Abstract
We identified a cysteine residue, conserved near the N terminus of Pex5p- and Pex20p-like proteins, that is essential for the cytosolic relocation of peroxisomal Pex20p. Surprisingly, this residue is not completely essential for the function of the protein; its point mutation into a serine in Pex20p(C8S) causes the accumulation of the protein at the peroxisome membrane, but this is quickly followed by its subsequent degradation by an ubiquitin-dependent quality control pathway called RADAR (receptor accumulation and degradation in the absence of recycling). This degradative pathway allows partial growth of the Pex20p(C8S) mutant on peroxisome-requiring medium. Mutation of cysteine 8 (C8S) and lysine 19 (K19R), the target residue of the RADAR pathway within Pex20p, leads to a stable but non-functional protein because it fails to recycle to the cytosol. This suggests a role for Cys-8 in Pex20p recycling and that constitutive degradation of peroxisomal receptors can be a partially functional alternative to receptor recycling. In addition, expression of this mutant protein in wild-type cells confers a dominant-negative, oleate-specific growth defect, which is a useful tool for a better understanding of peroxisomal receptor recycling.
Peroxisomal matrix proteins synthesized in the cytosol must be translocated post-translationally across the peroxisomal membrane (1). Matrix protein import into peroxisomes requires many peroxins involved in the recognition, targeting, and translocation of cargoes carrying a peroxisome targeting signal (PTS).3 The PTS on the cargo is bound by soluble, cytosolic receptors before the cargo:receptor complex is targeted to, and imported inside, the peroxisome. During matrix protein import, the PTS receptors/co-receptors, Pex5p, Pex7p, and Pex20p, cycle between the cytosol and the peroxisome as part of an “extended shuttle,” which implies the “recycling” of the receptors/co-receptors from the peroxisome back to the cytosol (2).
Many reports suggest a role for ubiquitin in receptor recycling, and indeed, several peroxins appear to be linked to ubiquitin metabolism (3). Pex4p is an ubiquitin-carrier protein (or E2) peripheral to the peroxisomal membrane and is required in the late steps of import, likely receptor recycling (4, 5). Also, three peroxins of the peroxisomal membrane possess a RING domain (Pex2p, Pex10p, Pex12p), often found in ubiquitin-protein isopeptide ligases (or E3s). These proteins may be involved in the retrotranslocation of the co-receptor, named Pex20p (6). Finally, a complex of AAA ATPases (Pex1p and Pex6p) is also required for the recycling of Pex5p and Pex20p (6–8). The situation resembles that at the endoplasmic reticulum, where E2s, E3s, and the AAA ATPase Cdc48p collaborate to extract and degrade misfolded or aberrant proteins from the lumen or membrane in a process called endoplasmic reticulum-associated degradation (9).
So far, the only ubiquitylated peroxins identified are Pex5p (PTS1 receptor (10, 11–13)) and Pex18p/Pex20p (PTS2 co-receptors (6, 14)). Ubiquitylated species of Pex5p were first identified in receptor recycling mutants, suggesting that ubiquitylation might be a prerequisite in the receptor shuttling process (10). However, this ubiquitylation now appears to be linked to a quality control mechanism that is triggered in response to receptor accumulation at the peroxisomal membrane as a consequence of their inability to recycle (13). By analogy to endoplasmic reticulum-associated degradation, we termed this mechanism the peroxisomal RADAR pathway (for receptor accumulation and degradation in the absence of recycling, (6)). On the other hand, work on Pex18p (14) and on Pex5p (11) identified monoubiquitylated species of these proteins, which may be indicative of a role for this modification in recycling. Indeed, monoubiquitylation is often associated with signaling or protein trafficking rather than proteolysis (15). Therefore, ubiquitin may play dual roles in peroxisome biogenesis. Consistent with this hypothesis, polyubiquitylation was found to be essential for the recycling step itself, independently of the RADAR (6).
Pex5p and Pex20p possess a conserved N-terminal domain (described here as “NtD”) (6). Deletion of the NtD in Pex20p abolishes its function and leads to a protein that accumulates at the peroxisome instead of being mostly cytosolic, suggesting an involvement of this region in recycling to the cytosol.
We studied in greater detail this NtD and observed that one residue, Cys-8, is essential for the recycling of Pex20p to the cytosol. However, we show that this residue is not completely essential for the function of the protein because similar to what happens in the recycling mutants, Pex20p(C8S) is constitutively degraded by the RADAR pathway, leading to a partially functional protein and therefore an alternative to receptor recycling.
EXPERIMENTAL PROCEDURES
Yeast Strains, Cultures, and Growth Assay
Strains and culture media were described previously (Ref. 6 and references cited therein). For growth assays, cells were grown overnight in rich (YPD) medium and inoculated again for 8 h prior to transfer in oleate (YNO) medium. Cultures were started at 0.3 A600/ml. After growth for the indicated time, cells were washed free of oleate medium and resuspended in the same volume of water. Growth was assayed by measurement of A600, and the initial A600 was subtracted from this value.
Constructs
To construct truncations and mutations within the Pex20p NtD, mutagenesis was performed on pSEB48 (6). Oligonucleotides are listed in Supplemental Table 1. Vectors were linearized with SalI and inserted at the HIS4 locus in the Δpex20, Δpex14, or wild-type (PPY12) strains. Other constructs were described previously (6).
Fluorescence Microscopy
Cells were grown on YPD and switched to YNO during exponential phase. The construct pPEX3::PEX3-mRFP (pJCF215) was a kind gift of Dr. J-C Farré, UCSD-Biology. Images were captured on a Zeiss Axioskop fluorescence microscope (AxioSkop 2 Plus, motorized) coupled to a cooled CCD monochrome camera (AxioCam MRM, Zeiss) and processed using AxioVision software.
Crude Extracts
Oleate-grown cells (8 ODs) were collected and resuspended in 200 μl of ice-cold immunoprecipitation lysis buffer (50 mm HEPES-KOH, pH 7.5, 0.5 m NaCl, 0.5% Nonidet P-40, 10% glycerol, 1 mm EDTA) containing the following inhibitors (Sigma): yeast protease inhibitor mixture, NaF (50 mm), leupeptin (12.5 μg/ml), aprotinin (50 μg/ml), phenylmethylsulfonyl fluoride (10 mm), N-ethylmaleimide (100 mm), and MG-132 (100 μm). Cells were broken with glass beads for 10 min at 4 °C. SDS sample buffer was then added, and samples were denatured for 5 min at 65 °C. Prior to loading, the sample was denatured again and centrifuged at 18,000 × g for 5 min at room temperature. 10 μl were subjected to SDS-PAGE and immunoblot analysis.
Subcellular Fractionation
Cells were grown overnight on YPD medium, precultured on YPD for 10 h, and transferred overnight into YNO. Subcellular fractionation has been described previously (6).
Co-immunoprecipitations
Cells (30 ODs) were prepared as for crude extracts (above) except that after the lysis with glass beads, the extracts were centrifuged at 14,000 × g for 10 min at 4 °C. Co-immunoprecipitations were performed on the supernatant, using a mixture of monoclonal GFP antibodies (clones 7.1 and 13.1; Roche Applied Sciences) at a concentration of 2.5 μg/ml crude extract. The samples were mixed by rotation for 3 h at 4 °C, and then 50 μl of Gammabind G-Sepharose beads (GE Healthcare Bio-Sciences Co.) were added for another 1.5 h. The beads were washed six times by rotating the beads for 5 min at 4 °C with 1 ml of immunoprecipitation lysis buffer (once) and then 1 ml of immunoprecipitation wash buffer (50 mm HEPES-KOH, pH 7.5, 150 mm NaCl, 1 mm EDTA) (5 times). The beads were finally boiled in SDS loading buffer (30 μl). The equivalent of 0.2 OD (input and unbound) or 2.5 ODs (immunoprecipitate) were loaded on SDS-PAGE gel and subjected to immunoblot analysis.
RESULTS AND DISCUSSION
A Conserved Cysteine Residue Is Required for Pex20p Function in the Absence of RADAR
We showed previously that the NtD of Pex20p is essential for its function; Pex20p(Δ1–19) is non-functional and accumulates in or on peroxisomes (6). The conserved lysine present in the NtD (Lys-19 in PpPex20p, Lys-22 in PpPex5p) is a target for polyubiquitylation and degradation by the RADAR pathway but is not directly involved in the recycling step since its substitution to arginine led to functional proteins (6, 13). Thus, the RADAR pathway is not essential for peroxisome biogenesis.
In contrast, deletion of the first 16 residues of Pex20p leads to a partially functional protein. We wondered how a growth defect could arise in the absence of residues 1–19, whereas residues 1–16 are not fully required and Lys-19 mutation has no effect on the protein function. We hypothesized that the NtD and the Lys-19 residue possess a redundant function that leads to no, or mild, phenotypes upon mutation of either of them.
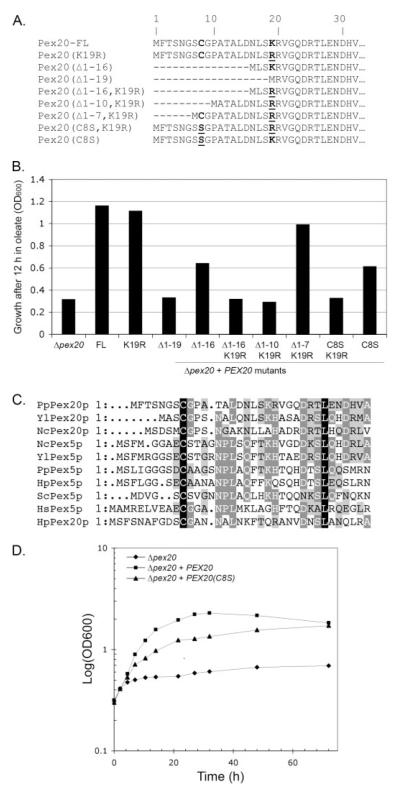
In agreement with this idea, we first observed that Lys-19 was required for Pex20p function in the absence of the 16 N-terminal residues (Pex20p(Δ1–16,K19R), Fig. 1, A and B). We then tried to identify which region/residue was required when Lys-19 is missing by performing N-terminal truncations in a K19R mutant background, namely, Pex20p(Δ1–7,K19R) and Pex20p(Δ1–10,K19R) (Fig. 1A). As shown in Fig. 1B, Pex20p(Δ1–7,K19R) fully restored the growth of the Δpex20 strain in oleate medium, as opposed to Pex20p(Δ1–10,K19R), Pex20p(Δ1–16,K19R), or Pex20p(Δ1–19), which failed to complement the Δpex20 growth defect. We concluded that residues 8–10 are required for Pex20p function in the absence of Lys-19. In particular, residue Cys-8 is present in all known or predicted Pex20p and Pex5p proteins (Fig. 1C), supporting the idea that it serves an important function. Although the C8S mutation (Pex20p(C8S)) altered only partially the function of Pex20p, the combination of both C8S and K19R mutations (Pex20p(C8S,K19R)) led to a non-functional protein, suggesting that these two residues have a redundant role regarding Pex20p function. Interestingly, a more detailed look at the intermediate growth of the Δpex20 strain complemented with the Pex20p(C8S) construct (Fig. 1D) indicated that this mutant protein is likely to be functional but may be less efficient, leading to a slower but constant growth on oleate medium that eventually reaches the same OD as wild-type cells after 3 days. Interestingly, similar results were obtained when the same experiment was performed with Pex5p(C10S) mutant, mutated in the conserved cysteine residue (data not shown).
FIGURE 1. A conserved cysteine residue is essential for Pex20p function when its degradation by RADAR is compromised.
A, schematic of the constructs used in this study. FL, full-length Pex20p. B, Δpex20 cells transformed with the constructs shown in A were assayed for growth on oleate medium. C, multiple alignment of the NtD of Pex20p and Pex5p proteins from various organisms. D, the effect of Cys-8 mutation on growth on oleate.
These results show that the function of Pex20p relies on the presence of the Cys-8 or Lys-19 residues. Given that the K19R mutation renders Pex20p insensitive to the RADAR pathway (6), we conclude that residue Cys-8 is essential when the degradation of Pex20p by RADAR is compromised. Conversely, when Cys-8 is mutated, Lys-19 is essential for Pex20p function.
Pex20p(C8S) Is Constitutively Degraded by Polyubiquitylation on Its Lys-19 Residue
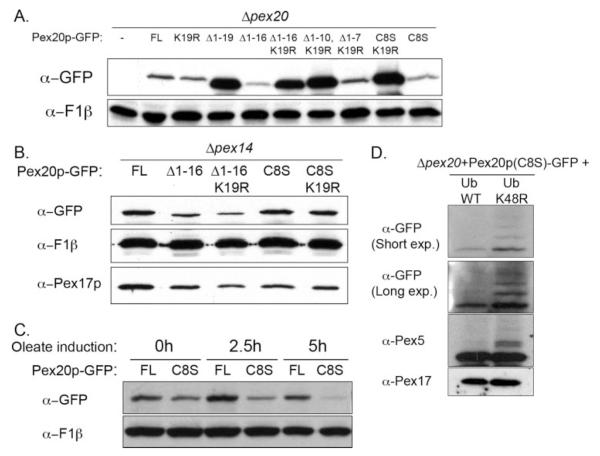
Since the RADAR pathway is involved in the degradation of receptors, we checked the steady-state levels of truncated/mutant proteins expressed in the Δpex20 background to see whether they differed from that of the wild-type protein. When crude extracts of oleate-grown cells were analyzed, it appeared that the (Δ1–16) truncation or C8S mutation led to lower steady-state levels of Pex20p (Fig. 2A, see also Fig. 3B), although both complemented the mutant strain (Fig. 1, B and D). This difference was more noticeable when the proteasome inhibitor MG132 was omitted in the lysis buffer (data not shown).
FIGURE 2. Low steady-state level of Pex20p lacking Cys-8 and dependence on the integrity of the peroxisome import machinery.
A, steady-state levels of the indicated proteins expressed in Δpex20 in the crude extracts of oleate-induced cells grown overnight. FL, full-length Pex20p. B, steady-state levels of the indicated proteins expressed in Δpex14 in the crude extracts of oleate-induced cells grown overnight. C, kinetics of Pex20p(C8S) steady-state levels on oleate medium. D, the effect of the overexpression of Ub or Ub(K48R) on Pex20p(C8S) steady-state levels. WT, wild type.
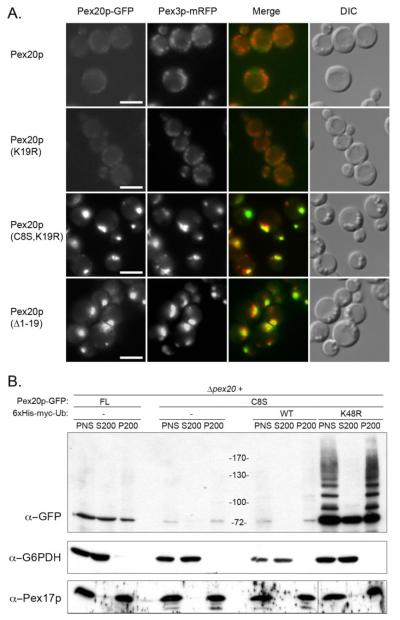
FIGURE 3. Pex20p(C8S) fails to relocate to the cytosol and as a result, is degraded at the peroxisome.
A, subcellular localization of Pex20p, Pex20p(K19R), and Pex20p(C8S,K19R) by fluorescence microscopy and co-localization with a peroxisomal marker (Pex3p-mRFP). Δpex20 cells expressing the various constructs were grown overnight on oleate medium. Bar = 5 μm. DIC, differential interference contrast. B, subcellular localization of Pex20p and Pex20p(C8S) proteins by subcellular fractionation and effect of overexpression of Ub and Ub-K48R on the localization and steady-state level of Pex20p(C8S). Δpex20 cells expressing the various constructs were grown overnight on oleate medium. FL, full-length Pex20p; WT, wild type.
This lower steady-state level was not observed when the peroxisomal docking factor, Pex14p, was missing (Fig. 2B). In this case, the stabilities of the C8S and C8S,K19R mutants were equivalent, as were those of Pex20p(Δ1–16) and Pex20p(Δ1–16, K19R), suggesting that the instabilities of C8S and (Δ1–16) are not intrinsic to the mutant proteins but rather require an intact peroxisomal import machinery. The Pex20p(C8S) steady-state levels decreased progressively with time after transfer into oleate medium, whereas wild-type Pex20p accumulated (Fig. 2C). Again, this supports the idea that this protein is not naturally unstable but that the lower steady-state level of the C8S mutant is linked with oleate utilization and therefore peroxisome biogenesis.
We addressed the origin of this low steady-state level by studying whether it is affected by overexpression of the ubiquitin mutant Ub(K48R), which interferes with Lys-48-branched polyubiquitylation and subsequent degradation by the proteasome and therefore inhibits the RADAR pathway. Overexpression of Ub(K48R) not only prevented Pex20p(C8S) degradation in Δpex20 cells but also allowed the detection of polyubiquitylated species of Pex20p(C8S) (Fig. 2D; see also Fig. 3B), indicating that the drop in Pex20p(C8S) steady-state level is likely due to an active degradation after polyubiquitylation via the ubiquitin-proteasome system. Overexpression of ubiquitin alone had no discernable effect.
An obvious candidate for the target residue of this polyubiquitylation within Pex20p(C8S) and Pex20p(Δ1–16) is Lys-19. Indeed, the steady-state levels of Pex20p(C8S,K19R) and Pex20p(Δ1–16,K19R) proteins were dramatically increased, showing an expression level that was even higher that that of the wild-type protein (Fig. 2A). In summary, these results demonstrate that Pex20p(C8S) mutant protein is actively degraded in wild-type cells by polyubiquitylation on residue Lys-19 and subsequent degradation by the ubiquitin-proteasome system pathway, in agreement with the hypothesis that Pex20p(C8S) is a target of the RADAR pathway.
Pex20p(C8S) Is Degraded by the RADAR Pathway at the Surface of the Peroxisome after a Round of Import because It Fails to Recycle
The above observations indicate that Pex20p(C8S) is constitutively degraded after the import cycle. Since its degradation happens by the RADAR pathway, which functions at the surface of the peroxisome, we checked whether its stabilization by the additional K19R mutation leads to the exclusive localization of the protein at the peroxisome. By fluorescence microscopy, we observed that although Pex20p or Pex20p(K19R) have a mostly cytosolic localization when transformed into the Δpex20 strain, Pex20p(C8S,K19R) behaved like Pex20p(Δ1–19) and was mostly peroxisomal (Fig. 3A). It should be noted that, as stated above, the K19R mutation alone does not alter the location of Pex20p, but by preventing access to the RADAR pathway and stabilizing the protein, it allowed us to determine that the Pex20p(C8S,K19R) protein accumulates at the peroxisome, indicative of a recycling defect. These data suggest the presence of two essential residues, Cys-8 and Lys-19, within the NtD, for recycling and RADAR, respectively.
We studied the subcellular localization of Pex20p(C8S) mutant protein when expressed in Δpex20 cells. As described previously, wild-type Pex20p was present mostly in the cytosol and partially in the organelle membrane fraction (6). However, despite being difficult to detect because of its low steady-state level (Fig. 2A), Pex20p(C8S) was mostly associated with the organelle pellet (Fig. 3B). This was a surprising observation since these constructs are partially functional, and proper function of Pex20p is thought to involve its shuttling between the cytosol and the peroxisome (6).
We hypothesized that Pex20p(C8S) is constitutively degraded at the surface of the peroxisomal membrane. To verify this, post-nuclear supernatants of Δpex20 cells co-expressing Pex20p(C8S) and Ub(K48R) or Ub (as a control) were subjected to differential centrifugation. Although strong, constitutive overexpression of Ub did not affect Pex20p(C8S) localization, overexpression of Ub(K48R) led to the appearance of abundant polyubiquitylated species of Pex20p in the pellet fraction (Fig. 3B). This observation is consistent with constitutive degradation of the mutant protein at the surface of the peroxisome, likely after the completion of an import cycle, since some import occurs with this partially functional construct. This would explain why Pex20p(C8S) has a low steady-state level but is yet functional and also why this protein is functional although it is mostly organelle-associated. This idea is also supported by the fact that stabilization of Pex20p(C8S) by the K19R mutation leads to a mostly peroxisomal protein (Fig. 3A).
Pex20p(C8S) is thus constitutively degraded by the RADAR pathway at the surface of the peroxisome, and prevention of this degradation by the K19R mutation leads to a stable, peroxisome-associated, and non-functional protein, presumably because it fails to recycle to the cytosol. Therefore, it is likely that the RADAR pathway can rescue the cells by degrading receptors that fail to recycle and that degradation at the peroxisome can be used as an alternative for recycling. In addition, our data point out to an essential role of residue Cys-8 in Pex20p recycling, and presumably, to a role for Cys-10 Pex5p recycling.
Mutations/Truncations in the NtD of Pex20p Do Not Prevent Its Association with the Docking and the RING Subcomplexes
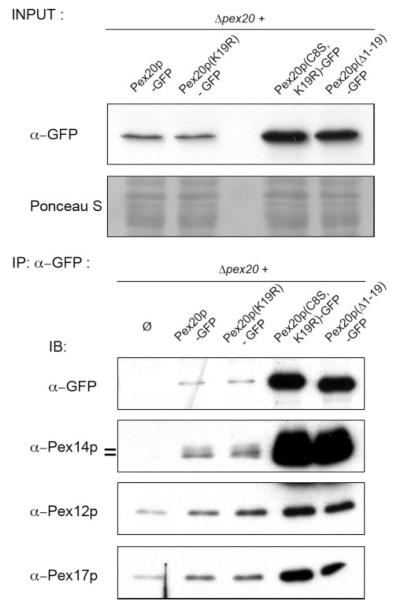
We investigated whether point mutations in, or even the complete absence of, the NtD of Pex20p changed its ability to interact with protein complexes of the peroxisomal membrane. Since Pex20p(Δ1–19) and Pex20p(C8S,K19R) are present at very high levels in the cells (Figs. 2A and 4, top panel), presumably because they are trapped at the peroxisome and cannot be degraded by the RADAR pathway, their immunoprecipitation led to a higher amount of recovered proteins that should be considered when analyzing the co-immunoprecipitation data. However, the results clearly indicate no qualitative change in the ability to interact with the docking proteins such as Pex14p or Pex17p (Fig. 4), in good agreement with the fact that the NtD is not required for the peroxisomal localization of Pex20p (Fig. 3A).
FIGURE 4. Mutations or deletion in the NtD of Pex20p do not lead to a qualitative change in its interaction with representative members of the docking or the RING subcomplexes.
Co-immunoprecipitations were performed on cells from oleate-induced, overnight-grown cells, and immunoprecipitates (IP) were analyzed by SDS-PAGE and immunoblotting (IB) with the indicated antibodies. WT, wild type.
We also observed a novel interaction between Pex20p and a member of the RING peroxin subcomplex, as shown by the co-immunoprecipitation of Pex12p with Pex20p (Fig. 4). Once again, mutation of the residue Cys-8, or deletion of residues 1–19 of Pex20p, did not lead to a change in this interaction. This result was expected since the RING peroxins play a role in the retrotranslocation of Pex20p from the peroxisome to the cytosol (6), and we report here that Pex20p(C8S) is degraded by the ubiquitin-proteasome system, suggesting that this retrotranslocation has occurred.
Previous results indicate a role for the AAA ATPase complex (Pex1p and Pex6p) in dislocation of Pex5p and Pex20p out of the peroxisomal membrane (7). Therefore, we addressed the interaction of Pex20p with Pex1p and Pex6p. However, none was detected at a significant level in the co-immunoprecipitate, neither with the wild-type Pex20p nor with the truncated proteins (data not shown). Similarly, with the yeast two-hybrid technique, no interactions were detected between Pex20p and Pex1p or Pex6p (data not shown), similarly to what was observed for Pex5p (16). This interaction may happen only in the presence of the full AAA ATPase complex and perhaps only when bound to ATP, a situation that probably was not mimicked in the yeast-two hybrid system. Therefore, the question remains open as to the effect of the C8S mutation on the interaction of Pex20p with the recycling complex.
Pex20p(C8S,K19R) Displays a Dominant-negative Phenotype
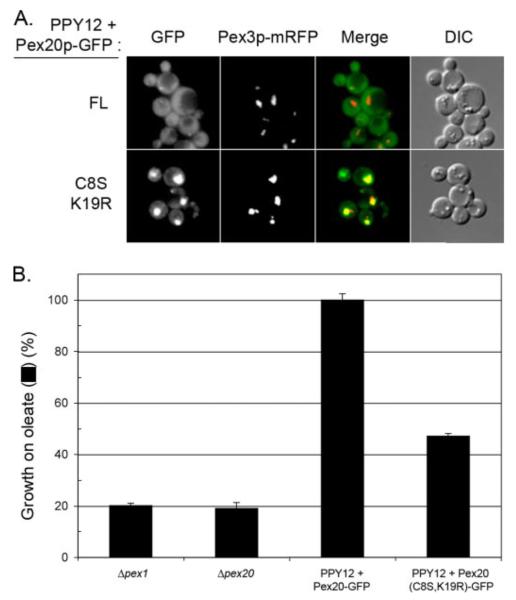
Since Pex20p(C8S,K19R) accumulates at high steady-state levels in the cells and since it is trapped in or on the peroxisome, we asked whether this situation can have deleterious consequences on peroxisome biogenesis in wild-type cells. First, we observed that when expressed in oleate-grown wild-type PPY12 cells, the Pex20p(C8S,K19R) mutant protein displayed the same subcellular localization as in the Δpex20 strain (Fig. 5A). In addition, we observed that expression of Pex20p(C8S,K19R) in wild-type cells led to an inhibition of growth on oleate (Fig. 5B). This phenotype was not observed when cells were grown on methanol as a carbon source (data not shown), a situation where peroxisomes are required for survival, but where the PTS2 import pathway, and therefore, Pex20p, is not required (6, 17). This indicates that instead of a general import defect, Pex20p(C8S,K19R) expression leads to a specific defect in the PTS2 pathway.
FIGURE 5. Pex20p(C8S,K19R) acts as a dominant-negative mutant.
A, subcellular localization of Pex20p-GFP and Pex20p(C8S,K19R)-GFP in wild-type cells grown on oleate for 6 h. Bar = 5 μm. DIC, differential interference contrast. B, growth on oleate medium (in the percentage of positive control, PPY12+Pex20p-GFP) of wild-type cells expressing Pex20p-GFP or Pex20p(C8S,K19R)-GFP.
A Conserved Cysteine Is Required for Pex20p Relocation to the Cytosol
We have shown that two routes can be used for the removal of Pex20p from the peroxisomal membrane. First, the recycling to the cytosol requires in cis a conserved cysteine residue in the NtD, and in trans, some specific peroxins, such as Pex1p, Pex6p, and Pex4p (6). Second, in the absence of any of these cis or trans recycling components, the RADAR pathway is activated and leads to the degradation of Pex20p by polyubiquitylation of the conserved Lys-19 residue present in the NtD. This is summarized in our current working model (Supplemental Fig. 1).
Although neither pathway is strictly essential per se, recycling appears to be the favored route since its impairment has a major effect on peroxisome biogenesis, whereas the RADAR pathway is not essential (6). However, our data show that the RADAR pathway, whose existence, but not function, has been known for some time in many organisms (for a review, see Ref. 18) can be used as a back-up system when recycling is compromised. Although we have not been able to observe a difference in the known protein-protein interactions between Pex20p and Pex20p(C8S,K19R), it is still possible that the C8S mutation modifies the affinity of the Pex20p toward the recycling machinery or a potential adaptor.
Our initial observations on the role of Cys-8 of Pex20p in peroxisome biogenesis were made with a cysteine-to-alanine mutant (data not shown). However, cysteine-to-serine mutations, such as the one described herein, are traditionally used because these two amino acids differ only in that the former carries a sulfur atom, whereas the latter has an oxygen, leading to little or no conformational change upon mutation. However, the sulfur-based chemistry, such as redox modifications (disulfide bond formation), thioether (prenylation), or thiolester linkages (S-acylation, palmitoylation, or ubiquitylation), cannot be supported by a serine. Therefore, another intriguing possibility is that Cys-8 is modified during the import cycle, and this modification is essential for recycling.
This idea was first supported by the fact that the C8K mutation has a less deleterious effect than its mutation to a serine, an alanine, or even an arginine,4 which led us to the tantalizing possibility that this cysteine might be a target for ubiquitylation. Indeed, ubiquitylation plays a clear role in receptor recycling (reviewed in Ref. 19). However, (i) we have been unable to demonstrate that this cysteine is ubiquitylated, and (ii) both Pex20p(C8K) and Pex20p(C8K,K19R) mutations restore the RADAR-mediated degradation by creating a new target site for ubiquitylation at the mutated position.5 Therefore, whether or not this cysteine is ubiquitylated remains unknown. No other types of modification are described for either Pex20p or Pex5p, but this is still a possibility that should be investigated. Identification of the components and interactions mediating the recycling and RADAR pathways will help in the global understanding of peroxisome biogenesis.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Drs. Xiumei Cao and Farida Khan (UCSD) for technical help and for unpublished data. We thank Dr. Ben Distel (Amsterdam Medical Center, University of Amsterdam, Netherlands) for helpful suggestions and for sharing unpublished data.
Footnotes
The abbreviations used are: PTS, peroxisome targeting signal; AAA, ATPases associated with various cellular activities; GFP, green fluorescent protein; NtD, N-terminal domain; RADAR, receptor accumulation and degradation in the absence of recycling; RING, really interesting new gene; Ub, ubiquitin.
F. Khan, S. Léon, and S. Subramani, unpublished data.
F. Khan and S. Subramani, unpublished data.
This work was supported by National Institutes of Health Grant DK41737 (to S. S.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains a supplemental figure and supplemental table.
REFERENCES
- 1.Purdue PE, Lazarow PB. Annu. Rev. Cell Dev. Biol. 2001;17:701–752. doi: 10.1146/annurev.cellbio.17.1.701. [DOI] [PubMed] [Google Scholar]
- 2.Kunau WH. Curr. Biol. 2001;11:R659–662. doi: 10.1016/s0960-9822(01)00386-4. [DOI] [PubMed] [Google Scholar]
- 3.Erdmann R, Schliebs W. Nat. Rev. Mol. Cell Biol. 2005;6:738–742. doi: 10.1038/nrm1710. [DOI] [PubMed] [Google Scholar]
- 4.Wiebel FF, Kunau WH. Nature. 1992;359:73–76. doi: 10.1038/359073a0. [DOI] [PubMed] [Google Scholar]
- 5.Collins CS, Kalish JE, Morrell JC, McCaffery JM, Gould SJ. Mol. Cell Biol. 2000;20:7516–7526. doi: 10.1128/mcb.20.20.7516-7526.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Léon S, Zhang L, McDonald WH, Yates J, III, Cregg JM, Subramani S. J. Cell Biol. 2006;172:67–78. doi: 10.1083/jcb.200508096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Platta HW, Grunau S, Rosenkranz K, Girzalsky W, Erdmann R. Nat. Cell Biol. 2005;7:817–822. doi: 10.1038/ncb1281. [DOI] [PubMed] [Google Scholar]
- 8.Miyata N, Fujiki Y. Mol. Cell Biol. 2005;25:10822–10832. doi: 10.1128/MCB.25.24.10822-10832.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romisch K. Annu. Rev. Cell Dev. Biol. 2005;21:435–456. doi: 10.1146/annurev.cellbio.21.012704.133250. [DOI] [PubMed] [Google Scholar]
- 10.Platta HW, Girzalsky W, Erdmann R. Biochem. J. 2004;384:37–45. doi: 10.1042/BJ20040572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kragt A, Voorn-Brouwer TM, Van den Berg M, Distel B. J. Biol. Chem. 2005;280:7867–7874. doi: 10.1074/jbc.M413553200. [DOI] [PubMed] [Google Scholar]
- 12.Kiel JA, Emmrich K, Meyer HE, Kunau WH. J. Biol. Chem. 2005;280:1921–1930. doi: 10.1074/jbc.M403632200. [DOI] [PubMed] [Google Scholar]
- 13.Kiel JA, Otzen M, Veenhuis M, van der Klei IJ. Biochim. Biophys. Acta. 2005;1745:176–186. doi: 10.1016/j.bbamcr.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 14.Purdue PE, Lazarow PB. J. Biol. Chem. 2001;276:47684–47689. doi: 10.1074/jbc.M106823200. [DOI] [PubMed] [Google Scholar]
- 15.Hicke L. Nat. Rev. Mol. Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 16.Faber KN, Heyman JA, Subramani S. Mol. Cell Biol. 1998;18:936–943. doi: 10.1128/mcb.18.2.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elgersma Y, Elgersma-Hooisma M, Wenzel T, McCaffery JM, Farquhar MG, Subramani S. J. Cell Biol. 1998;140:807–820. doi: 10.1083/jcb.140.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leon S, Goodman JM, Subramani S. Biochim. Biophys. Acta. 2006;1763:1552–1564. doi: 10.1016/j.bbamcr.2006.08.037. [DOI] [PubMed] [Google Scholar]
- 19.Thoms S, Erdmann R. Biochim. Biophys. Acta. 2006;1763:1621–1628. doi: 10.1016/j.bbamcr.2006.08.046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.