Background: To improve the effectiveness of chemo- and radiotherapy only in cancer tissue is important for avoiding side effects.
Results: Emi1 depletion enhanced the sensitivity of anticancer reagents and X-ray irradiation in cancer cells.
Conclusion: Emi1 siRNA would be a useful new modality for enhancing the effect of chemo- and radiotherapy in various tumors.
Significance: This work provides new insights regarding synergistic effect of Emi1 knockdown in combination therapies.
Keywords: Anticancer Drug, Cancer, Cancer Therapy, Cell Cycle, DNA Damage, DNA Synthesis, Radiation Biology
Abstract
Chemotherapy and radiation in addition to surgery has proven useful in a number of different cancer types, but the effectiveness in normal tissue cannot be avoided in these therapies. To improve the effectiveness of these therapies selectively in cancer tissue is important for avoiding side effects. Early mitotic inhibitor 1 (Emi1) is known to have the function to inhibit anaphase-promoting complex/cyclosome ubiquitin ligase complex, which ubiquitylates the cell cycle-related proteins. It recently has been shown that Emi1 knockdown prevents transition from S to G2 phase by down-regulating geminin via anaphase-promoting complex/cyclosome activation. At present, anticancer drugs for targeting DNA synthesis to interfere with rapidly dividing cells commonly are used. As Emi1 depletion interferes with completion of DNA synthesis in cancer cells, we thought that Emi1 knockdown might enhance the sensitivity for anticancer agents. Here, we confirmed that Emi1 siRNA induced polyploidy for preventing transition from S to G2 phase in several cancer cell lines. Then, we treated Emi1 depleted cells with doxorubicin. Interestingly, increased apoptotic cells were observed after doxorubicin treatment in Emi1 siRNA-treated cancer cells. In addition, Emi1 depletion enhanced the sensitivity of x-ray irradiation in cancer cells. Importantly, synergistic effect of Emi1 knockdown in these combination therapies was not observed in normal cells. These results suggest that Emi1 siRNA can be a useful tool for enhancing of sensitivity of cancer cells to anticancer reagents and radiation.
Introduction
Approximately 12.7 million cancers were diagnosed and 7.6 million people died of cancer worldwide in 2008 (1). This makes cancer the leading cause of death in the developed world and the second leading cause of death in the developing world (1). Cancer can be treated by various means such as surgery, chemotherapy, radiation therapy, immunotherapy, and monoclonal antibody therapy. The choice of therapy depends upon the location and grade of the tumor and the stage of the disease, as well as the physical condition of the patient. Complete inactivation of the cancer without damage to the rest of the body is the goal of treatment. Sometimes this can be accomplished by surgery, but the propensity of cancers to invade adjacent tissues or to spread to distant sites by microscopic metastasis often limits its effectiveness. Chemotherapy alone or in combination with other treatments such as surgery or radiation has proven useful in a number of different cancer types, but the effectiveness of chemotherapy is often limited by the toxicity to non-targeted tissues in the body. The majority of chemotherapeutic drugs can be divided into alkylating agents, antimetabolites, anthracyclines, plant alkaloids, topoisomerase inhibitors, and other anticancer agents (2, 3). All of these drugs affect cell division or DNA synthesis and act by killing cells that divide rapidly, one of the main properties of most cancer cells. Anticancer agents also harm cells that divide rapidly under normal circumstances: cells in the bone marrow, digestive tracts, and hair follicles. These effects result in the common side effects of chemotherapy, including myelosuppression, mucositis, and alopecia. Therefore, newer anticancer drugs, including monoclonal antibodies and the new tyrosine kinase inhibitors, e.g. imatinib mesylate (Gleevec or Glivec), which directly targets a molecular abnormality in certain types of cancer (chronic myelogenous leukemia, gastrointestinal stromal tumors), have been used (4). Targeted therapy is a type of medication that blocks the growth of cancer cells by interfering with specific targeted molecules needed for carcinogenesis and tumor growth, rather than by simply interfering with rapidly dividing cells. Targeted cancer therapies thus are expected to be more effective than conventional treatments and less harmful to normal cells. Many oncologists believe that targeted therapies are the chemotherapy of the future. At present, however, many traditional anticancer drugs for targeting DNA synthesis to interfere with rapidly dividing cells are commonly used. Radiation therapy is the medical application of ionizing radiation to suppressing tumor growth. Ionizing radiation works by damaging DNA to control tumor cell growth/division, but the effect of radiation in normal tissues cannot be avoided in these therapies.
The cell cycle is regulated by cell cycle regulation factors and many of these are degraded via ubiquitylation (5). Abnormality of ubiquitylation in degradation of proteins induces various diseases such as cancer (6–8). It is known that SCF (Skp1-Cullin-F-box) and anaphase-promoting complex/cyclosome (APC/C),4 ubiquitin ligases, are involved in ubiquitylation of cell cycle regulating factors (9, 10). In particular, APC/C is associated with the degradation of proteins in the M-G1 phase and plays a role in the regulation of spindle checkpoint and procession from the M to G1 phase. APC/C is composed of several dozen subunits, and its activity is regulated by co-activators Cdc20 or Cdh1 and phosphorylation of constitutive subunits (9). Activity of APC/CCdc20 increases from the prophase to prometaphase, and decrease in the anaphase by Cdc20 degradation (9). However, activity of APC/CCdh1 is maintained from the anaphase to G1/S phase, after which the activity is inhibited by Emi1 (11). Emi1 was identified as a factor inhibiting the function of APC/CCdh1 and is degraded by SCFβTrcp at early M phase (12–15). It recently has been reported that an abnormally high expression of Emi1 protein can be observed in various cancers (14, 16, 17). Moreover, inhibition of Emi1 interferes with progression to M phase by degradation of geminin, which is necessary for the completion of DNA synthesis (18, 19). Emi1-depleted cells show polyploidy and large nuclei because these cells cannot complete DNA synthesis (18, 19). These results suggest that Emi1-depleted cells remain in S phase. As Emi1 depletion interferes with completion of DNA synthesis in cancer cells (18, 19), we speculated that inhibition of Emi1 in cancer cells might enhance the sensitivity of anticancer agents. Moreover, cells lacking Emi1 undergo DNA damage, likely explained by replication stress (20). Therefore, we examined the combined effect of Emi1 knockdown and x-rays. In this study, we also examined the combined effects by one of the major anticancer agents, doxorubicin, and Emi1 depletion in various tumor cells.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Doxorubicin hydrochloride, camptothecin, etoposide, taxol (paclitaxel), and cobalt chloride (CoCl2) were obtained from Sigma. Commercial antibodies were from the following companies: anti-Emi1 and anti-Cul1 antibodies (Zymed Laboratories Inc.); anti-Aurora-A antibody and anti-fypoxia-inducible factor-1α (HIF-1α) (Transduction Laboratories); anti-cyclin A and anti-cyclin B antibodies (Santa Cruz Biotechnology); anti-Cdh1 and anti-Cdc20 antibodies, MBL; anti-E2F1 antibody (Cell Signaling Technology); and anti-β-actin antibody (Sigma). Anti-geminin polyclonal antibody was gift from Dr. Nishitani (University of Hyogo), and anti-TPX2 monoclonal antibody was a gift from Dr. Hans-Jürgen Heidebrecht (University of Kiel). For detection of the immunocomplex, the ECL Western blotting detection system (Amersham Biosciences) was used.
Tissue Samples
Sixty tissue samples of human head and neck squamous cell carcinoma were retrieved from the Surgical Pathology Registry of Hiroshima University Hospital, after approval by the Ethical Committee of Hiroshima University Hospital. Sixty head and neck squamous cell carcinoma cases were surgically resected before radiochemotherapy. Clinical information was gathered from surgical records of the patients. The histological grade of tumor was classified according to the criteria of the Japan Society for Head and Neck Cancer. Tissues were fixed in 10% buffered formalin and embedded in paraffin.
Cell Lines and Culture Conditions
Six head and neck cancer cell lines (HSC2, HSC3, HSC4, Ca9–22, Ho-1-N-1, and Ho-1-U-1), glioma cell line (T98G), and normal human lung fibroblasts (HFLIII) were provided by the Japanese Collection of Research Bioresources Cell Bank. HOC119, HOC313, HOC621, HOC719-PE, HOC719-NE, TSU, OM-1, and ZA cells have been described previously (21). Breast cancer cell line (MCF-7), colon cancer cell line (RKO and HCT116), osteosarcoma cell line (SaOS-2), and human breast epithelial cell line (MCF-10A) were purchased from ATCC. HaCaT cell line was obtained from N. E. Fusenig (German Cancer Research Center). The p53-null HCT116 cell line was obtained from B. Vogelstein (The Johns Hopkins University). Cells were maintained in RPMI 1640 medium or Dulbecco's modified Eagle's medium (Nissui Pharmaceutical Co., Tokyo, Japan) supplemented with 10% heat-inactivated FBS (Invitrogen) and 100 units/ml penicillin-streptomycin (Invitrogen) under conditions of 5% CO2 in air at 37 °C. Upon receiving the cell lines, they were immediately cultured and expanded to prepare frozen ampule stocks. Cells were passaged for no more than 2 to 3 months before establishing new culture from early-passage frozen ampules.
RT-PCR
Using the RNeasy mini kit (Qiagen, Hilden, Germany), total RNA from cultures of confluent cells and tissues was isolated. These isolates were quantified and their purity was evaluated by spectrophotometer. The cDNA was synthesized from 1 μg of total RNA according to ReverTra Dash (Toyobo Biochemicals, Tokyo, Japan). We used the following primers: human Emi1, 5′-tctttcgaaggggactcaga-3′ (forward) and 5′-tctggttgaagcatgaggtg-3′ (reverse); human GAPDH, 5′-tccaccaccctgttgctgta-3′ (forward) and 5′-accacagtccatgccatcac-3′ (reverse). Aliquots of total cDNA were amplified with 1.25 units of rTaq-DNA polymerase (Qiagen), and this amplification was done in a thermal cycler (MyCyler, Bio-Rad) for 30 cycles after initial 30 s of denaturation at 94 °C, annealing for 30 s at 60 °C, and extension for 1 min at 72 °C in all primers used. The amplification reaction products were resolved on 1.2% agarose/TAE gels (Nacalai Tesque), electrophoresed at 100 mV, and then finally visualized by using ethidium bromide.
Immunohistochemistry
Tumor tissues were fixed in 10% formalin, embedded in paraffin, and cut 4 μm thick. For immunohistochemical staining, tissue sections were deparaffinized in xylene and rehydrated in descending grades of ethanol. Endogenous peroxidase activity was blocked with methanol containing 0.3% H2O2 for 30 min. Antigen retrieval was done by the microwaving using a citrate phosphate buffer (pH 6.0), and then the sections were incubated with the primary antibody at 4 °C overnight. Immunohistochemical staining was carried out by a polyclonal anti-Emi1 antibody (Zymed Laboratories, Inc.). For detection of the reaction after incubation with secondary antibodies, we used diaminobenzidine (DAKO, Glostrup, Denmark). The sections were counterstained by hematoxylin and dehydrated in ascending grades of ethanol, and the slides were mounted. For evaluation of staining intensity, we graded + (weak/focal immunopositivity), ++ (strong/focal immunopositivity and weak/diffuse immunopositivity), +++ (strong/diffuse immunopositivity). Then, the expression of Emi1 was graded as high (>30% of tumor cells showing ++ or +++ intensity) or low (no staining or <30% of tumor cells showing + intensity).
Western Blot Analysis
Western blotting was carried out as described previously (15). The protein concentrations were measured by Bradford protein assay (Bio-Rad). Twenty μg of protein was subjected to 10% polyacrylamide gel electrophoresis, followed by electroblotting onto a nitrocellulose filter. For detection of the immunocomplex, the ECL Western blotting detection system (Amersham Biosciences) was used.
Cell Proliferation
For proliferation assay, 5000 cells were plated onto a 24-well plate, and the cells were allowed to grow and expand. The cells were then trypsinized and counted at 0, 2, 4, and 6 days by using cell counter (Coulter Z1, Beckman-Coulter). Doubling time was calculated by rate of cell proliferation.
Mitotic Index
Cells were seeded on glass coverslips. Forty eight hours after incubation, cells were fixed, permeabilized, and stained with pSer-10 histone H3 (Millipore) and DAPI. pSer-10 histone H3-positive and condensed chromatin cells were counted as mitotic cells. More than 400 cells were scored per condition.
RNA Interference
Logarithmically growing cells were seeded at a density of 105 cells/6-cm dish and transfected with oligonucleotides at 24 h after replating by using Oligofectamine (Invitrogen). Forty-eight hours after transfection, lysates were prepared and analyzed by SDS-PAGE and immunoblotting. The siRNA is a 19-bp duplex oligoribonucleotide with a sense strand corresponding to nucleotides 1446–1464 of the human Emi1 mRNA sequence; 5′-GAUUGUGAUCUCUUAUUAA-3′ (Dharmacon, Chicago, IL). Human cyclin A siRNA (sc-29282) and human E2F1 siRNA (sc-61861) were obtained from Santa Cruz Biotechnology. The negative control (siTrio negative control siRNA) was obtained from Cosmo Bio, Inc. (Tokyo, Japan).
Immunofluorescence
Cells grown on cover slips were fixed in 4% paraformaldehyde for 10 min at room temperature, rinsed three times with ice-cold PBS, and then permeabilized in 0.1% Triton X-100 in PBS for 15 min at room temperature. After rinsing three times with PBS, coverslips were incubated with DAPI staining. Immunostaining of cell preparations was recorded using an epifluorescence Zeiss Axioplan 2 (Zeiss, Inc., Thorwood, NY) microscope attached to a charge-coupled device camera.
For staining with γH2AX, cells were fixed in 4% paraformaldehyde for 10 min. Cells were then permeabilized for 2 min in 0.2% Triton X-100 (Sigma) in PBS and washed twice in PBS. Antibodies were diluted with 4% w/v BSA in PBS. Cells were incubated with mouse anti-γH2AX antibody (Millipore) for 1 h at 37 °C, washed three times in PBS, and incubated with FITC-conjugated rabbit anti-mouse IgG antibody (Sigma) for 1 h at room temperature. Slides were incubated in PBS containing DAPI for 5 min to stain the DNA and mounted using Vectashield (Vector Laboratories). Images were captured using an Olympus DP70 fluorescence microscope. Cell scoring was carried out blindly with >200 cells/sample (× two individual experiments).
Flow Cytometry
Cell cycle distribution was determined by DNA content analysis after propidium iodide (PI) staining (Sigma). Cells were cultured as described above, fixed in 70% ethanol, and stored at 4 °C before analysis. Flow cytometric determination of DNA content was performed on a FACS Calibur (Becton-Dickinson, San Jose, CA) flow cytometer. For each sample, 20,000 events were stored.
For flow cytometric analysis of γH2AX, cells were trypsinized, washed with PBS, and fixed for 10 min in 4% (w/v) paraformaldehyde and post-fixed in 70% ethanol for at least 2 h at −20 °C. After two washes with PBS, cells were incubated for 10 min on ice in hybridization buffer (PBS containing 0.1% BSA and 0.1% Triton X-100). After centrifugation, cells were hybridized with mouse anti-γH2AX antibody (Millipore) diluted at a ratio of 1:400 in hybridization buffer and incubated for 2 h in the dark at room temperature. Cells were washed with hybridization buffer and incubated with FITC-conjugated rabbit anti-mouse IgG antibody (Sigma) for 1 h at room temperature. Cells were then rinsed twice with PBS containing 0.1% Triton X-100 and stained with PI solution (PBS containing 5 μg/ml PI, 100 μg/ml RNase A) for 30 min before flow cytometry.
5-Ethynyl-2′-deoxyuridine (EdU) Labeling
For the labeling of DNA synthesis, cells were cultured with 10 μm EdU for 1 h. EdU-incorporated cells were detected by using the Click-it® EdU imaging kit (Invitrogen). EdU staining was conducted following the manufacturer's instructions. Briefly, cells were fixed with paraformaldehyde for 15 min, washed once with PBS, permeabilized with 0.2% Triton X-100 buffer for 10 min, washed once, treated with the click-reaction mixture for 30 min, washed once, and resuspended in buffer before staining of DNA content with PI solution.
Annexin V and PI Dual-staining Assay
After doxorubicin treatment, the cells were then stained with phycoerythrin-conjugated annexin V and 7-aminoactinomycin D, using the phycoerythrin-annexin V apoptosis detection kit (BD Pharmingen) according to the manufacturer's instructions. Apoptotic cells were identified by dual-staining with recombinant phycoerythrin-conjugated with annexin V and 7-aminoactinomycin D. Data acquisition and analysis were done in a FACS Calibur (Becton-Dickinson) flow cytometer using CellQuest software.
Radiation Exposure and Colony Formation Assay
Sensitivity to radiation was evaluated by colony formation assays. Cells were irradiated with x-rays (1.2 gray/min) generated by a TITAN x-ray irradiator (Shimadzu, Japan) at room temperature. After irradiation, cells were trypsinized and plated onto 100-mm culture dishes at appropriate cell density. After incubated for 10–14 days, cells were rinsed with PBS, fixed with 100% ethanol, and stained by crystal violet. Colonies consisting of >50 cells were scored as survivor.
Statistical Analysis
Statistical analyses were performed using STATVIEW (Abacus Concepts, Piscataway, NJ), and a Fidher protected least significant difference multiple-comparison test was used for all of the analyses in this study. Overall differences between the groups were determined by one-way analysis of variance, and Fisher protected least significant difference multiple comparison test was applied when the results of one-way analysis of variance were significant (p < 0.05).
RESULTS
High Expression of Emi1 Is Frequently Observed in Cancer Tissues
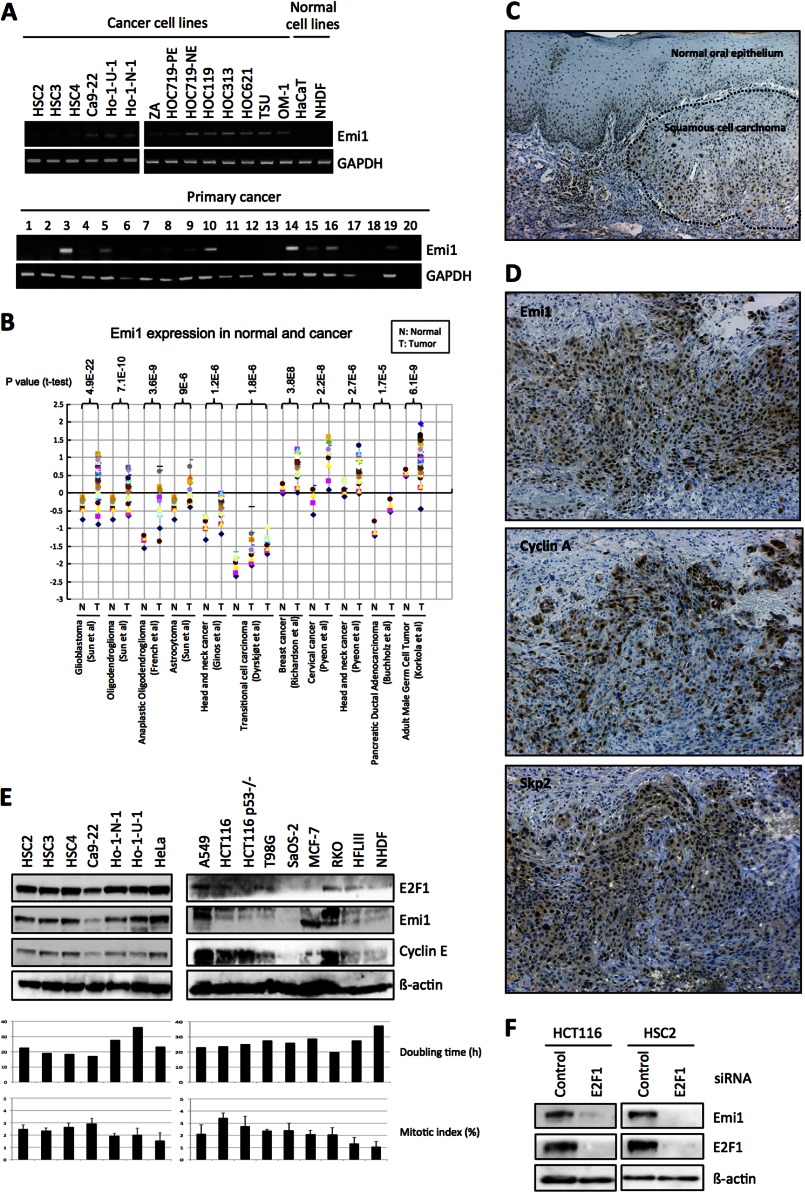
Recent reports show that high expression of Emi1 protein can be observed in various cancers (14, 16, 17). We also found Emi1 mRNA overexpression in head and neck cancer cells (Fig. 1A). Moreover, we investigated Emi1 expression in various tumor tissues by using the Oncomine database. As compared with normal counterparts, Emi1 expression was significantly increased in many types of tumors, such as head and neck, breast, prostate, cervix, pancreas, and brain tumors (Fig. 1B). Then, we examined the expression of Emi1 protein in head and neck cancer cases by immunohistochemistry. Emi1 expression was graded as high (over 30% of tumor cells showing ++ or +++ intensity) or low (no staining or less than 30% of tumor cells showing + intensity). Indeed, most cases with high expression of Emi1 showed strong and diffuse immunopositivity and most cases with low expression of Emi1 showed no or weak immunopositivity. Expression level of Emi1 in cancer cells was much higher than that in non-neoplastic cells (Fig. 1C). High Emi1 expression was observed in 53.3% of head and neck cancer cases and was well correlated with histological differentiation and lymph node metastasis (Table 1). Moreover, we compared the expression of Emi1 with p53 and APC/C substrates, cyclin A, Skp2, and Aurora-A in cancer tissues (Fig. 1D and Table 1). Interestingly, Emi1 expression was well correlated with p53 expression and APC/C substrates (Table 1).
FIGURE 1.
Emi1 overexpression in head and neck cancer. A, Emi1 mRNA expression was examined in 14 head and neck cancer cell lines, 2 normal cells, and 20 head and neck cancer tissues by RT-PCR. GAPDH was used as a control. B, mRNA level of Emi1 in different human tumors. All data were provided by the Oncomine database. Data from Refs. 43–50 were reanalyzed to show expression level of Emi1 in normal brain, glioblastoma, oligodendroglioma, anaplastic oligodendroglioma, astrocytoma, head and neck cancer, breast cancer, pancreatic ductal adenocarcinoma, cervical cancer, transitional cell carcinoma, and adult male germ cell tumor (43–50). N, normal tissues; T, tumor tissues. C, immunohistochemical expression of Emi1 in head and neck cancer. D, correlation between Emi1 expression and APC substrates, cyclin A, and Skp2 expression. We examined the expression of Emi1, cyclin A, and Skp2 in head and neck cancer cases. A representative head and neck cancer case is shown. E, high expression of Emi1 is regulated by E2F1 in cancer cells. Expression of Emi1, E2F1, and cyclin E was examined by Western blot analysis in cancer cell lines (HSC2, HSC3, HSC4, Ca9–22, Ho-1-N-1, Ho-1-U-1, HeLa, A549, HCT116, HCT116 p53−/−, T98G, SaOS-2, MCF-7, and RKO) and normal cells (HFL III and NHDF). β-Actin expression was used as a loading control. The graph shows the mitotic index (%) and doubling time (h) in each cell. F, E2F1 siRNA was transfected into HCT116 and HSC2 cells. After 48 h of E2F1 siRNA treatment, cells were collected. Emi1 and E2F1 expression was examined by Western blot analysis. β-Actin expression was used as a loading control.
TABLE 1.
Summary of Emi1 expression in head and neck cancer
| Total | Emi1 expression |
p value | ||
|---|---|---|---|---|
| Low | High | |||
| Head and neck cancer | 60 | 28 (46.7%) | 32 (53.3%) | |
| Histological differentiation | ||||
| Well | 11 | 8 (72.7%) | 3 (27.3%) | 0.02 |
| Moderate | 37 | 18 (48.6%) | 19 (51.4%) | 0.02 |
| Poor | 12 | 2 (16.7%) | 10 (83.3%) | 0.02 |
| Lymph node metastasis | ||||
| Negative | 40 | 23 (57.5%) | 17 (42.5%) | 0.02 |
| Positive | 20 | 5 (25.0%) | 15 (75.0%) | 0.02 |
| Cyclin A expression | ||||
| Low | 27 | 21 (77.8%) | 6 (22.2%) | 0.00001 |
| High | 33 | 7 (21.2%) | 26 (78.8%) | 0.00001 |
| Skp2 expression | ||||
| Low | 25 | 15 (60.0%) | 10 (40.0%) | 0.08 |
| High | 35 | 13 (37.1%) | 22 (62.9%) | 0.08 |
| Aurora-A expression | ||||
| Low | 27 | 19 (70.3%) | 8 (29.7%) | 0.0008 |
| High | 33 | 9 (27.3%) | 24 (72.7%) | 0.0008 |
| p53 expression | ||||
| Low | 19 | 13 (68.4%) | 6 (31.6%) | 0.02 |
| High | 41 | 15 (36.6%) | 26 (63.4%) | 0.02 |
As Emi1 itself is an E2F target gene, Emi1 mRNA overexpression might be caused by activated E2F in cancer cells (20). Indeed, Emi1 expression was well correlated with the E2F1 and E2F1 target gene, cyclin E in cancer cell lines, and normal cells (Fig. 1E). However, Emi1 expression was not correlated with doubling time and mitotic index (Fig. 1E). Accordingly, E2F1 knockdown reduced Emi1 expression in cancer cells (Fig. 1F).
Emi1 Knockdown Induces Polyploidy in Cancer Cells
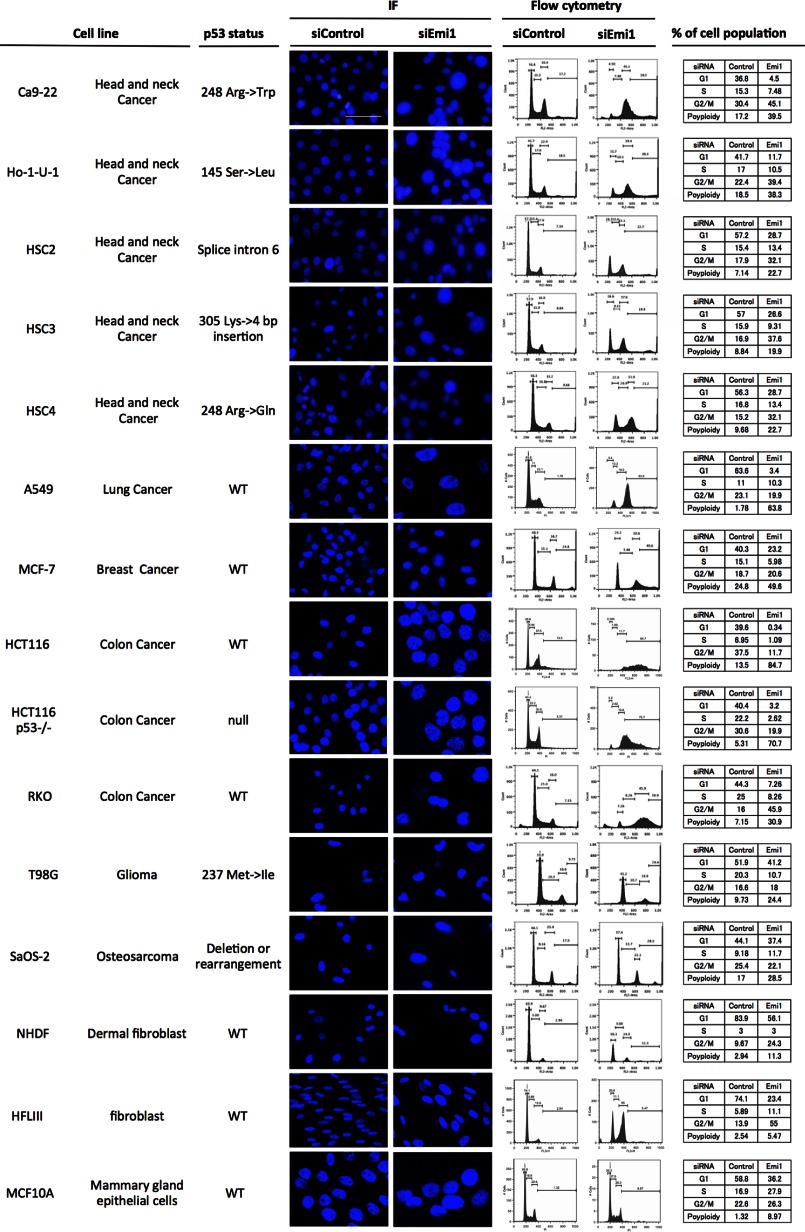
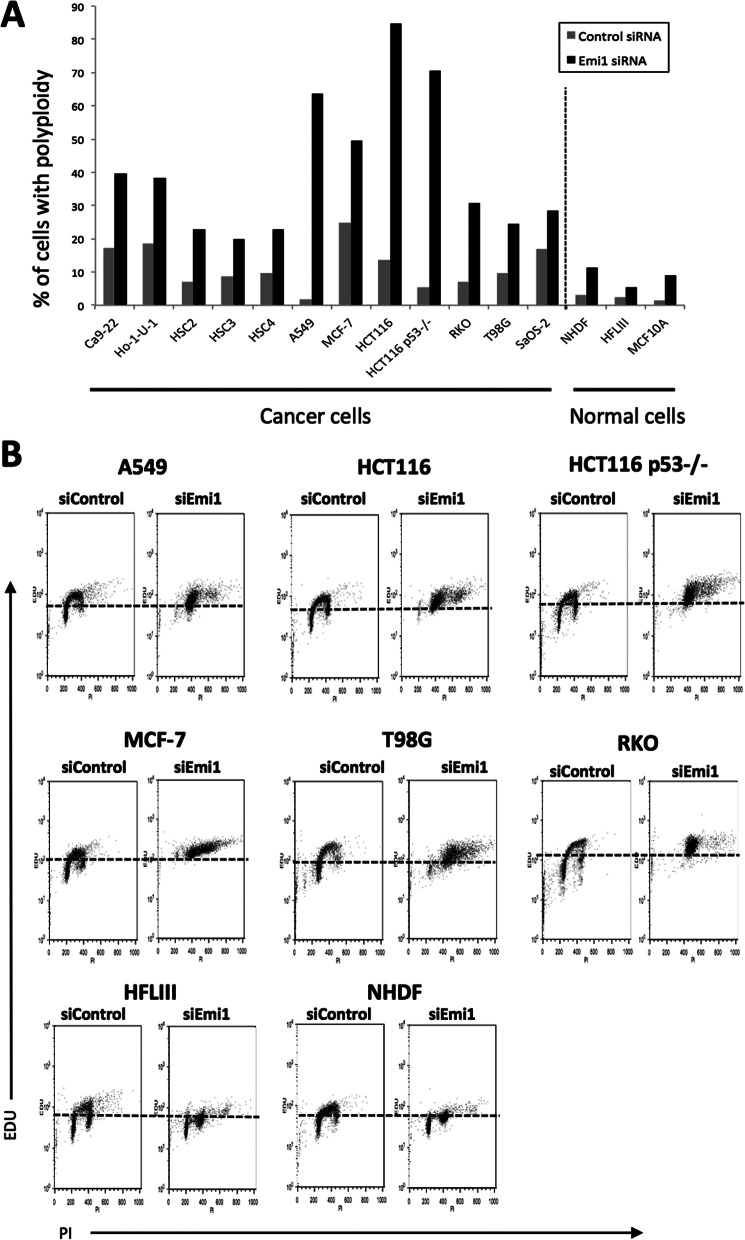
As shown in Fig. 1, Emi1 was frequently overexpressed in various types of cancer. Next, we examined the effect of Emi1 knockdown by using siRNA in various types of cancer cell lines. Previous reports showed that Emi1 depletion induced polyploidy and large nuclei because cells cannot complete DNA synthesis (18, 19). Similar to previous reports, we confirmed the effect of Emi1 knockdown on polyploidy and large nuclei in various cancer cell lines, including head and neck cancer (Ca9–22, Ho-1-U-1, HSC2, HSC3, and HSC4), lung cancer (A549), breast cancer (MCF-7), colon cancer (HCT116, HCT116 p53−/−, and RKO), glioma (T98G), and osteosarcoma (SaOS-2) (Fig. 2). Larger nuclei and increased number of cells with polyploidy were observed in all cancer cells after Emi1 siRNA treatment. Moreover, the efficiency of Emi1 knockdown was independent of p53 mutation status (Fig. 2). As shown in Fig. 3A, higher percentage of cells with polyploidy was observed in cancer cells, in comparison with that in normal cells. Indeed, Emi1 siRNA-treated cancer cells but not normal cells showed positive EDU (Fig. 3B), indicating that Emi1 depletion induced polyploidy through rereplication in cancer cells.
FIGURE 2.
Emi1 knockdown in various cancer cell lines. Emi1 siRNA was transfected into various cancer cell lines, including head and neck cancer (Ca9–22, Ho-1-U-1, HSC2, HSC3, and HSC4), lung cancer (A549), breast cancer (MCF-7), colon cancer (RKO, HCT116, and HCT116 p53−/−), glioma (T98G), and osteosarcoma (SaOS-2). Em1i siRNA was also transfected into normal cells, including NHDF (dermal fibroblast), HFL III (fibroblasts), and MCF10A (mammary gland epithelial cells). The status of p53 was shown in each cell line. Cells were stained with DAPI to visualize the nuclei. Cell cycle distribution was determined by DNA content analysis after PI staining using a flow cytometer. Scale bar, 100 μm. IF, immunofluorescence.
FIGURE 3.
Emi1 knockdown induced polyploidy in various cancer cell lines. A, the graph shows the percentage of cells with polyploidy in various cells. Percentage of cells with polyploidy was analyzed by a flow cytometer as shown in the figure. B, EdU staining was examined in various cells (A549, HCT116, HCT116 p53−/−, MCF-7, T98Gm RKO, HFLIII, and NHDF) by a flow cytometer after control siRNA or Emi1 siRNA treatment.
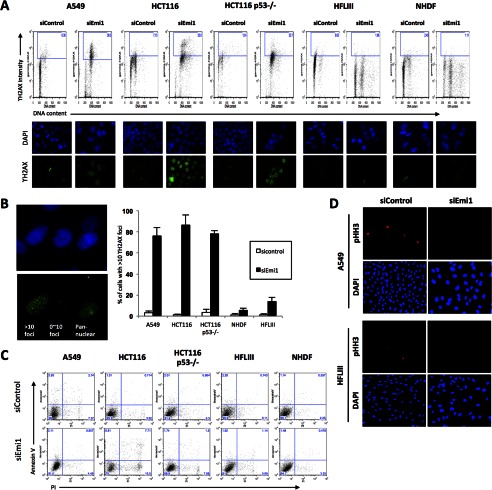
It has been shown that cells lacking Emi1 undergo DNA damage, likely explained by replication stress upon deregulated cyclin E- and A-associated kinase activities (20). Here, we examined the nuclear foci containing phosphorylated histone H2AX (γ-H2AX), a marker of DNA damage foci in Emi1 depleted cancer cells (A549, HCT116, and HCT116 p53−/−) and normal cells (HFLIII and neonatal normal human dermal fibroblast (NHDF)). Increased γ-H2AX foci and DNA content were observed in cancer cells, but not in normal cells (Fig. 4A). In particular, frequency of high levels of γH2AX foci (more than 10 foci per cell) in cancer cells was significantly higher than that in normal cells (Fig. 4B). Most cells did not show annexin V after Emi1 siRNA treatment (Fig. 4C). As the expression of phosphohistone H3 was not observed in Emi1 siRNA-treated cells (Fig. 4D), the elevated level of γH2AX was not caused by arresting at M phase or induction of apoptosis. These findings show that replication stress caused by Emi1 knockdown might lead to DNA damage.
FIGURE 4.
DNA damage in Emi1 depleted cancer cells. A, Emi1 knockdown induce γH2AX foci in tumor cells. Tumor cells (A549, HCT116, and HCT116 p53−/−) and normal cells (HFLIII and NHDF) were transfected with the indicated siRNA. After 48 h the transfection, cells were fixed and stained with anti-γH2AX antibodies. The levels of H2AX phosphorylation were detected using by the flow cytometer. Bivariate distributions representing expression of γH2AX versus DNA content of tumor and normal cells are shown. The levels of H2AX phosphorylation were also detected using by the fluorescence microscope. Representative images of γH2AX immunostaining (green) and nuclear staining (blue) in tumor or normal cells are shown. B, images on the left show typical images of cells presenting low (0∼10 foci/cell) or high levels of γH2AX foci or showing pan-nuclear staining γH2AX. The right graph indicates percentages of cells with high level (>10 foci/cell) of γH2AX foci. Results presented are the means of two independent experiments ± S.D. C, apoptosis was examined by annexin V staining using a flow cytometer in various cells (A549, HCT116, HCT116 p53−/−, HFLIII, and NHDF) after control siRNA or Emi1 siRNA treatment. D, expression of phosphohistone H3 (pHH3) was examined by the fluorescence microscope in A549 and HFLIII cells after control siRNA or Emi1 siRNA treatment. Cells were also stained with DAPI to visualize the nuclei.
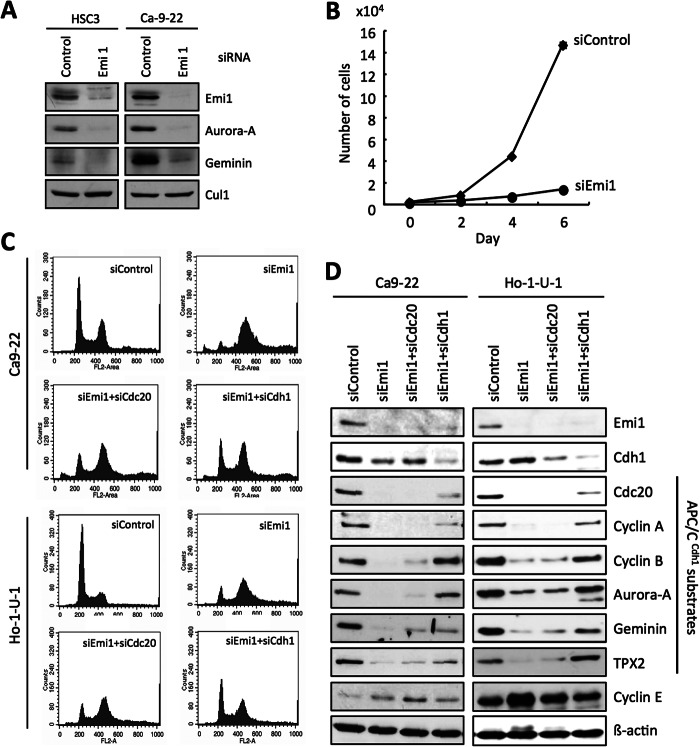
Next, we examined the detailed mechanism of Emi1 knockdown in Ca9–22 and Ho-1-U-1 cells. Emi1 is known as an inhibitor of APC/CCdc20 and APC/CCdh1 (12–15) and inhibition of Emi1 interferes with progression to the M phase by the APC/CCdh1-dependent proteolysis of geminin protein (18, 19). Indeed, expression of Emi1 and APC/CCdh1 substrates such as Aurora-A and geminin was down-regulated by Emi1 siRNA treatment in both cells (Fig. 5A). Moreover, Emi1 knockdown significantly suppressed proliferation (p < 0.05) (Fig. 5B). Then, we examined the co-depletion of Emi1 and Cdh1 or Cdc20 in Ca9–22 and Ho-1-U-1 cells. Cdh1 depletion corrected Emi1 knockdown-induced polyploidy to a certain extent (Fig. 5C). As expected, Emi1 knockdown induced down-regulation of APC/CCdh1 substrates such as Cdc20, cyclin A, cyclin B, Aurora-A, geminin, and TPX2 (Fig. 5D). Cdh1 depletion up-regulated these APC/CCdh1 substrates (Fig. 5D). Thus, as shown in previous reports (18, 19), Emi1 depletion induced rereplication because of geminin down-regulation via APCCdh1-dependent proteolysis.
FIGURE 5.
DNA damage and polyploidy by Emi1 knockdown via activation of APC/CCdh1. A, Emi1 siRNA was transfected into HSC3 or Ca9–22 cells. After 48 h of Emi1 siRNA transfection, cells were collected. The indicated proteins were examined by Western blotting. B, Emi1 siRNA inhibits cell growth in head and neck cancer cells. Cell growth of Ho-1-U-1 cells after Emi1 siRNA transfection. After 48 h of transfection, cells were plated in 24-well plates. Cells were counted by Cell Counter at 0, 2, 4, and 6 days. We assayed three times. C, Emi1 siRNA induces polyploidy through activation of APC/C. Co-depletion of Cdh1 or Cdc20 with Emi1 by siRNA in Ca9–22 and Ho-1-U-1 cells. These cells were transfected with the indicated siRNAs. Cell cycle distribution was determined by DNA content analysis after PI staining using a flow cytometer. D, co-depletion of Cdh1 or Cdc20 with Emi1 by siRNA in Ca9–22 and Ho-1-U-1 cells. The indicated proteins including APC/CCdh1 substrates in siRNA-treated cells were examined by Western blotting.
Emi1 Knockdown Enhances the Induction of Apoptosis by Doxorubicin in Human Cancer Cells, but Not in Normal Cells
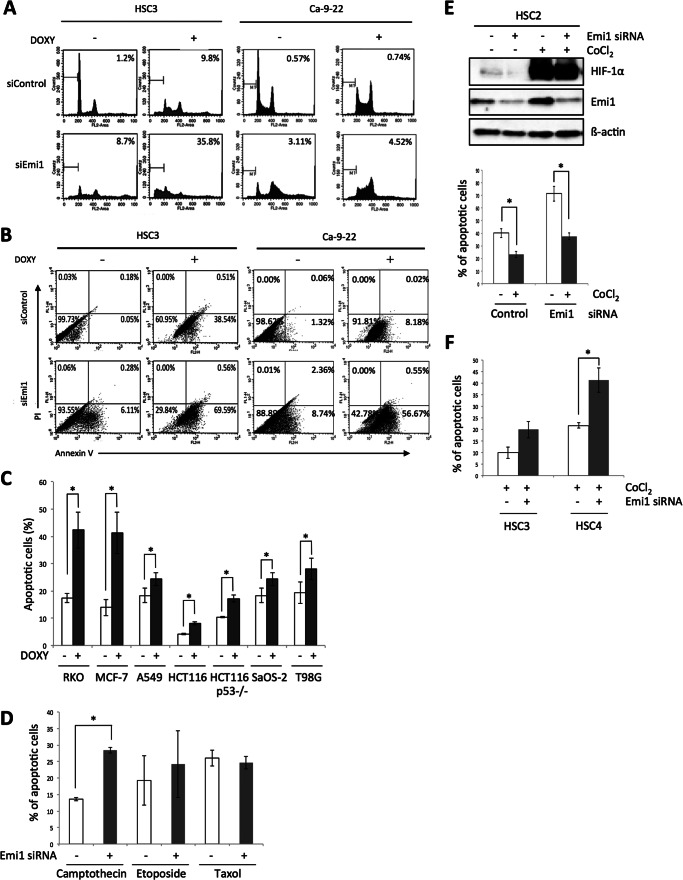
Many anticancer agents used in cancer therapy are intended to control DNA synthesis. Our observations made us hypothesize that Emi1 depletion in cancer cells might enhance the induction of apoptosis by anticancer agents. We treated doxorubicin after Emi1 knockdown in HSC3 and Ca9–22 cells. Doxorubicin treatment induced higher number of subG1 cells in Emi1 depleted cells in comparison with control cells (Fig. 6A). Annexin V-positive cells were observed in 38.5% of control (doxorubicin treatment alone) HSC3 cells and 69.59% of Emi1-depleted HSC3 cells (Fig. 6B). Annexin V-positive cells were observed in 8.18% of control (doxorubicin treatment alone) Ca9–22 cells and in 56.67% of Emi1-depleted Ca9–22 cells (Fig. 6B). We also examined the induction of apoptosis by doxorubicin treatment in various cancer cell lines (RKO, MCF-7, A549, HCT116, HCT116 p53−/−, SaOS-2, and T98G) after Emi1 depletion. In all cancer cells, the rate of apoptosis was significantly increased by Emi1 knockdown in combination with doxorubicin treatment (Fig. 6C). In this study, we used a common anticancer drug, doxorubicin. We first examined the induction of apoptosis by other anticancer drugs, camptothecin, etoposide, and Taxol in HSC3 cells after Emi1 depletion (Fig. 6D). Camptothecin is the prototypic DNA topoisomerase I inhibitor, and etoposide is the topoisomerase II inhibitor. Synergistic effect of Emi1 knockdown in combination with camptothecin and etoposide was observed in HSC3 cells (Fig. 6D). Taxol stabilizes microtubules and, as a result, interferes with the normal breakdown of microtubules during cell division. Distinct from other anticancer agents, Taxol does not affect DNA synthesis. As we expected, synergistic effect of Emi1 knockdown in combination with Taxol was not observed (Fig. 6D).
FIGURE 6.
Synergistic anticancer effect of Emi1 knockdown in combination with doxorubucin in cancer cells. A, after 48 h of Emi1 siRNA transfection, cells were treated with doxorubicin (0.5 μg/ml) for 12 h. Cells were fixed in 70% ethanol. Cell cycle distribution was determined by DNA content analysis after PI staining using a flow cytometer. For each sample, 20,000 events were stored. Percentage of the sub-G1 population is indicated. Representative data are shown. We performed three independent experiments. B, flow cytometric analysis of annexin V and PI staining in control and Emi1 siRNA-treated HSC3 or Ca9–22 cells after treatment with doxorubicin (0.5 μg/ml) for 12 h. We performed three independent experiments. C, synergistic anticancer effect of Emi1 knockdown in combination with doxorubucin (DOXY) in various cancer cells. Graph shows the average percentage of apoptotic cells in various cancer cells after treatment with doxorubicin (0.5 μg/ml) for 12 h. In this study, various cancer cell lines including lung cancer (A549), breast cancer (MCF-7), colon cancer (RKO, HCT116, and HCT116 p53−/−), glioma (T98G), and osteosarcoma (SaOS-2) were used. We performed three independent experiments. *, p < 0.05. D, synergistic anticancer effect of Emi1 knockdown in combination with camptothecin and etoposide but not with Taxol in HSC3 cells. The graph shows the average percentage of apoptotic cells in HSC3 cells after treatment with camtothecin (2 μm), etoposide (1 μm), or Taxol (1 nm) for 12–24 h. We performed two independent experiments. *, p < 0.05. E, hypoxic condition induced drug resistance. CoCl2 (100 μm) was treated for 6 h in HSC2 cells after 24 h of control or Emi1 siRNA transfection. Cells were collected and expression of HIF-1α and Emi1 was examined by Western blot analysis. β-Actin expression was used as a loading control. After 6 h of CoCl2 treatment, doxorubicin (DOXY; 0.5 μg/ml) was treated for 12 h in control or Emi1-depleted cells. Graph shows the average percentage of apoptotic cells (annexin V-positive cells) in HSC2 cells after treatment with doxorubicin for 12 h under normal or hypoxia condition (CoCl2 treatment). We performed two independent experiments. *, p < 0.05. F, synergistic anticancer effect of Emi1 knockdown in combination with doxorubicin in HSC3 and HSC4 cells under hypoxia conditions. The graph shows the average percentage of apoptotic cells (annexin V-positive cells) in HSC3 and HSC4 cells after treatment with doxorubicin for 12 h under normal or hypoxia conditions (CoCl2 treatment). We performed two independent experiments. *, p < 0.05.
Recent study showed that the hypoxic condition induced multidrug resistance (22). To mimic hypoxia conditions, we used cobalt chloride (CoCl2). CoCl2 treatment up-regulated HIF-1α in HSC2 cells (Fig. 6E). Under hypoxia conditions, we treated doxorubicin after Emi1 knockdown. Indeed, decreased number of annexin V-positive cells was observed in both control siRNA and Emi1 siRNA-treated cells under hypoxia conditions (Fig. 6E). Interestingly, a higher number of annexin V-positive cells was significantly observed in Emi1-depleted cells, in comparison with control cells under hypoxia conditions (Fig. 6, E and F).
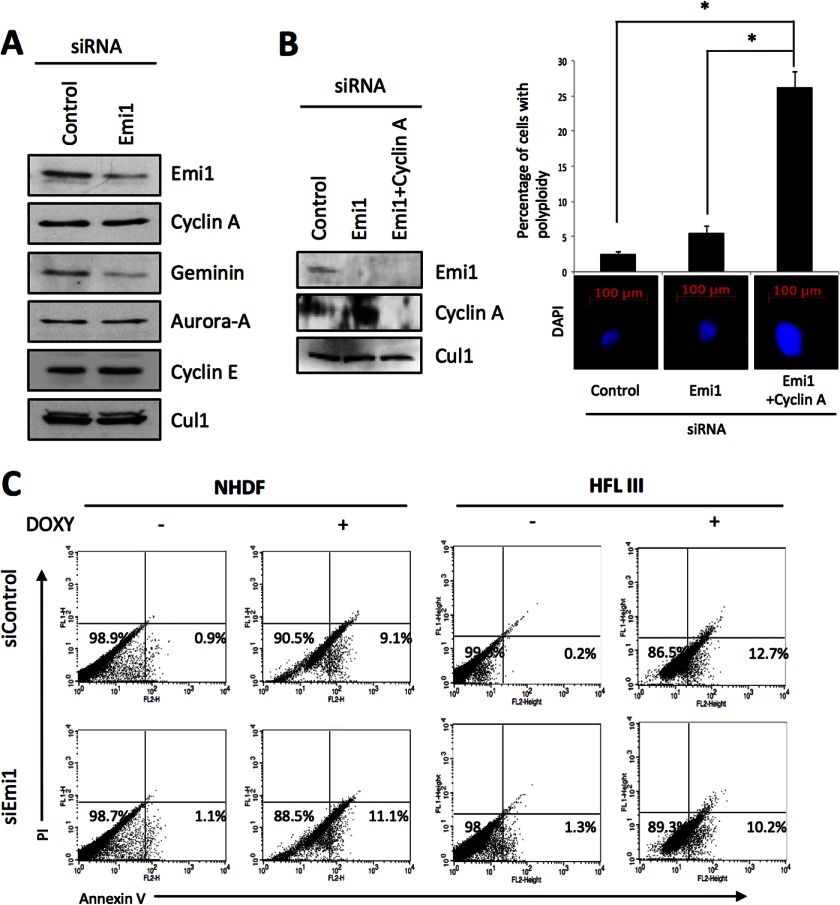
To find out the effect of Emi1 depletion in doxorubicin-treated normal cells, we examined the induction of apoptosis by doxorubicin treatment in normal fibroblasts (NHDF and HFL III) and mammary gland epithelial cells (MCF10A) after Emi1 depletion. Large nuclei were observed in HFL III cells, but not in NHDF and MCF10A cells (Fig. 2). Increased G2/M cell population was observed in HFL III cells (Fig. 2). In all normal cells, the effect of Emi1 knockdown on polyploidy was not observed (Fig. 2). As we checked the reduced expression of Emi1 after Emi1 knockdown in these cells by Western blot analysis, less effect of Emi1 knockdown on polyploidy and large nuclei was not caused by the transfection efficiency of Emi1 siRNA. In normal cells, rereplication is known to be induced by co-depletion of geminin and cyclin A (23). Moreover, both Cyclin A and geminin are required for suppression of over-replication and for genome stability in Drosophila cells (24). In this study, down-regulation of cyclin A was not observed in NHDF cells after Emi1 depletion (Fig. 7A). Although previous reports showed that accumulation of cyclin E was induced by Emi1 knockdown through activation of the APC/C-independent pathway (18, 19), up-regulation of cyclin E was not observed in NHDF cells (Fig. 7A). Then, we depleted both Emi1 and cyclin A in NHDF cells. As shown in previous reports (23, 24), knockdown of both Emi1 and cyclin A induced polyploidy and large nuclei (Fig. 7B). Interestingly, the synergistic effect of Emi1 knockdown on the induction of apoptosis by doxorubicin treatment was not observed in NHDF and HFL III cells (Fig. 7C).
FIGURE 7.
Synergistic anticancer effect of Emi1 knockdown in combination with doxorubucin in normal cells. A, Emi1 or control siRNA was transfected into NHDF cells, and cells were collected after 48 h. The indicated proteins in siRNA-treated NHDF cells were examined by Western blotting. B, Emi1 siRNA and/or cyclin A siRNA were transfected into NHDF cells. The left panel shows the expression of Emi1 and cyclin A examined by Western blot analysis after 48 h of siRNA transfection. β-Actin expression was used as a loading control. The right panel shows DAPI staining and percentage of cells with polyploidy after Emi1 siRNA and/or cyclin A siRNA transfection. Cells were stained with DAPI to visualize the nuclei, and percentage of cells with polyploidy was determined by DNA content analysis after PI staining using a flow cytometer. *, p < 0.05. C, flow cytometric analysis of annexin V and PI staining in control and Emi1 siRNA treated NHDF or HFL III cells after treatment with doxorubucin (DOXY; 0.5 μg/ml) for 12 h. We performed three independent experiments.
Emi1 Knockdown Enhances Radiosensitivity in Human Cancer Cells, but Not in Normal Cells
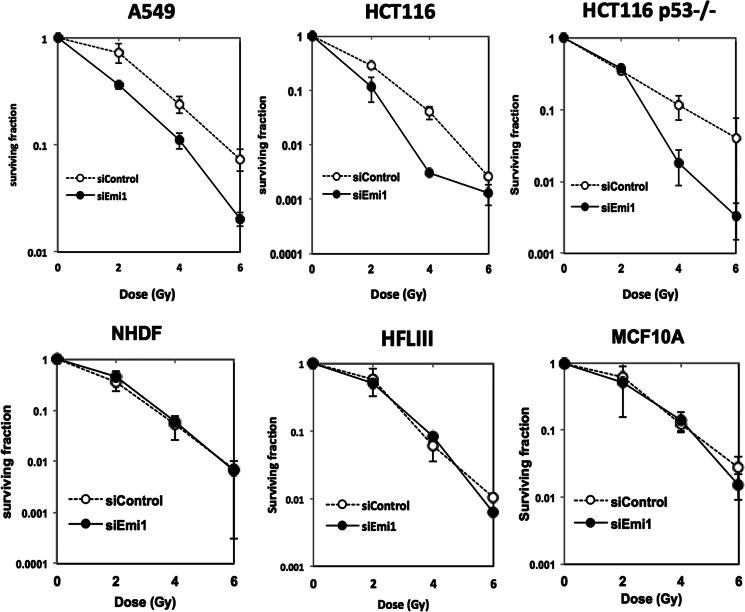
As shown in Fig. 4, Emi1 depletion induced DNA damage in cancer cells, but not in normal cells. Ionizing radiation works by damaging DNA of exposed tissue, leading to cellular death. Therefore, we examined whether Emi1 knockdown affects radiosensitivity in cancer and normal cells. We treated Emi1 siRNA in cancer cells (A549, HCT116 wild type, and HCT116 p53−/−) and normal cells (NHDF, HFL III, and MCF10A) and then exposed these cells to x-rays. Emi1 knockdown markedly increased the cellular sensitivity to ionizing radiation in these tumor cell lines (A549 and HCT116 p53 wild type) (Fig. 8). In HCT116 p53−/− cells, the survival ratio at low dose (2 gray) showed no significant change after Emi1 knockdown, whereas a clear radiosensitization effect was observed by Emi1 knockdown at 4–6 gray (Fig. 8). On the other hand, Emi1 knockdown did not affect radiosensitivity at all in normal cells (Fig. 8).
FIGURE 8.
Synergistic anticancer effect of Emi1 knockdown in combination with radiation. The effects of Emi1 knockdown on cell survival of A549, HCT116 wild type, and HCT116 p53−/−, HFLIII, NHDF, and MCF10A cells. Cells were transfected with Emi1 siRNA or negative control siRNA. After 48 h, cells were exposed to 0–6 gray x-rays. Cell survival was determined by a colony formation assay. Data shown are mean and S.E. of three independent experiments.
DISCUSSION
We found that Emi1 knockdown inhibited cell proliferation through induction of rereplication and enhanced the sensitivity of anticancer reagents and x-ray-treated cancer cells, but normal cells were not sensitized with Emi1 depletion. Interestingly, Emi1 was found to be up-regulated in several human tumors (11, 16, 17). APC/C inactivation by Emi1 leads to accumulation of APC/C substrates for progression of S phase and early mitosis (11, 14). Lehman et al. (16) found frequent overexpression of APC/C substrates (securin, Plk1, Aurora-A, and Skp2) and Emi1 in a large number of malignant tumors by immunohistochemical staining of tissue microarrays. Importantly, several or all of the APC/C substrates clustered together in various tumors with elevated Emi1 protein (16). Moreover, Emi1 overexpression leads to unscheduled cell proliferation, tetraploidy, and chromosomal instability in p53-deficient cells (25). Notably, Emi1 is the target of E2F transcription factors (11), potentially linking the frequent deregulation of the E2F pathway to APC/C deregulation. This observation is supported by our findings that (i) Emi1 expression was well correlated with E2F1 expression in cancer cells (Fig. 1E), and (ii) E2F1 knockdown down-regulated Emi1 expression (Fig. 1F). We also confirmed that Emi1 is frequently overexpressed in various cancers, including head and neck cancers, and that Emi1 expression is well correlated with Aurora-A and Skp2 expression. These findings suggest that deregulation of Emi1 may contribute to APC/C inactivation in cancer. Moreover, head and neck cancer cases with high expression of Emi1 showed malignant behaviors, including poor differentiation and lymph node metastasis, suggesting that Emi1 can be a possible therapeutic target in cancer.
DNA replication depends on the APC/C-mediated degradation of geminin, which releases the inhibition of Cdt1 incorporation into prereplicative complexes and permits the proper assembly of these complex components (26, 27). Origin firing can only occur after the APC/C is inactivated and CDKs become active. To strictly inhibit the assembly of prereplicative complexes during S to G2 phase transition, APC/C is inactivated by several mechanisms: binding to its inhibitor Emi1, CDK-mediated phosphorylation of Cdh1 (which blocks its ability to activate APC/C), and degradation of both Cdh1 (28) and its major ubiquitin C, namely UbcH10 (29). Indeed, Emi1 depletion induced rereplication with large nuclei and polyploidy in various cancer cells (Fig. 2), and Cdh1 rescued the down-regulation of geminin and rereplication caused by Emi1 depletion (Fig. 5C). These findings are consistent with previous findings showing that (i) rereplication caused by Emi1 depletion is due to decreased geminin via APC/C-mediated proteolysis (18, 19), and (ii) geminin depletion causes rereplication in cancer cells (30). In addition, Emi1 depletion induces DNA damage, likely explained by replication stress upon deregulated cyclin E- and A-associated kinase activities (20). We also found that Emi1 knockdown induced the frequency of high levels of γH2AX foci (>10 foci per cell) in cancer cells (Fig. 4B). γH2AX foci are known to be an early response marker for double-stranded breaks (DSBs) (31). Moreover, premature termination of DNA replication can lead to fork collapse and consequent DSBs, eventually triggering robust activation of the DNA damage checkpoint response (32). Therefore, Emi1 depletion is likely to induce DSBs via replication stress. Interestingly, frequency of high levels of γH2AX foci by Emi1 depletion in cancer cells was higher than that in normal cells (Fig. 4B). Chemotherapy and/or radiation in addition to surgery have proven useful in a number of different cancer types. At present many anticancer drugs for targeting DNA synthesis to interfere with rapidly dividing cells are commonly used. Ionizing radiation works by damaging DNA of exposed tissue leading to cellular death. Therefore, the finding that Emi1 depletion induced rereplication and DNA damage made us hypothesize that Emi1 knockdown might enhance the sensitivity of anticancer drugs and ionizing radiation. Interestingly, in the present study, we demonstrated that Emi1 knockdown by using siRNA enhanced the sensitivity of anticancer reagents treatment and x-rays in cancer cells. Although we found synergistic effect of Emi1 knockdown in combination with doxorubicin, camptothecin, and etoposide (Fig. 6D), we are planning to examine the synergistic effect of Emi1 knockdown in combination with other anticancer drugs that interfere with DNA synthesis. Recent study showed that the hypoxic condition induced multidrug resistance (22). Interestingly, Emi1 siRNA enhanced the sensitivity of anticancer drugs under hypoxia condition (Fig. 6, E and F), indicating that Emi1 depletion can be a useful for clinical application.
It is likely that the radioresistance is strong in the S phase because the DNA repair enzymes that allow radiation-induced damage to appear in DNA are at full activity during S phase and less so at other stages (33). The G2/M phase of the cell cycle is when cells are most sensitive to radiation (33). Indeed, paclitaxel, which inhibits the formation of mitotic spindles and arrests the cells in G2/M phase, enhances radiation efficacy on cell killing and suppression of growth (34–36). Moreover, inhibition of Aurora kinase enhances the sensitivity of radiation (37–40). Thus, some drugs for inducing cell cycle arrest at M phase are used for enhancing the sensitivity of radiation. Although Emi1-depleted cells were arrested at S phase by rereplication decreased survival cells was observed after x-ray exposure. As Emi1 depletion leads to DSBs via replication stress, DSBs induced by Emi1 depletion may enhance the sensitivity of radiation in cancer cells.
Emi1 knockdown suppressed cancer cell growth with continuous DNA synthesis, but normal cells did not seem to be affected by the knockdown. Therefore, the synergistic effect of Emi1 knockdown in combination with doxorubicin treatment and radiation was not observed in normal cells. These findings are consistent with previous finding that none of the effects are detected either in normal cells by geminin depletion (23). In normal cells, both geminin and cyclin A suppress rereplication and genomic instability (23, 24). In this study, down-regulation of cyclin A was not observed in normal fibroblasts after Emi1 depletion (Fig. 7A), whereas down-regulation of cyclin A was observed in cancer cells (Fig. 5D). As shown in Fig. 1, overexpression of Emi1 and APC/C substrates, including cyclin A, was observed in cancer cells. Activated APC/C by Emi1 depletion may degrade overexpressed APC/C substrates at S-G2 phase in cancer cells but not in normal cells. In normal cells, inhibitory mechanism of degradation of APC/C substrates may exist at S-G2 in addition to proper phase for their protein degradation. Indeed, depletion of both Emi1 and cyclin A induced polyploidy and large nuclei (Fig. 7B). Therefore, cyclin A may suppress rereplication in normal cells. This observation is supported by previous finding that treatment with geminin siRNA and cyclin A siRNA induced DNA rereplication in normal cells in similar to cancer cells induced by geminin siRNA alone (23). Previous reports showed that accumulation of cyclin E was induced by Emi1 knockdown through the APC/C-independent pathway (18, 19). As cyclin E is not a substrate of APC/C, it is still unclear why cyclin E is up-regulated by Emi1 depletion. Interestingly, up-regulation of cyclin E was not observed in normal cells (Fig. 7A), suggesting that induction of cyclin E may be involved in rereplication only in cancer cells. These findings suggest that Emi1 may play an important role in initiating DNA replication in cancer cells, whereas normal cells may possess additional safeguards such as cyclin A for preventing DNA rereplication. Importantly, no effect of Emi1 knockdown and no synergistic effect of Emi1 knockdown in combination with doxorubicin treatment and radiation in normal cells are useful findings for clinical application.
The therapeutic utility of siRNAs is thought to be limited by the requirement for complex formulations to deliver them to tissues. Recently, however, it has been identified single-strand siRNAs for silencing gene expression in animals (41). Indeed, single-strand siRNAs achieved potencies and selectivity for inhibiting mutant huntingtin in Huntington disease model mice (42). It is anticipated that technologies of siRNA delivery will be developed for efficient gene silencing in clinical application. Moreover, we will look for the small compound or peptide for inhibiting Emi1 function in addition to siRNA. In conclusion, we suggest that inhibition of Emi1 function could be a useful for enhancing sensitivity of cancer cells to chemotherapeutic drugs and ionizing radiation.
Acknowledgments
We thank Dr. Michele Pagano (New York University) for helpful discussions, Michie Akaishizawa (National Institute of Radiological Sciences) for expert technical assistance, and Dr. Rieko Arakaki (Tokushima University) and Dr. Keiji Tanimoto (Hiroshima University) for technical advice. We also thank Dr. Nishitani (University of Hyogo), Dr. Heidebrecht (University of Kiel), Dr. Fusenig (German Cancer Research Center), and Dr. Vogelstein (The Johns Hopkins University) for providing materials. This work was carried out at the Joint Usage/Research Center (RIRBM), Hiroshima University.
This work was supported in part by Grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (23689074 and 25670778 (to Y. K.), 21249088 (to T. Takata), and 23390301 and 24249067 (to R. O.)), Grant-in-aid for Cancer Research from the Ministry of Education, Culture, Sports, Science and Technology (19-9, H23-A-43; to R. O.), and a Research Fellowship for Young Scientists from the Japan Society for the Promotion of Science (23-6562; to T. Tsunematsu).
- APC/C
- anaphase-promoting complex/cyclosome
- DSB
- double-stranded break
- PI
- propidium iodide
- EdU
- 5-ethynyl-2′-deoxyuridine
- NHDF
- neonatal normal human dermal fibroblast.
REFERENCES
- 1. Jemal A., Bray F., Center M. M., Ferlay J., Ward E., Forman D. (2011) Global cancer statistics. CA-Cancer J. Clin. 61, 69–90 [DOI] [PubMed] [Google Scholar]
- 2. Takimoto C. H., Calvo E. (2008) Principles of Oncologic Pharmacotherapy in Cancer Management: A Multidisciplinary Approach (Pazdur R., Wagman L. D., Camphausen K. A., Hoskins W. J., eds) 11th Ed pp. 1–9, Cmp United Business Media, Manhasset, NY [Google Scholar]
- 3. Pommier Y., Leteurtre F., Fesen M. R., Fujimori A., Bertrand R., Solary E., Kohlhagen G., Kohn K. W. (1994) Cellular determinants of sensitivity and resistance to DNA topoisomerase inhibitors. Cancer Invest. 12, 530–542 [DOI] [PubMed] [Google Scholar]
- 4. Dancey J. E., Bedard P. L., Onetto N., Hudson T. J. (2012) The genetic basis for cancer treatment decisions. Cell 148, 409–420 [DOI] [PubMed] [Google Scholar]
- 5. Reed S. (2003) Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4, 855–864 [DOI] [PubMed] [Google Scholar]
- 6. Pagano M., Benmaamar R. (2003) When protein destruction runs amok, malignancy is on the loose. Cancer Cell 4, 251–256 [DOI] [PubMed] [Google Scholar]
- 7. Yamasaki L., Pagano M. (2004) Cell cycle, proteolysis and cancer. Curr. Opin. Cell Biol. 16, 623–628 [DOI] [PubMed] [Google Scholar]
- 8. Nakayama K. I., Nakayama K. (2006) Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 9. Peters J. M. (2006) The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7, 644–656 [DOI] [PubMed] [Google Scholar]
- 10. Cardozo T., Pagano M. (2004) The SCF ubiquitin ligase: insights into a molecular machine. Nat. Rev. Mol. Cell Biol. 5, 739–751 [DOI] [PubMed] [Google Scholar]
- 11. Hsu J. Y., Reimann J. D., Sørensen C. S., Lukas J., Jackson P. K. (2002) E2F-dependent accumulation of hEmi1 regulates S phase entry by inhibiting APCCdh1. Nat. Cell Biol. 4, 358–366 [DOI] [PubMed] [Google Scholar]
- 12. Reimann J. D., Gardner B. E., Margottin-Goguet F., Jackson P. K. (2001) Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 15, 3278–3285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reimann J. D., Freed E., Hsu J. Y., Kramer E. R., Peters J. M., Jackson P. K. (2001) Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell 105, 645–655 [DOI] [PubMed] [Google Scholar]
- 14. Margottin-Goguet F., Hsu J. Y., Loktev A., Hsieh H. M., Reimann J. D., Jackson P. K. (2003) Prophase destruction of Emi1 by the SCFβTrCP/Slimb ubiquitin ligase activates the anaphase promoting complex to allow progression beyond prometaphase. Dev. Cell 4, 813–826 [DOI] [PubMed] [Google Scholar]
- 15. Guardavaccaro D., Kudo Y., Boulaire J., Barchi M., Busino L., Donzelli M., Margottin-Goguet F., Jackson P. K., Yamasaki L., Pagano M. (2003) Control of meiotic and mitotic progression by the F box protein β-Trcp1 in vivo. Dev. Cell 4, 799–812 [DOI] [PubMed] [Google Scholar]
- 16. Lehman N. L., Tibshirani R., Hsu J. Y., Natkunam Y., Harris B. T., West R. B., Masek M. A., Montgomery K., van de Rijn M., Jackson P. K. (2007) Oncogenic regulators and substrates of the anaphase promoting complex/cyclosome are frequently overexpressed in malignant tumors. Am. J. Pathol. 170, 1793–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gütgemann I., Lehman N. L., Jackson P. K., Longacre T. A. (2008) Emi1 protein accumulation implicates misregulation of the anaphase promoting complex/cyclosome pathway in ovarian clear cell carcinoma. Mod. Pathol. 21, 445–454 [DOI] [PubMed] [Google Scholar]
- 18. Machida Y. J., Dutta A. (2007) The APC/C inhibitor, Emi1, is essential for prevention of rereplication. Genes Dev. 21, 184–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Di Fiore B., Pines J. (2007) Emi1 is needed to couple DNA replication with mitosis but does not regulate activation of the mitotic APC/C. J. Cell Biol. 177, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Verschuren E. W., Ban K. H., Masek M. A., Lehman N. L., Jackson P. K. (2007) Loss of Emi1-dependent anaphase-promoting complex/cyclosome inhibition deregulates E2F target expression and elicits DNA damage- induced senescence. Mol. Cell. Biol. 27, 7955–7965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yokoyama K., Kamata N., Fujimoto R., Tsutsumi S., Tomonari M., Taki M., Hosokawa H., Nagayama M. (2003) Increased invasion and matrix metalloproteinase-2 expression by Snail-induced mesenchymal transition in squamous cell carcinomas. Int. J. Oncol. 22, 891–898 [PubMed] [Google Scholar]
- 22. Onozuka H., Tsuchihara K., Esumi H. (2011) Hypoglycemic/hypoxic condition in vitro mimicking the tumor microenvironment markedly reduced the efficacy of anticancer drugs. Cancer Sci. 102, 975–982 [DOI] [PubMed] [Google Scholar]
- 23. Zhu W., Depamphilis M. L. (2009) Selective killing of cancer cells by suppression of geminin activity. Cancer Res. 69, 4870–4877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mihaylov I. S., Kondo T., Jones L., Ryzhikov S., Tanaka J., Zheng J., Higa L. A., Minamino N., Cooley L., Zhang H. (2002) Control of DNA replication and chromosome ploidy by geminin and cyclin A. Mol. Cell. Biol. 22, 1868–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lehman N. L., Verschuren E. W., Hsu J. Y., Cherry A. M., Jackson P. K. (2006) Overexpression of the anaphase promoting complex/cyclosome inhibitor Emi1 leads to tetraploidy and genomic instability of p53-deficient cells. Cell Cycle 5, 1569–1573 [DOI] [PubMed] [Google Scholar]
- 26. Wohlschlegel J. A., Dwyer B. T., Dhar S. K., Cvetic C., Walter J. C., Dutta A. (2000) Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science 290, 2309–2312 [DOI] [PubMed] [Google Scholar]
- 27. Tada S., Li A., Maiorano D., Méchali M., Blow J. J. (2001) Repression of origin assembly in metaphase depends on inhibition of RLF-B/Cdt1 by geminin. Nat. Cell Biol. 3, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Listovsky T., Oren Y. S., Yudkovsky Y., Mahbubani H. M., Weiss A. M., Lebendiker M., Brandeis M. (2004) Mammalian Cdh1/Fzr mediates its own degradation. EMBO J. 23, 1619–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rape M., Kirschner M. W. (2004) Autonomous regulation of the anaphase-promoting complex couples mitosis to S-phase entry. Nature 432, 588–595 [DOI] [PubMed] [Google Scholar]
- 30. Melixetian M., Ballabeni A., Masiero L., Gasparini P., Zamponi R., Bartek J., Lukas J., Helin K. (2004) Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 165, 473–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 32. Stiff T., Walker S. A., Cerosaletti K., Goodarzi A. A., Petermann E., Concannon P., O'Driscoll M., Jeggo P. A. (2006) ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 25, 5775–5782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terashima T., Tolmach L. J. (1963) X-ray sensitivity and DNA synthesis in synchronous populations of HeLa cells. Science 140, 490–492 [DOI] [PubMed] [Google Scholar]
- 34. Tishler R. B., Geard C. R., Hall E. J., Schiff P. B. (1992) Taxol sensitizes human astrocytoma cells to radiation. Cancer Res. 52, 3495–3497 [PubMed] [Google Scholar]
- 35. Milas L., Hunter N. R., Mason K. A., Kurdoglu B., Peters L. J. (1994) Enhancement of tumor radioresponse of a murine mammary carcinoma by paclitaxel. Cancer Res. 54, 3506–3510 [PubMed] [Google Scholar]
- 36. Zhang A. L., Russell P. J., Knittel T., Milross C. (2007) Paclitaxel enhanced radiation sensitization for the suppression of human prostate cancer tumor growth via a p53 independent pathway. Prostate 67, 1630–1640 [DOI] [PubMed] [Google Scholar]
- 37. Guan Z., Wang X. R., Zhu X. F., Huang X. F., Xu J., Wang L. H., Wan X. B., Long Z. J., Liu J. N., Feng G. K., Huang W., Zeng Y. X., Chen F. J., Liu Q. (2007) Aurora-A, a negative prognostic marker, increases migration and decreases radiosensitivity in cancer cells. Cancer Res. 67, 10436–10444 [DOI] [PubMed] [Google Scholar]
- 38. Tao Y., Zhang P., Frascogna V., Lecluse Y., Auperin A., Bourhis J., Deutsch E. (2007) Enhancement of radiation response by inhibition of Aurora-A kinase using siRNA or a selective Aurora kinase inhibitor PHA680632 in p53-deficient cancer cells. Br. J. Cancer 97, 1664–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moretti L., Niermann K., Schleicher S., Giacalone N. J., Varki V., Kim K. W., Kopsombut P., Jung D. K., Lu B. (2011) MLN8054, a small molecule inhibitor of aurora kinase a, sensitizes androgen-resistant prostate cancer to radiation. Int. J. Radiat. Oncol. Biol. Phys. 80, 1189–1197 [DOI] [PubMed] [Google Scholar]
- 40. Niermann K. J., Moretti L., Giacalone N. J., Sun Y., Schleicher S. M., Kopsombut P., Mitchell L. R., Kim K. W., Lu B. (2011) Enhanced radiosensitivity of androgen-resistant prostate cancer: AZD1152-mediated Aurora kinase B inhibition. Radiation Res. 175, 444–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lima W. F., Prakash T. P., Murray H. M., Kinberger G. A., Li W., Chappell A. E., Li CS, Murray S. F., Gaus H., Seth P. P., Swayze E. E., Crooke S. T. (2012) Single-stranded siRNAs activate RNAi in animals. Cell 150, 883–894 [DOI] [PubMed] [Google Scholar]
- 42. Yu D., Pendergraff H., Liu J., Kordasiewicz H. B., Cleveland D. W., Swayze E. E., Lima W. F., Crooke S. T., Prakash T. P., Corey D. R. (2012) Single-stranded RNAs use RNAi to potently and allele-selectively inhibit mutant Huntingtin expression. Cell 150, 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Buchholz M., Braun M., Heidenblut A., Kestler H. A., Klöppel G., Schmiegel W., Hahn S. A., Lüttges J., Gress T. M. (2005) Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions. Oncogene 24, 6626–6636 [DOI] [PubMed] [Google Scholar]
- 44. Dyrskjøt L., Kruhøffer M., Thykjaer T., Marcussen N., Jensen J. L., Møller K., Ørntoft T. F. (2004) Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 64, 4040–4048 [DOI] [PubMed] [Google Scholar]
- 45. French P. J., Swagemakers S. M., Nagel J. H., Kouwenhoven M. C., Brouwer E., van der Spek P., Luider T. M., Kros J. M., van den Bent M. J., Sillevis Smitt P. A. (2005) Gene expression profiles associated with treatment response in oligodendrogliomas. Cancer Res. 65, 11335–11344 [DOI] [PubMed] [Google Scholar]
- 46. Ginos M. A., Page G. P., Michalowicz B. S., Patel K. J., Volker S. E., Pambuccian S. E., Ondrey F. G., Adams G. L., Gaffney P. M. (2004) Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 64, 55–63 [DOI] [PubMed] [Google Scholar]
- 47. Korkola J. E., Houldsworth J., Chadalavada R. S., Olshen A. B., Dobrzynski D., Reuter V. E., Bosl G. J., Chaganti R. S. (2006) Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 66, 820–827 [DOI] [PubMed] [Google Scholar]
- 48. Pyeon D., Newton M. A., Lambert P. F., den Boon J. A., Sengupta S., Marsit C. J., Woodworth C. D., Connor J. P., Haugen T. H., Smith E. M., Kelsey K. T., Turek L. P., Ahlquist P. (2007) Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 67, 4605–4619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Richardson A. L., Wang Z. C., De Nicolo A., Lu X., Brown M., Miron A., Liao X., Iglehart J. D., Livingston D. M., Ganesan S. (2006) X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell 9, 121–132 [DOI] [PubMed] [Google Scholar]
- 50. Sun L., Hui A. M., Su Q., Vortmeyer A., Kotliarov Y., Pastorino S., Passaniti A., Menon J., Walling J., Bailey R., Rosenblum M., Mikkelsen T., Fine H. A. (2006) Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell 9, 287–300 [DOI] [PubMed] [Google Scholar]