Abstract
N6-(2-Deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine (MeFapy-dGuo) has been identified as a stable DNA adduct that arises from the reaction of DNA with a variety of methylating agents. Since this lesion persists in DNA and may contribute to the overall mutagenesis from electrophilic methylating agents, the MeFapy-dGuo lesion was incorporated into oligonucleotides, and its replication bypass was examined in vitro with a panel of eukaryotic high fidelity (hPols α, β, and δ/PCNA) and translesion (hPols η, κ, ι, Rev1, ν, and yPol ζ) polymerases to address its miscoding potential. The MeFapy-dGuo was found to be a strong block to the high fidelity polymerases at either the insertion or the extension step. Efficient translesion synthesis was observed for hPols η and κ, and the combined activities of hRev1 and yPol ζ. The nucleotide sequences of the extension products were determined by mass spectrometry. The error-free extension product was the most abundant product observed for each polymerase. Misreplication products, which included misinsertion of Thy, Gua, and Ade opposite the MeFapy-dGuo lesion, as well as an interesting one-nucleotide deletion product, were observed when hPols η and κ were employed; these events accounted for 8–29% of the total extension products observed. The distribution and abundance of the misreplication products were dependent on the polymerases and local sequence context of the lesion. Collectively, these data suggest that although MeFapy-dGuo adducts represent a relatively minor proportion of the total alkylated lesions, their miscoding potentials could significantly contribute to genomic instability.
INTRODUCTION
Electrophilic methylating agents have been widely used to study the mechanism of DNA damage, mutagenesis, and carcinogenesis of alkylating agents. Such laboratory reagents include methyl methanesulfonate (MMS), N-methyl-N-nitrosourea (NMU), N-methyl-N-nitroso-N-nitroguanidine (MNNG), as well as others.1 Potential endogenous, environmental, or occupational sources of DNA methylating agents include S-adenosylmethionine from the nonenzymatic methylation of DNA, the tobacco carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK), the food-associated nitrosamine and industrial byproduct N,N-dimethylnitrosamine (DMN), and the widely used agricultural fumigant methyl bromide.2 Methylation of DNA is also proposed to be the mechanism of action of the chemotherapeutic agents temozolomide, dacarbazine, and procarbazine.3,4
The reaction of DNA with electrophilic methylating agents gives rise to at least 12 different DNA adducts involving alkylation of the O- and N-atoms of the bases, in addition to the phosphate O-atoms.1 The distribution of these adducts is largely dependent on the nature of the methylating agent with so-called “soft” electrophilic species reacting preferentially at N7-dGuo (Figure 1), while “hard” electrophiles afford larger proportions of O6-Me-dGuo and O4-Me-dThd adducts.1,5–7 The minor O6-Me-dGuo and O4-Me-dThd were found to be much more mutagenic and cytotoxic than other adducts examined.8,9 Consequently, significant attention has been devoted to studying the occurrence, repair, mutagenicity, and carcinogenicity of O6-Me-dGuo since it is more abundant than O4-Me-dThd.10
Figure 1.
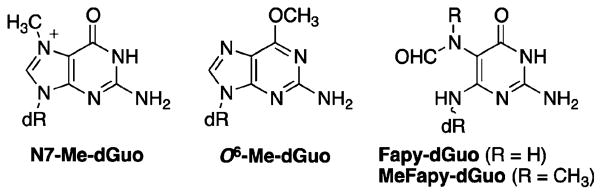
Structures of N7-Me-dGuo, O6-Me-dGuo, Fapy-dGuo, and MeFapy-dGuo.
The cationic N7-Me-dGuo adduct can undergo a secondary reaction with a hydroxide ion to afford the ring-opened MeFapy-dGuo lesion (Figure 1),11,12 which is significantly less studied than O6-Me-dGuo. A related lesion in which the N5-position is unsubstituted (Fapy-dGuo) is a major product of oxidative DNA damage.13–15 The MeFapy-dGuo lesion has been identified as the persistent lesion in rats treated with DMN, MNU, or 1,2-dimethylhydrazine, suggesting that MeFapy-dGuo may be important to the long-term carcinogenicity of methylating agents;16,17 however, an independent study failed to detect the MeFapy-dGuo lesion in rats treated with DMN.18 Additionally, N7-Me-dGuo has been reported to be stable toward ring-opening in calf thymus DNA.19 The deformylated MeFapy-Gua base, 2,6-diamino-4-hydroxy-5-methylaminopyrimidine, a proposed metabolite of MeFapy-dGuo, has been observed in the urine of healthy humans.20 Fapy-dGuo has been observed in human brain, lung, stomach, colon, breast, and ovarian cancer tissues.14,21 Fapy-dGuo and MeFapy-dGuo are substrates for the fpg/nei family of DNA glycosylases.14 Various glycosylases excise Fapy-dGuo, including E. coli formamidopyrimidine glycosylase (FPG), hNEIL1, yeast, mouse, and human oxoguanine glycosylase (OGG1), as well as others.14,22 MeFapy-dGuo has been shown to be a substrate for hOGG1,23,24 yOGG1,25 and hNEIL1;26 excision with mouse and human NTH1 was described as “fair to good”.26,27 Alternatively, MeFapy-dGuo is a poor substrate for hNEIL2 and mNEIL3, and E. coli Endo III and Endo VIII.26,28
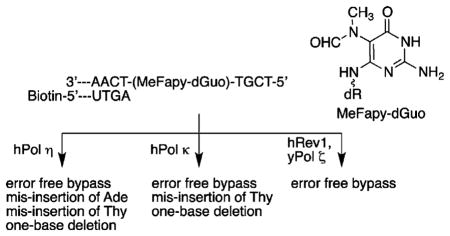
We have developed chemistry for the site-specific incorporation of the MeFapy-dGuo lesion into oligonucleotides and examined its replication by Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) and exonuclease-deficient Klenow fragment of Escherichia coli DNA polymerase I (Kf−).29,30 In vitro replication studies suggested that the MeFapy-dGuo has miscoding and DNA polymerase blocking properties. We now report the in vitro replication of two oligonucleotides that contain the MeFapy-dGuo lesion with human DNA polymerases (hPol) η, κ, ι, Rev1, ν, α, β, δ/PCNA, and yeast DNA polymerase (yPol) ζ. Of these polymerases, hPols η and κ and the combination of hRev1 and yPol ζ could efficiently bypass and extend the MeFapy-dGuo containing templates. The full-length extension products were sequenced by LC-ESI-MS-MS. The major extension products resulted from error-free bypass and extension; however, ~10–30% of the extension products identified resulted from error-prone translesion synthesis (TLS).
EXPERIMENTAL PROCEDURES
Materials
hPols η, κ, ι, Rev1, β, and yPol ζ were purchased from Enzymax (Lexington, KY). hPol α was purchased from CHIMERx (Milwaukee, WI). Recombinant hPols δ and ν were prepared as previously reported.31,32 Uracil DNA glycosylase (UDG) was obtained from Sigma Chemical Co. (St. Louis, MO). Piperidine was purchased from Aldrich and used as received. The dNTP solutions (100 mM) were purchased from New England Biolabs (Ipswich, MA), and streptavidin-coated beads (Streptavidin Sepharose High Performance) were purchased from GE Healthcare (Piscataway, NJ). Unmodified oligonucleotides and primers were purchased from Midland Certified Reagents (Midland, TX).
Oligonucleotide Synthesis, Labeling, and Annealing
The synthesis of site-specifically modified oligonucleotides containing the MeFapy-dGuo lesion were previously reported.29 Oligonucleotides were >98% pure as judged by capillary gel electrophoresis. The oligonucleotides were labeled and annealed as previously described.30
Steady-State Kinetics
These assays were performed according to a published procedure29 with the following modifications. All reactions (10 μL final volume) were run at nine dNTP concentrations and quenched with a solution of (70 μL) of EDTA (20 mM) in formamide (5:95, v/v) containing xylene cyanol and bromophenol blue dyes and heated at 95 °C for 10 min. Aliquots (6 μL) were separated by electrophoresis on denaturing gels. Each analysis was performed in duplicate (Table 1).
Table 1.
Steady-State Kinetic Parameters for Single-Nucleotide Incorporation Opposite the MeFapy-dGuo Adduct in a Local 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a) Sequence, as Well as Unmodified Control Templates (1b and 2b) by Human DNA Polymerases η, κ, and Rev1
| polymerase | template | dNTP | Km (μM) | kcat (s−1·10−3) | kcat/Km (μM −1·s−1·10−3) | fa |
|---|---|---|---|---|---|---|
| hPol η | MeFapy-Gua (1a) | C | 0.8 ± 0.01 | 26 ± 1 | 33 | 1 |
| T | 23 ± 3 | 17 ± 2 | 0.7 | 0.02 | ||
| G | 12 ± 1 | 10 ± 1 | 0.8 | 0.03 | ||
| A | 50 ± 8 | 14 ± 1 | 0.3 | 0.01 | ||
| Gua (1b) | C | 0.2 ± 0.05 | 40 ± 3 | 200 | 1 | |
| T | 40 ± 10 | 20 ± 3 | 0.5 | 0.003 | ||
| G | 75 ± 40 | 13 ± 1 | 0.2 | 0.001 | ||
| A | 80 ± 30 | 20 ± 2 | 0.3 | 0.002 | ||
| hPol κ | MeFapy-Gua (1a) | C | 4 ± 0.4 | 49 ± 2 | 12 | 1 |
| T | 45 ± 9 | 27 ± 5 | 0.6 | 0.05 | ||
| G | 135 ± 40 | 27 ± 8 | 0.2 | 0.02 | ||
| A | 138 ± 21 | 35 ± 9 | 0.3 | 0.03 | ||
| Gua (1b) | C | 0.08 ± 0.01 | 18 ± 2 | 225 | 1 | |
| T | 50 ± 12 | 32 ± 4 | 0.6 | 0.003 | ||
| G | 120 ± 18 | 22 ± 8 | 0.2 | 0.001 | ||
| A | 140 ± 30 | 25 ± 6 | 0.2 | 0.001 | ||
| hRev1 | MeFapy-Gua (1a) | C | 1.98 ± 0.3 | 5.2 ± 0.1 | 2.6 | |
| Gua (1b) | C | 0.85 ± 0.1 | 2.9 ± 0.1 | 3.4 | ||
| hPol η | MeFapy-Gua (2a) | C | 1.5 ± 0.5 | 22 ± 4 | 15 | 1 |
| T | 30 ± 12 | 10 ± 2 | 0.3 | 0.02 | ||
| G | 50 ± 21 | 15 ± 3 | 0.3 | 0.02 | ||
| A | 80 ± 27 | 13 ± 4 | 0.2 | 0.01 | ||
| Gua (2b) | C | 0.3 ± 0.01 | 38 ± 5 | 127 | 1 | |
| T | 60 ± 5 | 25 ± 3 | 0.4 | 0.003 | ||
| G | 45 ± 8 | 20 ± 4 | 0.4 | 0.003 | ||
| A | 57 ± 6 | 15 ± 2 | 0.3 | 0.002 | ||
| hPol κ | MeFapy-Gua (2a) | C | 2 ± 0.6 | 26 ± 2 | 13 | 1 |
| T | 65 ± 15 | 32 ± 8 | 0.5 | 0.04 | ||
| G | 80 ± 10 | 35 ± 4 | 0.4 | 0.03 | ||
| A | 143 ± 20 | 30 ± 6 | 0.2 | 0.02 | ||
| Gua (2b) | C | 0.1 ± 0.06 | 16 ± 1 | 160 | 1 | |
| T | 100 ± 18 | 35 ± 8 | 0.4 | 0.003 | ||
| G | 140 ± 22 | 45 ± 10 | 0.3 | 0.002 | ||
| A | 150 ± 20 | 22 ± 6 | 0.1 | 0.001 | ||
| hRev1 | MeFapy-Gua (2a) | C | 2.3 ± 0.3 | 3.0 ± 0.1 | 1.3 | |
| Gua (2b) | C | 0.56 ± 0.1 | 1.5 ± 0.08 | 2.7 |
f = misincorporation frequency = (Kcat/Km)incorrect dNTP/(Kcat/Km)correct dNTP (dCTP).
Single-Nucleotide Incorporation Assays
These assays were performed as previously described30 with the following modifications. The polymerase extension reactions were initiated by the addition of dNTP with final concentrations of 25, 50, and 100 μM to preincubated polymerase/DNA mixtures giving a final reaction volume of 10 μL. The final concentrations for the DNA polymerases were 2 nM when hPols η, κ, and ι were used individually or in combination. The final concentrations of the components for the bypass reactions were 100 nM DNA duplex, 50 mM Tris-HCl (pH 7.8), 1 mM dithiothreitol of (DTT), 50 μg/mL bovine serum albumin (BSA), 50 mM NaCl, and 5 mM MgCl2. The DNA polymerase reactions were run at 37 °C for 30 min. Control reactions with unmodified templates were carried out at half the polymerase concentration but otherwise under identical conditions. The polymerase reactions were quenched and analyzed as described above (Figure S1 of the Supporting Information).30
Full-Length Polymerase Extension Assays
These assays were performed as previously described30 with the following modifications. The extension reactions were initiated by the addition of the dNTP mixture (final concentrations of 50, 150, and 250 μM of each dNTP) to a preincubated solution of polymerase and DNA. The conditions were 100 nM DNA duplex, 50 mM Tris-HCl (pH 7.8), 1 mM DTT, 50 μg/mL BSA, 50 mM NaCl, and 5 mM MgCl2, giving a final reaction volume of 10 μL. The polymerase concentration was 2 nM when used individually or in combination. The polymerase extension reactions were carried out at 37 °C for 30 min. Control reactions with unmodified template were run at half the polymerase concentration but otherwise under identical conditions. The polymerase reactions were quenched and analyzed as described above (Figures 3 and 4, and S2 and S3 of the Supporting Information).30
Figure 3.
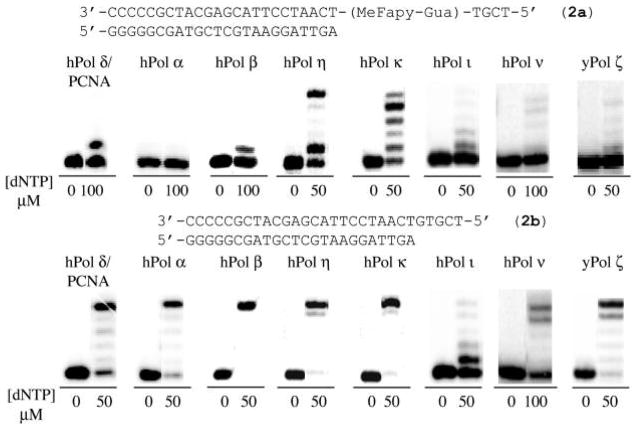
Gel analyses for the bypass and extension of 5′-T-(MeFapy-Gua)-T-3′ (2a, top) and unmodified control (2b, bottom) templates by eukaryotic DNA bypass polymerases. Reactions were carried out with the indicated concentration of each dNTPs for 30 min, except for the pol ν extension, which was run from 15 min. Results for the 5′-TXG-3′ (1) sequence are part of the Supporting Information (Figure S2).
Figure 4.

Replication of the 5′-T-(MeFapy-Gua)-T-3′ (2a, top) and unmodified control (2b, bottom) templates by hRev1, the combined activities of hRev1 and yPol ζ with the −1 primer, and extension by yPol ζ with the 0 primer. The dNTP concentrations are for dCTP alone for the hRev1 insertion and for each dNTP for the full-length extension reactions. Results for the 5′-TXG-3′ (1) sequence are part of the Supporting Information (Figure S3).
LC-ESI-MS-MS Sequencing of the Extension Reactions
These assays were performed as previously described30 with the following modifications. The extension reactions with hPols η and κ, and the combinations of hPols η/ι and κ/ι reactions were performed for 8 h in 50 mM Tris-HCl (pH 7.8), 1.55 nmol of DNA duplex containing the biotinylated primer, 1 mM DTT, 50 μg of BSA mL−1, 50 mM NaCl, and 5 mM MgCl2 (200 μL of total reaction mixture). The polymerase concentrations were 40 nM (Figures S4–S16 and Tables S1–S3 of the Supporting Information).
Construction of the Calibration Curves
The standard calibration curves were constructed as previously described (Figure S17 of the Supporting Information).30
RESULTS
Oligonucleotide Synthesis
We have recently described the synthesis of a phosphoramidite reagent of MeFapy-dGuo and its incorporation into oligonucleotides by solid-phase techniques.29 Polymerase extension reactions were carried out using templates in which the lesion was incorporated in local 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a) sequences (Figure 2).30 The MeFapy-dGuo containing templates were annealed to their complementary −1 primer strand for single-nucleotide insertion and full-length polymerase extension assays.
Figure 2.
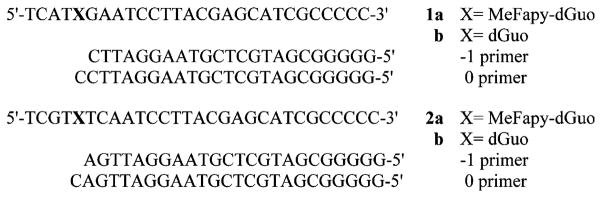
Sequences of the MeFapy-dGuo-containing and unmodified control template and primer oligonucleotides.
Polymerase Bypass of the MeFapy-dGuo Adduct
The in vitro replication bypass and extension of the MeFapy-dGuo adduct was examined using template oligonucleotides 1a (Figures S2 and S3 of the Supporting Information) and 2a (Figures 3 and 4) with hPols η, κ, ι, ν, Rev1, α, β, δ/PCNA, and yPol ζ. Gel analyses suggested that nucleotide insertions were highly error-prone for hPols η, and κ, and insertions of all four dNTPs opposite the MeFapy-dGuo were observed (Figure S1 of the Supporting Information). In contrast, only error-free insertion of dCTP was observed for human pols ι, ν, Rev1, β, and δ/PCNA, as well as yPol ζ (Figures 4, and S3 of the Supporting Information for the Rev1 results). However, the insertion efficiencies for Pols ι, ν, ζ, β, and δ/PCNA appeared low, and extension past the lesion in the presence of all four dNTPs was blocked or very inefficient, and no dNTP insertion was observed for hPol α (Figures 3 and S2 of the Supporting Information). As a result, replication of the MeFapy-dGuo lesion by these polymerases was not examined further.
The steady-state kinetic parameters for nucleotide incorporation opposite the MeFapy-dGuo lesion for hPols η, κ, and Rev1 are tabulated in Table 1. Error-free insertion of dCTP was favored for both sequences for each enzyme. The efficiencies of dCTP insertion opposite the lesion in the 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a) templates by hPol κ was only ~6- and ~9-fold lower than those for the unmodified templates (1b and 2b). Misincorporation frequencies (f) for the other three dNTPs ranged from 1 to 3% for the 5′-T-(MeFapy-Gua)-G-3′ (1a) template and 1–2% that of dCTP for the 5′-T-(MeFapy-Gua)-T-3′ (2a). The reduced catalytic efficiencies for the misinsertions were largely due to higher Km values.
The efficiencies for dCTP insertion opposite the lesion in the 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a) templates by hPol κ were 19 and 12 times lower than opposite the unmodified templates (1b and 2b), respectively (Table 1). The misincorporation frequencies (f) for the other dNTPs ranged from 2 to 4% in the 5′-T-(MeFapy-Gua)-G-3′ (1a) template and 2–5% in the 5′-T-(MeFapy-Gua)-T-3′ (2a) template. Human Rev1 was proficient at inserting dCTP opposite the MeFapy-Gua lesion, with catalytic efficiencies only 1.3- and 2.1-fold lower than the unmodified control templates for the 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a), respectively.
Sequencing Analysis of the Extension Products
To better understand the mechanism of translesion synthesis (TLS), the full-length extension products were sequenced using an LC-ESI-MS-MS method previously reported (Figure S4 of the Supporting Information).30,33–35 Briefly, a dUrd is positioned near the template–primer junction of a 5′-biotinylated primer strand. After extension with a DNA polymerase and all four dNTP’s, the extension products are immobilized on streptavidin-coated beads, which allows them to be separated from the reagents. Treatment with uracil DNA glycosylase (UDG) to generate an abasic site, followed by piperidine, releases a 3′-fragment of the extension products from the solid support, which are subsequently sequenced by LC-ESI-MS-MS. The masses of the extension products are measured in the first mass sector. Upon collision-induced dissociation (CID), DNA will fragment in a regular manner, and analysis of the fragment ion masses provides sequence information.36 This method allows for the rapid evaluation of all products from the extension reaction. A known quantity of a 13-mer oligonucleotide standard was added prior immediately to the mass analysis. Calibration curves were individually developed between the standard and each of the extension products in order to estimate the chemical yield of each product from the polymerization reactions. The error in the yield measurement was estimated to be within 10%. The remainder of the mass balance was assumed to be partially or unextended primers, which were not observed. The results from the sequencing are summarized in Table 2.
Table 2.
Summary of Bypass and Extension Products of the 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a) Templates As Determined by LC-ESI-MS-MS Sequencing and the Chemical (Relative) Yields of Each Product
| extension product | hPol η | hPol κ | hRev1/yPol ζ | comment |
|---|---|---|---|---|
| 3′--AGXTACT-5′ (template 1a) | ||||
| 5′-pTCCATGA-3′ (m/z 1078.8) | 57 (81)% | 58 (92)% | 55 (100)% | error-free |
| 5′-pTCTATGA-3′ (m/z 1086.9) | 8 (11)% | 5 (8)% | misinsertion of Thy | |
| 5′-pTCGATGA-3′ (m/z 1098.8) | 5 (7)% | misinsertion of Gua | ||
| 3′--ACTXTGCT-5′ (template 2a) | ||||
| 5′-pTGACACGA-3′ (m/z 1248.0) | 51 (71)% | 38 (78)% | 48 (100)% | error-free |
| 5′-pTGATACGA-3′ (m/z 1256.0) | 6 (8)% | misinsertion of Thy | ||
| 5′-pTGAAACGA-3′ (m/z 1259.5) | 5 (7)% | 4 (8)% | misinsertion of Ade | |
| 5′-pTGAC–CGA-3′ (m/z 1091.0) | 10 (14)% | 7 (14)% | one-nucleotide deletion |
Replication of the 5′-T-(MeFapy-Gua)-G-3′ (1a) template by hPol η resulted in three extension products with m/z 1078.8, 1086.9, and 1098.8 (Figure S5 of the Supporting Information). Since the isotopic distribution indicated that they were [M-2H]−2 ions, the masses of the extension products were calculated to be 2159.6, 2175.8, and 2199.6 Da, respectively. Given the sequence of the template, the nucleotide composition of the product with a mass of 2159.6 ± 2 Da was calculated to be pC2T2A2G. The CID spectrum of the m/z 1078.8 ion matched well with the predicted CID spectrum for the sequence 5′-pTCCATGA-3′, which is the product from error-free TLS.30 Additionally, the CID spectrum from the extension reaction was nearly identical to that of a commercially obtained, authentic standard, further confirming its sequence. The remaining two products were identified as resulting from the misincorporation of Thd (5′-pTCTATGA-3′, m/z 1086.9) and Gua (5′-pTCGATGA-3′, m/z 1098.8) by a similar analysis (Figures S8–S11 and Tables S1–S2 of the Supporting Information). Four extension products were observed for the 5′-T-(MeFapy-Gua)-T-3′ (2a) template (Figure S12 of the Supporting Information). These were identified as products resulting from error-free replication (5′-pTGACACGA-3′, m/z 1248.0), misincorporation of Thd (5′-pTGATACGA-3′, m/z 1256.0, Figure S15–16 and Table S3 of the Supporting Information), misincorporation of Ade (5′-pTGAAACGA-3′, m/z 1259.5), and an interesting one-nucleotide deletion product (5′-pTGAC–CGA-3′, m/z 1091.0). We have previously characterized the error-free extension products from both templates and the products derived from misinsertion of dATP and one-nucleotide deletion from the 5′-T-(MeFapy-Gua)-T-3′ (2a) template.30
The error-free extension products were the most abundant product observed. We estimate that the yield of error-free TLS catalyzed by hPol η was ~57% and ~51% for replication of the 5′-T-(MeFapy-Gua)-G-3′ (1a) and 5′-T-(MeFapy-Gua)-T-3′ (2a) templates, respectively. Products arising from misincorporation of dTTP and dGTP were observed with frequencies of ~8% and ~5% using the 5′-T-(MeFapy-Gua)-G-3′ (1a) template. The three misreplication products of the 5′-T-(MeFapy-Gua)-T-3′ (2a) template were observed in ~6–10% yield. Specifically, misincorporation of Thd (5′-pTGATACGA-3′) and Ade (5′-pTGAAACGA-3′) were observed in ~6% and ~5% yield, respectively. The most abundant misreplication product from the 5′-T-(MeFapy-Gua)-T-3′ (2a) template was a one-nucleotide deletion product (5′-pTGAC–CGA-3′), which was obtained in ~10% yield.
The error-free extension product was again the most abundant product from replication of both templates with hPol κ. The yield of the error-free product was ~58% for the 5′-T-(MeFapy-Gua)-G-3′ (1a) template and ~38% for the 5′-T-(MeFapy-Gua)-T-3′ (2a) template. Only one misreplication product was characterized from the 5′-T-(MeFapy-Gua)-G-3′ (1a) template (Figure S6 of the Supporting Information), which was the misincorporation of Thd (~5%). Products arising from misincorporation of Ade (~4%) and the same one-nucleotide deletion product (~7%) described above were obtained from the 5′-T-(MeFapy-Gua)-T-3′ (2a) template (Figure S13 of the Supporting Information).
It has been hypothesized that TLS can occur by the sequential action of two DNA polymerases. In such a scenario, one polymerase inserts a dNTP opposite the lesion (i.e., hPol ι or hRev1), then a second polymerase (hPols η, κ, or ζ) could extend from the lesion·N template–primer terminus, where N is the nucleotide inserted by the first polymerase.37–41 We examined the TLS of MeFapy-Gua containing templates 1a and 2a with combinations of hPols ι/κ, ι/η, and hRev1/yPol ζ. We did not observe a significant change in the product distribution when hPol ι was used in combination with either hPols η and κ from that of hPols η or κ alone, although the total yield of extension products was about half (data not shown). It is likely that hPol ι does not contribute significantly to the extension reaction but is effectively competing for the substrate binding and thereby accounting for the diminished yield of extension products. As noted earlier, dNTP insertion opposite the MeFapy-dGuo lesion by yPol ζ was inefficient. However, a significant level of the error-free extension product was observed when the human deoxycytidyl transferase Rev1 was used in combination with yPol ζ. These results suggest that yPol ζ effectively extends from a MeFapy-Gua·Cyt template–primer terminus. This was confirmed by gel analysis of the yPol ζ-catalyzed extension using the 0-primers in which dCyd was paired with the MeFapy-dGuo lesion; full-length extension of the 0-primers was observed to be efficient (Figure 4 and Figures S7 and S14 of the Supporting Information).
DISCUSSION
The replication of undamaged, genomic DNA in eukaryotic cells is performed by Pols α, δ, and ε, which are high-fidelity DNA polymerases. However, chain elongation by these polymerases is usually severely inhibited when encountering structural distortions that result from damaged nucleobases. In contrast, TLS polymerases are low fidelity, exonuclease-deficient enzymes that are often able to replicate through DNA lesions and alternative DNA structures.42–45 The eukaryotic TLS polymerases are classified in three major groups; the Y-family polymerases consist of hPols η, κ, ι, and Rev1, the B-family consists of yPol ζ, and the A-family polymerase consists of hPol ν, which has also been hypothesized to have TLS function.32,46,47 The functional role of these polymerases is assumed to be rescuing stalled replication forks at sites of base damage, such as those from DNA alkylation, as well as a possible role in gap-filling reactions associated with nucleotides excision repair.48,49
In this regard, methylation of DNA is hypothesized to be the cytotoxic mechanism of dacarbazine, temozolomide, and related compounds.4,50,51 These agents are of significant clinical interest; in particular, temozolomide shows promising oral activity against malignant gliomas.3,52 Temozolomide and dacarbazine are prodrugs and are precursors to methyl diazonium ion, which is the reactive DNA modifying agent.53 The predominant adduct from the nonenzymatic methylation of DNA is N7-Me-dGuo regardless of the electrophile.1,2 N7-Me-dGuo has been generally regarded as nonmiscoding since the modification does not alter the Watson–Crick hydrogen-bonding face of the Gua.54 Additionally, N7-modified dGuo lesions are not predicted to pose a significant block to replicative DNA polymerases because the modification resides in the major groove, and the major contact between such polymerases and the DNA helix is from the minor groove.55,56 This is in contrast to the minor groove adduct N3-Me-dAdo, which is a strong block to replication.55,57,58
TLS is a mechanism that contributes to resistance to chemotherapeutic DNA damaging agents. Understanding the function of polymerases that replicate DNA damage derived from chemotherapeutic agents may identify new cellular targets for combination therapies.59,60 In this study, we examined the ability of a panel of eukaryotic DNA polymerases, alone and in combination, to bypass the MeFapy-dGuo lesion in two sequence contexts in vitro. MeFapy-dGuo is not only a possible minor DNA lesion from electrophilic methylating agents but also a possibly more persistent one than the initially more abundant adducts.16,17 The MeFapy-dGuo lesion has previously been reported to block replicative prokaryotic polymerases.61–65 However, replication of the MeFapy-dGuo lesion by the Klenow fragment of E. coli DNA polymerase I resulted in error-free products when bypassed.30,65
All polymerases examined preferentially or exclusively inserted dCTP opposite the MeFapy-dGuo lesion. This result can be readily understood since the Watson–Crick hydrogen-bonding face of MeFapy-dGuo is not altered from dGuo. The insertion of dCTP was inefficient for the eukaryotic highfidelity polymerases including hPol δ/PCNA and hPol β, and further extension was completely inhibited in these cases; hPol α was also completely inhibited by the lesion. This is in contrast to O6-Me-dGuo, which was only a modest block to hPol δ/PCNA.66,67 These observations suggest that the MeFapy-dGuo may be a cytotoxic lesion or that TLS polymerases may be recruited as part of a damage tolerance mechanism of MeFapy-dGuo lesions.
In the current study, in vitro TLS of the MeFapy-dGuo containing templates (1a and 2a) was observed for hPols η and κ, and the combined activities of hRev1 and yPol ζ. Mass spectrometry was used to identify the replication products and estimate their chemical yield (Table 2). The major replication product observed for hPols η and κ resulted from error-free bypass and extension, accounting for >70% of the total replication products observed, and was the only product observed for hRev1/yPol ζ for both templates. The misinsertion of dTTP was the only misreplication product observed when hPol κ was employed. In contrast, two misreplication products were identified when the 5′-T-(MeFapy-Gua)-G-3′ (1a) template was replicated by hPol η, resulting from misinsertion of dGTP and dTTP opposite the MeFapy-dGuo. The total yields of bypass products were comparable for both polymerases (70% for hPol η and 63% for hPol κ based on the starting concentration of the template). Interestingly, the TLS polymerases used in this study were more error-prone than Kf− and Dpo4, which only produced the error-free product in the 5′-T-(MeFapy-Gua)-G-3′ (1a) template.30,65
We previously reported that replication of the MeFapy-dGuo lesion in the 5′-T-(MeFapy-Gua)-T-3′ (2a) template was more error-prone than the 5′-T-(MeFapy-Gua)-G-3′ (1a) template with Kf− or Dpo4.30 The misreplication products that were identified in this study were misinsertion of dATP and an unusual one-nucleotide deletion product that can be rationalized as arising from the correct insertion of dCTP opposite the MeFapy-dGuo followed by deletion of the 5′-dThd. In addition to these two misreplication products, we observed the misinsertion of dTTP when hPol η bypassed the 5′-T-(MeFapy-Gua)-T-3′ (2a) template. The two misinsertion products were observed in about equal yield (~7–8% relative yield), while the one-nucleotide deletion was the major misreplication product comprising ~14% of the products identified. One- and two-nucleotide deletions alter the reading frame of a DNA sequence and have the potential to be serious mutations. The deletion product observed in the present and previous replication studies occurred in the 5′-T-(MeFapy-Gua)-T-3′ (2a) template but not the 5′-T-(MeFapy-Gua)-G-3′ (1a) template. The sequence dependence of the deletion is not obvious from these two examples. The deletion is unusual since the misaligned template and primer would not involve the adduct, but rather the 5′-dThd; the 5′-flanking dThd is common to both sequences. The sequences differ in the 3′-flanking base (Gua vs Thy), and it is possible that the weaker Thy·Ade pair (2 hydrogen bonds) of the 5′-T-(MeFapy-Gua)-T-3′ (2a) template, as compared to the Gua–Cyt pair (3 hydrogen bonds) of the 5′-T-(MeFapy-Gua)-G-3′ (1a) template, allows for misalignment of the template and primer. The full-length extension assay showed a notable difference between the two MeFapy-dGuo containing templates. The insertion and extension steps by hPols η and κ were efficient for the 5′-T-(MeFapy-Gua)-G-3′ (1a, Figure 3S of the Supporting Information), and little or no pauses were observed. However, intermediate products were observed for the replication of the 5′-T-(MeFapy-Gua)-T-3′ (2a, Figure 3) template, suggesting that extension past the MeFapy-dGuo lesion may be rate-limiting. It is possible that the pausing of the hPols η and κ during the extension past the MeFapy-Gua·Cyt template–primer terminus allows for misalignment of the template and primer, leading to the one-nucleotide deletion.
The misinsertion of dATP and the one-nucleotide deletion products were the only misreplication products observed when the 5′-T-(MeFapy-Gua)-T-3′ (2a) template was bypassed by hPol κ; the yields of these two minor products were nearly identical to that of the hPol η reaction. The catalytic efficiency for misinsertion of a dNTP opposite the lesion in the 5′-T-(MeFapy-Gua)-T-3′ (2a) template was between 2 and 3% that of the dCTP insertion efficiency for hPol η and 2–5% for hPol κ; the misinsertion of dATP was the least efficient for both polymerases. The observation of an extension product resulting from initial misinsertion of dATP suggests that hPols η and κ can effectively extend from a MeFapy-Gua·Ade template–primer terminus in this sequence context.
Previous mutagenesis studies of oligonucleotides containing Fapy-dGuo have been reported in E. coli and mammalian COS-7 cells.68,69 The bacterial mutagenicity of Fapy-dGuo was examined in a 5′-T-(Fapy-Gua)-N-3′ local sequence and found to be ≤1.9%; the mutagenicity was relatively insensitive to the 3′-flanking base (mutagenic frequencies ranging from 1.2 to 1.9%) or SOS induction. In COS-7 cells, the mutagenicity was ~8% in a local 5′-T-(Fapy-Gua)-A-3′ sequence and ~30% in a local 5′-T-(Fapy-Gua)-T-3′ sequence. The bacterial and mammalian mutations resulted from misincorporation of dAdo opposite the Fapy-dGuo lesion (G→T transitions). Models for the instructive insertion of dATP opposite Fapy-dGuo have been proposed where the formyl N–H of Fapy-dGuo contributes a nascent hydrogen-bonding interaction with dATP.70 This critical hydrogen bond is missing when the formyl nitrogen is substituted as in MeFapy-dGuo, and therefore, the lower catalytic efficiency for dATP insertion opposite MeFapy-dGuo versus Fapy-dGuo can be rationalized.30
Overall, the panel of polymerases examined in this study showed a range of activities with the MeFapy-dGuo containing templates from efficient insertion and extension to complete inhibition. It is difficult to hypothesize a model that explains all of the activities in the absence of structural information. HPLC analysis of the MeFapy-dGuo containing templates showed four distinct species, which equilibrated over time, suggesting that the MeFapy-dGuo nucleotide exists in at least four interconverting conformations consisting of α/β-anomers and geometric isomers of the formamide group.71,72 The open active site of TLS polymerases may allow them to bind a variety of these conformations. It is possible, however, that only certain conformations can be bypassed, while others are strongly inhibitory. We previously established that the MeFapy-dGuo lesion exists initially as a ~60:40 ratio of β:α anomers.29,71 The total chemical yield of the pols η, κ, and Rev1/ζ bypass products range from approximately 48–72%. One interpretation of these results is that the bypass and extension is largely of the β-anomer, while the α-anomer’s is a reasonably strong block to replication. It is also noteworthy that the nucleotide insertion opposite the MeFapy-dGuo for the high-fidelity hPols δ/PCNA and β, as well as Kf−,30 appears to be better than that of the TLS pols ι, ν, and ζ. The tighter active sites of the high-fidelity polymerases may selectively bind and/or enforce a conformation that resembles that of dGuo, thereby facilitating correct insertion of dCTP. However, structural distortions prevent further extension of the MeFapy-dGuo·dCyt template–primer terminus. Previous structural and biochemical studies suggest that hPol ι utilizes the Hoogsteen face of the template for dNTP selection.73–76 The MeFapy-dGuo lesion can be viewed as having a chemically modified Hoogsteen face, and it is therefore not surprising that this lesion is a poor substrate for hPol ι. Since MeFapy-dGuo can be viewed as a major groove lesion, we were somewhat surprised that it was a poor substrate for hPol ν, which was shown to bypass other major groove lesions.47 MeFapy-dGuo is also a poor substrate for yPol ζ. Although yPol ζ has been shown to insert opposite DNA lesions,77 its primary role is believed to be the extension from distorted template–primer pairs such as those involving DNA lesions or mismatches.78 Consistent with this role, yPol ζ efficiently extends the primer after insertion of dCTP opposite the MeFapy-dGuo.
CONCLUSIONS
The cationic N7-Me-dGuo adduct, the major DNA lesion from nonenzymatic methylation of DNA, can undergo a secondary reaction to the ring-opened MeFapy-dGuo lesion. MeFapy-dGuo is likely to be a minor product from the reaction of DNA with genotoxic and cytotoxic methylating agents. We demonstrated that the MeFapy-dGuo lesion is a strong block to replicative eukaryotic DNA polymerases (hPol δ/PCNA, α, and β) in vitro, suggesting that MeFapy-dGuo has cytotoxic potential. These observations are in contrast to published reports showing that O6-Me-dGuo is only a modest block to hPol δ/PCNA.66,67 In vitro replication assays show that the MeFapy-dGuo lesion can be bypassed by hPols η and κ and the sequential activity of hRev1 and yPol ζ. Error-free bypass and extension was the major replication product observed for all three polymerase systems in both sequences examined. This is in accord with steady-state kinetic data showing that the insertion of dCTP opposite the MeFapy-dGuo lesion is highly favored compared to the other dNTPs. While TLS by Pols η, κ, and Rev1/Pol ζ are potentially important damage tolerance mechanisms, misreplication products were observed with hPols η and κ. These included the misinsertion of Thy, Gua, and Ade, as well as an unusual one-nucleotide deletion product. G→A transitions are commonly observed in cells and animals treated with methylating agents and are ascribed to the misinsertion of Thy opposite O6-Me-Gua.8,9,66,67 Our results suggest that a small percentage of G→A transitions may come from MeFapy-dGuo.
Supplementary Material
Acknowledgments
Funding
NIH Grants P01 ES05355 and P01 CA160032 (to C.J.R. and R.S.L.), R01 ES10375 (to F.P.G.), and R01 CA097175 (to R.D.W. and K.T.), center grant P30 ES00267 (to C.J.R. and F.P.G.), and a pilot project from center grant P30 ES007784 (to R.D.W. and K.T.), the M. D. Anderson Research Trust (to R.D.W. and K.T.), and NRF Grant 2010-0006538 from MEST Korea (to J.-Y.C.) supported this work.
ABBREVIATIONS
- O6-Me-dGuo
O6-methyl-2′-deoxyguanosine
- O4-Me-dThd
O4-methyl-thymidine
- MeFapy-dGuo
N6-(2-deoxy-D-erythro-pen-tofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylforma-midopyrimidine
- Dpo4
Sulfolobus solfataricus P2 DNA polymerase (Dpo4)
- Kf−
exonuclease-deficient Klenow fragment of Escherichia coli DNA polymerase I
- hpol
human DNA polymerase
- yPol
yeast DNA polymerase
- dNTPs
deoxynucleotide triphosphates
- CID
collision-induced dissociation
- TLS
translesion synthesis
Footnotes
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Additional tables and figures as described in the text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Beranek DT. Distribution of methyl and ethyl adducts following alkylation with monofunctional alkylating agents. Mutat Res. 1990;231:11–30. doi: 10.1016/0027-5107(90)90173-2. [DOI] [PubMed] [Google Scholar]
- 2.Kyrtopoulos SA. DNA adducts in humans after exposure to methylating agents. Mutat Res. 1998;405:135–143. doi: 10.1016/s0027-5107(98)00130-4. [DOI] [PubMed] [Google Scholar]
- 3.Friedman HS, Kerby T, Calvert H. Temozolomide and treatment of malignant glioma. Clin Cancer Res. 2000;6:2585–2597. [PubMed] [Google Scholar]
- 4.Newell D, Gescher A, Harland S, Ross D, Rutty C. N-methyl antitumour agents. A distinct class of anticancer drugs? Cancer Chemother Pharmacol. 1987;19:91–102. doi: 10.1007/BF00254559. [DOI] [PubMed] [Google Scholar]
- 5.Lown J, Chauhan S, Koganty R, Sapse A. Alkyldinitrogen species implicated in the carcinogenic, mutagenic, and anticancer activities of N-nitroso compounds: Characterization by 15N NMR of 15N-enriched compounds and analysis of DNA base situselectivity by ab initio calculations. J Am Chem Soc. 1984;106:6401–6408. [Google Scholar]
- 6.Loechler EL. A violation of the Swain-Scott principle, and not SN1 versus SN2 reaction mechanisms, explains why carcinogenic alkylating agents can form different proportions of adducts at oxygen versus nitrogen in DNA. Chem Res Toxicol. 1994;7:277–280. doi: 10.1021/tx00039a001. [DOI] [PubMed] [Google Scholar]
- 7.Wyatt MD, Pittman DL. Methylating agents and DNA repair responses: Methylated bases and sources of strand breaks. Chem Res Toxicol. 2006;19:1580–1594. doi: 10.1021/tx060164e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Newbold RF, Warren W, Medcalf AS, Amos J. Mutagenicity of carcinogenic methylating agents is associated with a specific DNA modification. Nature. 1980;283:596–599. doi: 10.1038/283596a0. [DOI] [PubMed] [Google Scholar]
- 9.Mitra G, Pauly GT, Kumar R, Pei GK, Hughes SH, Moschel RC, Barbacid M. Molecular analysis of O6-substituted guanine-induced mutagenesis of ras oncogenes. Proc Natl Acad Sci USA. 1989;86:8650–8654. doi: 10.1073/pnas.86.22.8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu AK, Essigmann JM. Site-specifically alkylated oligodeoxynucleotides: Probes for mutagenesis, DNA repair and the structural effects of DNA damage. Mutat Res. 1990;233:189–201. doi: 10.1016/0027-5107(90)90162-w. [DOI] [PubMed] [Google Scholar]
- 11.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 12.Tudek B. Imidazole ring-opened DNA purines and their biological significance. J Biochem Mol Biol. 2003;36:12–19. doi: 10.5483/bmbrep.2003.36.1.012. [DOI] [PubMed] [Google Scholar]
- 13.Greenberg MM. In vitro and in vivo effects of oxidative damage to deoxyguanosine. Biochem Soc Trans. 2004;32:46–50. doi: 10.1042/bst0320046. [DOI] [PubMed] [Google Scholar]
- 14.Dizdaroglu M, Kirkal iG, Jaruga P. Formamidopyrimidines in DNA: Mechanisms of formation, repair, and biological effects. Free Radical Biol Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Greenberg MM. The formamidopyrimidines: Purine lesions formed in competition with 8-oxopurines from oxidative stress. Acc Chem Res. 2012;45:588–597. doi: 10.1021/ar2002182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beranek DT, Weis CC, Evans FE, Chetsanga CJ, Kadlubar FF. Identification of N5-methyl-N5-formyl-2,5,6-triamino-4-hydroxypyrimidine as a major adduct in rat liver DNA after treatment with the carcinogens, N,N-dimethylnitrosamine or 1,2-dimethylhydrazine. Biochem Biophys Res Commun. 1983;110:625–631. doi: 10.1016/0006-291x(83)91195-6. [DOI] [PubMed] [Google Scholar]
- 17.Kadlubar FF, Beranek DT, Weis CC, Evans FE, Cox R, Irving CC. Characterization of the purine ring-opened 7-methylguanine and its persistence in rat bladder epithelial DNA after treatment with the carcinogen N-methylnitrosourea. Carcinogenesis. 1984;5:587–592. doi: 10.1093/carcin/5.5.587. [DOI] [PubMed] [Google Scholar]
- 18.Den Engelse L, Menkveld GJ, De Brij RJ, Tates AD. Formation and stability of alkylated pyrimidines and purines (including imidazole ring-opened 7-alkylguanine) and alkylphosphotriesters in liver DNA of adult rats treated with ethylnitrosourea or dimethylnitrosamine. Carcinogenesis. 1986;7:393–403. doi: 10.1093/carcin/7.3.393. [DOI] [PubMed] [Google Scholar]
- 19.Hendler S, Furer E, Srinivasan PR. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial deoxyribonucleic acid or ribonucleic acid polymerases. Biochemistry. 1970;9:4141–4153. doi: 10.1021/bi00823a017. [DOI] [PubMed] [Google Scholar]
- 20.Barak R, Vincze A, Bel P, Dutta SP, Chedda GB. Mass spectrometric investigation of the presence of 7-methyl ring-opened guanine derivatives in urine. Chem-Biol Interact. 1993;86:29–40. doi: 10.1016/0009-2797(93)90109-c. [DOI] [PubMed] [Google Scholar]
- 21.Evans MD, Dizdaroglu M, Cooke MS. Oxidative DNA damage and disease: Induction, repair and significance. Mutat Res. 2004;567:1–61. doi: 10.1016/j.mrrev.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 22.Hu J, de Souza-Pinto NC, Haraguchi K, Hogue BA, Jaruga P, Greenberg MM, Dizdaroglu M, Bohr VA. Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. J Biol Chem. 2005;280:40544–40551. doi: 10.1074/jbc.M508772200. [DOI] [PubMed] [Google Scholar]
- 23.Asagoshi K, Yamada T, Terato H, Ohyama Y, Monden Y, Arai T, Nishimura S, Aburatani H, Lindahl T, Ide H. Distinct repair activities of human 7,8-dihydro-8-oxoguanine DNA glycosylase and formamidopyrimidine DNA glycosylase for formamidopyrimidine and 7,8-dihydro-8-oxoguanine. J Biol Chem. 2000;275:4956–4964. doi: 10.1074/jbc.275.7.4956. [DOI] [PubMed] [Google Scholar]
- 24.Dherin C, Radicella JP, Dizdaroglu M, Boiteux S. Excision of oxidatively damaged DNA bases by the human α-hOgg1 protein and the polymorphic α-hOgg1(Ser326Cys) protein which is frequently found in human populations. Nucleic Acids Res. 1999;27:4001–4007. doi: 10.1093/nar/27.20.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karahalil B, Girard PM, Boiteux S, Dizdaroglu M. Substrate specificity of the Ogg1 protein of Saccharomyces cerevisiae: Excision of guanine lesions produced in DNA by ionizing radiation- or hydrogen peroxide/metal ion-generated free radicals. Nucleic Acids Res. 1998;26:1228–1233. doi: 10.1093/nar/26.5.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katafuchi A, Nakano T, Masaoka A, Terato H, Iwai S, Hanaoka F, Ide H. Differential specificity of human and Escherichia coli endonuclease III and VIII homologues for oxidative base lesions. J Biol Chem. 2004;279:14464–14471. doi: 10.1074/jbc.M400393200. [DOI] [PubMed] [Google Scholar]
- 27.Asagoshi K, Yamada T, Okada Y, Terato H, Ohyama Y, Seki S, Ide H. Recognition of formamidopyrimidine by Escherichia coli and mammalian thymine glycol glycosylases. Distinctive paired base effects and biological and mechanistic implications. J Biol Chem. 2000;275:24781–24786. doi: 10.1074/jbc.M000576200. [DOI] [PubMed] [Google Scholar]
- 28.Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christov PP, Brown KL, Kozekov ID, Stone MP, Harris TM, Rizzo CJ. Site-specific synthesis and characterization of oligonucleotides containing an N6-(2-deoxy-D-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem Res Toxicol. 2008;21:2324–2333. doi: 10.1021/tx800352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Christov PP, Angel KC, Guengerich FP, Rizzo CJ. Replication past the N5-methyl-formamidopyrimidine lesion of deoxyguanosine by DNA polymerases and an improved procedure for sequence analysis of in vitro bypass products by mass spectrometry. Chem Res Toxicol. 2009;22:1086–1095. doi: 10.1021/tx900047c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi JY, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J Mol Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 32.Takata K-i, Shimizu T, Iwai S, Wood RD. Human DNA polymerase N (POLN) is a low fidelity enzyme capable of error-free bypass of 5S-thymine glycol. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 33.Zang H, Goodenough AK, Choi JY, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J Biol Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 34.Christov PP, Petrova KV, Shanmugam G, Kozekov ID, Kozekova A, Guengerich FP, Stone MP, Rizzo CJ. Comparison of the in vitro replication of the 7-(2-oxoheptyl)-1,N2-etheno-2′-deoxyguanosine and 1,N2-etheno-2′-deoxyguanosine lesions by Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4) Chem Res Toxicol. 2010;23:1330–1341. doi: 10.1021/tx100082e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chowdhury G, Guengerich FP. Liquid chromatography-mass spectrometry analysis of DNA polymerase reaction products. Curr Prot Nucleic Acid Chem. 2011;47:7.16.11–17.16.11. doi: 10.1002/0471142700.nc0716s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McLuckey SA, Van Berker GJ, Glish GL. Tandem mass spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectrom. 1992:60–70. doi: 10.1016/1044-0305(92)85019-G. [DOI] [PubMed] [Google Scholar]
- 37.Yasui M, Laxmi Y, Ananthoju S, Suzuki N, Kim S, Shibutani S. Translesion synthesis past equine estrogen-derived 2′-deoxyadenosine DNA adducts by human DNA polymerases η and κ. Biochemistry. 2006;45:6187–6194. doi: 10.1021/bi0525324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 39.Washington MT, Minko IG, Johnson RE, Haracska L, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove N2-guanine adduct by the sequential action of yeast Rev1 and DNA polymerase ζ. Mol Cell Biol. 2004;24:6900–6906. doi: 10.1128/MCB.24.16.6900-6906.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, Prakash S, Prakash L. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and ζ. Mol Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA ι and κ. Mol Cell Biol. 2006;26:381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:296–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Loeb LA, Monnat RJ. DNA polymerases and human disease. Nat Rev Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 44.Bétous R, Rey L, Wang G, Pillaire MJ, Puget N, Selves J, Biard DSF, Shinya K, Vasquez KM, Cazaux C, Hoffmann JS. Role of TLS DNA polymerases eta and kappa in processing naturally occurring structured DNA in human cells. Mol Carcinog. 2009;48:369–378. doi: 10.1002/mc.20509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rey L, Sidorova JM, Puget N, Boudsocq F, Biard DSF, Monnat RJ, Cazaux C, Hoffmann JS. Human DNA polymerase η is required for common fragile site stability during unperturbed DNA replication. Mol Cell Biol. 2009;29:3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Arana ME, Takata K-i, Garcia-Diaz M, Wood RD, Kunkel TA. A unique error signature for human DNA polymerase ν. DNA Repair. 2007;6:213–223. doi: 10.1016/j.dnarep.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamanaka K, Minko IG, Takata K-i, Kolbanovskiy A, Kozekov ID, Wood RD, Rizzo CJ, Lloyd RS. Novel enzymatic function of DNA polymerase ν in translesion DNA synthesis past major groove DNA-peptide and DNA-DNA cross-links. Chem Res Toxicol. 2010;23:689–695. doi: 10.1021/tx900449u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ogi T, Lehmann AR. The Y-family DNA polymerase κ (pol κ) functions in mammalian nucleotide-excision repair. Nat Cell Biol. 2006;8:640–642. doi: 10.1038/ncb1417. [DOI] [PubMed] [Google Scholar]
- 49.Ogi T, Limsirichaikul S, Overmeer RM, Volker M, Takenaka K, Cloney R, Nakazawa Y, Niimi A, Miki Y, Jaspers NG, Mullenders LH, Yamashita S, Fousteri MI, Lehmann AR. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol Cell. 2010;37:714–727. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 50.Tentori L, Graziani G. Pharmacological strategies to increase the antitumor activity of methylating agents. Curr Med Chem. 2002;9:1285–1301. doi: 10.2174/0929867023369916. [DOI] [PubMed] [Google Scholar]
- 51.Kyrtopoulos SA, Anderson LM, Chhabra SK, Souliotis VL, Pletsa V, Valavanis C, Georgiadis P. DNA adducts and the mechanism of carcinogenesis and cytotoxicity of methylating agents of environmental and clinical significance. Cancer Detect Prev. 1997;21:391–405. [PubMed] [Google Scholar]
- 52.Quinn JA, Jiang SX, Reardon DA, Desjardins A, Vredenburgh JJ, Rich JN, Gururangan S, Friedman AH, Bigner DD, Sampson JH, McLendon RE, Herndon JE, Walker A, Friedman HS. Phase II trial of Temozolomide plus O6-benzylguanine in adults with recurrent, Temozolomide-resistant malignant glioma. J Clin Oncol. 2009;27:1262–1267. doi: 10.1200/JCO.2008.18.8417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug Temozolomide and its interaction with DNA. Biochemistry. 1994;33:9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 54.Boysen G, Pachkowski BF, Nakamura J, Swenberg JA. The formation and biological significance of N7-guanine adducts. Mutat Res. 2009;678:76–94. doi: 10.1016/j.mrgentox.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larson K, Sahm J, Shenkar R, Strauss B. Methylation-induced blocks to in vitro DNA replication. Mutat Res. 1985;150:77–84. doi: 10.1016/0027-5107(85)90103-4. [DOI] [PubMed] [Google Scholar]
- 56.Hogg M, Wallace SS, Doublié S. Bumps in the road: How replicative DNA polymerases see DNA damage. Curr Opin Struct Biol. 2005;15:86–93. doi: 10.1016/j.sbi.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 57.Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson LD. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 58.Fronza G, Gold B. The biological effects of N3-methyladenine. J Cell Biochem. 2004;91:250–257. doi: 10.1002/jcb.10698. [DOI] [PubMed] [Google Scholar]
- 59.Dorjsuren D, Wilson DM, Beard WA, McDonald JP, Austin CP, Woodgate R, Wilson SH, Simeonov A. A real-time fluorescence method for enzymatic characterization of specialized human DNA polymerases. Nucleic Acids Res. 2009;37:e128. doi: 10.1093/nar/gkp641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berdis AJ. DNA polymerases as therapeutic targets. Biochemistry. 2008;47:8253–8260. doi: 10.1021/bi801179f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boiteux S, Laval J. Imidazole open ring 7-methylguanine: An inhibitor of DNA synthesis. Biochem Biophys Res Commun. 1983;110:552–558. doi: 10.1016/0006-291x(83)91185-3. [DOI] [PubMed] [Google Scholar]
- 62.O’Connor TR, Boiteux S, Laval J. Ring-opened 7-methylguanine residues in DNA are a block to in vitro DNA synthesis. Nucleic Acids Res. 1988;16:5879–5894. doi: 10.1093/nar/16.13.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tudek B, Boiteux S, Laval J. Biological properties of imidazole ring-opened N7-methylguanine in M13mp18 phage DNA. Nucleic Acids Res. 1992;20:3079–3084. doi: 10.1093/nar/20.12.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tudek B, Graziewicz M, Kazanova O, Zastawny TH, Obtulowicz T, Laval J. Mutagenic specificity of imidazole ring-opened 7-methylpurines in M13mp18 phage DNA. Acta Biochim Pol. 1999;46:785–799. [PubMed] [Google Scholar]
- 65.Asagoshi K, Terato H, Ohyama Y, Ide H. Effects of a guanine-derived formamidopyrimidine lesion on DNA replication: Translesion DNA synthesis, nucleotide insertion, and extension kinetics. J Biol Chem. 2002;277:14589–14597. doi: 10.1074/jbc.M200316200. [DOI] [PubMed] [Google Scholar]
- 66.Choi JY, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J Biol Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 67.Haracska L, Prakash S, Prakash L. Replication past O6-methylguanine by yeast and human DNA polymerase η. Mol Cell Biol. 2000;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Genetic effects of oxidative DNA damages: Comparative mutagenesis of the imidazole ring-opened formamidopyrimidines (Fapy lesions) and 8-oxo-purines in simian kidney cells. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Patro JN, Wiederholt CJ, Jiang YL, Delaney JC, Essigmann JM, Greenberg MM. Studies on the replication of the ring opened formamidopyrimidine, Fapy•dG in Escherichia coli. Biochemistry. 2007;46:10202–10212. doi: 10.1021/bi700628c. [DOI] [PubMed] [Google Scholar]
- 70.Wiederholt CJ, Greenberg MM. Fapy•dG instructs Klenow exo− to misincorporate deoxyadenosine. J Am Chem Soc. 2002;124:7278–7279. doi: 10.1021/ja026522r. [DOI] [PubMed] [Google Scholar]
- 71.Christov PP, Banerjee S, Stone MP, Rizzo CJ. Selective incision of the α-N5-methyl formamidopyrimidine anomer by Escherichia coli Endonuclease IV. J Nucleic Acids. 2010 doi: 10.4061/2010/850234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomasz M, Lipman R, Lee MS, Verdine GL, Nakanishi K. Reaction of acid-activated mitomycin C with calf thymus DNA and model guanines: Elucidation of the base-catalyzed degradation of N7-alkylguanine nucleosides. Biochemistry. 1987;26:2010–2027. doi: 10.1021/bi00381a034. [DOI] [PubMed] [Google Scholar]
- 73.Johnson RE, Prakash L, Prakash S. Biochemical evidence for the requirement of Hoogsteen base pairing for replication by human DNA polymerase ι. Proc Natl Acad Sci USA. 2005;102:10466–10471. doi: 10.1073/pnas.0503859102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Human DNA polymerase ι incorporates dCTP opposite template G via a G•C+ Hoogsteen base pair. Structure. 2005;13:1569–1577. doi: 10.1016/j.str.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 75.Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat Struct Mol Biol. 2006;13:619–625. doi: 10.1038/nsmb1118. [DOI] [PubMed] [Google Scholar]
- 76.Kirouac KN, Ling H. Unique active site promotes error-free replication opposite an 8-oxo-guanine lesion by human DNA polymerase ζ. Proc Natl Acad Sci USA. 2011;108:3210–3215. doi: 10.1073/pnas.1013909108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stone JE, Kumar D, Binz SK, Inase A, Iwai S, Chabes A, Burgers PM, Kunkel TA. Lesion bypass by S. cerevisiae Pol ζ alone. DNA Repair. 2011;10:826–834. doi: 10.1016/j.dnarep.2011.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Waters LS, Minesinger BK, Wiltrout ME, D’Souza S, Woodruff RV, Walker GC. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol Mol Biol Rev. 2009;73:135–154. doi: 10.1128/MMBR.00034-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.