Background: The functions of the deubiquitinating enzymes JosD1 and JosD2, related to ATXN3, are unknown.
Results: JosD1 is activated by ubiquitination, localizes to plasma membrane, and affects membrane dynamics, cell motility, and endocytosis.
Conclusion: JosD1 and JosD2 possess divergent properties, with JosD1 regulating membrane-related functions.
Significance: Our findings provide insight into diverse functions of a disease-linked family of deubiquitinating enzymes.
Keywords: Cell Motility, Deubiquitination, Endocytosis, Ubiquitin, Ubiquitination, Ataxin-3, Deubiquitinating Enzyme, Josephin Domain-containing Protein 1, Membrane Dynamics
Abstract
The functional diversity of deubiquitinating enzymes (DUBs) is not well understood. The MJD family of DUBs consists of four cysteine proteases that share a catalytic “Josephin” domain. The family is named after the DUB ATXN3, which causes the neurodegenerative disease Machado-Joseph disease. The two closely related Josephin domain-containing (JosD) proteins 1 and 2 consist of little more than the Josephin domain. To gain insight into the properties of Josephin domains, we investigated JosD1 and JosD2. JosD1 and JosD2 were found to differ fundamentally in many respects. In vitro, only JosD2 can cleave ubiquitin chains. In contrast, JosD1 cleaves ubiquitin chains only after it is monoubiquitinated, a form of posttranslational-dependent regulation shared with ATXN3. A significant fraction of JosD1 is monoubiquitinated in diverse mouse tissues. In cell-based studies, JosD2 localizes to the cytoplasm whereas JosD1 preferentially localizes to the plasma membrane, particularly when ubiquitinated. The membrane occupancy by JosD1 suggests that it could participate in membrane-dependent events such as cell motility and endocytosis. Indeed, time-lapse imaging revealed that JosD1 enhances membrane dynamics and cell motility. JosD1 also influences endocytosis in cultured cells by increasing the uptake of endocytic markers of macropinocytosis while decreasing those for clathrin- and caveolae-mediated endocytosis. Our results establish that two closely related DUBs differ markedly in activity and function and that JosD1, a membrane-associated DUB whose activity is regulated by ubiquitination, helps regulate membrane dynamics, cell motility, and endocytosis.
Introduction
Deubiquitinating enzymes (DUBs)3 are specialized proteases that cleave the chemical bond between ubiquitin (Ub) and another protein, including Ub-Ub linkages (1). By controlling this critical post-translational modification, DUBs regulate cellular processes and pathways from gene transcription to protein degradation (2). The importance of DUBs to cellular health and homeostasis is underscored by links to malignancies and neurological diseases (3, 4). According to a recent study, most DUBs in the fruit fly Drosophila melanogaster were required developmentally or in adults (5). Nearly 100 potential DUBs are encoded by the human genome, classified into five distinct families based on similarities in the catalytic domain: UCHs, USPs, MJDs, OTUs, and JAMMs (1). Except for JAMM DUBs, which are zinc-dependent metalloproteases, all other DUB families are cysteine proteases. DUB activity can be regulated at several levels including translation, degradation, post-translational modification, substrate-induced conformational change, and functional interactions with other proteins. Although some DUBs have been studied extensively (2), little is known about the activity, expression, and function of most DUBs.
The smallest family of DUBs is the MJD class, which includes four proteins: ataxin-3 (ATXN3), ataxin-3-like (ATXN3L) and Josephin domain-containing proteins 1 and 2 (JosD1 and JosD2) (1, 4). All share a highly conserved “Josephin” catalytic domain, named after the neurodegenerative disease Machado-Joseph disease (MJD), which is caused by a polyglutamine expansion mutation in ATXN3. Because of its link to disease, ATXN3 is the best studied member of the MJD family. ATXN3 contains an N-terminal catalytic Josephin domain, which is globular and conserved among MJD family members, and a flexible C-terminal region that includes three Ub-interacting motifs (UIMs) flanking the polyglutamine domain (6). ATXN3 displays several remarkable properties. For example, its activity is directly regulated by ubiquitination, which enhances AXTN3-mediated cleavage of Ub chains (7, 8). It also partners with specific E3 ubiquitin ligases to regulate the extent of substrate ubiquitination (9, 10). These properties suggest that ATXN3 functions in cellular protein quality control (6).
Approximately 180 amino acids long, the Josephin catalytic domain is the unifying feature of the MJD DUBs. ATXN3 and ATXN3L also share the UIMs and the polyglutamine domain, whereas JosD1 and JosD2 contain essentially only the catalytic Josephin domain (Fig. 1A) (1). The catalytic residues within the Josephin domain are relatively conserved among MJD DUBs (Fig. 1B). Whereas JosD1 and JosD2 represent distinct orthologs in vertebrates, only one JosD1/2-related gene is identified in Caenorhabditis elegans and Drosophila (5, 11).
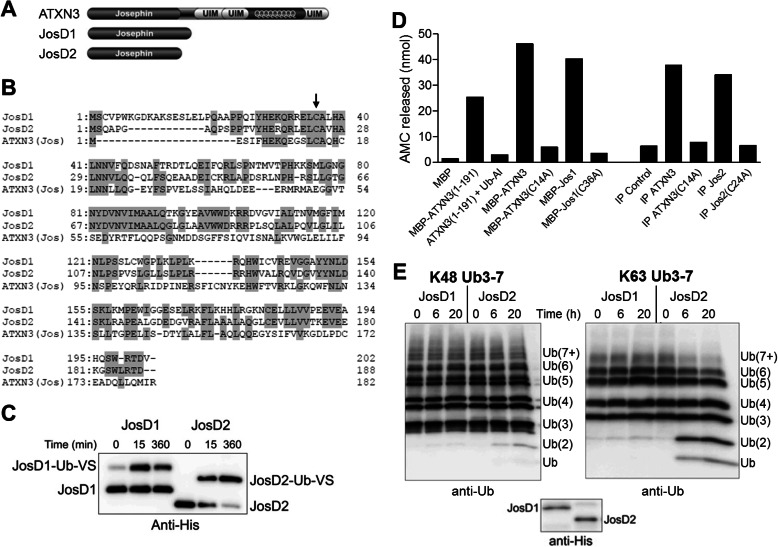
FIGURE 1.
JosD1 and JosD2 differ in DUB activity in vitro. A, domain structure composition of ATXN3, JosD1, and JosD2. In addition to the Josephin domain, ATXN3 has three UIMs flanking a polyglutamine repeat. B, sequence comparison of JosD1, JosD2, and the Josephin domain of ATXN3. Amino acid residues conserved between two or more proteins are highlighted with gray. Arrow indicates the cysteine residue essential for catalytic activity (JosD1, Cys36; JosD2, Cys24; ATXN3, Cys14). C, catalytic activity of JosD1 and JosD2 assessed by reaction with Ub-VS. His-tagged JosD1 and JosD2 were incubated with Ub-VS for the indicated times and detected by immunoblotting with anti-His tag antibody. Ub-VS-modified JosD1 and JosD2 electrophorese at higher molecular mass. D, DUB activity of ATXN3, JosD1, and JosD2 determined by Ub-AMC. DUB activity was measured by fluorescence AMC released from Ub-AMC. Recombinant JosD1 fused to maltose-binding protein (MBP) has DUB activity similar to full-length ATXN3 and the isolated Josephin domain (1–191) of ATXN3. Immunoprecipitated (IP) JosD2 also has DUB activity. DUB activity of ATXN3, JosD1, and JosD2 is eliminated by mutating the catalytic cysteine residue (Cys14 in ATXN3, Cys36 in JosD1, and Cys24 in JosD2). E, divergent DUB activity of JosD1 and JosD2 assessed in vitro by cleavage of polyubiquitin chains. His-tagged JosD1 and JosD2 were incubated with Lys48-linked (left) or Lys63-linked (right) poly-Ub chains (3–7 Ubs in length) for the indicated times. Ub chains were detected by immunoblotting with anti-ubiquitin antibody. Lower immunoblot shows the amount of JosD1 and JosD2 used in the assay, detected with anti-His antibody. Whereas JosD2 cleaves poly-Ub chains, JosD1 shows little to no cleavage.
The functions of JosD1 and JosD2 are unknown. To expand general understanding of DUB activity and to explore the potential roles of the MJD family DUBs, we chose to investigate JosD1 and JosD2. Defining the properties of these two closely related proteins that seemingly comprise only the Josephin domain also may shed light on why the disease protein ATXN3 possesses a C-terminal extension with multiple UIMs and polyglutamine domain. Our results show that, despite their structural similarity, JosD1 and JosD2 differ with respect to base-line catalytic activity, modification by ubiquitin, and subcellular localization. We also find that, similar to ATXN3 (7, 8), the catalytic activity of JosD1 is regulated by ubiquitination, suggesting that ubiquitination-dependent regulation may be a more common regulatory mechanism for DUBs than previously anticipated. Finally, we show that JosD1 tends to localize to the plasma membrane, especially when ubiquitinated, where it influences membrane dynamics, cell motility and endocytosis.
EXPERIMENTAL PROCEDURES
DNA Constructs
For recombinant protein expression, ATXN3, JosD1, and JosD2 constructs were subcloned into pGEX6P1 (for GST-tagged, GE Healthcare) and pET28a (for His-tagged; Millipore). For mammalian cell expression, JosD1 and JosD2 constructs were subcloned into pcDNA3.1/V5-His TOPO (Invitrogen) to fuse with V5 tag, into pEGFP-C1 (Clontech) to fuse with GFP, and into pFLAG-CMV-6c (Sigma) to fuse with FLAG tag. The HA-ubiquitin construct was subcloned into pRcCMV. The QuikChange II XL site-directed mutagenesis kit (Agilent) was used to generate catalytically inactive mutants. All constructs were confirmed by sequencing.
Ub-VS and Ub-AMC Assay
Recombinant proteins were prepared as described previously (12, 13). Ub-VS (Boston Biochem, Cambridge, MA) is a potent inhibitor of cysteine protease DUBs. Recombinant His-tagged JosD1 and JosD2 (2 μm) were incubated with 2 μm Ub-VS at 37 °C for 15 or 360 min, and the reaction was stopped by adding Laemmli sample buffer and boiling. Samples were subjected to SDS-PAGE, followed by immunoblotting with anti-His tag antibody. Because Ub-VS forms a covalent bond resistant to Laemmli buffer, the Ub-VS-modified proteins electrophorese as higher molecular mass proteins.
DUB activity was also assessed by the cleavage of Ub-AMC, a fluorescent probe, as described previously (14). Briefly, recombinant ATXN3 (full-length or 1–191(Josephin domain alone)), JosD1 and JosD2 (100 nm) were incubated with Ub-AMC (500 nm) at 37 °C for 15 min. Fluorescence of AMC released from Ub-AMC by DUB activity was measured using a Biotek Synergy HT fluorometer.
Deubiquitination Reactions
Deubiquitination reactions were conducted as described previously (7). Briefly, Ub chains (300–400 nm; Boston Biochem) were incubated with recombinant JosD1 or JosD2 (50–100 nm) in DUB reaction buffer (50 mm HEPES, 0.5 mm EDTA, 1 mm DTT, 0.1 mg/ml ovalbumin, pH 7.5) at 37 °C for the indicated times or overnight. Reactions were stopped by adding 2% SDS buffer and boiling. Samples were subjected to SDS-PAGE followed by immunoblotting with anti-Ub rabbit polyclonal antibody (Dako).
In Vitro Ubiquitination
In vitro ubiquitination of recombinant JosD1 and JosD2 was conducted as described previously (7). Briefly, GST-JosD1 or GST-JosD2 was incubated with 1 μm CHIP, 8 μm UbcH5c, 0.16 μm E1, 50 μm ubiquitin (Boston Biochem), 4.5 μm MgCl2, and 4.5 μm ATP in kinase reaction buffer (50 mm Tris, 50 mm KCl, 0.2 mm DTT, pH 7.5) for 2 h at 37 °C.
JosD1 Expression in Various Mouse Tissues
Various mouse tissues were lysed in RIPA lysis buffer (20 mm Tris, 150 mm NaCl, 0.1% SDS, 0.5% deoxycholic acid, 1% Nonidet P-40, pH 7.5) supplemented with protease inhibitors (Sigma-Aldrich) using a Dounce homogenizer. Equal amounts of proteins from these tissues were subjected to SDS-PAGE, followed by immunoblotting with anti-JosD1 rabbit polyclonal antibody, generated by Sigma Genosys using recombinant JosD1 peptide.
Cell Culture, Transfections, and Lysate Preparation
HEK-293 and COS-7 cells were cultured and maintained as described previously (15). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instruction. Cells were harvested in SDS (2%) buffer supplemented with 100 mm DTT 2–3 days after transfection. Lysates were boiled for 15 min, brought to room temperature, centrifuged, and loaded onto 10, 12, or 15% SDS-PAGE gels.
To examine subcellular localization of JosD1 and JosD2, proteins of transfected HEK-293 cells were fractionated into cytosolic (S), membrane (M), nuclear (N), and cytoskeletal (CS) fractions using the ProteoExtract subcellular proteome extraction kit (Millipore) according to the manufacturer's instruction. The protein concentration of each fraction was quantified using Protein Quantification Assay kit (Macerey-Nagel, Düren, Germany)
Immunopurification
For experiments examining ubiquitination of JosD1 in cells, we conducted stringent denature/renature immunopurification as described previously (12). Briefly, cells transfected to express JosD1-V5 and HA-Ub were lysed in RIPA buffer with protease inhibitors, denatured for 30 min by adding SDS to 1%, and then renatured in additional 4.5% Triton X-100 for 30 min. Lysates were then incubated with anti-V5 mouse monoclonal antibody (Sigma-Aldrich) for 2 h at 4 °C, rinsed four times with RIPA + protease inhibitors, eluted with Laemmli buffer, and boiled for 5 min. Ubiquitination of JosD1 was determined by immunoblotting with anti-V5 (Invitrogen) and anti-HA (Santa Cruz Biotechnology antibodies).
Pulldown with Nickel-Agarose Beads
To detect interactions of JosD1/JosD2 with other proteins, we pulled down V5-tagged JosD1/JosD2 with nickel-agarose beads, made possible because the vector used to fuse V5 to JosD1 or Jos D2 also contains a His tag next to the V5 tag. Transfected cells expressing JosD1-V5 or JosD2-V5 were lysed with lysis buffer (150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100, 20 mm HEPES, pH 7.5) containing protease inhibitors and 5 mm imidazole to minimize nonspecific binding to the beads. Lysates were incubated with nickel-agarose beads (EZview red HIS-select HC nickel affinity gel; Sigma) for 4 h at 4 °C, rinsed four times with lysis buffer with protease inhibitors, and eluted with lysis buffer containing 1 m imidazole. Pulldown products and input lysates (5–10%) were immunoblotted with anti-β-actin (Sigma), anti-α-tubulin (Cell Signaling Technologies), and anti-V5 antibodies.
Immunofluorescence
Immunofluorescence was conducted as described previously (16). Briefly, cells were plated onto glass-bottom dishes 1 day after transfection. After further cultivation for 1–2 days, cells were fixed with 4% paraformaldehyde followed by membrane permeabilization with 0.3% Triton X-100 for 10 min. Cells were incubated with primary antibodies (mouse anti-V5 (Invitrogen; 1:1000) or rabbit anti-early endosomal antigen 1 (EEA1) (Affinity BioReagents; 1:500)) for >1 h, followed by incubation with secondary antibody (Alexa Fluor 488 anti-mouse IgG (1:500), Alexa Fluor 568 anti-mouse IgG (1:500), or Alexa Fluor 647 anti-rabbit IgG (1:500); Invitrogen) for at least 1 h. For F-actin staining, phalloidin-TRITC (100 nm; Invitrogen) was incubated with fixed cells expressing GFP-JosD1 after membrane permeabilization. Single optical plane images were obtained with an A1 confocal microscope (Nikon).
Time-lapse Imaging
One day after transfection of GFP or GFP-JosD1, HEK-293 cells were detached and plated onto glass-bottom dishes (MatTek, Ashland, MA) at 0.5–1 × 104 cells/dish. After another day of cultivation, GFP fluorescence and bright field images were monitored in living cells using Deltavison-RT live imaging system (Applied Precision, Issaquah, WA). Images were obtained every 2 min for 60 min.
To quantify changes in cell morphology, we drew a circle to fit the outermost cell edges at every 10-min interval. We estimated morphological changes by the changes in circle diameter in each 10-min interval. Cell motility was evaluated by the distance that the center of the nucleus (a proxy for the cell center) moved in 10 min. The mean value of six different time intervals (0–10, 10–20, 20–30, 30–40, 40–50, 50–60 min) represents an estimate of morphological change or cell motility for each cell. Duration of blebbing was evaluated by the number of sequential images in which a cell had any detectable blebs.
Uptake of Fluorescent Endocytic Markers
JosD1-V5-transfected cells were plated onto glass-bottom dishes as described above. Two days after transfection, cells were incubated with Lucifer Yellow (LY, 1 mg/ml; Sigma) for 1 h, with transferrin (Tf) fused with Alexa Fluor 568 (Invitrogen; 10 μg/ml for clathrin-mediated endocytosis) for 5 min, or cholera toxin subunit B (CTxB) fused with Alexa Fluor 555 (Invitrogen; 1 μg/ml for caveolae-mediated endocytosis) for 5 min to visualize fluid-phase, clathrin-mediated, or caveolae-mediated endocytosis, respectively. After incubation, cells were fixed with 4% paraformaldehyde and immunostained with anti-V5 antibody. To visualize macropinocytosis, GFP-JosD1 was transfected instead of JosD1-V5, and cells were incubated for 1 h with dextran 70-kDa protein (Dex) conjugated to tetramethylrhodamine (TMR) (0.5 mg/ml; Invitrogen). Single optical plane fluorescence images were obtained using the A1 confocal microscope (Nikon, Tokyo, Japan). Each fluorescence image contains cells both expressing and not expressing JosD1 to compare the uptake of fluorescence markers. Uptake of these markers was quantified by computer-based analysis using ImageJ software. We evaluated the mean fluorescence intensity from whole cells and the number and mean intensity of fluorescent puncta per cell.
Quantification and Statistical Analysis
To analyze immunoblot results, immunoblots were scanned using a scanner, and images were collected in Adobe Photoshop. Densitometry was measured using ImageJ software. To analyze results from immunofluorescence and fluorescence marker uptake quantitatively, single optical plane fluorescence images obtained by a confocal microscopy were analyzed using ImageJ software, as described above. All data are represented as the percentage of the average values from nonexpressing cells. Unpaired Student's t test was used to determine statistical differences between two groups indicated in figure legends. Prism 4 software (GraphPad Software, La Jolla, CA) was used to conduct Student's t test and to generate graphs.
RESULTS
Catalytic Activity of JosD1 and JosD2
We employed two common methods to assess in vitro the activity of recombinant JosD1 and JosD2. First, we used the suicide probe Ub-VS, an inhibitor of cysteine protease DUBs that reacts with and inactivates the catalytic cysteine. Recombinant JosD1 and JosD2 both reacted with Ub-VS (Fig. 1C). Although we commonly observed complete or nearly complete reaction of JosD2 with Ub-VS, not all JosD1 was modified by Ub-VS regardless of the tag used (GST, maltose-binding protein, or His6). Thus, a portion of bacterially expressed JosD1 is inactive, which could result from the propensity of bacterially expressed JosD1 to aggregate.4 Second, we tested the ability of both proteins to cleave Ub from a fluorescent probe (Ub-AMC). As shown in Fig. 1D, both JosD1 and JosD2 cleaved Ub from the probe. Mutating the catalytic cysteine of each DUB abolished this activity (C14A for ATXN3, C36A for JosD1, and C24A for JosD2, Fig. 1B). These results confirm that JosD1 and JosD2 are active DUBs in vitro.
We next assessed the ability of recombinant JosD1 and JosD2 to cleave Ub chains of different lengths in vitro. We tested Lys48-linked Ub chains, which commonly target proteins for degradation, and Lys63-linked Ub chains, most often linked to nondegradative pathways. Interestingly, we observed only minimal activity from JosD1 toward either type of Ub chain (Fig. 1E) even though JosD1 is clearly active against mono-Ub probes (Fig. 1, C and D). In contrast, JosD2 cleaves both types of chains, although it appears to have a preference for Lys63 linkages (Fig. 1E).
JosD1 Expression and Ubiquitination
In efforts to generate antibodies against JosD1 and JosD2, we succeeded in obtaining an antibody reactive against JosD1 but not JosD2 (Fig. 2A). This antibody allowed us to examine JosD1 expression in mice. As shown in Fig. 2B, JosD1 is expressed in all tissues tested. In addition to the main JosD1-reactive band, more slowly migrating JosD1-positive bands were also seen. These could represent ubiquitinated forms of JosD1, as described for ATXN3 (7, 8). Indeed, stringent immunopurification of JosD1 from cells expressing tagged forms of JosD1 and ubiquitin established that a substantial fraction of JosD1 is ubiquitinated in cells (Fig. 2C). In contrast, JosD2 does not appear to be ubiquitinated in cells under similar experimental conditions (data not shown). These data demonstrate that JosD1 is expressed widely and is prone to be ubiquitinated in cells.
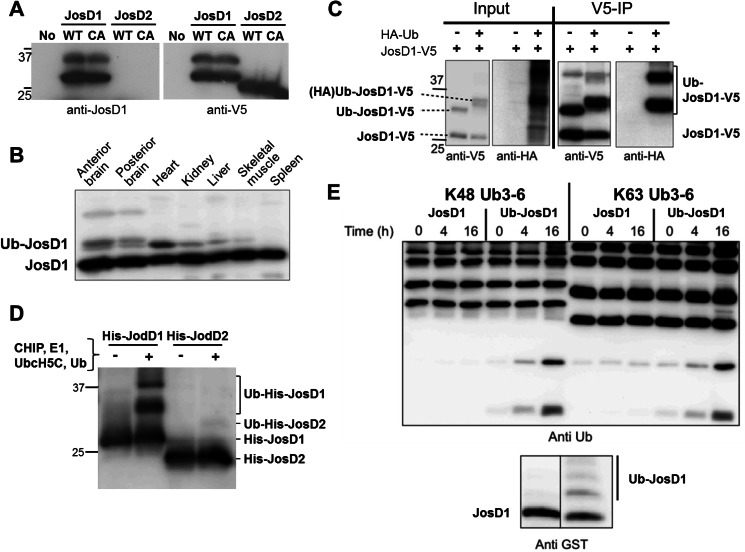
FIGURE 2.
Ubiquitination of JosD1 enhances DUB activity. A, specificity of generated anti-JosD1 antibody. JosD1-V5 (wild-type (WT) and C36A (CA) mutant) and JosD2 (WT and C24A (CA) mutant), when transiently expressed in HEK-293 cells, were detected by immunoblotting with anti-JosD1 (left) and anti-V5 (right) antibodies. The JosD1 antibody detected overexpressed JosD1-V5, but not JosD2-V5. This antibody could not detect endogenous JosD1 in HEK-293 cells. B, JosD1 expression in mouse tissues. Protein lysates from the indicated tissues were immunoblotted with anti-JosD1 antibody. A strong band is seen in all tissues, with some tissues also showing a higher molecular mass band consistent with monoubiquitinated JosD1 (Ub-JosD1). C, confirmation of JosD1 monoubiquitination cells. JosD1-V5 and HA-Ub were co-expressed in COS-7 cells, and cell lysates were immunoprecipitated (IP) with anti-V5 antibody. HA-ubiquitinated JosD1 was strongly detected in immunoprecipitates. D, ubiquitination of JosD1 in vitro. Recombinant His-JosD1 and His-JosD2 were incubated with CHIP, UbcH5c, E1, and Ub to drive ubiquitination. JosD1, but not JosD2, is ubiquitinated in these reactions. E, ubiquitination of JosD1 enhances DUB activity in vitro. GST-JosD1 and ubiquitinated GST-JosD1 (Ub-JosD1) were incubated with Lys48-linked (left) or Lys63-linked (right) poly-Ub chains for the indicated periods of time. Lower immunoblot shows the levels of JosD1 and Ub-JosD1 used in the assay, detected with anti-GST antibody.
Ubiquitination Activates JosD1
Monoubiquitination of ATXN3 directly enhances its DUB activity in vitro and modulates function in cultured cells (7, 8). Because a fraction of JosD1 is ubiquitinated in cells (Fig. 2, A–C), we tested whether ubiquitination similarly regulates the DUB activity of JosD1. To generate ubiquitinated JosD1, we employed the E3 ligase CHIP because the Josephin domain of ATXN3 is known to be ubiquitinated in vitro by this ligase (8). Consistent with our results in transfected cells, CHIP ubiquitinated JosD1 in vitro, but failed to ubiquitinate JosD2 (Fig. 2D). Purified JosD1 from in vitro ubiquitination reactions was then tested for ability to cleave Ub chains. Compared with unmodified JosD1, a mixture of ubiquitinated and unmodified JosD1 showed enhanced catalytic activity toward Ub chains (Fig. 2E). Thus, we conclude that ubiquitination positively regulates the DUB activity of JosD1 in vitro as is true for at least one other MJD family member, ATXN3.
Differential Subcellular Localization of JosD1, JosD1-Ub, and JosD2
Because our JosD1 antibodies do not reliably detect endogenous JosD1 in fixed tissue or cells, we expressed V5-tagged JosD1 and JosD2 (JosD1-V5 and JosD2-V5) in various cell lines to examine their subcellular localization. For our analysis, we excluded cells expressing epitope-tagged JosD1 or JosD2 with very high levels because they can give spurious results regarding subcellular localization. As shown in Fig. 3A, JosD1 localizes to the plasma membrane and cytoplasmic puncta in HEK-293 cells, whereas JosD2 is diffusely cytoplasmic. Neither protein localized to the nucleus, distinguishing them from ATXN3 which shuttles in and out of the nucleus (6). Divergent patterns of subcellular localization for JosD1 and JosD2 were also observed in COS-7 and SH-SY5Y cells and when employing different epitope tags (myc, or GFP, data not shown). These results suggest that JosD1 and JosD2, despite extensive sequence similarity, possess sequence or structural differences that mediate distinct cellular localizations for the two proteins.
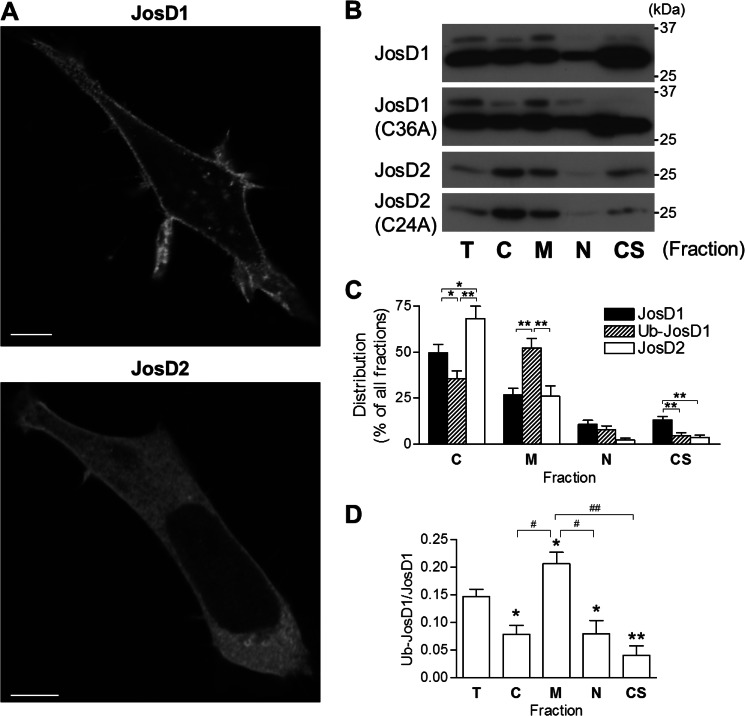
FIGURE 3.
JosD1 and JosD2 differ in subcellular localization. A, HEK-293 cells expressing JosD1-V5 (upper panel) and JosD2-V5 (lower panel) immunostained with anti-V5 antibody. JosD1 localizes to the plasma membrane and cytoplasmic puncta, whereas JosD2 localizes diffusely throughout the cytoplasm and is largely excluded from the nucleus. Scale bars, 10 μm. B, subcellular fractionation of cells expressing JosD1 or JosD2. Transfected cells were fractionated into cytosolic (C), membrane (M), nuclear (N), and cytoskeletal (CS) fractions. Equal amount of protein from total cell lysates (T) and the four indicated fractions were subjected to SDS-PAGE, followed by immunoblotting with anti-V5 antibody. Images are representative of five independent experiments. Catalytically inactive mutants of JosD1 and JosD2 show distribution similar to wild-type JosD1 and JosD2. C, quantification of subcellular distribution of JosD1, Ub-JosD1, and JosD2. Ub-JosD1 preferentially localizes to the membrane fraction over nonubiquitinated JosD1. Shown are mean values from five independent experiments. Error bars, S.E. *, p < 0.05; **, p < 0.01 (unpaired t test). D, quantification of the Ub-JosD1/JosD1 ratio in each fraction. The ratio is significantly higher in the membrane fraction and significantly lower in the other three fractions compared with the ratio for the total cellular pool. Shown are mean values from five independent experiments. Error bars, S.E. *, p < 0.05 versus total cell lysate (T); #, p < 0.05; ##, p < 0.01 (unpaired t test).
Next, we examined whether ubiquitinated JosD1 (Ub-JosD1) with its increased DUB activity preferentially localizes to certain cellular regions (Fig. 3B). Subcellular fractionation confirmed with marker proteins for various fractions (data not shown), indicated that unmodified JosD1 resides in the cytoskeletal (CS), cytosolic (C) and membrane (M) fractions but is nearly absent from the nuclear (N) fraction when equal protein amounts were assessed for each fraction. In contrast, Ub-JosD1 was more strongly detected in the membrane fraction. JosD2 mainly localized to the cytosolic fraction (Fig. 3B). Subcellular distribution of unmodified JosD1, Ub-JosD1, and JosD2 was calculated based on the total protein for each fraction (Fig. 3C). Ub-JosD1 partitioned more significantly to the membrane fraction than did unmodified JosD1 or JosD2; conversely, unmodified JosD1 localized more significantly to the cytoskeletal fraction. Correspondingly, the Ub-JosD1/JosD1 ratio was highest in the membrane fraction and lowest in cytoskeletal fraction (Fig. 3D). Thus, Ub-JosD1 preferentially localizes to the plasma membrane, whereas unmodified JosD1 associates primarily with the cytoskeleton. The presence or absence of the catalytic cysteine did not alter the relative cellular distribution of JosD1, Ub-JosD1, or JosD2 (Fig. 3B), implying that DUB activity per se does not drive this preferential cellular localization of either protein.
JosD1 Regulates Membrane Dynamics and Cell Motility
Because JosD1 partially localizes to the plasma membrane and ubiquitin-dependent pathways regulate membrane dynamics at the cell surface (17), we next tested the hypothesis that JosD1 regulates membrane dynamics. We expressed GFP-fused JosD1 (GFP-JosD1) in HEK-293 cells and observed cellular dynamics by time-lapse microscopy (see supplemental Movies 1–3). Compared with control GFP-expressing cells, cells expressing GFP-JosD1 showed increased membrane dynamics. We quantitatively analyzed morphological changes by measuring over time the change in the circle diameter circumscribing cells (Fig. 4A). Average changes of circle diameter in 10-min intervals were significantly elevated in cells expressing GFP-JosD1 (Fig. 4B). This elevation was also observed in cells expressing catalytically inactive GFP-JosD1(C36A).
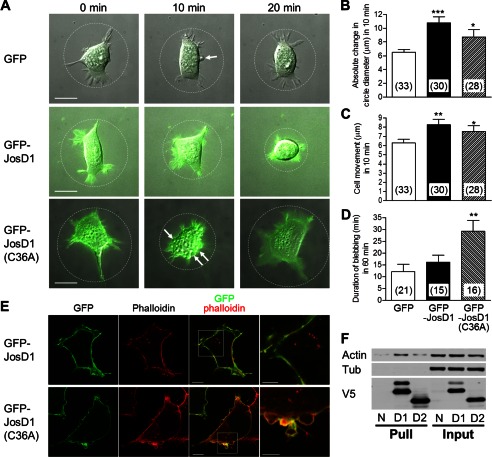
FIGURE 4.
JosD1 affects membrane dynamics and cell motility, independent of DUB activity. A, time-lapse imaging of HEK-293 cells expressing GFP and or GFP-JosD1. Sequential images of GFP fluorescence and differential interference contrast microscopy were obtained at 2-min intervals for 60 min. Representative merged images of GFP fluorescence and differential interference contrast microscopy are shown at 0, 10, and 20 min. Cells expressing GFP-JosD1 and GFP-JosD1(C36A) more readily changed morphology than cells expressing GFP alone. Circles drawn to fit the outer cell edges were used to evaluate changes in cell morphology. Arrows indicate blebs observed in time-lapse imaging. Scale bars, 20 μm. B, quantification of changes in cell morphology. Absolute changes in cell diameter (as shown in A) were measured at 10-min intervals for a total of 60 min, and the mean value of change per 10-min epoch in a given cell serves as a measure of change in cell morphology. Numbers in parentheses indicate the cell number analyzed in each group. GFP-JosD1 or GFP-JosD1(C36A) expression significantly increases morphological changes. C, quantification of changes in cell motility, estimated from the mean distance a cell nucleus moves in 10-min intervals for a total of 60 min. GFP-JosD1 and GFP-JosD1(C36A) significantly increase motility. D, quantification of cell blebbing which was frequently observed during 60-min time-lapse imaging. We determined the duration that cells blebbed during the 60-min observation period. Catalytically inactive GFP-JosD1 significantly prolonged the extent of blebbing. Means ± S.E. (error bars) are shown. Numbers in parentheses indicate the number of cells with blebs. *, p < 0.05; **, p < 0.01; ***, p < 0.001 versus GFP-expressing cells (unpaired t test). E, F-actin staining of cells expressing wild-type or catalytically inactive JosD1. Representative fluorescence images (GFP, phalloidin-TRITC, and merged) of blebbing cells expressing GFP-JosD1 (upper panels) or GFP-JosD1(C36A) (lower panels) are shown. Far right images are higher power magnification views of boxed areas in merged images. Scale bars of merged and high magnification images are 10 and 5 μm, respectively. F, interaction between JosD1 and β-actin. JosD1-V5 and JosD2-V5, both of which also have a His tag, were pulled down with nickel-agarose beads to test for an interaction with β-actin. Pulldown product (Pull) and 10% input were immunoblotted with anti-β-actin (Actin), anti-α-tubulin (Tub), and anti-V5 antibodies. N, D1, and D2 represent nonexpressing cells or transfected cells expressing JosD1-V5 or JosD2-V5, respectively. Immunoblot with anti-β-actin antibody reveals interaction of β-actin with JosD1. Neither JosD1 nor JosD2 interacts with α-tubulin.
Next, we analyzed cell motility by measuring the distance that cells moved in 10-min intervals. Cells expressing GFP-JosD1 and GFP-JosD1(C36A) moved significantly greater distances than controls expressing GFP (Fig. 4C). In addition, blebs of cell membrane were frequently observed during time-lapse observation (arrows in Fig. 4A). Although the frequency of cells showing blebs was similar among cells expressing GFP alone (64%, 21/33 cells), GFP-JosD1 (50%, 15/30 cells), and GFP-JosD1(C36A) (57%, 16/28 cells), the duration of blebs was significant longer in cells expressing GFP-JosD1(C36A) (Fig. 4D). Blebbing can be triggered by the local destabilization of actin cytoskeleton (18). JosD1 potentially could affect the stability and arrangement of the actin cytoskeleton, thereby altering membrane dynamics and cell motility. In cells expressing GFP-JosD1 and stained with phalloidin-TRITC to visualize F-actin, JosD1 and F-actin co-localized near the plasma membrane (Fig. 4E, upper panels). In blebbing cells expressing GFP-JosD1(C36A), JosD1 lined the blebs whereas F-actin remained in the cell proper, just below the blebs (Fig. 4E, lower panels). Consistent with a functional interaction between JosD1 and F-actin and in keeping with the cellular fractionation shown above, JosD1 co-immunoprecipitated with β-actin (Fig. 4F). We conclude that JosD1 influences membrane dynamics and cell motility independent of its DUB activity, possibly through an interaction with the actin cytoskeleton.
JosD1 Modulates Endocytosis
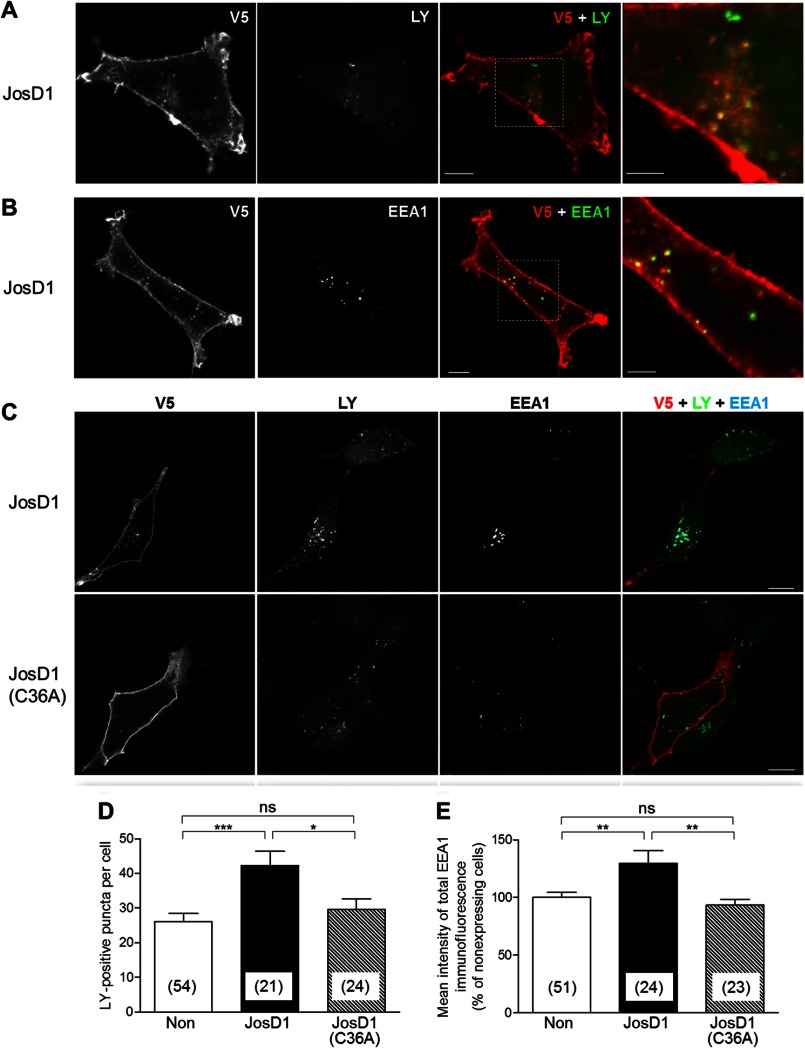
Because JosD1 affects membrane dynamics, this membrane-associated DUB could contribute to the regulation of endocytosis. To explore this possibility, we visualized fluid-phase endocytosis by incubating HEK-293 cells expressing JosD1-V5 with extracellular LY for 1 h (19). Cytoplasmic puncta of JosD1-V5 frequently, but not always, co-localized with LY-positive puncta (Fig. 5A) and EEA1, an early endosome marker (Fig. 5B). JosD1 also increased the number of LY-positive puncta (Fig. 5, C and D) and the immunofluorescence intensity of EEA1-positive puncta (Fig. 5, C and E). Importantly, these increases depended on the catalytic activity of JosD1 (Fig. 5, C–E).
FIGURE 5.
JosD1 enhances endocytosis in an activity-dependent manner. A and B, partial localization of JosD1 to early endosomes is shown. HEK-293 cells expressing JosD1-V5 were incubated with LY, a marker of fluid-phase endocytosis, for 1 h. After fixation, cells were immunostained with anti-V5 and anti-EEA1 antibodies. Cytoplasmic puncta of JosD1 partially co-localize with LY-positive (A) and EEA1-positive (B) puncta. Scale bars in merged and higher power images are 10 and 5 μm, respectively. C, JosD1 increases LY- and EEA1-positive puncta compared with nonexpressing cells (upper cell in upper panel), whereas catalytically inactive JosD1 does not. Scale bar, 20 μm. D and E, LY-positive puncta (D) and EEA1 immunoreactivity are quantified (E). Both are significantly increased in cells expressing JosD1 but not JosD1(C36A). Means ± S.E. (error bars) are shown. Numbers in parentheses indicate the number of cells analyzed in each group. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ns, not significant; unpaired t test.
Several endocytic pathways have been identified including macropinocytosis, clathrin-mediated endocytosis, and caveolae-mediated endocytosis (20). To determine which pathways JosD1 affects, we examined the uptake of fluorescence markers specific for these three pathways: Dex-TMR (macropinocytosis), Tf-Alexa Fluor 568 (clathrin-mediated endocytosis), and CTxB-Alexa Fluor 555 (caveolae-mediated endocytosis) (21, 22).
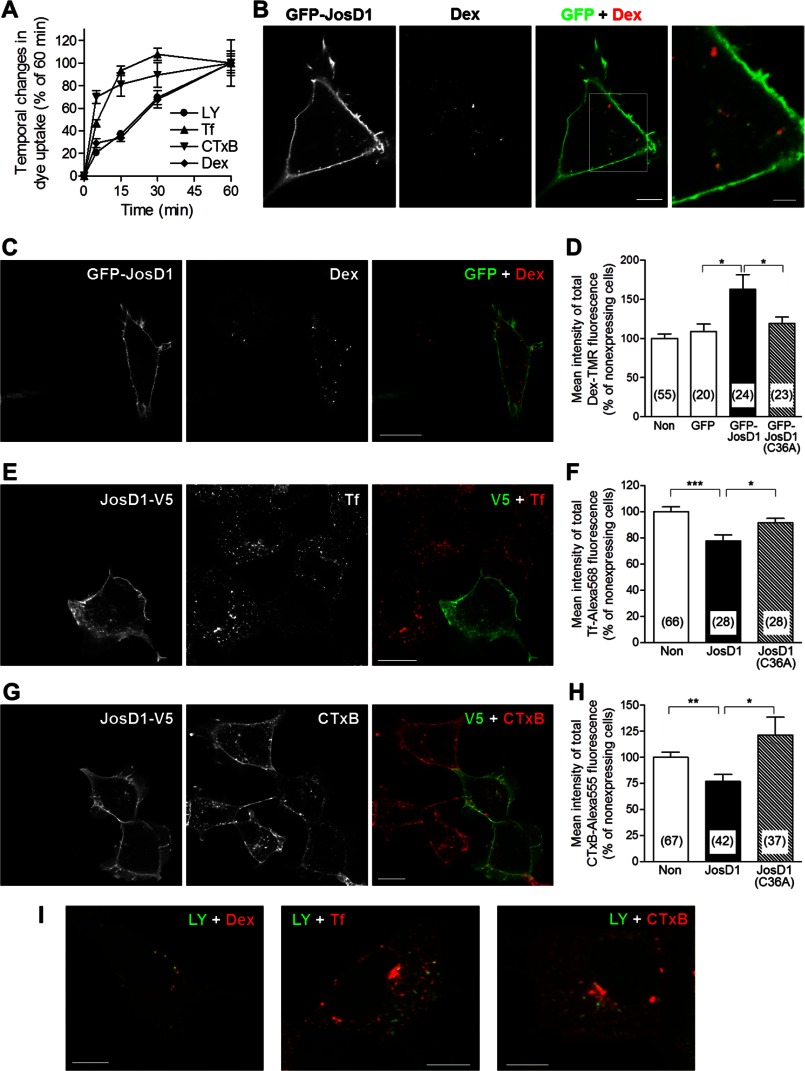
Uptake of Dex continuously increased over 1 h, similar to LY uptake (Fig. 6A). Therefore, the effect of JosD1 on Dex uptake was evaluated after 1-h uptake. We used GFP-JosD1 to assess the effect on Dex uptake to avoid the repetitive washes required for immunostaining because Dex-TMR fluorescence rapidly disappears during washes (data not shown). GFP-JosD1 significantly increased Dex uptake over 1 h (Fig. 6, C and D). Cytoplasmic puncta containing GFP-JosD1 partially co-localized with Dex-positive puncta (Fig. 6B) and F-actin (Fig. 4E). Because macropinosomes are surrounded by F-actin (23), JosD1 would be expected to co-localize to macropinosomes. In addition, LY-positive puncta strongly overlapped with Dex-positive puncta after 1 h uptake, but not with Tf- and CTxB-positive puncta Fig. 6I). These findings indicate that JosD1 enhances macropinocytosis, resulting in increased uptake of LY and Dex.
FIGURE 6.
JosD1 overexpression enhances macropinocytosis but suppresses clathrin-mediated and caveolae-mediated endocytosis. A, time course of uptake of fluorescent dyes used to detection various types of endocytosis in HEK-293 cells. Mean fluorescence intensity of each dye was plotted after 5, 15, 30, and 60 min. Uptake of Tf and CTxB nears saturation at around 15 min, whereas LY and Dex uptake increases continuously over 60 min. B, partial co-localization of GFP-JosD1- and Dex-positive puncta. Representative fluorescence images (GFP, Dex, and merged) of cells expressing GFP-JosD1 are shown. Far right image is higher power magnification views of boxed area in merged image. Scale bars of merged and high magnification images are 10 and 5 μm, respectively. C and D, uptake of Dex-TMR, a marker of macropinocytosis, in HEK-293 cells expressing GFP-JosD1. C, representative cell images visualizing GFP (left), Dex-TMR (center), and merged (right). Scale bar, 20 μm. D, quantification of mean intensity of total Dex-TMR fluorescence showing that Dex uptake is significantly increased in cells expressing JosD1, but not JosD1(C36A). E and F, uptake of Tf-Alexa Fluor 568, a marker of clathrin-mediated endocytosis, in cells expressing JosD1-V5. E, representative cell images, viewed by V5 tag immunofluorescence (left), Tf-Alexa Fluor 568 (center), and merged (right). Scale bar, 20 μm. F, quantification of mean intensity of total Tf-Alexa Fluor 568 fluorescence showing that Tf uptake is significantly decreased in cells expressing JosD1-V5 but not JosD1(C36A)-V5. G and H, uptake of CTxB-Alexa Fluor 555, a marker of caveolae-mediated endocytosis, in cells expressing JosD1-V5. G, representative cell images, viewed by V5 tag immunofluorescence (left), CTxB-Alexa Fluor 555 (center), and merged (right). Scale bar, 20 μm. H, quantification of mean intensity of total CTxB-Alexa Fluor 555 fluorescence showing that CTxB uptake is significantly decreased in cells expressing JosD1-V5 but not JosD1(C36A)-V5. For D, F, and H, means ± standard errors (error bars) are shown. Numbers in parentheses indicate the number of cells analyzed in each group. *, p < 0.05; **, p < 0.01; ***, p < 0.001, unpaired t test. I, Uptake of LY with Dex (left), Tf (center), or CTxB (right) in the same cell for 60 min. LY puncta strongly co-localized with Dex puncta, but only partially co-localized with Tf and CTxB puncta. Scale bars, 10 μm.
In contrast to LY and Dex, uptake of Tf or CTxB was nearly saturated within 15 min of incubation (Fig. 6A). Thus, we evaluated the effects of JosD1 on Tf and CTxB uptake during a 5-min period. JosD1 significantly decreased the uptake of Tf or CTxB (Fig. 6, E–H), suggesting that JosD1 suppresses clathrin- and caveolae-mediated endocytosis. These divergent effects of JosD1 on separate endocytic pathways were not observed in cells expressing catalytically inactive JosD1 (Fig. 6, C–H). Taken together, these results suggest that JosD1 differentially regulates three endocytic pathways in a manner that depends on its DUB activity.
DISCUSSION
Our study provides insight into the properties and functions of a poorly understood class of DUBs, the MJD family, and particularly of JosD1 and JosD2. Despite the close sequence similarity of JosD1 and JosD2, our results show that they differ in fundamental ways including basal catalytic activity in vitro, capacity for ubiquitination in vitro and in cells, and subcellular localization. We also demonstrate that the DUB activity of JosD1 is positively regulated by its own ubiquitination, similar to another MJD DUB, ATXN3. Finally, our data reveal that JosD1 regulates membrane dynamics, including endocytosis, in cultured cells.
Our result that recombinant JosD2 has greater in vitro DUB activity than JosD1 is consistent with previous reports (11, 24), but our observation that ubiquitination directly activates JosD1, similar to the disease protein ATXN3 (7, 8), is novel. Ubiquitination-dependent regulation of ATXN3 likely depends on a conformational change in the Josephin domain, because none of the five identified Ub binding sites on ATXN3 is necessary for this activation (8). Ubiquitination of ATXN3 at a specific lysine residue near the catalytic groove, Lys117, is sufficient to enhance DUB activity. We speculate that the DUB activity of JosD1 is similarly regulated by ubiquitination through a conformational change at or near the catalytic site. In fact, sequence alignments (Fig. 1B) identify a lysine residue in JosD1 that closely aligns with Lys117 of ATXN3. The possibility that ubiquitination alters the conformation of JosD1 is indirectly supported by our finding that unmodified, recombinant JosD1 is active against a mono-Ub probe but incapable of cleaving Ub chains. Perhaps the catalytic cleft of unmodified JosD1 cannot interact productively with larger Ub fusions, exemplified by Ub-Ub linkages, unless ubiquitination alters the conformation of the catalytic groove, opening it sufficiently to accommodate Ub-Ub linkages. Structural studies are needed to address this possibility.
Our finding that ubiquitination activates JosD1 also suggests that ubiquitination may be a more common regulator of DUB catalytic activity than has been appreciated. Whether this phenomenon is limited to the MJD DUB family or applies to other DUBs is an open question. A recent study demonstrated that many DUBs are monoubiquitinated in vitro by several different E3 ligases (25). Another study presented evidence that ubiquitination of the DUB USP25 increased its activity in cell culture (26). We should also emphasize that, even though we do not observe JosD1 activity against Ub chains in vitro, we cannot exclude the possibility that unmodified JosD1 possesses in vivo activity either toward specific substrates or that is mediated by specific protein interactions.
At the subcellular level JosD1 appears most concentrated at the plasma membrane, whereas JosD2 is diffusely localized in the cytosol. Because JosD1 does not have hydrophobic motifs to localize it to the membrane, it could be targeted to plasma membrane by interacting with membrane proteins or lipids. Although JosD1 and JosD2 share a high similarity in amino acid sequence, the most divergent sequence between JosD1 and JosD2 is at the N terminus, where there are 12 additional amino acids in JosD1 compared with JosD2 (Fig. 1B). We deleted these additional amino acids from JosD1, but subcellular localization of JosD1 was not affected by this deletion (data not shown). Therefore, subcellular localization of JosD1 may be determined by one or more post-translational modifications or protein-protein interactions that are specific to JosD1.
Subcellular fractionation revealed that ubiquitinated JosD1 preferentially partitions to the plasma membrane. JosD1 with enhanced DUB activity could thus regulate plasma membrane-related functions, such as membrane dynamics, cell motility, and endocytosis. Indeed, our study revealed that JosD1 differentially regulates three types of endocytosis in a manner that depends on its DUB activity: it enhances macropinocytosis and suppresses clathrin- and caveolae-mediated endocytosis. Clathrin- and caveolae-mediated endocytosis processes are important for the internalization of membrane receptors and membrane proteins (20). Many receptors are ubiquitinated in response to agonists, and ubiquitinated receptors are internalized into endocytic vesicles (27). Membrane-localized JosD1 could deubiquitinate membrane receptors after agonist stimulation, leading to a decrease in clathrin- and caveolae-mediated endocytosis.
Macropinocytosis mediates the nonselective uptake of extracellular molecules, nutrients, and antigens (28, 29). Macropinocytosis depends on actin organization and is regulated by various molecules including PI3-kinase and many small GTPases (29). Most of these regulators are also implicated in membrane dynamics and cell motility (29). Thus, JosD1 could affect the activity of such regulators, thereby activating macropinocytosis as well as regulating membrane dynamics and cell motility. Some of these regulators are ubiquitinated and degraded by the proteasome (30, 31). One or more of these proteasome-sensitive regulators may be targets of JosD1.
Macropinocytosis participates in antigen capture and presentation in immune cells (29), but its role in other cell types is not fully elucidated. Recently, macropinocytosis was shown to be important for axon guidance by inducing growth cone collapse through plasma membrane retrieval (32, 33). Because JosD1 is strongly expressed in brain (Fig. 2B), it will be interesting to determine whether it alters neuronal macropinocytosis or neuronal network formation during development or in adults.
The significance of our findings would be strengthened by experiments in which the level of endogenous JosD1 is reduced by siRNA. Unfortunately, endogenous JosD1 was not detected in HEK-293 cells by immunoblotting with our antibody (Fig. 2A). Although we could detect low level expression of endogenous JosD1 in HeLa and COS-7 cells, we were unable to achieve effective knockdown of JosD1 using several commercially available shRNA or siRNA (data not shown). The fact that we can readily detect JosD1 in various mouse tissues (Fig. 2B) suggests that construction of JosD1 knock-out mice would be useful in providing further physiological insight into the functional roles of JosD1.
In summary, we have shown that two closely related members of the MJD family of DUBs differ markedly in a variety of functional properties. Because of the high structural similarity of JosD1 and ATXN3 in their catalytic domain, these functions might also be regulated by ATXN3 and be involved in MJD disease pathogenesis. Our findings offer new insights into the functions of the MJD family DUBs.
Acknowledgments
We thank Randall Pittman, Barrington Burnett, and Fushing Li (University of Pennsylvania) for early contributions and thoughtful insight into this work. This work was carried out using equipment at Microscopy and Image Analysis Laboratory, University of Michigan.
This work was supported, in whole or in part, by National Institutes of Health Grants R00 NS064097 (to S. V. T.) and R01 NS038712 (to H. L. P.).

This article contains supplemental Movies 1–3.
T. Seki, L. Gong, A. J. Williams, N. Sakai, S. V. Todi, and H. L. Paulson, unpublished observations.
- DUB
- deubiquitinating enzyme
- ATXN3
- ataxin-3
- CHIP
- C terminus of Hsc70-interacting protein
- CTxB
- cholera toxin subunit B
- Dex
- dextran
- EEA1
- early endosomal antigen 1
- JosD1/2
- Josephin domain-containing protein 1/2
- LY
- Lucifer Yellow
- MJD
- Machado-Joseph disease
- RIPA
- radioimmunoprecipitation assay
- Tf
- transferrin
- TMR
- tetramethylrhodamine
- TRITC
- tetramethylrhodamine isothiocyanate
- Ub
- ubiquitin or ubiquitinated
- Ub-AMC
- ubiquitin-7-amino-4-methylcoumarin
- Ub-VS
- ubiquitin-vinylsulfone
- UIM
- ubiquitin-interacting motif.
REFERENCES
- 1. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 2. Burrows J. F., Johnston J. A. (2012) Regulation of cellular responses by deubiquitinating enzymes: an update. Front. Biosci. 17, 1184–1200 [DOI] [PubMed] [Google Scholar]
- 3. Sippl W., Collura V., Colland F. (2011) Ubiquitin-specific proteases as cancer drug targets. Future Oncol. 7, 619–632 [DOI] [PubMed] [Google Scholar]
- 4. Todi S. V., Paulson H. L. (2011) Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 34, 370–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tsou W. L., Sheedlo M. J., Morrow M. E., Blount J. R., McGregor K. M., Das C., Todi S. V. (2012) Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS One 7, e43112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costa Mdo C., Paulson H. L. (2012) Toward understanding Machado-Joseph disease. Prog. Neurobiol. 97, 239–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Todi S. V., Winborn B. J., Scaglione K. M., Blount J. R., Travis S. M., Paulson H. L. (2009) Ubiquitination directly enhances activity of the deubiquitinating enzyme ataxin-3. EMBO J. 28, 372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Todi S. V., Scaglione K. M., Blount J. R., Basrur V., Conlon K. P., Pastore A., Elenitoba-Johnson K., Paulson H. L. (2010) Activity and cellular functions of the deubiquitinating enzyme and polyglutamine disease protein ataxin-3 are regulated by ubiquitination at lysine 117. J. Biol. Chem. 285, 39303–39313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scaglione K. M., Zavodszky E., Todi S. V., Patury S., Xu P., Rodríguez-Lebrón E., Fischer S., Konen J., Djarmati A., Peng J., Gestwicki J. E., Paulson H. L. (2011) Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol. Cell 43, 599–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Durcan T. M., Kontogiannea M., Thorarinsdottir T., Fallon L., Williams A. J., Djarmati A., Fantaneanu T., Paulson H. L., Fon E. A. (2011) The Machado-Joseph disease-associated mutant form of ataxin-3 regulates parkin ubiquitination and stability. Hum. Mol. Genet. 20, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tzvetkov N., Breuer P. (2007) Josephin domain-containing proteins from a variety of species are active de-ubiquitination enzymes. Biol. Chem. 388, 973–978 [DOI] [PubMed] [Google Scholar]
- 12. Todi S. V., Laco M. N., Winborn B. J., Travis S. M., Wen H. M., Paulson H. L. (2007) Cellular turnover of the polyglutamine disease protein ataxin-3 is regulated by its catalytic activity. J. Biol. Chem. 282, 29348–29358 [DOI] [PubMed] [Google Scholar]
- 13. Winborn B. J., Travis S. M., Todi S. V., Scaglione K. M., Xu P., Williams A. J., Cohen R. E., Peng J., Paulson H. L. (2008) The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys-63 linkages in mixed linkage ubiquitin chains. J. Biol. Chem. 283, 26436–26443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burnett B., Li F., Pittman R. N. (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum. Mol. Genet. 12, 3195–3205 [DOI] [PubMed] [Google Scholar]
- 15. Berke S. J., Chai Y., Marrs G. L., Wen H., Paulson H. L. (2005) Defining the role of ubiquitin-interacting motifs in the polyglutamine disease protein, ataxin-3. J. Biol. Chem. 280, 32026–32034 [DOI] [PubMed] [Google Scholar]
- 16. Seki T., Yoshino K. I., Tanaka S., Dohi E., Onji T., Yamamoto K., Hide I., Paulson H. L., Saito N., Sakai N. (2012) Establishment of a novel fluorescence-based method to evaluate chaperone-mediated autophagy in a single neuron. PLoS One 7, e31232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schaefer A., Nethe M., Hordijk P. L. (2012) Ubiquitin links to cytoskeletal dynamics, cell adhesion and migration. Biochem. J. 442, 13–25 [DOI] [PubMed] [Google Scholar]
- 18. Fackler O. T., Grosse R. (2008) Cell motility through plasma membrane blebbing. J. Cell Biol. 181, 879–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Racoosin E. L., Swanson J. A. (1992) M-CSF-induced macropinocytosis increases solute endocytosis but not receptor-mediated endocytosis in mouse macrophages. J. Cell Sci. 102, 867–880 [DOI] [PubMed] [Google Scholar]
- 20. Doherty G. J., McMahon H. T. (2009) Mechanisms of endocytosis. Annu. Rev. Biochem. 78, 857–902 [DOI] [PubMed] [Google Scholar]
- 21. Al Soraj M., He L., Peynshaert K., Cousaert J., Vercauteren D., Braeckmans K., De Smedt S. C., Jones A. T. (2012) siRNA and pharmacological inhibition of endocytic pathways to characterize the differential role of macropinocytosis and the actin cytoskeleton on cellular uptake of dextran and cationic cell penetrating peptides octaarginine (R8) and HIV-Tat. J. Control. Release 161, 132–141 [DOI] [PubMed] [Google Scholar]
- 22. Prieto-Sánchez R. M., Berenjeno I. M., Bustelo X. R. (2006) Involvement of the Rho/Rac family member RhoG in caveolar endocytosis. Oncogene 25, 2961–2973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee E., Knecht D. A. (2002) Visualization of actin dynamics during macropinocytosis and exocytosis. Traffic 3, 186–192 [DOI] [PubMed] [Google Scholar]
- 24. Weeks S. D., Grasty K. C., Hernandez-Cuebas L., Loll P. J. (2011) Crystal structure of a Josephin-ubiquitin complex: evolutionary restraints on ataxin-3 deubiquitinating activity. J. Biol. Chem. 286, 4555–4565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Loch C. M., Strickler J. E. (2012) A microarray of ubiquitylated proteins for profiling deubiquitylase activity reveals the critical roles of both chain and substrate. Biochim. Biophys. Acta 1823, 2069–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Denuc A., Bosch-Comas A., Gonzàlez-Duarte R., Marfany G. (2009) The UBA-UIM domains of the USP25 regulate the enzyme ubiquitination state and modulate substrate recognition. PLoS One 4, e5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Haglund K., Dikic I. (2012) The role of ubiquitylation in receptor endocytosis and endosomal sorting. J. Cell Sci. 125, 265–275 [DOI] [PubMed] [Google Scholar]
- 28. Amyere M., Mettlen M., Van Der Smissen P., Platek A., Payrastre B., Veithen A., Courtoy P. J. (2002) Origin, originality, functions, subversions and molecular signalling of macropinocytosis. Int. J. Med. Microbiol. 291, 487–494 [DOI] [PubMed] [Google Scholar]
- 29. Lim J. P., Gleeson P. A. (2011) Macropinocytosis: an endocytic pathway for internalising large gulps. Immunol. Cell Biol. 89, 836–843 [DOI] [PubMed] [Google Scholar]
- 30. Orme M., Bianchi K., Meier P. (2012) Ubiquitin-mediated regulation of RhoGTPase signalling: IAPs and HACE1 enter the fray. EMBO J. 31, 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Weisz Hubsman M., Volinsky N., Manser E., Yablonski D., Aronheim A. (2007) Autophosphorylation-dependent degradation of Pak1, triggered by the Rho-family GTPase, Chp. Biochem. J. 404, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kolpak A. L., Jiang J., Guo D., Standley C., Bellve K., Fogarty K., Bao Z. Z. (2009) Negative guidance factor-induced macropinocytosis in the growth cone plays a critical role in repulsive axon turning. J. Neurosci. 29, 10488–10498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kabayama H., Nakamura T., Takeuchi M., Iwasaki H., Taniguchi M., Tokushige N., Mikoshiba K. (2009) Ca2+ induces macropinocytosis via F-actin depolymerization during growth cone collapse. Mol. Cell. Neurosci. 40, 27–38 [DOI] [PubMed] [Google Scholar]