and F1 generations of the entire life cycle; F0 miracidia → F0 cercariae → F0 adult worms generated in hamsters → F1 miracidia from F0 adults → F1 cercariae → F1 adults. Loss of transcript detection in F2 stages indicated that, although clearly being carried in germline cells, the GFP transgene was not stably integrated, if at all, in the germinal cell genome (Beckmann et al. 2007).
A successful alternative approach for delivery and expression of foreign or altered genes in the various schistosome stages explores the use of transposons, retrotransposons and pseudo-typed retroviruses as schistosome-transducing agents (Mann et al. 2008). Applications to asexually developing larval stages have been limited, although work reported by Kines et al. (2006) appears to show promise. In vitro-cultured S. mansoni sporocysts were infected with S. mansoni promoter-flanked eGFP sequence incorporated into a Moloney murine leukaemia retroviral vector packaged into a non-replicative VSV-pseudotyped retrovirus. Larval exposure of the retroviral construct in polybrene resulted in infection and viral transduction of sporocysts as evidenced by localization of virions at the larval surface, integration of proviral sequences into sporocyst genomic DNA, and detection of reporter gene expression in virally-infected, cultured larvae. Whether or not retroviral transformation will prove to be a practical method for producing genetically-modified schistosome lines has yet to be determined. However, early results show considerable promise.
RNA interference as a functional genomics tool in trematodes
The post-transcriptional silencing of specific messenger RNA expression by RNA interference (RNAi) in parasitic helminths, including trematodes, is a significant advance in parasite postgenomics, as it represents the only approach to date for experimentally manipulating expression of specifically-targeted endogenous genes, thereby providing insight into putative gene function. Although there is some controversy as to whether helminth parasites, in particular parasitic nematodes, possess a fully-functional RNAi mechanism (Geldhof et al. 2007; Knox et al. 2007; Viney and Thompson, 2008), accumulating evidence for trematodes strongly supports a functional system for the regulation of mRNA expression by small interfering (si) RNA and/or microRNA (Verjovski-Almeida et al. 2003; Xue et al. 2008; Krautz-Peterson and Skelly, 2008; Gomes et al. 2009). Because several recent reviews and primary reports cover the topic of RNAi applications in trematodes, mainly emphasizing the mammalian stages of schistosomes (Skelly, 2006; Geldhof et al. 2007; Kalinna and Brindley, 2007; Brindley and Pearce, 2007; Ndegwa et al. 2007; Morales et al. 2008; Pereira et al. 2008; Rinaldi et al. 2009; Krautz-Peterson et al. 2009 – in current special issue) and the liver fluke Fasciola hepatica (Rinaldi et al. 2008), we will focus our attention on its application to stages within the snail intermediate host; namely miracidia and primary sporocysts.
The possible existence of an RNAi-like mechanism in trematodes was first demonstrated in S. mansoni, initially in adult worms (Skelly et al. 2003) and then in the primary sporocyst (Boyle et al. 2003). In these studies, isolated cercariae and miracidia were treated in vitro with dsRNAs synthesized from transcripts of target genes by incubating (soaking) parasites for 6 days in dsRNA-containing media, during which time juvenile or larval stages transformed to their successive stages of development: cercariae → schistosomula and miracidia → sporocysts. These first attempts at silencing the cathepsin B gene in schistosomula (Skelly et al. 2003) and a glucose transporter (SGTP1) gene in primary sporocysts (Boyle et al. 2003) have now led to a number of follow-up studies investigating a variety of target transcripts, dsRNA/siRNA delivery systems and other experimental parameters aimed at optimizing the specificity and efficacy of gene knockdown (Skelly, 2006; Dinguirard and Yoshino, 2006; Krautz-Peterson et al. 2007; Ndegwa et al. 2007; Mourão et al. 2009). As a result of the convergence of technologies involving in vitro cultivation, stage-associated gene discovery/expression and gene manipulation through reverse-genetics approaches, we anticipate that functional genomic studies will not only continue in the schistosomes at an accelerating pace, but incorporate a diversity of other trematode species as well.
Assessing gene function in the development or activities of the miracidium of S. mansoni has been accomplished by introduction of gene-specific double-stranded (ds) RNA into eggs recovered from adult female worms or isolated from infected livers. Freitas et al. (2007) demonstrated that a S. mansoni Inhibin/Activin gene (SmInAct; member of the TGF-β signaling family) was a key regulator of miracidial embryogenesis by soaking eggs, newly deposited by cultured female worms, in SmInAct dsRNA and quantifying inhibition of larval development in ovo. A similar methodological approach was used to confirm previous pharmacological findings (Xu and Dresden, 1986) that S. mansoni leucine aminopeptidases (SmLAP) produced by miracidia in ovo were required for egg hatching. Rinaldi et al. (2009) exposed liver-isolated eggs to dsRNA of the two SmLAP isoforms (1 and 2) for 7 days at 37 C, after which time they were induced to hatch. Results demonstrated that egg hatching in the presence of SmLAP1 dsRNA, SmLAP2 dsRNA or both, was reduced by ~80% compared to both irrelevant dsRNA-treated and untreated control groups, and was comparable to groups treated with the LAP inhibitor bestatin. Concomitant with this hatching phenotype were significant reductions in specific LAP transcripts and enzymatic activity in dsRNA-treated eggs, indicating a clear relationship between LAP gene expression and hatching (Rinaldi et al. 2009). These studies not only provide a valuable method for screening potential drug targets for chemotherapeutic intervention, but also can be used for investigating the potential functions of genes involved in miracidial development, behaviour (e.g. ciliary activity, phototaxis), and larval transformation.
Application of RNAi methods to primary (mother) sporocyst stages of Schistosoma spp. also has taken advantage of in vitro culture methods that have permitted development of the miracidial to cercarial stages (Ivanchenko et al. 1999). As noted above, the first application of RNAi to sporocysts involved incubation (soaking) of freshly-isolated miracidia in snail ringers (CBSS) containing dsRNA and allowing them to transform to sporocysts followed by cultivation for 6 days in the presence of dsRNA (Boyle et al. 2003). Using dsRNA for the S. mansoni glucose transporter (SGTP1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), they demonstrated specific knockdown of SGTP1 transcripts upon exposure to SGTP1 dsRNA, using GAPDH dsRNA exposure as a specificity control. The gene knockdown effect was dependent on the presence of dsRNA during miracidial transformation and persisted in larvae maintained for up to 28-days in culture. Importantly, tritiated-glucose uptake by sporocysts treated with SGTP1 dsRNA, but not those exposed to GAPDH dsRNA, was significantly reduced, indicating a functional role of SGTP1, or a structurally related homologue (Skelly et al., 1994), in sporocyst glucose acquisition. A second study, using essentially the same protocols as Boyle et al. (2003), examined the role of a sporocyst-expressed CD36-like scavenger receptor (SR) gene in modified (acetylated) low-density lipoprotein (LDL) binding to the sporocyst tegument and larval growth (Dinguirard and Yoshino, 2006). Consistent with previous findings, exposure of S. mansoni miracidia to SR dsRNA during in vitro transformation to sporocysts and incubation for 6-days, resulted in a significant decrease in SR transcripts, decreases in the prevalence and intensity of acetylated-LDL binding to sporocysts, and reduction in larval length. These examples illustrate the types of functional information that may be gained by applying RNAi-type approaches to in vitro developing schistosome larvae.
Universality of gene-specific RNAi-like effects in cultured larval schistosomes
In our experience investigating dsRNA-mediated gene silencing we noted that not all genes employed in RNAi experiments yielded consistent and/or specific knockdowns. This prompted us to examine a wider range of different genes for their abilities to produce RNAi-like effects both at the transcriptional and phenotypic levels (Mourão et al. 2009). Briefly, in vitro transforming miracidia of S. mansoni were treated with dsRNAs (Boyle et al. 2003) generated from 32 genes expressed in primary sporocysts, including those encoding anti-oxidants, signaling proteins, transcription factors, metabolic enzymes and the like, and screened for morphological changes in larval phenotype. After 7 days in culture, a reduction in sporocyst size (length), similar to that reported by Dinguirard and Yoshino (2006), was the only morphological phenotype noted, and this was observed for 11 of the 32 genes examined. Of these 11 genes associated with the ‘size’ phenotype, 6 exhibited consistent knockdown in their steady-state transcript levels by real-time quantitative PCR, one (SOD) was highly elevated compared to the irrelevant GFP dsRNA-treatment control, while the remaining 4 phenotype-presenting genes were unchanged in transcript levels. The remaining 21 genes tested did not produce a consistent ‘size’ phenotype, although significant transcript knockdown was demonstrated for half of these genes (Mourão et al. 2009).
Results of this profiling study were enlightening as it pointed out several aspects of RNAi-type experiments incorporating larval schistosomes that require careful consideration: (1) The choice of gene to be examined – For reasons yet unknown transcript knockdown appears to be depend on the target gene or the specific gene segment being used to template dsRNA synthesis. Genes of interest should be pre-tested to ensure consistent, significant knockdown effects (Mourão et al. 2009). (2) Off-target effects – Some genes or gene segments may produce non-specific or off-target effects that are not consistent with the predicted interaction with and/or degradation of specific target transcripts, but instead appear to affect phenotype-associated non-target genes. Off-target effects are one of the common unwanted side-effects of RNAi manipulation (Kulkarni et al. 2006), although steps can be taken to minimize these effects (Reynolds et al. 2004). (3) Optimizing dsRNA delivery and timing – Although we use miracidial incubation (soaking) in dsRNA for 6 days as part of our standard delivery protocol for gene knockdown in sporocysts, others have incorporated square-wave electroporation as effective alternative dsRNA delivery methods for mammalian stages of schistosomes (Correnti et al. 2005; Osman et al. 2006; Sayed et al. 2006; Krautz-Peterson et al. 2007; Morales et al. 2008; Zhao et al. 2008) and fasciolids (Rinaldi et al. 2008). In preliminary electroporation experiments we have found that over a range of delivered voltages (65–230 V) and capacitances (25–100 mF), 65–70% maximum sporocysts survival rates were attained at 72 h post-treatment resulting in an apparent consistency of dsRNA delivery (Fig. 4). Follow-up studies are now needed to determine if, in fact, specific gene silencing in surviving larvae is achieved by this approach and how knockdown rates and long-term larval viability compare to soaking or other delivery methods. Consistent with previous finding in schistosomula (Skelly et al. 2003), in general, sporocysts did not tolerate lipofection-type reagents used within recommended concentration ranges.
Fig. 4.
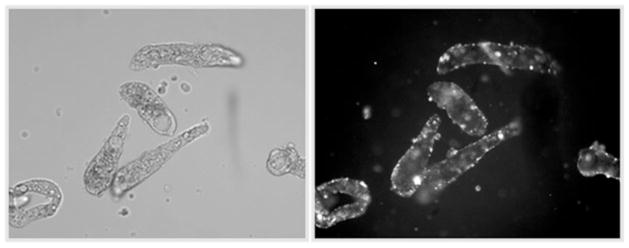
Example of Schistosoma mansoni primary sporocysts subjected to square-wave electroporation in the presence of rhodamine-labeled double-stranded (ds)RNA. Localization of electroporated dsRNA is mainly tegumental in discrete ‘patches’ (right panel; epifluorescence). Intact sporocysts are shown in the left panel.
SUMMARY AND CONCLUDING REMARKS
With the dramatic advancements in DNA and amino acid sequencing technologies made over the last decade, combined with the powerful analytical tools of bioinformatics, identification of genes and their products in diverse organisms, including model parasitic helminths, has become almost routine. However, it should come as no surprise that despite the rapid accumulation of gene/protein databases to date, our knowledge, and therefore understanding, of the role or function of identified genes/proteins is lagging far behind. This is especially evident in parasitic metazoans such as the digenetic trematodes, in which the entire genomes of selected species are only now completed. Although parasite expression profiling leading to the identification and activities of specific genes or gene networks can have immediate practical applications involving drug targeting or protective immune interventions (Brindley and Pearce, 2007; Pearce and Freitas, 2008; Han et al. 2009), experimental approaches for addressing fundamental questions of how genes may be functioning in the biology of parasites undergoing complex life cycle changes are urgently needed. As summarized in this review, part of this need is being met by the establishment of larval isolation and in vitro cultivation methods capable of generating and maintaining the major intramolluscan stages for trematodes of several prominent families (Schistosomatidae, Fasciolidae, Echinostomatidae), in combination with gene discovery efforts that have already yielded completed draft genomes of S. mansoni and S. japonicum, and ever-growing stage-associated transcriptomic and proteomic databases for these and other digenean species. The next major challenge for trematode biologists is to effectively combine these different approaches to investigating the functional role of expressed genes and their products in the maintenance, growth and development these parasites as they undergo morphological and physiological changes from one stage to the next. Recent exciting and promising research into gene transfer techniques (in vitro transgenesis and viral transduction) with the prospect of germline transfection, and reverse-genetic approaches using RNAi to manipulate endogenous gene function offer novel approaches for meeting this challenge as we truly advance into the post-genomics era.
Supplementary Material
Acknowledgments
Selected published and unpublished data cited in this review were supported by NIH grants AI061436 and AI015503 (TPY). MMM was supported by a CAPES (Process no. 4753/06-2) and MCT/CNPq 02/2006 Universal (Process no. 475151). We also thank Maria Castillo, Andrew Taft and 2 anonymous reviewers for their valuable input.
References
- Adema CM, Léonard PM, DeJong RJ, Day HL, Edwards DJ, Burgett G, Hertel LA, Loker ES. Analysis of messages expressed by Echinostoma paraensei miracidia and sporocysts, obtained by random EST sequencing. Journal of Parasitology. 2000;86:60–65. doi: 10.1645/0022-3395(2000)086[0060:AOMEBE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Ataev G, Fournaier A, Coustau C. Comparison of Echinostoma caproni mother sporocyst development in vivo and in vitro using Biomphalaria glabrata snails and a B. glabrata embryonic cell line. Journal of Parasitology. 1998;84:227–235. [PubMed] [Google Scholar]
- Barakat A, DiLoreto DS, Zhang Y, Smith C, Baier K, Powell WA, Wheeler N, Sederoff R, Carlson JE. Comparison of the transcriptomes of American chestnut (Castanea dentata) and Chinese chestnut (Castanea mollissima) in response to the chestnut blight infection. BMC Plant Biology. 2009;9:51. doi: 10.1186/1471-2229-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch PF. Schistosomes – Development, Reproduction and Host Relations. Oxford University Press; New York and Oxford: 1991. p. 248. [Google Scholar]
- Beckmann S, Wippersteg V, El-Bahay A, Hirzmann J, Oliveria G, Grevelding CG. Schistosoma mansoni: germ-line transformation approaches and actin-promoter analysis. Experimental Parasitology. 2007;117:292–303. doi: 10.1016/j.exppara.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Bender RC, Bixler LM, Lerner JP, Bayne CJ. Schistosoma mansoni sporocysts in culture: host plasma hemoglobin contributes to in vitro oxidative stress. Journal of Parasitology. 2002;88:14–18. doi: 10.1645/0022-3395(2002)088[0014:SMSICH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Berriman M, Haas BJ, Loverde PT, Wilson RA, Dillon GP, Cerqueira GC, Mashiyama ST, Al-Lazikani B, Andrade LF, Ashton PD, Aslett MA, Bartholomeu DC, Blandin G, Caffrey CR, Coghlan A, Coulson R, Day TA, Delcher A, Demarco R, Djikeng A, Eyre T, Gamble JA, Ghedin E, Gu Y, Hertz-Fowler C, Hirai H, Hirai Y, Houston R, Ivens A, Johnston DA, Lacerda D, Macedo CD, McVeigh P, Ning Z, Oliveira G, Overington JP, Parkhill J, Pertea M, Pierce RJ, Protasio AV, Quail MA, Rajandream MA, Rogers J, Sajid M, Salzberg SL, Stanke M, Tivey AR, White O, Williams DL, Wortman J, Wu W, Zamanian M, Zerlotini A, Fraser-Liggett CM, Barrell BG, El-Sayed NM. The genome of the blood fluke Schistosoma mansoni. Nature. 2009;460:352–358. doi: 10.1038/nature08160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle JP, Wu XJ, Shoemaker CB, Yoshino TP. Using RNA interference to manipulate endogenous gene expression in Schistosoma mansoni sporocysts. Molecular and Biochemical Parasitology. 2003;128:205–215. doi: 10.1016/s0166-6851(03)00078-1. [DOI] [PubMed] [Google Scholar]
- Brindley PJ, Pearce EJ. Genetic manipulation of schistosomes. International Journal for Parasitology. 2007;37:465–473. doi: 10.1016/j.ijpara.2006.12.012. [DOI] [PubMed] [Google Scholar]
- Campbell WC, Todd AC. In vitro metamorphosis of the miracidium of Fascioloides magna (Bassi, 1851) Ward, 1917. Transactions of the Americian Microscopical Society. 1955;74:225–228. [Google Scholar]
- Chernin E. Observations on hearts explanted in vitro from the snail Australorbis glabratus. Journal of Parasitology. 1963;49:353–364. [PubMed] [Google Scholar]
- Coppin JF, Lefebvre C, Caby S, Cocquerelle C, Vicogne J, Coustau C, Dissous C. Gene expression changes in Schistosoma mansoni sporocysts induced by Biomphalaria glabrata embryonic cells. Parasitology Research. 2002;89:113–119. doi: 10.1007/s00436-002-0643-2. [DOI] [PubMed] [Google Scholar]
- Correnti JM, Brindley PJ, Pearce EJ. Long-term suppression of cathepsin B levels by RNA interference retards schistosome growth. Molecular and Biochemical Parasitology. 2005;143:209–215. doi: 10.1016/j.molbiopara.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Coustau C, Ataev G, Jourdane J, Yoshino TP. Schistosoma japonicum: in vitro cultivation of miracidium to daughter sporocyst using a Biomphalaria glabrata embryonic cell line. Experimental Parasitology. 1997;87:77–87. doi: 10.1006/expr.1997.4184. [DOI] [PubMed] [Google Scholar]
- Coustau C, Yoshino TP. Flukes without snails: advances in the in vitro cultivation of intramolluscan stages of trematodes. Experimental Parasitology. 2000;94:62–66. doi: 10.1006/expr.1999.4462. [DOI] [PubMed] [Google Scholar]
- Curwen RS, Ashton PD, Johnston DA, Wilson RA. The Schistosoma mansoni soluble proteome: a comparison across four life-cycle stages. Molecular and Biochemistry Parasitology. 2004;138:57–66. doi: 10.1016/j.molbiopara.2004.06.016. [DOI] [PubMed] [Google Scholar]
- Curwen RS, Ashton PD, Sundaralingam S, Wilson RA. Identification of novel proteases and immunomodulators in the secretions of schistosome cercariae that facilitate host entry. Molecular and Cellular Proteomics. 2006;5:835–844. doi: 10.1074/mcp.M500313-MCP200. [DOI] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proceedings of the National Academy of Sciences, USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon GP, Illes JC, Isaacs HV, Wilson RA. Patterns of gene expression in schistosomes: localization by whole mount in situ hybridization. Parasitology. 2007;134:1589–1597. doi: 10.1017/S0031182007002995. [DOI] [PubMed] [Google Scholar]
- Dinguirard N, Yoshino TP. Potential role of a CD36-like class B scavenger receptor in the binding of modified low-density lipoprotein (acLDL) to the tegumental surface of Schistosoma mansoni sporocysts. Molecular and Biochemical Parasitology. 2006;146:219–230. doi: 10.1016/j.molbiopara.2005.12.010. [DOI] [PubMed] [Google Scholar]
- El-Sayed NM, Bartholomeu D, Ivens A, Johnston DA, LoVerde PT. Advances in schistosome genomics. Trends in Parasitolology. 2004;20:154–157. doi: 10.1016/j.pt.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, Johnston DA, Williams GW, Williams DJ, Freeman TC, Dunne DW, Hoffmann KF. An oligonucleotide microarray for transcriptome analysis of Schistosoma mansoni and its application/use to investigate gender-associated gene expression. Molecular and Biochemical Parasitology. 2005;141:1–13. doi: 10.1016/j.molbiopara.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick JM, Protasio AV, McArdle AJ, Williams GA, Johnston DA, Hoffmann KF. Use of genomic DNA as an indirect reference for identifying gender-associated transcripts in morphologically identical, but chromosomally distinct, Schistosoma mansoni cercariae. PLoS Negected Tropical Diseases. 2008;2:e323. doi: 10.1371/journal.pntd.0000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco GR, Adams MD, Soares MB, Simpson AJ, Venter JC, Pena SD. Identification of new Schistosoma mansoni genes by the EST strategy using a directional cDNA library. Gene. 1995;23:141–147. doi: 10.1016/0378-1119(94)00747-g. [DOI] [PubMed] [Google Scholar]
- Franco GR, Rabelo EM, Azevedo V, Pena HB, Ortega JM, Santos TM, Meira WS, Rodrigues NA, Dias CM, Harrop R, Wilson A, Saber M, Abdel-Hamid H, Faria MS, Margutti ME, Parra JC, Pena SD. Evaluation of cDNA libraries from different developmental stages of Schistosoma mansoni for production of expressed sequence tags (ESTs) DNA Research. 1997;30:231–240. doi: 10.1093/dnares/4.3.231. [DOI] [PubMed] [Google Scholar]
- Franco GR, Valadão AF, Azevedo V, Rabelo EM. The Schistosoma gene discovery program: state of the art. International Journal of Parasitology. 2000;30:453–463. doi: 10.1016/s0020-7519(00)00020-5. [DOI] [PubMed] [Google Scholar]
- Freitas TC, Jung E, Pearce EJ. TGF-β signaling controls embryo development in the parasitic flatworm Schistosoma mansoni. PLoS Pathogens. 2007;3:e52. doi: 10.1371/journal.ppat.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldhof P, Visser A, Clark D, Saunders G, Britton C, Gilleard J, Berriman M, Knox D. RNA interference in parasitic helminths: current situation, potential pitfalls and future prospects. Parasitology. 2007;134:609–619. doi: 10.1017/S0031182006002071. [DOI] [PubMed] [Google Scholar]
- Gobert GN, Moertel L, Brindley PJ, McManus DP. Developmental gene expression profiles of the human pathogen Schistosoma japonicum. BMC Genomics. 2009;10:128. doi: 10.1186/1471-2164-10-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes MS, Cabral FJ, Jannotti-Passos LK, Carvalho O, Rodriques V, Baba EH, Sa RG. Preliminary analysis of miRNA pathway in Schistosoma mansoni. Parasitology International. 2009;58:61–68. doi: 10.1016/j.parint.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Gourbal BEF, Guillou F, Mitta G, Sibille P, Theron A, Pointier JP, Coustau C. Excretory–secretory products of larval Fasciola hepatica investigated using a two-dimensional proteomic approach. Molecular and Biochemical Parasitology. 2008;161:63–66. doi: 10.1016/j.molbiopara.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Grevelding CG. Transgenic flatworms. In: Maule AG, Marks NJ, editors. Parasitic Flatworms: Molecular Biology, Biochemistry, Immunology, and Physiology. CABI; Oxfordshire, UK: 2006. pp. 149–173. [Google Scholar]
- Guillou F, Roger E, Mone Y, Rognon A, Grunau C, Theron A, Mitta G, Coustau C, Gourbal BEF. Excretory–secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Molecular and Biochemical Parasitology. 2007;155:45–56. doi: 10.1016/j.molbiopara.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Gunasekera AM, Patankar S, Schug J, Eisen G, Kissinger J, Roos D, Wirth DF. Widespread distribution of antisense transcripts in the Plasmodium falciparum genome. Molecular and Biochemical Parasitology. 2004;136:35–42. doi: 10.1016/j.molbiopara.2004.02.007. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Berriman M, Hirai H, Cerqueira GG, LoVerde PT, El-Sayed NM. Schistosoma mansoni genome: closing in on a final gene set. Experimental Parasitology. 2007;117:225–228. doi: 10.1016/j.exppara.2007.06.005. [DOI] [PubMed] [Google Scholar]
- Han ZG, Brindley PJ, Wang SY, Chen Z. Schistosome genomics: new perspectives on schistosome biology and host-parasite interactions. Annual Review of Genomics and Human Genetics. 2009;10:10.1–10.30. doi: 10.1146/annurev-genom-082908-150036. [DOI] [PubMed] [Google Scholar]
- Hastings ML, Ingle HA, Lazar MA, Munroe SH. Post-transcriptional regulation of thyroid hormone receptor expression by cis-acting sequences and a naturally occurring antisense RNA. Journal of Biological Chemistry. 2000;275:11507–11153. doi: 10.1074/jbc.275.15.11507. [DOI] [PubMed] [Google Scholar]
- Heyers O, Walduck AK, Brindley PJ, Bleizß W, Lucius R, Dorbic T, Wittig B, Kalinna BH. Schistosoma mansoni miracidia transformed by particle bombardment infect Biomphalaria glabrata snails and develop into transgenic sporocysts. Experimental Parasitology. 2003;105:174–178. doi: 10.1016/j.exppara.2003.11.001. [DOI] [PubMed] [Google Scholar]
- Hokke CH, Fitzpatrick JM, Hoffmann KF. Integrating transcriptome, proteome and glycome analyses of Schistosoma biology. Trends in Parasitology. 2007;23:165–174. doi: 10.1016/j.pt.2007.02.007. [DOI] [PubMed] [Google Scholar]
- Hu W, Yan Q, Shen DK, Liu F, Zhu ZD, Song HD, Xu XR, Wang ZJ, Rong YP, Zeng LC, Wu J, Zhang X, Wang JJ, Xu XN, Wang SY, Fu G, Zhang XL, Wang ZQ, Brindley PJ, McManus D, Zue CL, Feng Z, Chen Z, Han ZG. Evolutionary and biomedical implications of a Schistosoma japonicum complementary DNA resource. Nature Genetics. 2003;35:139–147. doi: 10.1038/ng1236. [DOI] [PubMed] [Google Scholar]
- Ivanchenko MG, Lerner JP, McCormick RS, Toumadje A, Allen B, Fischer K, Hedstrom O, Helmrich A, Barnes DW, Bayne CJ. Continuous in vitro propagation and differentiation of cultures of the intramolluscan stages of the human parasite Schistosoma mansoni. Proceedings of the National Academy of Sciences, USA. 1999;96:4965–4970. doi: 10.1073/pnas.96.9.4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolly ER, Chin CS, Miller S, Baghat MM, Lim KC, DeRisi J, McKerrow JH. Gene expression patterns during adaptation of a helminth parasite to different environmental niches. Genome Biology. 2007;8:R65. doi: 10.1186/gb-2007-8-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp K, Coustau C, Wippersteg V, Jourdane J, Kunz W, Grevelding CG. Transplantation of in vitro-generated Schistosoma mansoni mother sporocysts into Biomphalaria glabrata. Parasitology Research. 2003;91:482–485. doi: 10.1007/s00436-003-0951-1. [DOI] [PubMed] [Google Scholar]
- Kalinna BH, Brindley PJ. Manipulating the manipulators: advances in parasitic helminth transgenesis. Trends in Parasitology. 2007;23:197–204. doi: 10.1016/j.pt.2007.03.007. [DOI] [PubMed] [Google Scholar]
- Kines KJ, Mann VH, Morales ME, Shelby BD, Kalinna BH, Gobert GN, Chirgwin SR, Brindley PJ. Transduction of Schistosoma mansoni by vesicular stomatitis virus glycoprotein-pseudotyped Moloney murine leukemia retrovirus. Experimental Parasitology. 2006;112:209–220. doi: 10.1016/j.exppara.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Knox DP, Geldhof P, Visser A, Britton C. RNA interference in parasitic nematodes of animals: a reality check? Trends in Parasitology. 2007;23:105–107. doi: 10.1016/j.pt.2007.01.007. [DOI] [PubMed] [Google Scholar]
- Knudsen GM, Medzihradszky KF, Lim KC, Hansell E, McKerrow JH. Proteomic analysis of Schistosoma mansoni cercarial secretions. Molecular and Cellular Proteomics. 2005;4:1862–1875. doi: 10.1074/mcp.M500097-MCP200. [DOI] [PubMed] [Google Scholar]
- Krautz-Peterson G, Bhardwaj R, Faghiri Z, Tararam CA, Skelly PJ. RNA interference in schistosomes: machinery and methodology. Parasitology. 2009;137 doi: 10.1017/S0031182009991168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krautz-Peterson G, Radwanska M, Ndegwa D, Shoemaker CB, Skelly PJ. Optimizing gene suppression in schistosomes using RNA interference. Molecular and Biochemical Parasitology. 2007;153:194–202. doi: 10.1016/j.molbiopara.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Krautz-Peterson G, Skelly PJ. Schistosoma mansoni: the dicer gene and its expression. Experimental Parasitology. 2008;118:122–128. doi: 10.1016/j.exppara.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nature Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- Laursen JR, Yoshino TP. Biomphalaria glabrata embryonic (Bge) cell line supports in vitro miracidial transformation and early larval development of the deer liver fluke, Fascioloides magna. Journal of Parasitology. 1999;118:187–194. doi: 10.1017/s0031182098003710. [DOI] [PubMed] [Google Scholar]
- Lehr T, Beuerlein K, Doenhoff MJ, Grevelding CG, Geyer R. Localization of carbohydrate determinants common to Biomphalaria glabrata as well as to sporocysts and miracidia of Schistosoma mansoni. Parasitology. 2008;135:931–942. doi: 10.1017/S0031182008004514. [DOI] [PubMed] [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Liu F, Lu J, Hu W, Wang SY, Cui SJ, Chi M, Yan Q, Wang XR, Song HD, Xu XN, Wang JJ, Zhang XL, Zhang X, Wang ZQ, Xue CL, Brindley PJ, McManus DP, Yang PY, Feng Z, Chen Z, Han ZG. New perspectives on host-parasite interplay by comparative transcriptomic and proteomic analyses of Schistosoma japonicum. PLoS Pathogens. 2006;2:e29. doi: 10.1371/journal.ppat.0020029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loker ES, Cimino DF, Hertel LA. Excretory-secretory products of Echinostoma paraensei sporocysts mediate interference with Biomphalaria glabrata hemocyte function. Journal of Parasitology. 1992;78:104–115. [PubMed] [Google Scholar]
- LoVerde PT, Hirai H, Merrick JM, Lee NH, El-Sayed N. Schistosoma mansoni genome project: an update. Parasitology International. 2004;53:183–192. doi: 10.1016/j.parint.2004.01.009. [DOI] [PubMed] [Google Scholar]
- Luther HP, Haase H, Hohaus A, Beckmann G, Reich J, Morano I. Characterization of naturally occurring myosin heavy chain antisensemRNA in rat heart. Journal of Cellular Biochemistry. 1998;70:110–120. [PubMed] [Google Scholar]
- Mann VH, Morales ME, Kines KJ, Brindley PJ. Transgenesis of schistosomes: approaches employing mobile genetic elements. Parasitology. 2008;135:141–153. doi: 10.1017/S0031182007003824. [DOI] [PubMed] [Google Scholar]
- McManus DP, Dalton JP. Vaccines against the zoonotic trematodes Schistosoma japonicum, Fasciola hepatica and Fasciola gigantica. Parasitology. 2006;133:S43–61. doi: 10.1017/S0031182006001806. [DOI] [PubMed] [Google Scholar]
- McManus DP, Hu W, Brindley PJ, Feng Z, Han ZG. Schistosome transcriptome analysis at the cutting edge. Trends in Parasitology. 2004;20:301–304. doi: 10.1016/j.pt.2004.04.011. [DOI] [PubMed] [Google Scholar]
- Morales ME, Rinaldi G, Gobert GN, Kines KJ, Tort JF, Brindley PJ. RNA interference of Schistosoma mansoni cathepsin D, the apical enzyme of the hemoglobin proteolysis cascade. Molecular and Biochemical Parasitology. 2008;157:160–168. doi: 10.1016/j.molbiopara.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourão MM, Dinguirard N, Franco GR, Yoshino TP. Phenotypic screen of early-developing larvae of the blood fluke, Schistosoma mansoni, using RNA interference. PLoS Neglected Tropical Diseases. 2009;3:e502. doi: 10.1371/journal.pntd.0000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndegwa D, Krautz-Peterson G, Skelly PJ. Protocols for gene silencing in schistosomes. Experimental Parasitology. 2007;117:284–291. doi: 10.1016/j.exppara.2007.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W. Unconventional secretory routes: direct protein export across the plasma membrane of mammalian cells. Traffic. 2005;6:607–614. doi: 10.1111/j.1600-0854.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- Nordstrom T, Gharizadeh B, Pourmand N, Nyren P, Ronaghi M. Method enabling fast partial sequencing of cDNA clones. Analytical Biochemistry. 2001;15:266–271. doi: 10.1006/abio.2001.5094. [DOI] [PubMed] [Google Scholar]
- Nowak TS, Loker ES. Echinostoma paraensei: differential gene transcription in the sporocyst stage. Expimental Parasitology. 2005;109:94–105. doi: 10.1016/j.exppara.2004.11.005. [DOI] [PubMed] [Google Scholar]
- Osman A, Niles EG, Verjovski-Almeida S, LoVerde PT. Schistosoma mansoni TGF-β receptor II: role in host ligand-induced regulation of a schistosome target gene. PLoS Pathogen. 2006;2:e54. doi: 10.1371/journal.ppat.0020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira GC, Franco G, Verjovski-Almeida S. The Brazilian contribution to the study of the Schistosoma mansoni transcriptome. Acta Tropica. 2008;108:179–182. doi: 10.1016/j.actatropica.2008.04.022. [DOI] [PubMed] [Google Scholar]
- Oliveira GC, Rodrigues NB, Romanha A, Bahia D. Genome and genomics of schistosomes. Canadian Journal of Zoology. 2004;82:375–390. [Google Scholar]
- Patankar S, Munasinghe A, Shoaibi A, Cummings LM, Wirth DF. Serial analysis of gene expression in Plasmodium falciparum reveals the global expression profile of erythrocytic stages and the presence of anti-sense transcripts in the malarial parasite. Molecular Biology of the Cell. 2001;12:3114–1325. doi: 10.1091/mbc.12.10.3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EJ, Freitas TC. Reverse genetics and the study of the immune response to schistosomes. Parasite Immunology. 2008;30:215–221. doi: 10.1111/j.1365-3024.2007.01005.x. [DOI] [PubMed] [Google Scholar]
- Peng HJ, Chen XG, Wang XZ, Lun ZR. Analysis of the gene expression profile of Schistosoma japonicum cercariae by a strategy based on expressed sequence tags. Parasitology Research. 2003;90:287–293. doi: 10.1007/s00436-003-0843-4. [DOI] [PubMed] [Google Scholar]
- Pereira TC, Pascoal VDB, Marchesini RB, Maia IG, Magalhaes LA, Zanotti-Magalhaes EM, Lopes-Cendes I. Schistosoma mansoni: Evaluation of an RNAi-based treatment targeting HGPRTase gene. Experimental Parasitology. 2008;118:619–623. doi: 10.1016/j.exppara.2007.11.017. [DOI] [PubMed] [Google Scholar]
- Petalidis L, Bhattacharyya S, Morris GA, Collins VP, Freeman TC, Lyon PA. Global amplification of mRNA by template-switching PCR: linearity and application to microarray analysis. Nucleic Acids Research. 2003;25:402–408. doi: 10.1093/nar/gng142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson NA, Hokke CH, Deelder AM, Yoshino TP. Glycotope analysis in miracidia and primary sporocysts of Schistosoma mansoni: differential expression during the miracidium-to-sporocyst transformation. International Journal for Parasitology. 2009;39:1331–1344. doi: 10.1016/j.ijpara.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy K, Salafsky B, Potluri S, He YX, Li JW, Shibuya T. Secretion of an anti-inflammatory, immunomodulatory factor by Schistosomulae of Schistosoma mansoni. Journal of Inflammation. 1995;46:13–22. [PubMed] [Google Scholar]
- Radke JR, Behnke MS, Mackey AJ, Radke JB, Roos DS, White MW. The transcriptome of Toxoplasma gondii. BMC biology. 2005;2:3–26. doi: 10.1186/1741-7007-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nature Biotechnology. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rinaldi G, Morales ME, Alrefaei YN, Cancela M, Castillo E, Dalton JP, Tort JF, Brindley PJ. RNA interference targeting leucine aminopeptidase blocks hatching of Schisotosoma mansoni eggs. Molecular and Biochemical Parasitology. 2009;167:118–126. doi: 10.1016/j.molbiopara.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaldi G, Morales ME, Cancela M, Castillo E, Brindley PJ, Jose F, Tort JF. Development of functional genomic tools in trematodes: RNA interference and luciferase reporter gene activity in Fasciola hepatica. PLoS Neglected Tropical Diseases. 2008;2:e260. doi: 10.1371/journal.pntd.0000260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijn ML, Wujrer M, Kornelis D, Deelder AM, Geyer R, Hokke CH. Mapping fucosylated epitopes on glycoproteins and glycolipids of Schistosoma mansoni cercariae, adult worms and eggs. Parasitology. 2005;130:67–77. doi: 10.1017/s0031182004006390. [DOI] [PubMed] [Google Scholar]
- Roger E, Gourbal B, Grunau C, Pierce RJ, Galinier R, Mitta G. Expression analysis of highly polymorphic mucin proteins (Sm PoMuc) from the parasite Schistosoma mansoni. Molecular and Biochemical Parasitology. 2008b;157:217–227. doi: 10.1016/j.molbiopara.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Roger E, Grunau C, Pierce RJ, Hirai H, Gourbal B, Galinier R, Emans R, Cesari IM, Cosseau C, Mitta G. Controlled chaos of polymorphic mucins in a metazoan parasite (Schistosoma mansoni) interacting with its invertebrate host (Biomphalaria glabrata) PLoS Neglected Tropical Diseases. 2008c;2(11):e330. doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger E, Mitta G, Mone Y, Bouchut A, Rognon A, Grunau C, Boissier J, Theron A, Gourbal BEF. Molecular determinants of compatibility polymorphism in the Biomphalaria glabrata/Schistosoma mansoni model: New candidates identified by a global comparative proteomics approach. Molecular and Biochemical Parasitology. 2008a;157:205–216. doi: 10.1016/j.molbiopara.2007.11.003. [DOI] [PubMed] [Google Scholar]
- Saha S, Sparks AB, Rago C, Akmaev V, Wang CJ, Vogelstein B, Kinzler KW, Velculescu VE. Using the transcriptome to annotate the genome. Nature Biotechnology. 2002;20:508–512. doi: 10.1038/nbt0502-508. [DOI] [PubMed] [Google Scholar]
- Santos TM, Johnston DA, Azevedo V, Ridgers IL, Martinez MF, Marotta GB, Santos RL, Fonseca SJ, Ortega JM, Rabelo EM, Saber M, Ahmed HM, Romeih MH, Franco GR, Rollinson D, Pena SD. Analysis of the gene expression profile of Schistosoma mansoni cercariae using the expressed sequence tag approach. Molecular and Biochemistry Parasitology. 1999;20:79–97. doi: 10.1016/s0166-6851(99)00100-0. [DOI] [PubMed] [Google Scholar]
- Sargent TD, Dawid IB. Differential gene expression in the gastrula of Xenopus laevis. Science. 1983;222:135–139. doi: 10.1126/science.6688681. [DOI] [PubMed] [Google Scholar]
- Sayed AH, Cook SK, Williams DL. Redox balance mechanisms in Schistosoma mansoni rely on peroxiredoxins and albumin and implicate peroxiredoxins as novel drug targets. Journal of Biological Chemistry. 2006;281:17001–17010. doi: 10.1074/jbc.M512601200. [DOI] [PubMed] [Google Scholar]
- Simpson AJ, Sher A, McCutchan TF. The genome of Schistosoma mansoni: isolation of DNA, its size, bases and repetitive sequences. Molecular and Biochemistry Parasitology. 1982;6:125–137. doi: 10.1016/0166-6851(82)90070-6. [DOI] [PubMed] [Google Scholar]
- Skelly PJ. Gene silencing in flatworms using RNA interference. In: Maule AG, Marks NJ, editors. Parasitic Flatworms: Molecular Biology, Biochemistry, Immunology, and Physiology. CABI; Oxfordshire, UK: 2006. pp. 423–434. [Google Scholar]
- Skelly PJ, Da’dara A, Harn DA. Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. International Journal for Parasitology. 2003;33:363–369. doi: 10.1016/s0020-7519(03)00030-4. [DOI] [PubMed] [Google Scholar]
- Skelly PJ, Kim JW, Cunningham J, Shoemaker CB. Cloning, characterization, and functional expression of cDNAs encoding glucose transporter proteins from the human parasite Schistosoma mansoni. Journal of Biological Chemistry. 1994;269:4247–4253. [PubMed] [Google Scholar]
- Taft AS, Vermeire JJ, Bernier J, Birkeland SR, Cipriano MJ, Papa AR, McArthur AG, Yoshino TP. Transcriptome analysis of Schistosoma mansoni larval development using serial analysis of gene expression (SAGE) Parasitology. 2009;136:469–485. doi: 10.1017/S0031182009005733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-seq whole-transcriptome analysis of a single cell. Nature Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Theodoropoulos G, Hicks SJ, Corfield AP, Miller BG, Carrington SD. The role of mucins in host-parasite interactions. Part II. Helminth parasites. Trends in Parasitology. 2001;17:130–135. doi: 10.1016/s1471-4922(00)01775-x. [DOI] [PubMed] [Google Scholar]
- The Schistosoma japonicum Genome Sequencing and Functional Analysis Consortium; Genome annotation and evolution analysis: Zhou, Y., Zheng, H., Chen, Y., Zhang, L., Wang, K., Guo, J., Huang, Z., Zhang, B., Huang, W., Jin, K., Dou, T., Hasegawa, M., Wang, L., Zhang, Y., Zhou, J., Tao, L., Cao, Z., Li, Y., Vinar, T., Brejova, B., Brown, D., Li, M., Miller, D. J., Blair, D., Zhong, Y., Chen, Z., Functional genomics analysis: Liu, F., Hu, W., Wang, Z. Q., Zhang, Q. H., Song, H. D., Chen, S., Xu, X., Xu, B., Ju, C., Huang, Y., Brindley, P. J., McManus, D. P., Feng, Z., Han, Z. G., Sequencing and assembly: Lu, G., Ren, S., Wang, Y., Gu, W., Kang, H., Chen, J., Chen, X., Chen, S., Wang, L., Yan, J., Wang, B., Li, X., Jin, L., Wang, B., Pu, S., Zhang, X., Zhang, W., Hu, Q., Zhu, G., Wang, J., Yu, J., Wang, J., Yang, H., Ning, Z., Berriman, M., Wei, C. L., Ruan, Y., Zhao, G., Wang, S., Paper writing: Liu, F., Zhou, Y., Wang, Z. Q., Lu, G., Zheng, H., Brindley, P. J., McManus, D. P., Blair, D., Zhang, Q. H., Zhong, Y., Wang, S., Han, Z. G, Chen, Z., Project leaders: Wang, S., Han, Z. G. and Chen, Z. The Schistosoma japonicum genome reveals features of host-parasite interplay. Nature. 2009;460:345–351. doi: 10.1038/nature08140.
- Valentim CL, LoVerde PT, Anderson TJ, Criscione CD. Efficient genotyping of Schistosoma mansoni miracidia following whole genome amplification. Molecular and Biochemistry Parasitology. 2009;166:81–84. doi: 10.1016/j.molbiopara.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Molecular Ecology. 2008;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Verjovski-Almeida S, DeMarco R, Martins EA, Guimarães PE, Ojopi EP, Paquola AC, Piazza JP, Nishiyama MY, Jr, Kitajima JP, Adamson RE, Ashton PD, Bonaldo MF, Coulson PS, Dillon GP, Farias LP, Gregorio SP, Ho PL, Leite RA, Malaquias LC, Marques RC, Miyasato PA, Nascimento AL, Ohlweiler FP, Reis EM, Ribeiro MA, Sá RG, Stukart GC, Soares MB, Gargioni C, Kawano T, Rodrigues V, Madeira AM, Wilson RA, Menck CF, Setubal JC, Leite LC, Dias-Neto E. Transcriptome analysis of the acoelomate human parasite Schistosoma mansoni. Nature Genetics. 2003;35:148–157. doi: 10.1038/ng1237. [DOI] [PubMed] [Google Scholar]
- Vermeire JJ, Taft AS, Hoffmann KF, Fitzpatrick JM, Yoshino TP. Schistosoma mansoni: DNA microarray gene expression profiling during the miracidium-to-mother sporocyst transformation. Molecular and Biochemical Parasitology. 2006;147:39–47. doi: 10.1016/j.molbiopara.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Vermeire JJ, Yoshino TP. Antioxidant gene expression and function in in vitro-developing Schistosoma mansoni mother sporocysts: possible role in self-protection. Parasitology. 2007;134:1369–1378. doi: 10.1017/S0031182007002697. [DOI] [PubMed] [Google Scholar]
- Viney ME, Thompson FJ. Two hypotheses to explain why RNA interference does not work in animal parasitic nematodes. International Journal for Parasitology. 2008;38:43–47. doi: 10.1016/j.ijpara.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Williams DL, Sayed AA, Bernier J, Birkeland SR, Cipriano MJ, Papa AR, McArthur AG, Taft A, Vermeire JJ, Yoshino TP. Profiling Schistosoma mansoni development using Serial Analysis of Gene Expression (SAGE) Experimental Parasitology. 2007;117:246–258. doi: 10.1016/j.exppara.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RA, Ashton PD, Braschi S, Dillion GP, Berriman M, Ivens A. ‘Oming in on schistosomes: prospects and limitations for post-genomics. Trends in Parasitology. 2006;23:14–20. doi: 10.1016/j.pt.2006.10.002. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Kapp K, Kunz W, Grevelding CG. Characterization of the cysteine protease ER60 in transgenic Schistosoma mansoni larvae. International Journal for Parasitology. 2002b;32:1219–1224. doi: 10.1016/s0020-7519(02)00092-9. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Kapp K, Kunz W, Jackstadt WP, Zahner H, Grevelding CG. HSP70-controlled GFP expression in transiently transformed schistosomes. Molecular and Biochemical Parasitology. 2002a;120:141–150. doi: 10.1016/s0166-6851(01)00446-7. [DOI] [PubMed] [Google Scholar]
- Wippersteg V, Ribeiro F, Liedtke S, Kunz W, Grevelding CG. The uptake of Texas red bovine serum albumin in the excretory system of schistosomes, and its colocalization with ER60 promoter-induced GFP in transiently transformed adult males. International Journal for Parasitology. 2003;33:1139–1143. doi: 10.1016/s0020-7519(03)00168-1. [DOI] [PubMed] [Google Scholar]
- Wu XJ, Sabat G, Brown JF, Zhang M, Taft A, Peterson NP, Harms A, Yoshino TP. Proteomic analysis of Schistosoma mansoni proteins released during in vitro miracidium-to-sporocyst transformation. Molecular and Biochemical Parasitology. 2009;164:32–44. doi: 10.1016/j.molbiopara.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YZ, Dresden MH. Leucine aminopeptidase and hatching of Schistosoma mansoni eggs. Journal of Parasitology. 1986;72:507–511. [PubMed] [Google Scholar]
- Xue X, Sun J, Zhang Q, Wang Z, Huang Y, Pan W. Identification and characterization of novel microRNAs from Schistosoma japonicum. PLoS One. 2008;3:e4034. doi: 10.1371/journal.pone.0004034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang LL, Lu ZY, Hu SM, He SJ, Li ZY, Zhang SM, Zheng HQ, Li MT, Yu XB, Fung MC, Wu ZD. Schistosoma japonicum: proteomics analysis of differentially expressed proteins from ultraviolet-attenuated cercariae compared to normal cercariae. Parasitology Research. 2009;105:237–248. doi: 10.1007/s00436-009-1387-z. [DOI] [PubMed] [Google Scholar]
- Yoshino TP, Boswell CA. Antigen sharing between larval trematodes and their snail hosts: how real a phenomenon in immune evasion? In: Lackie AM, editor. Immune Mechanisms in Invertebrate Vectors. Clarendon Press; Oxford: 1986. pp. 221–238. [Google Scholar]
- Yoshino TP, Laursen JR. Production of Schistosoma mansoni daughter sporocysts from mother sporocysts maintained in synxenic culture with Biomphalaria glabrata embryonic (Bge) cells. Journal of Parasitology. 1995;81:714–722. [PubMed] [Google Scholar]
- Zhao ZR, Lei L, Liu M, Zhu SC, Ren CP, Wang XN, Shen JJ. Schistosoma japonicum: inhibition of mago nashi gene expression by shRNA-mediated RNA interference. Experimental Parasitology. 2008;119:379–384. doi: 10.1016/j.exppara.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Zelck UE, Von Janowsky B. Antioxidant enzymes in intramolluscan Schistosoma mansoni and ROS-induced changes in expression. Parasitology. 2004;128:493–501. doi: 10.1017/s0031182004004895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.