Abstract
OBJECTIVE
Genomic copy number variations (CNVs) have been strongly implicated as important genetic factors for obesity. A recent genome-wide association study identified a novel variant, rs12444979, which is in high linkage disequilibrium with CNV 16p12.3, for association with obesity in Europeans. The aim of this study was to directly examine the relationship between the CNV 16p12.3 and obesity phenotypes, including body mass index (BMI) and body fat mass.
SUBJECTS
Subjects were a multi-ethnic sample, including 2286 unrelated subjects from a European population and 1627 unrelated Han subjects from a Chinese population. Body fat mass was measured using dual energy X-ray absorptiometry.
RESULTS
Using Affymetrix Genome-Wide Human SNP Array 6.0, we directly detected CNV 16p12.3, with the deletion frequency of 27.26 and 0.8% in the European and Chinese populations, respectively. We confirmed the significant association between this CNV and obesity (BMI: P = 1.38 × 10−2; body fat mass: P = 2.13 × 10−3) in the European population. Less copy numbers were associated with lower BMI and body fat mass, and the effect size was estimated to be 0.62 (BMI) and 1.41 (body fat mass), respectively. However, for the Chinese population, we did not observe significant association signal, and the frequencies of this deletion CNV are quite different between the European and Chinese populations (P<0.001).
CONCLUSION
Our findings first suggest that CNV 16p12.3 might be ethnic specific and cause ethnic phenotypic diversity, which may provide some new clues into the understanding of the genetic architecture of obesity.
Keywords: CNV, 16p12.3, BMI, body fat mass, association
INTRODUCTION
Obesity is a major public health problem, resulting in increased risk for multiple disorders, including type 2 diabetes, cardiovascular disease and some forms of cancer.1,2 Although excessive energy intake and diminished physical activity contribute to the increasing prevalence of obesity, previous studies have demonstrated a strong genetic component to obesity risk, with heritability estimated of ~40–70%.3,4 Body mass index (BMI) is the most widely used phenotype to evaluate obesity due to its ease of measurement. Besides BMI, body fat mass can also be used as a complementary phenotype to characterize obese status. Identifying genetic determinants of these phenotypes could lead to a better understanding of the pathogenesis of obesity.
In the past years, genome-wide association studies (GWASs) have successfully identified a number of loci for obesity, including FTO, MC4R and so on.5 – 8 Although most of the findings of GWASs are highly reproducible, they explain only a small portion of heritability (<10%) of obesity collectively.9 This situation suggests that the missing heritability might in part be explained by additional genes or different types of genetic variants, such as copy number variations (CNVs). CNV is a common type of genomic variability with size of 1 kb to several Mb. CNVs have been demonstrated to be related to a series of complex diseases, such as autism,10 schizophrenia11 and osteoporosis.12 Several CNV regions have also been reported for obesity and BMI, including 16p11.2,13,14 10q11.22(ref.15) and 11q11,16 which implicate that CNVs have an important role in predisposing to obesity. Recently, two large-scale GWASs on obesity have detected two SNPs, rs2815752 and rs12444979, which are in strong linkage disequilibrium with two CNVs, 1p31.1 (tagged by rs2815752)17 and 16p12.3 (tagged by rs12444979).18 CNV 1p31.1 has been successfully confirmed for obesity by another group.16 However, for CNV 16p12.3, the result for association with obesity is still controversial.16 Replication of genetic associations in additional populations by independent group is necessary to evaluate a positive finding and determine its generality. Therefore, the aim of this study was to directly examine the relationship between the CNV 16p12.3 and obesity phenotypes, including BMI and body fat mass. Our samples included not only Whites, but also Chinese, to see whether this CNV is common or ethnicity-specific.
MATERIALS AND METHODS
Subjects
The study was approved by the Institutional Review Board or Research Administration of Xi’an Jiaotong University, Hunan Normal University, Creighton University and University of Missouri-Kansas City. Signed informed consent documents were obtained from all study participants before entering the study.
European sample
The European sample consisted of 2286 unrelated adults living in the US Midwestern area. All of the subjects were normal healthy subjects of Northern European origin and defined by a comprehensive suite of exclusion criteria.19 Briefly, subjects with chronic diseases and conditions involving vital organs (heart, lung, liver, kidney and brain) and severe endocrine, metabolic or nutritional diseases that might affect fat metabolism were excluded from this study.
BMI was calculated as body weight (kg) divided by the square of height (m). Weight was measured in light indoor clothing without shoes, using a calibrated balance beam scale, and height was measured using a calibrated stadiometer. Body fat mass was measured using dual energy X-ray absorptiometry by Hologic 4500 DEXA machines (Hologic Inc., Bedford, MA, USA). The short-term reproducibility (coefficient of variation) of BMI and fat mass measurements is on average 0.2 and 1.1%, respectively. Measurement of body fat mass by DEXA is considered to be highly accurate and gold standard. The correlation between BMI and fat mass was 0.85 (P<0.01) in this sample.
Chinese sample
The Chinese sample consisted of 1627 unrelated subjects. The subjects were recruited from Midwestern Chinese Han adults living in Xi’an and Changsha cities. The exclusion criteria was same as with European sample. The measurements of BMI and body fat mass were obtained by the same approach as those adopted in the European sample.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using standard protocols. For all the samples, SNP genotyping was performed using Genome-Wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA, USA) according to the Affymetrix protocol, which has been described in our previous publication.20 Only samples with a minimum call rate of 95% were included. Because of efforts of repeat experiments, all samples met this criteria, and the final mean call rate reached a high level of 98.93% for the European sample and 98.96% for the Chinese sample, respectively.
CNV detection
Common CNVs were identified using the CANARY algorithm implemented in the Birdsuite software,21 which utilized a previously defined copy number polymorphism (CNP, namely CNV with frequency greater than 1%) map based on HapMap samples.22 CNV 16p12.3 was denoted by CNP 2150 according to the CNP map.
Statistical analyses
Before association analyses, principal component analysis implemented in EIGENSTRAT23 was used to correct for potential population stratification that may lead to spurious association results. The first 10 principal components emerging from the EIGENSTRAT analyses, along with sex and age, were used as covariates to adjust the raw BMI or fat mass in each sample. Association analyses were performed by linear regression for the adjusted residuals.
RESULTS
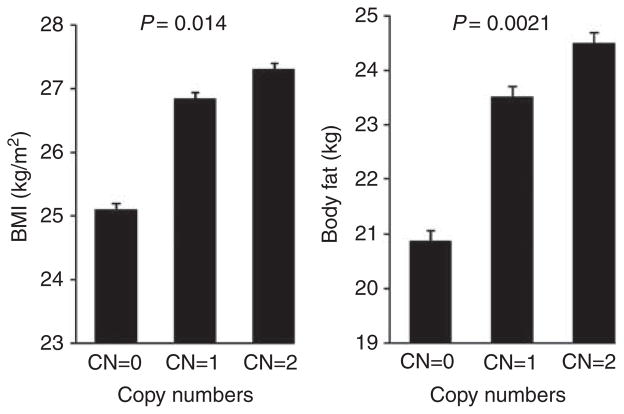
The basic characteristics of the study subjects are presented in Table 1. We summarized the association results for CNP 2150 with BMI and body fat mass in Table 2. According to the CNP map, CNP 2150 is located between 19 853 151 and 19 874 863 bp on chromosome 16 spanning ~21 kb (named CNV 16p12.3). For the European sample, we detected 46 subjects (2.02%) with a homozygous deletion, 577 subjects (25.24%) with a heterozygous deletion (1 copy) and 1663 subjects (72.74%) with wild type (2 copies). The prevalence of the deletion polymorphism was concordant with that expected from the Hardy–Weinberg equilibrium test (P = 0.676). We were able to confirm the association between this CNV and BMI (P = 1.38 × 10−2) in the European sample. We also found a more significant association signal between this CNV and body fat mass, with a P-value of 2.13 × 10−3. As shown in Figure 1, less copy numbers were significantly associated with lower BMI and body fat, and the effect size (β) was estimated to be 0.62 (BMI) and 1.41 (body fat mass) for each copy number, respectively. In addition, as this CNV is in high linkage disequilibrium with the tagging SNP rs12444979 (r2 = 0.96), we also examined associations between rs12444979 and obesity phenotypes. Significant associations were observed both for BMI (P = 0.010) and body fat mass (P = 0.002).
Table 1.
Basic characteristics of the study subjects
| Trait | European sample | Chinese sample |
|---|---|---|
| Number | 2286 | 1627 |
| Age (years) | 51.37 (13.76) | 34.49 (13.24) |
| Weight (kg) | 75.27 (17.54) | 60.12 (10.48) |
| Height (cm) | 166.35 (8.47) | 164.25 (8.16) |
| Female/male | 1727/558 | 825/802 |
| BMI (kg m−2) | 27.14 (5.75) | 22.21 (3.03) |
| Body fat (kg) | 24.17 (10.63) | 14.02 (5.44) |
Abbreviation: BMI, body mass index. Data are shown as mean (s.d.).
Table 2.
Association results for CNV 16p12.3 with obesity phenotypes in European and Chinese samples
| Sample |
Copy numbers (N)
|
HWE | P-value
|
|||
|---|---|---|---|---|---|---|
| N = 0 | N = 1 | N = 2 | BMI | Body fat mass | ||
| European | 46 | 577 | 1663 | 0.676 | 1.38 × 10−2 | 2.13 × 10−3 |
| Chinese | 0 | 13 | 1614 | 0.862 | 0.993 | 0.613 |
Abbreviations: BMI, body mass index; HWE, Hardy-Weinberg equilibrium.
Figure 1.
Phenotype differences for CNV 16p12.3 in the European sample.
For the Chinese sample, we found that the frequency of this deletion CNV was different from that in the European sample (P<0.001 by χ2-test). We only detected 13 subjects (0.8%) with a heterozygous deletion. Most of the subjects were wild type with two copy numbers (99.2%). We didn’t observe significant association between this CNV, and BMI and body fat mass (Table 2). For the tagging SNP rs12444979, there is no polymorphism in the Chinese sample.
DISCUSSION
A recent large-scale GWAS identified a novel variant, rs12444979, which is in high linkage disequilibrium with CNV 16p12.3, for association with obesity.18 The deletion allele was tagged by the T allele of rs12444979 with the allele frequency of 0.13. This study provided an indirect evidence for the association between CNV 16p12.3 and obesity, as the identification of CNV was based on the tagging SNP. In our study, we successfully detected CNV 16p12.3 by using the Affymetrix Genome-Wide Human SNP Array 6.0, which is designed for the detection of common CNVs, and directly confirmed the significant association between this CNV and obesity phenotypes in the European population. Our results demonstrated the validity of the initial finding and further supported the potential contribution of CNV 16p12.3 to the pathogenesis of obesity.
The statistical power of our study is estimated by using the program Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/qtlassoc.html). We set the significance level at P = 0.05. Assuming that a marker has a minor frequency of 0.05 and is in strong linkage disequilibrium (D′= 0.9) with a functional mutation that accounts for 1% variation of a phenotype, our European and Chinese samples can achieve >95% statistical power, which is large enough to detect a genetic variant. However, we could not replicate the association between CNV 16p12.3 and obesity in the Chinese population, which revealed an ethnic differentiation for this CNV. One possible explanation for such ethnic difference could be that the frequencies of this CNV are quite different between the European and Chinese populations (P<0.001). The copy number deletion of CNV 16p12.3 is much common in the European population with the deletion frequency of 27.26%, which may contribute to the overall risk of obesity in Europeans; whereas, only 0.8% of the Chinese subjects have this deletion CNV. Our findings supported that it is necessary to evaluate associations in populations of different ancestries, especially for CNV analyses, as the genomic variation is greater when compared across ethnicities.
CNV 16p12.3 is a 21-kb deletion that locates ~50 kb upstream of the gene GPRC5b (G protein-coupled receptor, family C, group 5, member B). The protein encoded by this gene is a member of the type-3 G protein-coupled receptor family. This protein may mediate the cellular effects of retinoic acid on the G protein signal transduction cascade. The direct connection between this protein and the biology of obesity phenotypes is still unknown. Our results may provide an important entry point for the future investigation.
In conclusion, we confirmed the recently found association between CNV 16p12.3 and obesity phenotypes in the European population, which highlights the importance of this CNV for obesity. Future functional studies are needed to elucidate its detailed role and uncover the new insight into the biology of obesity. We first suggest that this CNV is ethnic specific, as it has no effect on obesity in the Chinese population. The development and application of future therapies and interventions should consider the ethnicity of subjects.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81000363, 31000554), the Grants from NIH (R01 AR050496, R21 AG027110, R01 AG026564, P50 AR055081, R01 AR057049-01A1 and R21 AA015973). The study was also funded by the Fundamental Research Funds for the Central Universities, the PhD Programs Foundation of Ministry of Education of China (20100201120058), Shanghai Leading Academic Discipline Project (S30501), a Grant from Ministry of Education to ShangHai University of Science and Technology, and startup fund from University of Shanghai for Science and Technology, Xi’an Jiaotong University and the Ministry of Education of China. This work was also supported by Dr Hong-Wen Deng’s Dickson/Missouri Endowment at University of Missouri-Kansas City and the Edward G Schlieder Endowment at Tulane University.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 2.Lewis CE, McTigue KM, Burke LE, Poirier P, Eckel RH, Howard BV, et al. Mortality, health outcomes, and body mass index in the overweight range: a science advisory from the American Heart Association. Circulation. 2009;119:3263–3271. doi: 10.1161/CIRCULATIONAHA.109.192574. [DOI] [PubMed] [Google Scholar]
- 3.Maes HH, Neale MC, Eaves LJ. Genetic and environmental factors in relative body weight and human adiposity. Behav Genet. 1997;27:325–351. doi: 10.1023/a:1025635913927. [DOI] [PubMed] [Google Scholar]
- 4.Stunkard AJ, Foch TT, Hrubec Z. A twin study of human obesity. JAMA. 1986;256:51–54. [PubMed] [Google Scholar]
- 5.Chambers JC, Elliott P, Zabaneh D, Zhang W, Li Y, Froguel P, et al. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat Genet. 2008;40:716–718. doi: 10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 6.Dina C, Meyre D, Gallina S, Durand E, Korner A, Jacobson P, et al. Variation in FTO contributes to childhood obesity and severe adult obesity. Nat Genet. 2007;39:724–726. doi: 10.1038/ng2048. [DOI] [PubMed] [Google Scholar]
- 7.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loos RJ, Lindgren CM, Li S, Wheeler E, Zhao JH, Prokopenko I, et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat Genet. 2008;40:768–775. doi: 10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walley AJ, Asher JE, Froguel P. The genetic contribution to non-syndromic human obesity. Nat Rev Genet. 2009;10:431–442. doi: 10.1038/nrg2594. [DOI] [PubMed] [Google Scholar]
- 10.Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stefansson H, Rujescu D, Cichon S, Pietilainen OP, Ingason A, Steinberg S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang TL, Chen XD, Guo Y, Lei SF, Wang JT, Zhou Q, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet. 2008;83:663–674. doi: 10.1016/j.ajhg.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bochukova EG, Huang N, Keogh J, Henning E, Purmann C, Blaszczyk K, et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2010;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walters RG, Jacquemont S, Valsesia A, de Smith AJ, Martinet D, Andersson J, et al. A new highly penetrant form of obesity due to deletions on chromosome 16p11. 2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sha BY, Yang TL, Zhao LJ, Chen XD, Guo Y, Chen Y, et al. Genome-wide association study suggested copy number variation may be associated with body mass index in the Chinese population. J Hum Genet. 2009;54:199–202. doi: 10.1038/jhg.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarick I, Vogel CI, Scherag S, Schafer H, Hebebrand J, Hinney A, et al. Novel common copy number variation for early onset extreme obesity on chromosome 11q11 identified by a genome-wide analysis. Hum Mol Genet. 2011;20:840–852. doi: 10.1093/hmg/ddq518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willer CJ, Speliotes EK, Loos RJ, Li S, Lindgren CM, Heid IM, et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat Genet. 2009;41:25–34. doi: 10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng HW, Deng H, Liu YJ, Liu YZ, Xu FH, Shen H, et al. A genomewide linkage scan for quantitative-trait loci for obesity phenotypes. Am J Hum Genet. 2002;70:1138–1151. doi: 10.1086/339934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang TL, Guo Y, Liu YJ, Shen H, Liu YZ, Lei SF, et al. Genetic variants in the SOX6 gene are associated with bone mineral density in both Caucasian and Chinese populations. Osteoporos Int. 2012;23:781–787. doi: 10.1007/s00198-011-1626-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korn JM, Kuruvilla FG, McCarroll SA, Wysoker A, Nemesh J, Cawley S, et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]