Abstract
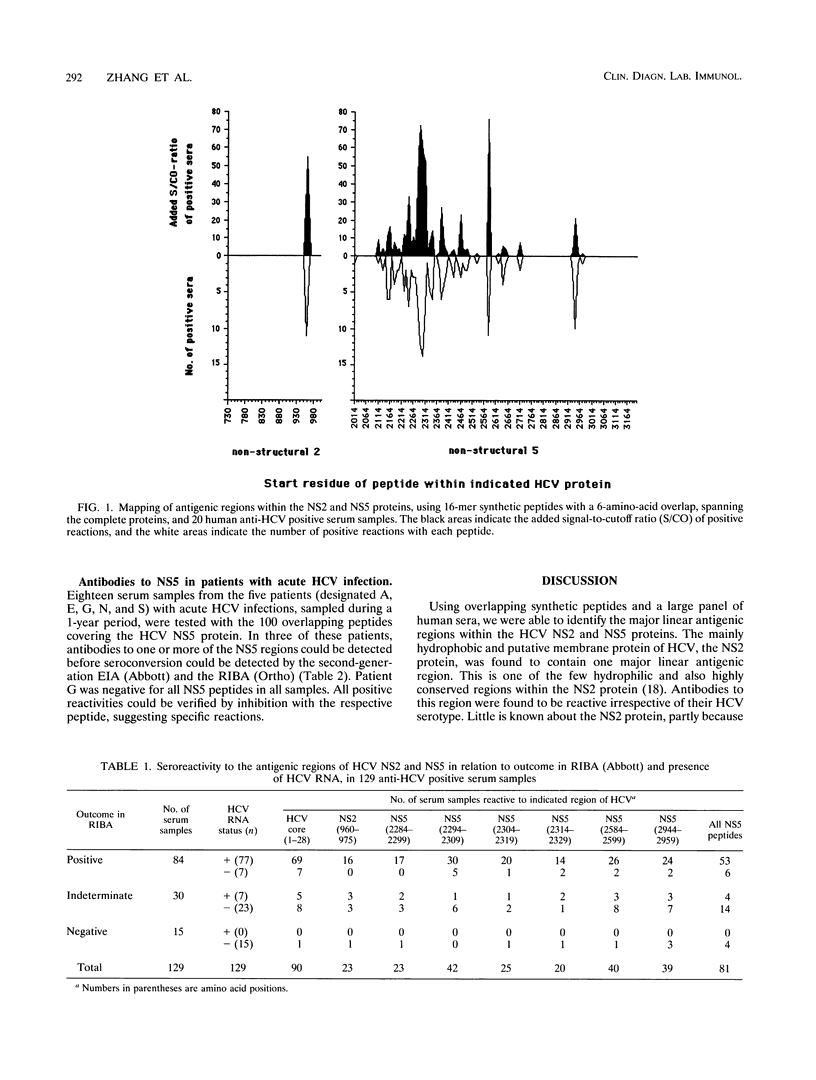
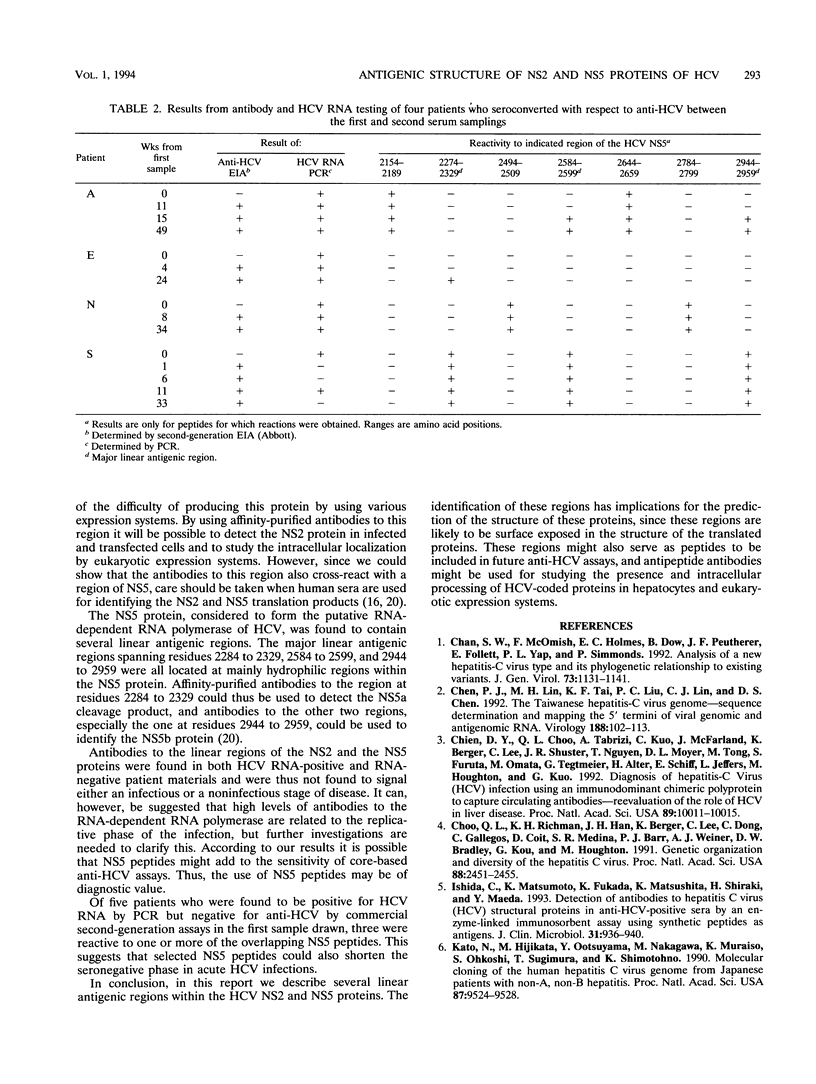
Antigenic regions within the nonstructural (NS) 2 and 5 proteins of hepatitis C virus (HCV) were identified and characterized by the use of 127 overlapping synthetic peptides and a serum panel consisting of 167 human serum samples from persons with antibodies to HCV. Initially, 20 anti-HCV-positive serum samples were used to screen the peptides covering the complete NS2 and NS5 proteins. Among the 27 overlapping peptides spanning the NS2 protein of HCV, only the peptide covering residues 960 to 975 was recognized by human sera. Within the 100 peptides covering the NS5 protein, major linear antigenic regions were located at residues 2284 to 2329 within the putative NS5a and at residues 2584 to 2599 and 2944 to 2959 within the putative NS5b. Additional minor linear antigenic regions were also identified within the NS5. The sequence of the antigenic region of the NS2 protein is, unlike most parts of the NS2 protein, highly conserved among the described types of HCV, whereas the sequence of the major antigenic region of NS5 shows variability among HCV types. The recognition of a peptide corresponding to a part of the major region of NS5 was found to be dependent on HCV type. In 129 anti-HCV-positive serum samples, the prevalence of antibodies to the NS2 protein was found to be 23% among HCV RNA-positive sera and 10% among HCV RNA-negative sera. In the same samples, reactivity to the major linear antigenic regions of HCV NS5 was found in 68% of the HCV RNA-positive sera and 67% of the HCV RNA-negative sera. Of 18 serum samples from five patients with acute HCV infections, and who seroconverted with respect to anti-HCV, 4 were found to be reactive to one or more of the 100 NS5 peptides and in three serum samples the NS5 reactivities were found to shorten the time for serodiagnosis of cross-reactive with a region form residues 2584 to 2599 of NS5, which has 67% homology with a six-residue sequence of NS2. In conclusion, in this study we have identified and evaluated the potential use of synthetic peptides corresponding to linear antigenic regions of the NS2 and NS5 proteins.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chan S. W., McOmish F., Holmes E. C., Dow B., Peutherer J. F., Follett E., Yap P. L., Simmonds P. Analysis of a new hepatitis C virus type and its phylogenetic relationship to existing variants. J Gen Virol. 1992 May;73(Pt 5):1131–1141. doi: 10.1099/0022-1317-73-5-1131. [DOI] [PubMed] [Google Scholar]
- Chen P. J., Lin M. H., Tai K. F., Liu P. C., Lin C. J., Chen D. S. The Taiwanese hepatitis C virus genome: sequence determination and mapping the 5' termini of viral genomic and antigenomic RNA. Virology. 1992 May;188(1):102–113. doi: 10.1016/0042-6822(92)90739-c. [DOI] [PubMed] [Google Scholar]
- Chien D. Y., Choo Q. L., Tabrizi A., Kuo C., McFarland J., Berger K., Lee C., Shuster J. R., Nguyen T., Moyer D. L. Diagnosis of hepatitis C virus (HCV) infection using an immunodominant chimeric polyprotein to capture circulating antibodies: reevaluation of the role of HCV in liver disease. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10011–10015. doi: 10.1073/pnas.89.21.10011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo Q. L., Richman K. H., Han J. H., Berger K., Lee C., Dong C., Gallegos C., Coit D., Medina-Selby R., Barr P. J. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida C., Matsumoto K., Fukada K., Matsushita K., Shiraki H., Maeda Y. Detection of antibodies to hepatitis C virus (HCV) structural proteins in anti-HCV-positive sera by an enzyme-linked immunosorbent assay using synthetic peptides as antigens. J Clin Microbiol. 1993 Apr;31(4):936–940. doi: 10.1128/jcm.31.4.936-940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato N., Hijikata M., Ootsuyama Y., Nakagawa M., Ohkoshi S., Sugimura T., Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal G. J., Baroudy B. M., Kuramoto I. K., McDonald F. F., Schiff G. M., Holland P. V., Zeldis J. B. Detection of acute hepatitis C virus infection by ELISA using a synthetic peptide comprising a structural epitope. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4486–4489. doi: 10.1073/pnas.89.10.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo G., Choo Q. L., Alter H. J., Gitnick G. L., Redeker A. G., Purcell R. H., Miyamura T., Dienstag J. L., Alter M. J., Stevens C. E. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989 Apr 21;244(4902):362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Kurai K., Okada S., Yamamoto K., Lizuka H., Tanaka T., Fukuda S., Tsuda F., Mishiro S. Full-length sequence of a hepatitis C virus genome having poor homology to reported isolates: comparative study of four distinct genotypes. Virology. 1992 May;188(1):331–341. doi: 10.1016/0042-6822(92)90762-e. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Okada S., Sugiyama Y., Kurai K., Iizuka H., Machida A., Miyakawa Y., Mayumi M. Nucleotide sequence of the genomic RNA of hepatitis C virus isolated from a human carrier: comparison with reported isolates for conserved and divergent regions. J Gen Virol. 1991 Nov;72(Pt 11):2697–2704. doi: 10.1099/0022-1317-72-11-2697. [DOI] [PubMed] [Google Scholar]
- Okamoto H., Tsuda F., Machida A., Munekata E., Akahane Y., Sugai Y., Mashiko K., Mitsui T., Tanaka T., Miyakawa Y. Antibodies against synthetic oligopeptides deduced from the putative core gene for the diagnosis of hepatitis virus infection. Hepatology. 1992 Feb;15(2):180–186. doi: 10.1002/hep.1840150203. [DOI] [PubMed] [Google Scholar]
- Selby M. J., Choo Q. L., Berger K., Kuo G., Glazer E., Eckart M., Lee C., Chien D., Kuo C., Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993 Jun;74(Pt 6):1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- Simmonds P., Rose K. A., Graham S., Chan S. W., McOmish F., Dow B. C., Follett E. A., Yap P. L., Marsden H. Mapping of serotype-specific, immunodominant epitopes in the NS-4 region of hepatitis C virus (HCV): use of type-specific peptides to serologically differentiate infections with HCV types 1, 2, and 3. J Clin Microbiol. 1993 Jun;31(6):1493–1503. doi: 10.1128/jcm.31.6.1493-1503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sällberg M., Rudén U., Magnius L. O., Norrby E., Wahren B. Rapid "tea-bag" peptide synthesis using 9-fluorenylmethoxycarbonyl (Fmoc) protected amino acids applied for antigenic mapping of viral proteins. Immunol Lett. 1991 Sep;30(1):59–68. doi: 10.1016/0165-2478(91)90090-w. [DOI] [PubMed] [Google Scholar]
- Sällberg M., Rudén U., Wahren B., Magnius L. O. Antigenic regions within the hepatitis C virus envelope 1 and non-structural proteins: identification of an IgG3-restricted recognition site with the envelope 1 protein. Clin Exp Immunol. 1993 Mar;91(3):489–494. doi: 10.1111/j.1365-2249.1993.tb05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sällberg M., Rudén U., Wahren B., Magnius L. O. Immune response to a single peptide containing an immunodominant region of hepatitis C virus core protein: the isotypes and the recognition site. Immunol Lett. 1992 Jun;33(1):27–33. doi: 10.1016/0165-2478(92)90089-7. [DOI] [PubMed] [Google Scholar]
- Sällberg M., Rudén U., Wahren B., Magnius L. O. Immunodominant regions within the hepatitis C virus core and putative matrix proteins. J Clin Microbiol. 1992 Aug;30(8):1989–1994. doi: 10.1128/jcm.30.8.1989-1994.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamizawa A., Mori C., Fuke I., Manabe S., Murakami S., Fujita J., Onishi E., Andoh T., Yoshida I., Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991 Mar;65(3):1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Kato N., Nakagawa M., Ootsuyama Y., Cho M. J., Nakazawa T., Hijikata M., Ishimura Y., Shimotohno K. Molecular cloning of hepatitis C virus genome from a single Japanese carrier: sequence variation within the same individual and among infected individuals. Virus Res. 1992 Apr;23(1-2):39–53. doi: 10.1016/0168-1702(92)90066-i. [DOI] [PubMed] [Google Scholar]
- Tomei L., Failla C., Santolini E., De Francesco R., La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993 Jul;67(7):4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner A. J., Geysen H. M., Christopherson C., Hall J. E., Mason T. J., Saracco G., Bonino F., Crawford K., Marion C. D., Crawford K. A. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]



