Abstract
Objective
Bone and muscle, two major tissue types of musculoskeletal system, have strong genetic determination. Abnormality in bone and/or muscle may cause musculoskeletal diseases such as osteoporosis and sarcopenia. Bone size phenotypes (BSPs), such as hip bone size (HBS), appendicular bone size (ABS), are genetically correlated with body lean mass (mainly muscle mass). However, the specific genes shared by these phenotypes are largely unknown. In this study, we aimed to identify the specific genes with pleiotropic effects on BSPs and appendicular lean mass (ALM).
Methods
We performed a bivariate genome-wide association study (GWAS) by analyzing ~690,000 SNPs in 1,627 unrelated Han Chinese adults (802 males and 825 females) followed by a replication study in 2,286 unrelated US Caucasians (558 males and 1728 females).
Results
We identified 14 interesting single nucleotide polymorphisms (SNPs) that may contribute to variation of both BSPs and ALM, with p values <10−6 in discovery stage. Among them, the association of three SNPs (rs2507838, rs7116722, and rs11826261) in/near GLYAT (glycine-N-acyltransferase) gene was replicated in US Caucasians, with p values ranging from 1.89×10−3 to 3.71×10−4 for ALM-ABS, from 5.14×10−3 to 1.11×10−2 for ALM-HBS, respectively. Meta-analyses yielded stronger association signals for rs2507838, rs7116722, and rs11826261, with pooled p values of 1.68×10−8, 7.94×10−8, 6.80×10−8 for ALB-ABS and 1.22×10−4, 9.85×10−5, 3.96×10−4 for ALM-HBS, respectively. Haplotype allele ATA based on these three SNPs were also associated with ALM-HBS and ALM-ABS in both discovery and replication samples. Interestingly, GLYAT was previously found to be essential to glucose metabolism and energy metabolism, suggesting the gene’s dual role in both bone development and muscle growth.
Conclusions
Our findings, together with the prior biological evidence, suggest the importance of GLYAT gene in co-regulation of bone phenotypes and body lean mass.
Keywords: Bivariate GWAS, Bone size, Lean mass, GLYAT
Introduction
Bone and muscle are two major tissue types of musculoskeletal system. Bones sustain mechanical loads and provide load points for muscles, and muscles keep bones in place and are responsible for major mechanical loading of bones (Handoll et al. 2007). Abnormality in bone and/or muscle may cause musculoskeletal diseases such as osteoporosis and sarcopenia. Osteoporosis is a major public disease characterized by decreased bone strength and increased fracture risk (McCloskey 2011; Orwig et al. 2011). Hip fracture is the most common and serious type of osteoporotic fracture, often leading to prolonged or permanent disability, or even death, for some patients (McCloskey 2011; Orwig et al. 2011). Bone mineral density (BMD) is considered to be an important, but not exclusive, determining factor for bone strength and fracture risk. Bone size, independent of BMD, is another important factor, that determines bone strength and is directly associated with osteoporotic fractures (Ahlborg et al. 2003; McCreadie and Goldstein 2000; Tan et al. 2008; Deng et al. 2002b; Deng et al. 2002a). Many recent studies have suggested that hip bone size (HBS) could be an useful measurement for assessment of hip fracture risk (Ahlborg et al. 2003; McCreadie and Goldstein 2000; Tan et al. 2008; Deng et al. 2002b; Deng et al. 2002a).
The muscular tissue, as characterized by body lean mass, is also closely associated with human health. Low body lean mass is related to a series of health problems, such as sarcopenia, impaired protein balance, obesity, and osteoporosis (Fry and Rasmussen 2011; Lee et al. 2011; Capozza et al. 2008). Body lean mass and bone size are closely related (Ferretti et al. 2003; Ferretti et al. 2001; Cointry et al. 2004). It has been demonstrated that lean mass can predict bone size in pre-pubertal children and can serve as a major determinant of bone size (Martin 2002; Micklesfield et al. 2011; Baptista et al. 2012). Bone size, in turn, has been shown to be adapted to the dynamic load imposed by muscle force. Dynamic strains provided by muscle may be an important stimulus of bone adaptation (Ferretti et al. 2003).
From the genetic perspective, both body lean mass and bone size are known to have strong genetic components, with heritability over 50% (Liu et al. 2009b; Lei et al. 2011; Karasik et al. 2009; Livshits et al. 2007; Deng et al. 2002b; Deng et al. 2002a). Genetic analyses also have shown that body lean mass are significantly correlated with bone size (Chumlea et al. 2002), thus these phenotypic traits might share some common genetic factors. However, the specific SNPs/genes linked with both BSPs and body lean mass are largely unknown.
Bivariate genome-wide association study (GWAS) is an effective approach to detect pleiotropic genes for complex traits (Liu et al. 2009a; Pei et al. 2009; Liu et al. 2009c; Zhang et al. 2009; Sun et al. 2011). To identify the specific pleiotropic SNPs/genes that contribute to both BSPs and body lean mass, we performed a bivariate GWAS in a large Chinese sample, and followed by a replication study in Caucasians. Since appendicular lean mass (ALM, sum of lean mass in the arms and legs) is a better proxy measure of body skeletal muscle mass than total body lean mass for assessing exercise capacity and predicting related diseases (Kim et al. 2002; Heymsfield et al. 1990), we utilized ALM as the phenotype for association analyses. In addition to bivariate analysis on ALM and HBS, we also analyzed ALM and appendicular bone size (ABS), as ALM is most naturally correlated with bone size of the appendicular skeleton.
Materials and Methods
Subjects and phenotypes
The study was approved by the involved Institutional Review Board. All study participants signed informed consent documents before they entered the project. Two independent cohorts were included in this study: a cohort of 1,627 unrelated adult Han Chinese (802 males and 825 females) recruited from Changsha and Xi’an and their surroundings areas, and another cohort of 2,286 unrelated homogeneous US Caucasians (including 558 males and 1,728 females) living in Kansas City and its surrounding areas. Both cohorts were recruited for studies aimed in searching for genes underlying variations in body compositions (bone mass, fat mass and lean mass). Anthropometric measures and a structured questionnaire including diet, lifestyle, medical history, family information and others were obtained for all subjects. Strict exclusion criteria were adopted to minimize any known and potential confounding effects on variation of body composition phenotypes. Generally, subjects with chronic diseases and conditions involving vital organs (heart, lung, liver, kidney, and brain) and severe endocrinological, metabolic, and nutritional diseases were excluded from this study.
Two BSPs and ALM were used in this study. HBS was areal bone size (cm2) at the total hip (femoral neck, trochanter and intertrochanteric region). ABS was the sum of areal bone size (cm2) of arms and legs. ALM (g) was the sum of lean soft tissue (nonfat, non-bone) mass in the arms and legs. In both cohorts, HBS, ABS, and ALM were measured by dual-energy Xray absorptiometry (DXA) with Hologic densitometers (Hologic Inc., Waltham, MA, USA) followed the standard protocol recommended by the manufacturer. The machines were calibrated daily. The coefficient of variation (CV) values of DXA measurements for Chinese subjects were comparable to those obtained for US Caucasians.
Genotyping
Genomic DNA was extracted from peripheral blood leukocytes using the Puregene DNA isolation kit (Gentra Systems, Minneapolis, MN, USA). All subjects were genotyped using the Human mapping SNP 6.0 assay kit (Affymetrix, Inc, Santa Clara, CA), following the standard protocol recommended by the manufacturer. For quality control (QC) of SNPs, we set the default value of greater than 0.4 as the contrast QC threshold. The final average contrast QC across the entire sample reached the high level of 2.62. In the initial stage, 909,622 SNPs were genotyped for the Chinese subjects. After excluding 17,888 SNP with allele frequencies deviating from Hardy-Weinberg equilibrium (p < 0.01) and 202,984 SNPs with minor allele frequencies (MAF) < 1% (618 SNP were included by both exclusion criteria), a final total of 689,368 SNPs were retained for subsequent analyses, yielding an average marker spacing of ~4 kb throughout the human genome.
Statistical Analyses
Previous studies suggested that bone phenotypes and body lean mass are two related phenotypes (Ahlborg et al. 2003; Chumlea et al. 2002; Capozza et al. 2008; Cointry et al. 2004; Szulc et al. 2005). We tested the phenotypic correlation between bone size and body lean mass in the two studied population by the bivariate correlation analysis with the function cor.test of R package (version 2.13.1).
We adopted similar statistical analyses in the initial GWAS and replication study. Before association analyses, raw phenotypes of BSPs and ALM were adjusted for age, sex and height. To ensure adequate normality of quantitative traits, we applied an inverse normal transformation to each trait prior to association analysis. The inverse normal transformation reduces the impact of outliers and deviations from normality on statistical analysis. The transformation involves ranking all available phenotypes, transforming these ranks into quantiles and, finally, converting the resulting quantiles into normal deviates (Scuteri et al. 2007; Uda et al. 2008). Principal component analysis (PCA) was performed by EIGENSTRAT to calculate the principal components, and the ten default main eigenvectors were used as covariates to adjust raw phenotypic data for correction of population stratification (Price et al. 2006). We first performed bivariate GWAS to detect associations between each SNP and two phenotypes in the discovery cohort, and we then conducted bivariate association analysis in the replication cohort. Haplotype association analyses of particular identified SNP groups were performed with both univariate and bivariate framework in the discovery and replication cohorts. An additive genetic model and a linear model were applied to both univariate and bivariate association analyses using the R software package (version 2.13.1). This method is expressed as the following formula, Yi = μ+ βXi + εi, for an individual i, Yi is a vector with length of 1 (for univariate analysis) or 2 (for bivariate analysis) coding the individual’s phenotype, μ is the ground mean vector, β’s are the corresponding effects of the locus (SNP or haplotype block) under test; Xi is the genotype score at the locus of interest for individual i, and εi is the vector of random error. We compared the likelihood of the model under the null hypothesis (locus effects are restricted to 0), with that under the alternative hypothesis (the locus effects are not 0), to test the alternative hypothesis. Then the likelihood ratio can convert to an F-statistic, which follows an F-distribution under the null hypothesis. The p value was calculated based on the F-statistic. To quantify overall evidence of association achieved in our discovery GWAS cohort and in the US replication cohort, we combined individual p values of the two cohorts with a Fisher’s method for meta-analysis. The calculation was performed using the MetaP web tool (http://people.genome.duke.edu/dg48/metap.php). The linkage disequilibrium (LD) [standardized D′(D/Dmax)] patterns of interesting SNPs and the haplotype block map was analyzed using Haploview software (available at http://www.broad.mit.edu/mpg/haploview/) (Barrett et al. 2005). Phased haplotype of each block were obtained by using Plink (version 1.07, http://pngu.mgh.harvard.edu/,purcell/plink/) in the discovery GWAS and the replication study (Purcell et al. 2007).
Results
The basic characteristics of the study subjects are summarized in Table 1. Apparently, ALM, ABS and HBS in women were lower than those in men. And all these three measurements were lower in Chinese than in Caucasian subjects.
Table 1.
Basic characteristics of study subjects.
| Traits | Chinese Sample | Caucasian Sample | ||
|---|---|---|---|---|
|
| ||||
| Male (N = 802) | Female (N = 825) | Male (N = 558) | Female (N = 1728) | |
| Age (years) | 31.27±12.78 | 37.46±13.77 | 50.72±16.05 | 51.58±12.92 |
| Height (cm) | 170.27±5.96 | 157.75±13.69 | 174.882±17.96 | 163.13±8.89 |
| Weight (kg) | 65.74±9.64 | 54.26±11.07 | 86.46±20.08 | 71.35±16.55 |
| HBS (cm2) | 37.66±7.93 | 29.88±7.67 | 45.58±8.46 | 35.86±5.42 |
| ABS (cm2) | 1145.85±114.24 | 954.83±107.49 | 1359.57±130.94 | 1132.03±110.78 |
| ALM (kg) | 23.94±3.20 | 15.73±2.15 | 29.89±4.85 | 20.21±3.53 |
Note: all the values are means ± SD.
Correlation analysis of our phenotype data showed that ALM was highly significantly correlated with BPS (ABS, HBS), with correlation coefficients over 0.50 (p < 0.001) in both Chinese and Caucasian cohorts. The significant correlations between phenotype pairs indicate the necessity of bivariate analysis to identify the underlying pleiotropic genes.
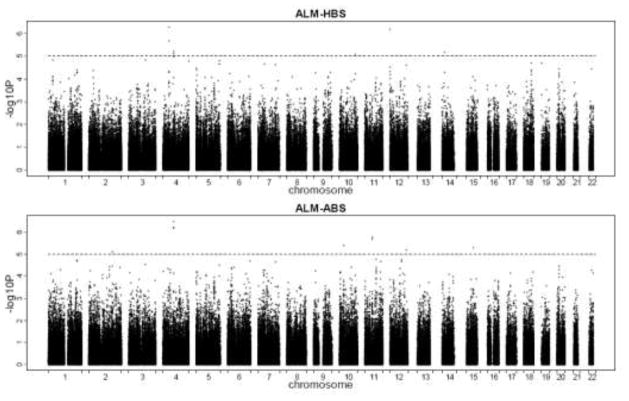
Fig. 1 shows the distribution of p values of bivariate GWAS across the genome in Chinese sample. We identified 14 interesting SNPs that were moderately bivariately associated with ALM and bone sizes (p at the level of 1.0×10−6 or less for ALM-HBS or/and ALM-ABS) (Table 2). Among them, there are two groups of closely neighboring SNPs. Group II was composed of rs961719, rs12648730 and rs1584984 of chromosome 4, which are the top 1–3 significant SNPs for the bivariate association of ALM-ABS (P values were 6.248×10−7, 6.70×10−7, 3.43×10−7, respectively). These three SNPs were in moderate LD (r 2 > 0.50). Group II was composed of SNPs rs2507838, rs7116722 and rs11826261 on chromosome 11 which were in strong LD (r 2 > 0.91) with each other. The bivariate association p values of the three SNPs in group II with ALM-ABS genotype pair ranged from 1.75×10−6 to 2.06×10−6.
Fig. 1.
Distribution of p values across the genome in bivariate association analyses in Chinese sample. The p values are plotted according to its physical position on successive chromosomes. The dashed lines represent association signals with p = 1.00E-05. ALM appendicular lean mass, HBS hip bone size, ABS appendicular bone size.
Table 2.
SNP results of bivariate association for ALM and BSPs (At least one p < 1.00E-05 for each SNP).
| SNP | Chromosome | Posision | Allele (minor/ major) | Gene | Predicted function | Discovery Chinese sample | Replication Caucasian sample | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| MAF | Bivariate p | MAF | Bivariate p | ||||||||
| ALM-HBS | ALM-ABS | ALM-HBS | ALM-ABS | ||||||||
| rs4325816 | 2 | 174517145 | C/T | SP3 | intronic | 0.02 | 2.55E-03 | 7.68E-06 | 0.22 | - | - |
| rs4145971 | 4 | 44537971 | T/G | RP11-55C6.1 | non-coding | 0.25 | 5.27E-07 | - | 0.25 | - | - |
| rs961719 | 4 | 80247809 | C/G | non-coding intronic | 0.10 | 1.01E-05 | 6.24E-07 | 0.45 | - | - | |
| rs12648730 | 4 | 80278147 | G/A | RP11-438E5.1 | non-coding intronic | 0.10 | 7.95E-06 | 6.70E-07 | 0.45 | - | - |
| rs1584984 | 4 | 80279231 | A/G | non-coding intronic | 0.10 | 6.24E-06 | 3.43E-07 | 0.29 | - | - | |
| rs11008640 | 10 | 32124485 | G/C | ARHGAP12 | 3′ donwstream | 0.50 | - | 3.93E-06 | 0.19 | - | - |
| rs2768331 | 10 | 119245685 | C/G | EMX2OS | 5′ upstream | 0.25 | 8.15E-06 | - | 0.22 | - | - |
| rs2507838 | 11 | 58229375 | A/C | intronic | 0.05 | 1.90E-03 | 2.06E-06 | 0.03 | 5.14E-03 | 3.71E-04 | |
| rs7116722 | 11 | 58263484 | T/G | GLYAT | 3′ donwstream | 0.05 | 7.52E-04 | 1.75E-06 | 0.02 | 1.03E-02 | 2.23E-03 |
| rs11826261 | 11 | 58271739 | A/G | 3′ donwstream | 0.05 | 3.16E-03 | 1.75E-06 | 0.02 | 1.11E-02 | 1.89E-03 | |
| rs10505721 | 12 | 1075963 | G/A | ERC1 | intronic | 0.18 | 6.44E-07 | - | 0.13 | - | - |
| rs11068990 | 12 | 117471566 | G/A | RP11-121J20.1-001 | 3′ donwstream | 0.03 | 2.52E-05 | 6.62E-06 | 0.11 | - | - |
| rs12891306 | 14 | 35967587 | G/A | SFTA3 | 3′ donwstream | 0.22 | 7.04E-06 | - | 0.39 | - | - |
| rs901130 | 15 | 72360960 | C/G | CCDC33 | intronic | 0.48 | 2.22E-03 | 5.10E-06 | 0.29 | - | - |
|
| |||||||||||
| Phenotype correlation coefficient | 0.50* | 0.88* | 0.54* | 0.84* | |||||||
Note: - p > 0.05,
p < 0.001,
ALM appendicular lean mass, BSP bone size phenotype, HBS hip bone size, ABS appendicular bone size.
Using Ensemble genome browser (e66), we obtained the information of position and effect of the 14 SNPs on specific genes and transcripts (Table 2). Four identified SNPs are located in protein coding gene region. Specifically, the SNP rs2507838 (one SNP in group II) is in the intron of glycine-N-acyltransferase gene (GLYAT), and SNP rs10505721 is in the intron of ELKS/RAB6-interacting/CAST family member 1 gene (ERC1), and rs4325816 is in the intron of Sp3 transcription factor (SP3), and SNP rs901130 is in the intron of coiled-coil domain containing 33 gene (CCDC33). The other two SNPs (rs7116722 and rs11826261) of group II are intergenic, about 7kb and 16kb away from the gene GLYAT. The three SNPs of group I locate in a non-coding antisense gene (RP11-438E5.1).
We further performed bivariate analysis for the 14 identified SNPs in the replication Caucasian sample. Replication analyses confirmed the association of the three consecutive SNPs of group II(rs2507838, rs7116722, rs11826261), with p values ranging from 1.89×10−3 to 3.71×10−4 for ALM-ABS, from 5.14×10−3 to 1.11×10−2 for ALM-HBS, respectively (Table 2). Meta-analyses yielded stronger association signals, with pooled p values of 1.68×10−8, 7.94×10−8, 6.80×10−8 for ALB-ABS and 1.22×10−4, 9.85×10−5, 3.96×10−4 for ALM-HBS, respectively.
We also performed bivariate association analyses for haplotypes constructed by the two identified SNP-groups. For SNP group I (rs961719, rs12648730 and rs1584984), as shown in Table 3, we detected that hayplotype allele CGA were bivariately associated with ALM-HBS (p = 6.92×10−6) and ALM-ABS (p = 6.92×10−7) in the discovery Chinese cohort. However, we failed to replicate the association in the replication cohort. For the SNP group II (rs2507838, rs7116722 and rs11826261), haplotype allele ATA were bivariately associated with ALM-HBS (p = 3.20×10−3) and ALM-ABS (p = 2.05×10−6) in the discovery Chinese cohort. Furthermore, haplotype allele ATA were also bivariately associated with ALM-HBS and ALM-ABS in replication Caucasian sample at the nominal significance level (p < 0.05) (Table 3). The results of univariate haplotype association were less significant but with consistent trend to those of bivariate haplotype association anlaysis (Table 3) in both populations.
Table 3.
Haplotype results of association for ALM and BSPs.
| Chromosome | SNPs | Tested Haplotype Allele | Sample | Bivariate p | Univariate p | ||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| ALM-HBS | ALM-ABS | ALM | ABS | HBS | |||||
| Group I | 4 | rs961719 | CGA | Chinese | 6.92E-06 | 6.92E-07 | 1.09E-06 | 1.65E-06 | 2.10E-02 |
| rs12648730 | |||||||||
| rs1584984 | Caucasian | 2.82E-01 | 3.06E-01 | 4.36E-01 | 4.57E-01 | 1.35E-01 | |||
| Group II | 11 | rs2507838 | ATA | Chinese | 3.20E-03 | 2.05E-06 | 1.30E-02 | 2.78E-06 | 2.10E-01 |
| rs7116722 | |||||||||
| rs11826261 | Caucasian | 4.36E-02 | 6.88E-04 | 1.89E-03 | 3.40E-03 | 8.00E-03 | |||
Note: ALM appendicular lean mass, BSP bone size phenotype, HBS hip bone size, ABS appendicular bone size.
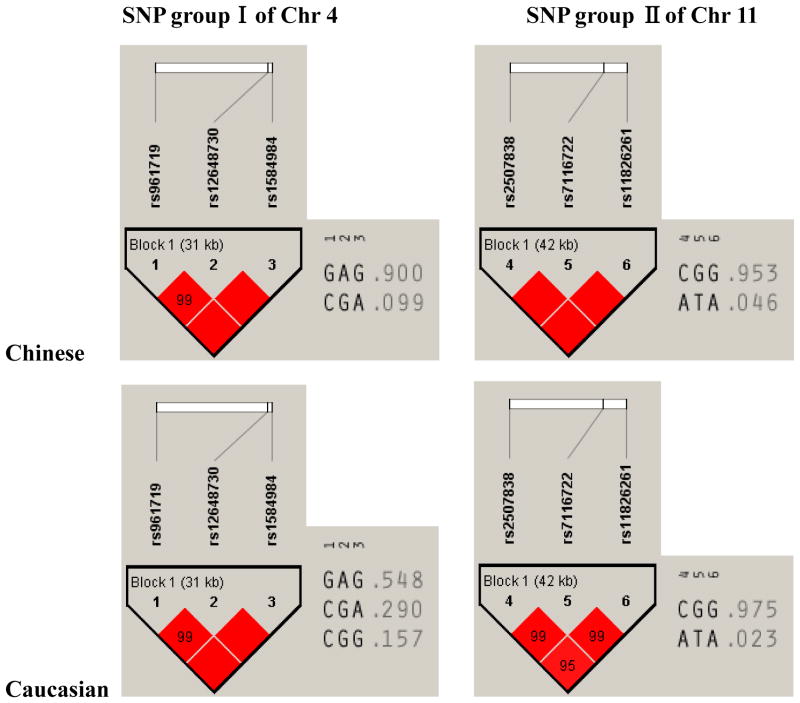
Fig. 2 shows the LD pattern and the haplotype frequency for the two consecutive SNP groups both in the Chinese and Caucasian samples. As shown in Fig. 2, these two SNP groups formed tightly combined haplotype block in both population. There is great difference in haplotype frequency for the block of SNP group I (rs961719, rs12648730, rs1584984) between the two populations, and haplotype allele CGG (frequency is 0.157) was only observed in Caucasian sample. While much similar haplotype frequencies were observed for the block of SNP group II (rs2507838, rs7116722, rs11826261) between the two populations.
Fig. 2.
Haplotype blocks for the two groups of consecutive SNPs located on chromosome 4 and 11. The LD pattern (D′) and haplotype frequency for each block were analyzed and plotted using the Haploview program.
Discussion
We performed bivariate GWAS analyses for ALM and BSPs to identify potential pleiotropic genes. We found that 14 SNPs (some located within/near four genes, GLYAT, ERC, SP31 and CCDC33) showed associations with both ALM and BSPs.
GWAS is a powerful tool for detecting novel genetic variants underlying common human diseases. To date, however, most published GWASs were largely performed using a univariate framework to analyze individual phenotypes separately. Although univariate GWASs have led to the discovery of novel genes for some specific diseases/traits, univariate analysis generally has insufficient power to detect genes with pleiotropic effects on two or more correlated phenotypes (such as lean mass and bone size). Recently developed methods for bivariate GWAS allow us to identify potential pleiotropic genes with much increased power than univariate approaches (Liu et al. 2009c; Zhang et al. 2009; Sun et al. 2011; Liu et al. 2009a; Pei et al. 2009).
The bivariate associations of three SNPs (rs2507838, rs7116722 and rs11826261) with ALM and BSPs in our discovery Chinese cohort were replicated in a Caucasian population. Furthermore, haplotype allele ATA based on these three SNPs were also bivariately associated with ALM and BSPs both in discovery Chinese sample and in replication Caucasian sample, and univariate haplotype analysis with ALM or BSPs showed association signals of the same trend to those of bivariate analyses. The consistent results suggested that the genomic region harboring these SNPs may contain genes with pleiotropic effects on ALM and BSPs in different ethnic groups.
The gene GLYAT, where rs2507838 is located, encodes the glycine-N-acyltransferase protein, a metabolic enzyme conjugating glycine with acyl-CoA substrates in the mitochondria. GLYAT is thought to be important in the detoxification of endogenous and xenobiotic acyl-CoA’s and in regulating glucose metabolism and energy metabolism (Yamamoto et al. 2009; Lino Cardenas et al. 2010). Reduced expression of GLYAT is associated to hyperglycemia diseases. Glucose metabolism, one of the most basic cellular biochemical reactions, provides energy and material for fundamental cellular activities such as protein metabolism, cell growth, and proliferation (Vander Heiden et al. 2001; Movcessian et al. 1972). These activities are essential for normal muscle growth, and may influence lean mass in human (Leidy et al. 2007). Glucose metabolism is also associated with bone development, as elevated glucose levels have been shown to inhibit calcium uptake and bone mineralization (Balint et al. 2001). Certain expression data from the public databases Gene Expression Omnibus and ArrayExpress also support the association of GLYAT to skeleton muscle system. GLYAT is over expressed in human muscle (p < 1.0E-10) in a global map of human gene expression (Lukk et al. 2010). GLYAT is also nominally differentially expressed in a range of muscle disease status, including Emery-Dreifuss muscular dystrophy (p = 1.2E-02), Becker muscular dystrophy (p = 8.0E-03), and juvenile dermatomyositis (p = 2.7E-02) (Bakay et al. 2006). Statistical evidences obtained in our study, together with the known biological functions and expressions of this gene, support the role of GLYAT in regulation of metabolism of both muscle and bone.
Our study found another group of 3 consecutive SNPs (rs961719, rs12648730 and rs1584984,) ranked at the top for bivariate association with ALM and BSPs in the Chinese cohort, though the findings were not replicated in Caucasians. Furthermore, no significant association signals were observed around the region in Caucasians. Detailed results are shown in the Appendix S1. In fact, differences in haplotype frequency for this three-SNPs block were observed between the Chinese population and the Caucasian population. It is possible that the failure to replicate the associations in Caucasians is due to the genome structure diversity in this region or even genuine diversity of the genetic effects between Chinese and Caucasians. The associations of the other 8 SNPs (rs4325816, rs4145971, rs11008640, rs2768331, rs10505721, rs11068990, rs12891306, rs901130) in Chinese sample have also not been replicated in Caucasian sample. Genetic heterogeneity has been wildly observed among different races (Schimmenti et al. 2008; Palmer et al. 2008; Kochi et al. 2009). The validity and generality of the identified genetic effects need to be evaluated in additional replication studies of different populations or even through laboratory functional studies.
Most interesting SNPs identified in the initial GWAS in Chinese locates within non-coding genomic regions. Particularly, the most significant 3 contiguous SNPs (rs1584984, rs961716 and rs12648730) locate within antisense RNA coding gene RP11-438E5.1. Over the past few years, GWASs have revealed a large number of genetic variants related to complex diseases/traits; however, at least one-third of the identified variants are not within protein-coding regions. Although enhancers in the non-coding regions have been anticipated to contain some of these risk variants, another possibility is that these risk variants reside in non-coding RNAs, which are evolutionally conserved across mammals and are biologically functional as cis- and/or trans-regulators of gene activity (Jin et al. 2011). For example, recent emerging evidence has indicated the important role of genetic variants of microRNAs in diseases (Mattick 2009). The current study provided some evidence that variants of non-coding RNAs may be important in etiology of bone and muscle related diseases.
In a previous GWAS study of BSPs in Caucasian subjects, we identified that PLCL1 (phospholipase c-like 1) is a novel gene associated with variation of HBS in female Caucasians (Liu et al. 2008). In current Chinese sample, SNPs of PLCL1 gene also showed association evidence at HBS, with most p values at the level of 1.0E-4. However, in the current study, we did not find any association signal at ABS, possibly suggesting the differences of genetic determination of bone size at skeletal sites. Meanwhile, SNPs of PLCL1 were not detected to associate with lean mass, with all p value over 0.05.
This study has limitations in sample matching between the two studied cohorts. First, the average ages between two cohorts are different (34.5 years in Chinese versus 51.6 years in Caucasian). Second, the sex composition is different, with almost half men and half women in Chinese, but many more women in Caucasian (558 vs 1728). These may be the two potential factors underlying discrepancy of the findings between Chinese and Caucasians.
In summary, we conducted a bivariate GWAS study Chinese, followed by a replication study in Caucasians. We identify GLYAT gene which appear to co-regulate BSPs and ALM. The study findings may improve our knowledge of genetic correlation between bone and muscle, and may shed light on pathophysiology of the related diseases such as osteoporosis and sarcopenia.
Supplementary Material
Acknowledgments
This work was partially supported by grants from the NIH (R01 AR050496, R21 AG027110, R01 AG026564, P50 AR055081, R01 AR057049 and R21 AA015973). The study was also supported by the National Natural Science Foundation of China (81000363, 31000554, and 31150006), the Foundation for Distinguished Young Talents in Higher Education of Guangdong (LYM11040), the Dean Fund from Southern Medical University (JC1103), the Fundamental Research Funds for the Central Universities of China (2011JBM134, 2009JBM099), Shanghai Leading Academic Discipline Project (S30501), and startup fund from University of Shanghai for Science and Technology, Xi’an Jiaotong University, and the Ministry of Education of China.
References
- Ahlborg HG, Johnell O, Turner CH, Rannevik G, Karlsson MK. Bone loss and bone size after menopause. N Engl J Med. 2003;349:327–334. doi: 10.1056/NEJMoa022464. [DOI] [PubMed] [Google Scholar]
- Bakay M, Wang Z, Melcon G, Schiltz L, Xuan J, Zhao P, Sartorelli V, Seo J, Pegoraro E, Angelini C, Shneiderman B, Escolar D, Chen YW, Winokur ST, Pachman LM, Fan C, Mandler R, Nevo Y, Gordon E, Zhu Y, Dong Y, Wang Y, Hoffman EP. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain. 2006;129:996–1013. doi: 10.1093/brain/awl023. [DOI] [PubMed] [Google Scholar]
- Balint E, Szabo P, Marshall CF, Sprague SM. Glucose-induced inhibition of in vitro bone mineralization. Bone. 2001;28:21–28. doi: 10.1016/s8756-3282(00)00426-9. [DOI] [PubMed] [Google Scholar]
- Baptista F, Barrigas C, Vieira F, Santa-Clara H, Homens PM, Fragoso I, Teixeira PJ, Sardinha LB. The role of lean body mass and physical activity in bone health in children. J Bone Miner Metab. 2012;30:100–108. doi: 10.1007/s00774-011-0294-4. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Capozza RF, Cure-Cure C, Cointry GR, Meta M, Cure P, Rittweger J, Ferretti JL. Association between low lean body mass and osteoporotic fractures after menopause. Menopause. 2008;15:905–913. doi: 10.1097/gme.0b013e318164ee85. [DOI] [PubMed] [Google Scholar]
- Chumlea WC, Wisemandle W, Guo SS, Siervogel RM. Relations between frame size and body composition and bone mineral status. Am J Clin Nutr. 2002;75:1012–1016. doi: 10.1093/ajcn/75.6.1012. [DOI] [PubMed] [Google Scholar]
- Cointry GR, Capozza RF, Negri AL, Roldan EJ, Ferretti JL. Biomechanical background for a noninvasive assessment of bone strength and muscle-bone interactions. J Musculoskelet Neuronal Interact. 2004;4:1–11. [PubMed] [Google Scholar]
- Deng HW, Deng XT, Conway T, Xu FH, Heaney R, Recker RR. Determination of bone size of hip, spine, and wrist in human pedigrees by genetic and lifestyle factors. J Clin Densitom. 2002a;5:45–56. doi: 10.1385/jcd:5:1:045. [DOI] [PubMed] [Google Scholar]
- Deng HW, Xu FH, Davies KM, Heaney R, Recker RR. Differences in bone mineral density, bone mineral content, and bone areal size in fracturing and non-fracturing women, and their interrelationships at the spine and hip. J Bone Miner Metab. 2002b;20:358–366. doi: 10.1007/s007740200052. [DOI] [PubMed] [Google Scholar]
- Ferretti JL, Cointry GR, Capozza RF, Capiglioni R, Chiappe MA. Analysis of biomechanical effects on bone and on the muscle-bone interactions in small animal models. J Musculoskelet Neuronal Interact. 2001;1:263–274. [PubMed] [Google Scholar]
- Ferretti JL, Cointry GR, Capozza RF, Frost HM. Bone mass, bone strength, muscle-bone interactions, osteopenias and osteoporoses. Mech Ageing Dev. 2003;124:269–279. doi: 10.1016/s0047-6374(02)00194-x. [DOI] [PubMed] [Google Scholar]
- Fry CS, Rasmussen BB. Skeletal Muscle Protein Balance and Metabolism in the Elderly. Curr Aging Sci. 2011 doi: 10.2174/1874609811104030260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoll HH, Gillespie WJ, Gillespie LD, Madhok R. Moving towards evidence-based healthcare for musculoskeletal injuries: featuring the work of the Cochrane Bone, joint and Muscle Trauma Group. J R Soc Promot Health. 2007;127:168–173. doi: 10.1177/1466424007079491. [DOI] [PubMed] [Google Scholar]
- Heymsfield SB, Smith R, Aulet M, Bensen B, Lichtman S, Wang J, Pierson RN., Jr Appendicular skeletal muscle mass: measurement by dual-photon absorptiometry. Am J Clin Nutr. 1990;52:214–218. doi: 10.1093/ajcn/52.2.214. [DOI] [PubMed] [Google Scholar]
- Jin G, Sun J, Isaacs SD, Wiley KE, Kim ST, Chu LW, Zhang Z, Zhao H, Zheng SL, Isaacs WB, Xu J. Human polymorphisms at long non-coding RNAs (lncRNAs) and association with prostate cancer risk. Carcinogenesis. 2011;32:1655–1659. doi: 10.1093/carcin/bgr187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasik D, Zhou Y, Cupples LA, Hannan MT, Kiel DP, Demissie S. Bivariate genome-wide linkage analysis of femoral bone traits and leg lean mass: Framingham study. J Bone Miner Res. 2009;24:710–718. doi: 10.1359/JBMR.081222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Wang Z, Heymsfield SB, Baumgartner RN, Gallagher D. Total-body skeletal muscle mass: estimation by a new dual-energy X-ray absorptiometry method. Am J Clin Nutr. 2002;76:378–383. doi: 10.1093/ajcn/76.2.378. [DOI] [PubMed] [Google Scholar]
- Kochi Y, Suzuki A, Yamada R, Yamamoto K. Genetics of rheumatoid arthritis: underlying evidence of ethnic differences. J Autoimmun. 2009;32:158–162. doi: 10.1016/j.jaut.2009.02.020. [DOI] [PubMed] [Google Scholar]
- Lee CG, Boyko EJ, Strotmeyer ES, Lewis CE, Cawthon PM, Hoffman AR, Everson-Rose SA, Barrett-Connor E, Orwoll ES. Association between insulin resistance and lean mass loss and fat mass gain in older men without diabetes mellitus. J Am Geriatr Soc. 2011;59:1217–1224. doi: 10.1111/j.1532-5415.2011.03472.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei SF, Shen H, Yang TL, Guo Y, Dong SS, Xu XH, Deng FY, Tian Q, Liu YJ, Liu YZ, Li J, Deng HW. Genome-wide association study identifies HMGN3 locus for spine bone size variation in Chinese. Hum Genet. 2011 doi: 10.1007/s00439-011-1093-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidy HJ, Carnell NS, Mattes RD, Campbell WW. Higher protein intake preserves lean mass and satiety with weight loss in pre-obese and obese women. Obesity(Silver Spring) 2007;15:421–429. doi: 10.1038/oby.2007.531. [DOI] [PubMed] [Google Scholar]
- Lino Cardenas CL, Bourgine J, Cauffiez C, Allorge D, Lo-Guidice JM, Broly F, Chevalier D. Genetic polymorphisms of glycine N-acyltransferase (GLYAT) in a French Caucasian population. Xenobiotica. 2010;40:853–861. doi: 10.3109/00498254.2010.519407. [DOI] [PubMed] [Google Scholar]
- Liu J, Pei Y, Papasian CJ, Deng HW. Bivariate association analyses for the mixture of continuous and binary traits with the use of extended generalized estimating equations. Genet Epidemiol. 2009a;33:217–227. doi: 10.1002/gepi.20372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XG, Tan LJ, Lei SF, Liu YJ, Shen H, Wang L, Yan H, Guo YF, Xiong DH, Chen XD, Pan F, Yang TL, Zhang YP, Guo Y, Tang NL, Zhu XZ, Deng HY, Levy S, Recker RR, Papasian CJ, Deng HW. Genome-wide association and replication studies identified TRHR as an important gene for lean body mass. Am J Hum Genet. 2009b;84:418–423. doi: 10.1016/j.ajhg.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YZ, Pei YF, Liu JF, Yang F, Guo Y, Zhang L, Liu XG, Yan H, Wang L, Zhang YP, Levy S, Recker RR, Deng HW. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS One. 2009c;4:e6827. doi: 10.1371/journal.pone.0006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YZ, Wilson SG, Wang L, Liu XG, Guo YF, Li J, Yan H, Deloukas P, Soranzo N, Chinappen-Horsley U, Cervino A, Williams FM, Xiong DH, Zhang YP, Jin TB, Levy S, Papasian CJ, Drees BM, Hamilton JJ, Recker RR, Spector TD, Deng HW. Identification of PLCL1 gene for hip bone size variation in females in a genome-wide association study. PLoS One. 2008;3:e3160. doi: 10.1371/journal.pone.0003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livshits G, Kato BS, Wilson SG, Spector TD. Linkage of genes to total lean body mass in normal women. J Clin Endocrinol Metab. 2007;92:3171–3176. doi: 10.1210/jc.2007-0418. [DOI] [PubMed] [Google Scholar]
- Lukk M, Kapushesky M, Nikkila J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin RB. Size, structure and gender: lessons about fracture risk. J Musculoskelet Neuronal Interact. 2002;2:209–211. [PubMed] [Google Scholar]
- Mattick JS. The genetic signatures of noncoding RNAs. PLoS Genet. 2009;5:e1000459. doi: 10.1371/journal.pgen.1000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey E. Preventing osteoporotic fractures in older people. Practitioner. 2011;255:19–3. [PubMed] [Google Scholar]
- McCreadie BR, Goldstein SA. Biomechanics of fracture: is bone mineral density sufficient to assess risk? J Bone Miner Res. 2000;15:2305–2308. doi: 10.1359/jbmr.2000.15.12.2305. [DOI] [PubMed] [Google Scholar]
- Micklesfield LK, Norris SA, Pettifor JM. Determinants of bone size and strength in 13-year-old South African children: the influence of ethnicity, sex and pubertal maturation. Bone. 2011;48:777–785. doi: 10.1016/j.bone.2010.12.032. [DOI] [PubMed] [Google Scholar]
- Movcessian SG, Boghosian AA, Kamalian RG, Urganjian MK. Effect of Krebs cycle components and glucose on the deamination of mono- and dinucleotides in brain mitochondrial fractions. Vopr Biokhim Mozga. 1972;7:11–17. [PubMed] [Google Scholar]
- Orwig DL, Chiles N, Jones M, Hochberg MC. Osteoporosis in men: update 2011. Rheum Dis Clin North Am. 2011;37:401–14. vi. doi: 10.1016/j.rdc.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Palmer CG, Martinez A, Fox M, Sininger Y, Grody WW, Schimmenti LA. Ethnic differences in parental perceptions of genetic testing for deaf infants. J Genet Couns. 2008;17:129–138. doi: 10.1007/s10897-007-9134-z. [DOI] [PubMed] [Google Scholar]
- Pei YF, Zhang L, Liu J, Deng HW. Multivariate association test using haplotype trend regression. Ann Hum Genet. 2009;73:456–464. doi: 10.1111/j.1469-1809.2009.00527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmenti LA, Martinez A, Telatar M, Lai CH, Shapiro N, Fox M, Warman B, McCarra M, Crandall B, Sininger Y, Grody WW, Palmer CG. Infant hearing loss and connexin testing in a diverse population. Genet Med. 2008;10:517–524. doi: 10.1097/gim.0b013e31817708fa. [DOI] [PubMed] [Google Scholar]
- Scuteri A, Sanna S, Chen WM, Uda M, Albai G, Strait J, Najjar S, Nagaraja R, Orru M, Usala G, Dei M, Lai S, Maschio A, Busonero F, Mulas A, Ehret GB, Fink AA, Weder AB, Cooper RS, Galan P, Chakravarti A, Schlessinger D, Cao A, Lakatta E, Abecasis GR. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Tan LJ, Lei SF, Chen XD, Li X, Pan R, Yin F, Liu QW, Yan XF, Papasian CJ, Deng HW. Bivariate genome-wide association analyses of femoral neck bone geometry and appendicular lean mass. PLoS One. 2011;6:e27325. doi: 10.1371/journal.pone.0027325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulc P, Beck TJ, Marchand F, Delmas PD. Low skeletal muscle mass is associated with poor structural parameters of bone and impaired balance in elderly men--the MINOS study. J Bone Miner Res. 2005;20:721–729. doi: 10.1359/JBMR.041230. [DOI] [PubMed] [Google Scholar]
- Tan LJ, Liu YZ, Xiao P, Yang F, Tang ZH, Liu PY, Recker RR, Deng HW. Evidence for major pleiotropic effects on bone size variation from a principal component analysis of 451 Caucasian families. Acta Pharmacol Sin. 2008;29:745–751. doi: 10.1111/j.1745-7254.2008.00806.x. [DOI] [PubMed] [Google Scholar]
- Uda M, Galanello R, Sanna S, Lettre G, Sankaran VG, Chen W, Usala G, Busonero F, Maschio A, Albai G, Piras MG, Sestu N, Lai S, Dei M, Mulas A, Crisponi L, Naitza S, Asunis I, Deiana M, Nagaraja R, Perseu L, Satta S, Cipollina MD, Sollaino C, Moi P, Hirschhorn JN, Orkin SH, Abecasis GR, Schlessinger D, Cao A. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci US A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Plas DR, Rathmell JC, Fox CJ, Harris MH, Thompson CB. Growth factors can influence cell growth and survival through effects on glucose metabolism. Mol Cell Biol. 2001;21:5899–5912. doi: 10.1128/MCB.21.17.5899-5912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, Nonen S, Fukuda T, Yamazaki H, Azuma J. Genetic polymorphisms of glycine N-acyltransferase in Japanese individuals. Drug Metab Pharmacokinet. 2009;24:114–117. doi: 10.2133/dmpk.24.114. [DOI] [PubMed] [Google Scholar]
- Zhang L, Pei YF, Li J, Papasian CJ, Deng HW. Univariate/multivariate genome-wide association scans using data from families and unrelated samples. PLoS One. 2009;4:e6502. doi: 10.1371/journal.pone.0006502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.